Abstract
Quality control and harmonization of data is a vital and challenging undertaking for any successful data coordination center and a responsibility shared between the multiple sites that produce, integrate, and utilize the data. Here we describe a coordinated effort between scientists and data managers in the Cancer Family Registries to implement a data governance infrastructure consisting of both organizational and technical solutions. The technical solution uses a rule-based validation system that facilitates error detection and correction for data centers submitting data to a central informatics database. Validation rules comprise both standard checks on allowable values and a crosscheck of related database elements for logical and scientific consistency. Evaluation over a 2-year timeframe showed a significant decrease in the number of errors in the database and a concurrent increase in data consistency and accuracy.
Keywords: Breast cancer, colon cancer, cancer registry, coordination, data governance, bioinformatics, epidemiology, ESAC, informatics
Introduction
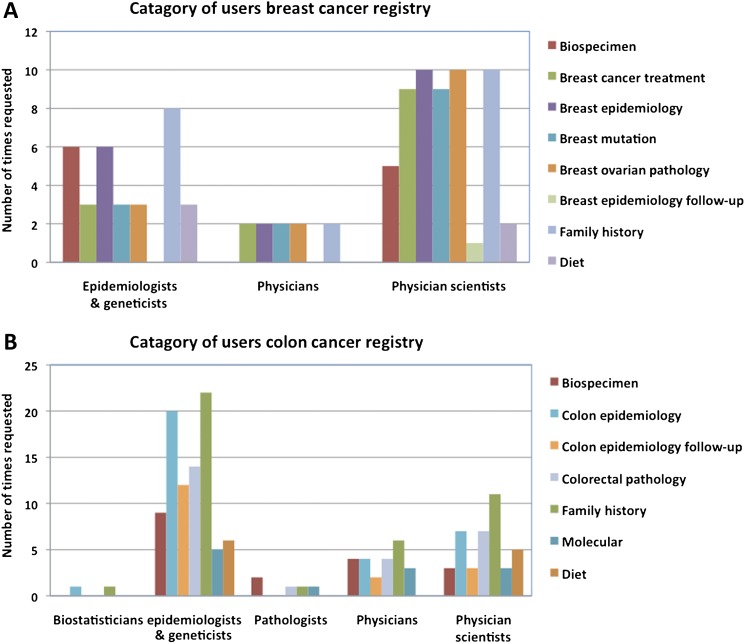
The Cancer Family Registries (CFR), consisting of the Breast Cancer Family Registry (B-CFR)1 and the Colon Cancer Family Registry (C-CFR),2 were established by the National Cancer Institute (NCI) to serve as a unique resource for conducting studies on the genetics and epidemiology of breast and colon cancer. To better enable the work of these registry sites, the NCI has funded a common informatics support center (ISC) to facilitate data collection and handle data requests from the scientific community. The Georgetown Data Coordination and Informatics Center (GDCIC) has served in this capacity since April 2009. The GDCIC maintains a centralized database, and facilitates data requests from the global scientific community via an application approved through the NCI. Figure 1A,B contains high-level summaries of the types of CFR ISC users and data they request. As of May 2011, the C-CFR database contained 468 037 records of individuals from 17 274 families collected from seven sites. Information including questionnaires and/or biological sample data is available for 42 403 individuals from 14 923 families. The B-CFR database contained 467 797 records of individuals from 16 245 families collected from six sites. In the latter database, questionnaires and/or biological sample data are available for 66 533 individuals from 15 436 families.
Figure 1.
Summary of the different backgrounds of Cancer Family Registries (CFR) data users and the types of CFR data they have most commonly requested since 2009 for (A) the Breast Cancer Family Registry (B-CFR) and (B) the Colon Cancer Family Registry (C-CFR).
Case description
A number of long-standing data management challenges were inherited by the GDCIC, including: (1) legacy systems from two previous ISCs and CFR sites for data storage and submission; their database, software, and practices had changed over time and were no longer adequately documented; (2) data gaps owing to incomplete data dictionaries used to standardize submissions across sites, which rendered some information inaccessible to data requestors; (3) poor technical communication and inadequate metadata since documents and modifications to data dictionaries were shared by email instead of a central repository, and were thus frequently lost or outdated; and (4) data quality problems—data profiling3 revealed inconsistencies, missing values, and errors in baseline epidemiology data that could be traced to several common sources of data quality problems4 including: multiple data sources, input rules being bypassed, and distributed heterogeneous systems.
