Abstract
Background
Although the penetration of electronic health records is increasing rapidly, much of the historical medical record is only available in handwritten notes and forms, which require labor-intensive, human chart abstraction for some clinical research. The few previous studies on automated extraction of data from these handwritten notes have focused on monolithic, custom-developed recognition systems or third-party systems that require proprietary forms.
Methods
We present an optical character recognition processing pipeline, which leverages the capabilities of existing third-party optical character recognition engines, and provides the flexibility offered by a modular custom-developed system. The system was configured and run on a selected set of form fields extracted from a corpus of handwritten ophthalmology forms.
Observations
The processing pipeline allowed multiple configurations to be run, with the optimal configuration consisting of the Nuance and LEADTOOLS engines running in parallel with a positive predictive value of 94.6% and a sensitivity of 13.5%.
Discussion
While limitations exist, preliminary experience from this project yielded insights on the generalizability and applicability of integrating multiple, inexpensive general-purpose third-party optical character recognition engines in a modular pipeline.
Keywords: Luke, bioinformatics
Introduction
In the realm of biomedical research, the electronic health record (EHR) is increasingly realized as a rich source of longitudinal medical information. One such effort leveraging this realization is the Electronic Medical Records and Genomics Network,1 which has a primary goal of developing and evaluating novel methods for extracting clinical information from the EHR.2 Although many institutions are implementing EHRs, the data collected are often not in an easily computable form—instead of discrete data elements, they are collected as unstructured text notes or even document images. The latter include digital forms collected via tablet-based computers, or paper documents that are later scanned as an attempt to ‘back load’ historical medical data.
Although optical character recognition (OCR) in the medical domain is not, in principle, different from OCR in other domains, the interpretation of physician handwriting has been notoriously challenging, even for humans. Studies on the use of OCR within the medical domain have been limited and have followed one of two approaches. The first is focused on designing forms to capture hand-printed data specifically for OCR processing,3–5 which facilitates prospective data collection to be more amenable to OCR processing by segmenting each individual character via controlled forms. However, this approach does not help with the detection of historical forms. These studies have utilized a complete third-party OCR system to manage definition of the form fields, extract the data from completed forms, and manually verify and correct errors. While there are benefits to commercial systems, they also suffer from higher costs and the risk of vendor ‘lock-in’. The second approach utilized custom-developed OCR engines to perform the handwriting recognition, including unconstrained handwriting where adjacent characters may touch. Some promising results have been reported applying these engines in the medical domain.6–8 A limitation of this approach is that such development requires a level of funding and expertise in OCR research that is absent at most institutions.
Recent work in the natural language processing domain has focused on the development of modular pipelines for analysis of biomedical documents.9 10 In the pipeline paradigm, individual software modules (such as data handling, preprocessing, postprocessing, and data presentation) are chained together such that the output from one module is the input to the next.11 Such pipelines support a ‘mix and match’ strategy that maximizes flexibility while minimizing the need for local expertise. To our knowledge, no work to date has explored a comparable pipeline approach to OCR of biomedical documents. This paper describes the development and preliminary evaluation of such a pipeline.
Methods
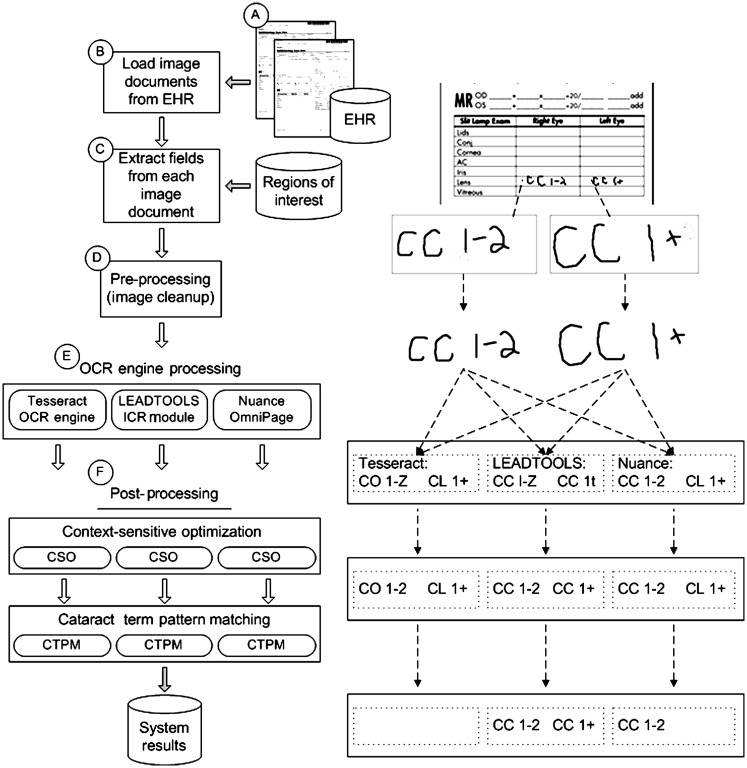
The specific task was to identify cataract type and severity for subjects using purely automated methods, which would be used for a genome-wide association study. Because the goal of the driving study was to find a limited number of ‘clean’ cases from among a large population, the primary performance goal was a high positive predictive value at the expense of sensitivity. A goal of the Electronic Medical Records and Genomics Project was to develop phenotyping tools and approaches that could be applied to multiple institutions.1 This ruled out not only most proprietary document management systems, but also systems that required deep OCR expertise to implement. The strategy selected was to focus on open-source and moderately priced, open application programming interface OCR engines. The development of a pipeline architecture allowed the OCR engines to be tested in multiple configurations, with or without supporting modules earlier or later in the pipeline. The utility of using a pipeline was evaluated through several configurations of the system overall in order to arrive at an optimal combination of modules and OCR engines. The overall architecture of the resulting OCR system is shown in figure 1, with the pipeline modules described below. Some pipeline modules required initial configuration and setup, specific to the documents that were being processed, but no manual intervention was required during the pipeline execution.
Figure 1.
The optical character-recognition (OCR) system pipeline (left) and sample output (right). Image documents from an electronic health record (EHR) were extracted, and specific fields of interest were selected on the basis of regions defined in an external configuration file. After the images had been preprocessed, they were sent through three separate OCR engines. The results of the engines were postprocessed using context-sensitive optimization rules, and the final results of cataract types and severities were found by pattern matching.
Corpus
The cohort for this study was based on a population of 20 000 subjects enrolled in the Marshfield Clinic Personalized Medicine Research Project.12 13 All participants provided written informed consent to participate in the biobank, and the project was reviewed and approved by the Marshfield Clinic Institutional Review Board. From the full cohort, a subset of subjects with International Classification of Diseases (ICD)9 codes related to senile cataract was selected to identify those most likely to contain cataract-related assessments in their medical record. A random sample of 949 ophthalmology forms stored as images in the tagged image file format was selected from a corpus of 49 753 relevant forms in the Marshfield Clinic EHR system, with the images extracted from the document repository in this first step.
Loading document images
All forms, both paper and electronic, contain a form identifier printed in the lower left portion of each page to assist with versioning and tracking of form templates. Within this study, the form identifier was used to identify region(s) of the document to be processed. For documents created on the tablet PC, the identifier was embedded in the associated metadata. Forms that were filled out on paper and later scanned in required an additional OCR processing step to extract the identifier from the form image. Given that the identifier was printed on the form within the same location on the page, it was amenable to processing by an OCR engine and the resulting output matched against a list of form identifiers of interest.
Extracting fields from document images
On the basis of a review by a domain expert and a trained research coordinator, certain fields from ophthalmology forms were identified as the most likely to contain data pertaining to cataracts. Using empty forms, bounding boxes were defined around the target fields to determine their coordinates and size. In addition, regions of some forms were identified as having data but were not clearly delineated form fields, such as areas designated for diagrams. The coordinates for each form field were added to a configuration file, including the unique identifier assigned to each form. Using these configuration files, fields were extracted from the full form and placed in a secure storage location for subsequent processing steps in the pipeline.
Preprocessing
Initial testing of the OCR engines showed issues with recognition if the handwriting overlapped the lines on the form, or contained additional artifacts such as speckles, which commonly appear as noise in scanned documents. To improve OCR performance, speckles were removed utilizing the capabilities of the LEADTOOLS image processing toolkit, and lines were removed using a custom-developed algorithm based on methods described in the literature.14 A separate approach was needed for form fields designed to contain diagrams, since the diagram template was not as easy to remove as bordering lines for traditional form fields. We utilized the diagram outline as a mask to be removed from the end image, while factoring in that characters would overlap the diagram. This step was executed each time the pipeline ran (as opposed to preprocessing images outside of the pipeline), which allowed us to adjust the preprocessing algorithms without having to re-extract all of the form fields.