Data governance
Prioritizing and addressing the various data governance and quality challenges of the CFR required both organizational and technical solutions. The organizational structure of the CFR includes a steering committee composed of the principal investigators of each site, and ISC and NCI personnel; a technical working group (TWG) composed of data coordinators and technical staff from each site and the ISC; and an analytical working group (AWG), which includes scientists from each site who use the data in their own research; as well as specialized working groups for biospecimens, epidemiology, and other topics.1 2 Members of the Georgetown ISC took over leadership of the TWG and began working closely with NCI management and the steering committee to prioritize activities and tackle the inherited data issues. The following sections provide details on the organizational and technical solutions we have adopted.
TWG process improvements
One of the first priorities was to create an improved charter for the TWG that delineated responsibilities in relation to the various other working groups and the overall operating process. The TWG is explicitly chartered to ensure the accuracy and completeness of the CFR data. The TWG contains at least two representatives from each site along with representatives from the ISC. The TWG meets online twice a month (more than any other working group in the CFR) and also tries to meet once a year for a face-to-face meeting. The TWG serves as an open forum for the discussion and resolution of all technical issues, both general and site-specific, relating to data submission, storage, and handling. Its main activity is to facilitate the creation of new or updated data dictionaries in response to the needs of the CFR community. Data dictionaries contain detailed descriptions of data tables and their respective data elements, including descriptions and permissible values, and each must be approved by the TWG prior to adoption. The ISC team helps facilitate this process, and provides staff and technical advice as needed. All data dictionary proposals in development or approved are posted on a public Wiki along with meeting information and implementation schedules. After a data dictionary is approved, the ISC software team begins the process of implementation. All data submitted by the CFR sites must conform to the approved dictionaries.
Data cleaning and improvements in the quality control framework
One cannot overestimate the importance of quality control (QC) for the successful operation of a data coordination center. However, it was recognized within the CFR that some of the baseline epidemiology data derived from questionnaires had issues including missing data, out of range values, or internal inconsistencies. To address the data issues, the ISC was tasked with developing a centralized system to help identify problems and assist in cleaning errors and inconsistencies where possible. Two areas were identified where technical solutions could help improve the QC framework: (1) improvements in the data dictionaries' format and data validations; and (2) new tools to move validation checks to earlier in the data submission process.
Data dictionaries
The original data dictionaries were maintained in MS Word and PDF files, which were difficult to keep current. We moved maintenance to XML that could be used to automatically produce HTML versions for review. (For an illustrative example of the HTML validations, see https://cfrisc.georgetown.edu/isc/dd.variablessummary.do?MODULE=breast-epi#breast-epi-AMENORRHEA_AGE.) To improve existing data validations, the ISC, in collaboration with the registry sites and the AWGs, implemented a process to review and rewrite existing validations, and add new rules to validate data elements. Initially, subject matter experts from the AWG were assigned to review various sections of the epidemiology data dictionaries and provide feedback. The TWG then used these comments to rewrite the validations into a logical set of independent sequentially executed if-then statements. Validations were written into a revised data dictionary, then reviewed and approved by the TWG.
Validation checks
The original data management system ran validation checks during data loading, generating log files that were returned to the registry sites for review. Previously, sites would get the logs late in the process and often could not make corrections within the time window for data submissions. In some cases, the only way to check the original data was to complete a manual inspection of the original paper questionnaire. In an improved system, the new XML data dictionaries are now used directly in a new QC process and framework consisting of the XML-based data dictionary and a rules engine to determine if the data submissions pass or fail each individual logic check. Two tiers of validation checks are used. First, a quality control tool (QCT), developed by the GDCIC, which runs as a Java WebStart application to download the current release of the XML data dictionaries, performs all of the validation checks on the registries' hardware prior to data submission. The QCT outputs a list of the validation errors and warnings to assist the data submitters in correcting their data submission file to match the data dictionary. This allows the sites to prepare and fix many data problems weeks before submitting the file to the ISC. Whenever rules are modified, a few may generate unexpected false fires where a discrepancy is found that turns out to be correct or explainable. This happens in spite of careful review of the data dictionaries and testing of the QCT. When these problems are identified, the validations are corrected and a new version of the QCT is built before the final submission. Second, once the data have been submitted, the corrected data dictionaries are used to validate the entire dataset yet again. The GDCIC provides training sessions to ensure that each registry's data manager can effectively use this resource.