OCR engine processing
The pipeline was executed with different configurations of three OCR engines. The engines generated plain text interpretations of the handwriting and were not used to generate image features or other intermediate results from the recognition process. Given this configuration, running engines in serial (where order of execution would be important) was not a consideration, so each preprocessed form field was sent through each engine in parallel, and the results recorded.
The first engine evaluated was version 3.0 of the Tesseract OCR engine, originally developed at Hewlett-Packard Labs from 1985 to 1995, and now available as an open-source project.15 Tesseract was an attractive option, as it was open source, is still under development (version 3.0 was released in late 2010), could be trained to recognize a specific set of characters, and be further constrained to understand a set lexicon. Within this study, the lexicon targeted three cataract types from the ophthalmology forms: nuclear sclerotic, posterior subcapsular, and cortical. The process of training the Tesseract engine is described in appendix A of the online data supplement, with the optimal configuration used in this study including a limited character set that represented only those characters expected to occur in a cataract description: A, B, C, D, E, I, L, N, O, P, R, S, T, U, 1, 2, 3, 4, +, −. Two commercial engines were also used (both of which were preconfigured to recognize all English alphabetic, numeric, and punctuation characters), the first of which was version 16 of the LEADTOOLS Intelligent Character Recognition (ICR) Module16 available from LEAD Technologies (Charlotte, NC, USA), which was developed specifically for handwriting recognition. The LEADTOOLS ICR Module is an add-on to a sophisticated document-imaging toolkit, which was also used for other image-processing steps within this system. The final engine was the OmniPage Capture SDK from Nuance Communications (Burlington, MA, USA),17 which also supports hand-printed character recognition through an ICR recognition module. Although the Nuance SDK also offered a toolkit of image-processing features, they were not used in this study.
Postprocessing
As expected, all OCR engines were susceptible to false-negative, false-positive, and character-substitution errors when applied to unconstrained text. Further evaluation revealed that these mis-recognitions tended to follow predictable patterns. A set of regular expressions was developed to correct the most common insertion, deletion, and substitution errors from all engines in an attempt to improve recognition rates. These context-sensitive optimization rules (a subset of which is shown in table 1) were created on the premise that a limited set of characters and combinations of those characters made sense within this domain. By using the context of surrounding characters and results from known misclassifications (similar in nature to the spellcheck in word-processing software), rules could be developed to infer a correct answer. The rules were derived from the analysis of a training set of 120 form regions that were manually validated by a human reader, by testing different scenarios to find regular expressions which improved accuracy and reduced the false-positive rate. The final rules were manually reviewed before being accepted for use by the system.
Table 1.
Subset of the context-sensitive optimization rules that were applied to the raw output from the optical character recognition engines
| Matching pattern | Corrected result | Description |
| R[1IL\\/]S | NS | A lowercase ‘r’ next to a character appearing as a vertical bar (‘1’, ‘I’, ‘L’, ‘/’ or ‘\’) is a common misclassification for an ‘N’. This rule would only be applied if an ‘S’ was subsequently found, which would indicate the abbreviation for nuclear sclerotic. |
| [LI\\/][T+] | 1+ | A common misclassification is with characters shaped as vertical bars, which means a 1 may be misclassified as an ‘L’, ‘I’ or slash (‘/’ or ‘\’). One of these characters followed by a ‘+’ (or a ‘T’ which is a common misclassification for a ‘+’) is most likely a ‘1’. A known documentation pattern for severity is ‘1+’. |
| [Z][T+] | 2+ | A ‘Z’ is a common misclassification for a ‘2’. Similarly to the previous rule mentioned for ‘1+’, it requires a ‘+’-like character to immediately follow. |
The optimization rules were applied to the results from each OCR engine, and additional regular expressions were then used to match documentation patterns of cataract type and severity in a final extraction step to create a dataset.
Gold standard and validation
Each form region was interpreted separately by two human readers, who reviewed them in random order. The readers recorded the cataract type and severity for each finding listed, or whether the region was blank or illegible. Thirteen regions were noted as illegible by one reader, but none by both. Initially, readers agreed on 92.4% of the regions. Disagreements were resolved by consensus of both readers following a review and discussion of each region for which codings did not match. One of the authors (LR) acted as a facilitator. The readers were able to reach agreement for all 949 regions. The final standard included 475 positive regions containing 682 mentions of cataract type (647 mentions of both type and severity) and 474 regions that were blank or contained no data of interest.
Statistical analysis
To evaluate the configurations that were run through the pipeline, the performance of each engine was evaluated singly and in combination with other engines using both conjunction (AND) and disjunction (OR) combining rules, which were applied in the postprocessing module. For each combination, positive predictive value (PPV), negative predictive value (NPV), sensitivity and specificity were computed. Details on the methods of validating against this gold standard are given in appendix B of the online supplement.
Observations
During construction of the pipeline, the use of preconfigured commercial OCR engines required less time to integrate into the OCR pipeline than the Tesseract engine, which required substantial training. However, once each engine was configured for use by the pipeline, subsequent use and exchange of each engine was negligible.
The pipeline architecture facilitated multiple configurations to be executed against the test corpus. For each execution, the pipeline was quickly configured to use any or all of the OCR engines, whether to use the context-sensitive optimization rules within the postprocessing step, and if results from multiple engines required full agreement in order to be included in the output. The results from each experiment were compared with the gold standard in order to assess the accuracy of that particular configuration. Once all experiments had been run, the results were evaluated to determine how the system performed under different scenarios.
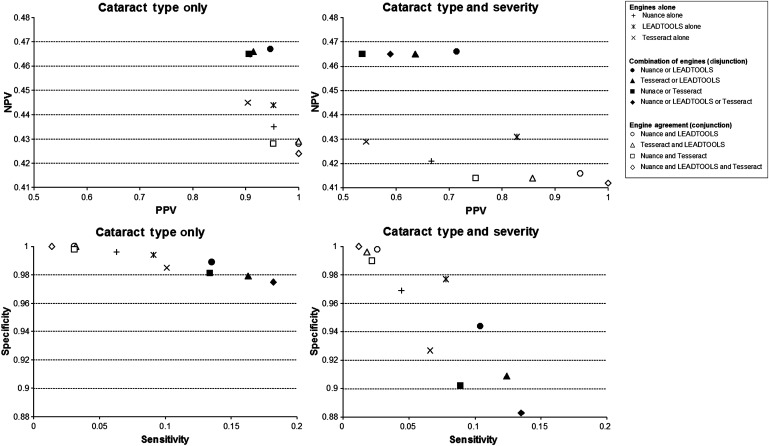
Summary statistics for all combinations of the three engines are shown in table 2, with complete experimental results listed in table 4 of appendix C in the online data supplement. Summary graphs showing the comparison of PPV with NPV and sensitivity with specificity are shown in figure 2, and include results for the detection of cataract type only as well as cataract type and severity. These results show that the pipeline allowed multiple configurations to be run, which also provided the ability to arrive at an optimal configuration—the Nuance and LEADTOOLS engines running in parallel, which demonstrated the highest PPV (94.6%) for cataract type and/or severity. As with all of the engine configurations, the PPV was higher than sensitivity (13.5% for the chosen configuration), which is consistent with the design criteria.
Table 2.
Summary statistics for all combinations of the three engines, including positive predictive value (PPV), negative predictive value (NPV), sensitivity (Sn) and specificity (Sp)
| Cataract type only | Cataract type and severity | |||||||
| PPV (%) | NPV (%) | Sn (%) | Sp (%) | PPV (%) | NPV (%) | Sn (%) | Sp (%) | |
| Nuance alone | 95.3 | 43.5 | 6.3 | 99.6 | 66.7 | 42.1 | 4.4 | 96.9 |
| LEADTOOLS alone | 95.2 | 44.4 | 9.1 | 99.4 | 82.8 | 43.1 | 7.8 | 97.7 |
| Tesseract alone | 90.4 | 44.5 | 10.1 | 98.5 | 54.3 | 42.9 | 6.6 | 92.7 |
| Nuance or LEADTOOLS | 94.6 | 46.7 | 13.5 | 98.9 | 71.4 | 46.6 | 10.4 | 94.4 |
| Tesseract or LEADTOOLS | 91.4 | 46.6 | 16.3 | 97.9 | 63.6 | 46.5 | 12.4 | 90.9 |
| Nuance or Tesseract | 90.6 | 46.5 | 13.4 | 98.1 | 53.6 | 46.5 | 8.9 | 90.2 |
| Nuance or LEADTOOLS or Tesseract | 90.8 | 46.5 | 18.2 | 97.5 | 58.9 | 46.5 | 13.5 | 88.3 |
| Nuance and LEADTOOLS | 100.0 | 42.8 | 3.1 | 100.0 | 94.7 | 41.6 | 2.6 | 99.8 |
| Tesseract and LEADTOOLS | 100.0 | 42.9 | 3.2 | 100.0 | 85.7 | 41.4 | 1.8 | 99.6 |
| Nuance and Tesseract | 95.2 | 42.8 | 3.1 | 99.8 | 75.0 | 41.4 | 2.2 | 99.0 |
| Nuance and LEADTOOLS and Tesseract | 100.0 | 42.4 | 1.4 | 100.0 | 100.0 | 41.2 | 1.2 | 100.0 |
Figure 2.