Results and observations
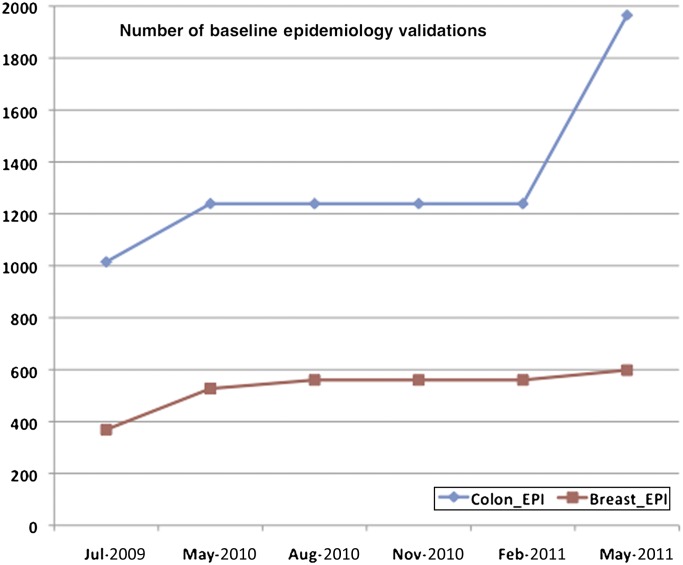
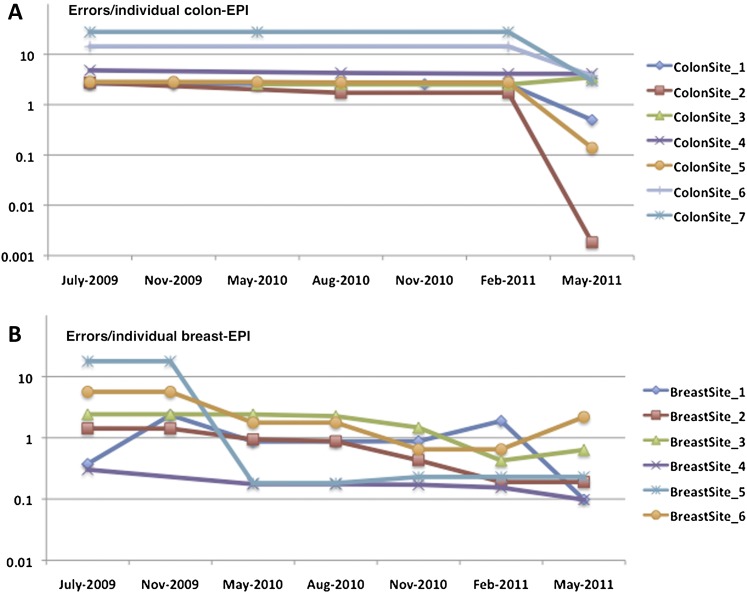
Overall, there is a clear sense that the implemented changes have improved the efficacy and operational effectiveness of the consortium, especially with respect to data quality. Figure 2 shows the growth in total number of validations implemented for the breast and colon epidemiology tables in the ISC database. The CFR data and the data dictionary validations have fluctuated over time, occasionally introducing false fires and thus making the strict counts of errors from each submission an inadequate measure of quality. To estimate aggregate quality improvements, we ran the most current version of the QCT and data dictionaries against all versions of the data submitted between July 2009 and May 2011. We normalized the errors by total number of individual participants (patient or relative) at each registry site. A plot of errors per individual for the epidemiology data from colon sites is shown in figure 3A. A plot of similar data from breast sites is shown in figure 3B. The graphs show a downward trend for errors per individual, often with steep declines, and a few increases coinciding with the implementation of new sets of validations. To asses the significance of this decline in errors per individual, we performed two Wilcoxon signed-rank tests (one for the B-CFR data and one for the C-CFR data) to test if the reduction in validation errors between time points was statistically significant. Reductions in errors per individual for both CFRs were significant, with p=0.031 for the B-CFR data and p=0.047 for the C-CFR data. The variability between individual registry sites is likely due to individual differences in the level of resources and effort available at each site to go back and fix errors. Reducing the number of errors at each of the registry sites is an ongoing effort.