Performance results for all combinations of optical character recognition (OCR) engines. Multiple engines separated by ‘OR’ reflect a logical disjunction of the engines, where each engine contributes results regardless of whether both engines agree. Multiple engines separated by ‘AND’ reflect where the engines both arrived at the same determination. The top row provides results for positive predictive value (PPV) and negative predictive value (NPV), while the bottom row provides results for sensitivity and specificity. The left column has results for the detection of cataract type only, and the right column has results for the detection of both cataract type and severity compared with the gold standard.
The context-sensitive optimization rules negatively affected results in multiple runs by increasing the number of falsely identified ‘results’ where none existed, contrary to the initially positive results during early system training and development. These rules also increased the number of correctly identified results, but not to a significant degree and not to a degree that offset the negative results. No analysis was performed to assess the differential impact of individual rules on the false-positive rate.
Configuration of the postprocessing module to require the OCR engines to agree markedly reduced the number of findings detected from 88 to 20 in the optimal configuration for cataract type only, and from 70 to 18 for cataract type and severity. However, this also had the effect of reducing false identifications, the largest reduction being 28 to 1 misclassifications for cataract type and severity. The use of any engine independently produced fewer correctly identified form fields than the use of both engines together, indicating that each engine would detect at least some handwriting that another engine could not.
Discussion
The study has two main findings. First, it is possible to integrate multiple, inexpensive general-purpose OCR engines into a single, modular pipeline. The use of a modular pipeline in this study demonstrated the ability to rapidly test multiple configurations of the system in order to arrive at a functional solution. Second, the best performance was achieved not by any individual OCR engine, but by the logical combination of the results of multiple engines. The rapid interchange of experimental modules, OCR engines or cleanup steps in the pipeline would accommodate institutions investigating methods to improve handwriting recognition. Institutions wishing to use a pipeline for applied recognition tasks, such as in medical research, may benefit from the ability to adopt new OCR engines as improvements are made in commercial offerings.
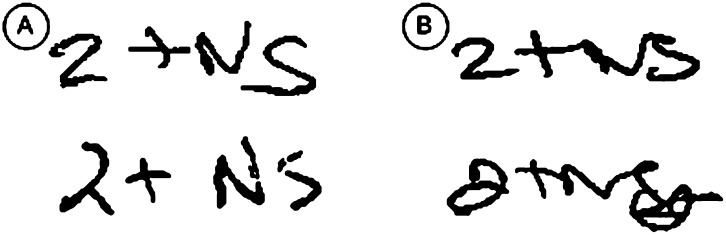
Detection rates found in this study were low compared with other studies applying OCR techniques on handwritten form fields.3 6 7 This is partially explained by the attempt to detect unconstrained handwriting, as opposed to hand-printed characters as in other studies, and the use of engines designed primarily for hand-printed characters. Figure 3 shows additional handwriting samples which provided varying levels of difficulty for detection by the OCR engines.
Figure 3.
Multiple handwriting samples of the same cataract type and severity, which demonstrate the challenges of optical character recognition (OCR). (A) Two examples of well-segmented characters that were detected by the system. (B) Two examples of handwriting samples that were not detected by the system.
This study had several limitations. We intentionally limited component choices to open-source or relatively inexpensive OCR components that could be integrated into our pipeline. Time and resource constraints prohibited an exhaustive comparison of all possible OCR options, and some OCR engine authors did not respond to a request to evaluate their engine. A purpose-built custom OCR engine would likely have produced higher recognition rates, as reported in other studies6 7; however, this would also likely have been at the expense of slower implementation and less generalizability than our modular approach. In addition, the system was only evaluated on documents created at one institution and focused on one specific disease, and used a very limited vocabulary.