Figure 2.
Total number of baseline epidemiology validations for the Breast and Colon Cancer Family Registries from July 2009 to May 2011. Breast validations rose from a total of 368 to 598 for the 214 data elements in the breast epidemiology baseline table, with most revisions being implemented in May 2010. Approximately two thirds of the validations are logical checks with the remaining one third being allowable values checks with a few format and other types of checks. Colon epidemiology validations rose from a total of 1015 to 1965 for the 625 data elements in the colon epidemiology baseline table with revisions being implemented in May 2010 and May 2011. Approximately half of the validations are logical checks with the rest being allowable values with some format and other types of checks.
Figure 3.
Logarithmic plot of the number of errors and warnings per individual family member from the colon and breast epidemiology baseline tables from July 2009 until May 2011 for each of the sites. (A) Colon data. Of the 1965 epidemiology validations, 278 new validations were added in May 2010 and 726 were added in May 2011. All validations were reviewed and some modified or deleted as well as new ones added. The number of individuals with epidemiology records increased by 3.7% during this time to 42 033. (B) Breast data. Of the 595 epidemiology validations, 158 new validations were added in May 2010 and 71 were added between May 2010 and May 2011. The original 369 validations were all reviewed, and some also modified or deleted. The number of individuals with epidemiology records increased by 3.4% during this time to 36 149.
To examine qualitative aspects of this improvement, we looked at samples of errors generated in 2009 but not in 2011. The earlier data were subject to inconsistent coding and missing values, causing difficulties for end user interpretation. For example, subjects were asked if they had taken a particular class of drugs and if so, they were subsequently asked about specific drugs. Much of this specific information was blank as subjects usually only took one drug, but the correct options were limited to yes, no, not asked, or unknown. Now these previously missing data are consistently and unambiguously coded most often as ‘unknown.’ Other more important changes were observed. For example, one site originally had 20 individuals with a diagnosis of primary amenorrhea (ie, ‘failure of menstrual periods to start naturally’). Rules that checked the age at diagnosis against allowable ranges and/or against the age of first menstruation flagged data that were internally inconsistent and where subjects seemed to have started menstrual periods within a normal time frame. After review at the site, 17 cases had the diagnosis removed from the database.
Discussion
Ensuring data quality is an essential undertaking for any successful data coordination center, yet often among the most difficult aspects to successfully address. This is especially true in research-oriented databases where data types and methods of data collection have evolved over time. The CFR programs have been in operation for 15 years, and have undergone iterative personnel and procedural changes. QC is a shared responsibility between data-generating and data-integrating sites, so data coordinators need to be involved not only during the initial study design and data collection phase, but also as processes, methods, and the databases evolve over time. The progress outlined in this work was only possible because the principal investigators of the CFR sites made the quality assurance review and validation efforts a priority for their scientists and staff, and the NCI funded the GDCIC for its efforts in this area. From a technical perspective, the ability of the CFR sites to verify their data locally, using the QCT before integration to the central database, has been a major factor in our success.
Further improvements in the QCT will be made. False fires and how to best handle valid biological exceptions to a rule remain a challenge. Although designed for this effort, the QC framework will be refactored for used in future projects.
Acknowledgments
The authors would like to thank the members of C-CFR and B-CFR, Sheri Schully and Scott Rogers of the National Cancer Institute, and Andrew Shinohara and Kevin Rosso of ESAC Inc.
Footnotes
Funding: This work was supported by NIH/NCI HHSN261200900010C.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.John EM, Hopper JL, Beck JC, et al. The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res 2004;6:R375–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newcomb PA, Baron J, Cotterchio M, et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomark Prev 2007;16:2331–43 [DOI] [PubMed] [Google Scholar]
- 3.Olson JE. Data Quality: The Accuracy Dimension. San Francisco, CA: Morgan Kaufmann, 2003 [Google Scholar]
- 4.Lee YW, Pipino LL, Funk JD, et al. Journey to Data Quality. Cambridge, MA: MIT Press, 2006 [Google Scholar]