Although the results are promising, it would be premature to attempt to generalize these results too broadly. The goal of this exploratory work was to evaluate the feasibility of the pipeline OCR strategy, rather than to develop a definitive architecture for such a pipeline or to define a detailed set of data representation or interconnect standards. One implication of this work is that OCR is more likely to be useful in instances where one needs to identify a limited number of patients from a large population, such as genomic studies or subject recruitment, as opposed to instances where all positive cases need to be identified, such as decision support. In spite of these limitations, this study demonstrates that a pipeline-based architecture may be a viable OCR strategy in certain domains and facilitates the rapid addition or removal of components in an attempt to improve recognition results. As continued advances are made in the detection of unconstrained handwriting, new engines may be plugged into a similarly configured pipeline for improved recognition results.
Acknowledgments
We thank Dr Lin Chen and Carol Waudby for guidance on the selection of form fields for the study. In addition, we acknowledge the anonymous reviewers whose recommendations about initial drafts and suggestion of the Nuance OCR engine have improved the resulting article.
Footnotes
Funding: This work was funded by NIH grant 5U01HG004608-02 from the National Human Genome Research Institute (NHGRI), and supported by grant 1UL1RR025011 from the Clinical and Translational Science Award (CTSA) program of the National Center for Research Resources, National Institutes of Health.
Competing interests: None.
Ethics approval: Ethics approval was provided by Marshfield Clinic Institutional Review Board.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.The eMERGE Network: electronic Medical Records & Genomics. http://www.gwas.org (accessed 19 May 2011).
- 2.Kho AN, Pacheco JA, Peissig PL, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med 2011;3:79re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biondich PG, Overhage JM, Dexter PR, et al. A modern optical character recognition system in a real world clinical setting: some accuracy and feasibility observations. Proc AMIA Symp 2002:56–60 [PMC free article] [PubMed] [Google Scholar]
- 4.Bussmann H, Wester CW, Ndwapi N, et al. Hybrid data capture for monitoring patients on highly active antiretroviral therapy (HAART) in urban Botswana. Bull World Health Organ 2006;84:127–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Titlestad G. Use of document image processing in cancer registration: how and why? Medinfo 1995:462. [PubMed] [Google Scholar]
- 6.Govindaraju V. Emergency medicine, disease surveillance, and informatics. Proc of the 2005 National Conference on Digital Government Research, Digital Government Society of North America, 2005:167–8 [Google Scholar]
- 7.Milewski R, Govindaraju V. Handwriting analysis of pre-hospital care reports. Proc CBMS, IEEE Computer Society, Los Alamitos, CA, 2004:428–33 [Google Scholar]
- 8.Piasecki M, Broda B. Correction of Medical Handwriting OCR Based on Semantic Similarity, Intelligent Data Engineering and Automated Learning—IDEAL 2007. Vol. 4881 Berlin, Heidelberg: Springer Berlin/Heidelberg, 2007:437–46 [Google Scholar]
- 9.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham H, Maynard D, Bontcheva K, et al. GATE: a framework and graphical development environment for robust NLP tools and applications. Proc of the 40th Assoc for Comp Ling (ACL'02); Philadelphia, July 2002 [Google Scholar]
- 11.Ghezzi C, Jazayeri M, Mandrioli D. Fundamentals of Software Engineering. 2nd edn Harlow: Prentice Hall, 2002 [Google Scholar]
- 12.McCarty CA, Wilke RA, Giampietro PF, et al. Marshfield Clinic Personalized Medicine Research Project (PMRP): design, methods and recruitment for a large population-based biobank. Per Med 2005;2:49–79 [DOI] [PubMed] [Google Scholar]
- 13.McCarty CA, Peissig P, Caldwell MD, et al. The Marshfield Clinic Personalized Medicine Research Project: 2008 scientific update and lessons learned in the first 6 years. Per Med 2008;5:529–42 [DOI] [PubMed] [Google Scholar]
- 14.Yoo JY, Kim MK, Han SY, et al. Line removal and restoration of handwritten characters on the form documents. ICDAR'97, IEEE Computer Society, Los Alamitos, CA, 1997:128–31 [Google Scholar]
- 15.Tesseract-OCR engine homepage Google Code. http://code.google.com/p/tesseract-ocr/ (accessed 19 May 2011).
- 16.LEADTOOLS Developer's Toolkit ICR Engine. http://www.leadtools.com/sdk/ocr/icr.htm (accessed 19 May 2011).
- 17.Nuance OmniPage CSDK. http://www.nuance.com/for-business/by-product/omnipage/csdk/index.htm (accessed 19 May 2011).