Abstract
Objective
Electronically linked datasets have become an important part of clinical research. Information from multiple sources can be used to identify comorbid conditions and patient outcomes, measure use of healthcare services, and enrich demographic and clinical variables of interest. Innovative approaches for creating research infrastructure beyond a traditional data system are necessary.
Materials and methods
Records from a large healthcare system's enterprise data warehouse (EDW) were linked to a statewide population database, and a master subject index was created. The authors evaluate the linkage, along with the impact of missing information in EDW records and the coverage of the population database. The makeup of the EDW and population database provides a subset of cancer records that exist in both resources, which allows a cancer-specific evaluation of the linkage.
Results
About 3.4 million records (60.8%) in the EDW were linked to the population database with a minimum accuracy of 96.3%. It was estimated that approximately 24.8% of target records were absent from the population database, which enabled the effect of the amount and type of information missing from a record on the linkage to be estimated. However, 99% of the records from the oncology data mart linked; they had fewer missing fields and this correlated positively with the number of patient visits.
Discussion and conclusion
A general-purpose research infrastructure was created which allows disease-specific cohorts to be identified. The usefulness of creating an index between institutions is that it allows each institution to maintain control and confidentiality of their own information.
Keywords: Master subject index, record linking, confidentiality, cancer cohort, population database, informatics, statistics, record linking, master subject index, population database
Background and significance
Considerable benefits are associated with the use of electronically linked datasets for clinical research.1 2 The use of linked datasets allows (1) researchers to examine relationships between variables not available in a single dataset and can provide more complete information on individual patients and populations,2 3 (2) long-term follow-up of patients and supports outcome research,4–8 and (3) the safety, effectiveness, and cost of care to be studied.9–11 For example, cancer registries and patient health records have been linked to vital statistic records, health record datasets, claims and administrative data, quality of life surveys, and outcome and mortality surveys.3 12–20
A collaborative approach among organizations or data resources can fill the gaps in existing data infrastructure.21 Record linking is typically performed to combine resources for investigating a specific disease or for a certain study objective. Many institutions create data warehouses where administrative and clinical data can be viewed together and some communities have expanded the data warehouse model to form research infrastructures that can support data requests for many different projects across institutions.22–24
Record linkage is an intensive process that requires an in-depth knowledge of the source datasets and an understanding of how linkage parameters may influence which records match. It can be time-consuming and costly to use human review. Consequently, studies that contain linkage often use small sets of records that can be verified for correctness or do not evaluate the linkage for accuracy or the introduction of bias. In addition to the technical issues associated with linking data, the creation of data infrastructure requires negotiation and agreement between data sources. Different types of organizations may have varying interests in research, and concerns about privacy may lead to policies that prevent records from being easily linked.21
This paper describes and evaluates the linkage between the Utah Population Database (UPDB) and the enterprise data warehouse (EDW) maintained by Intermountain Healthcare; this presents an example of a data infrastructure that extends beyond traditional data systems. We describe a solution for record linkage and long-term access to data that addresses the confidentiality of persons identified in the records and the concerns of the data contributors. This infrastructure contains records for patients of all age groups that had either inpatient or outpatients encounters. It represents a heterogeneous patient population that can be sampled for patients of interest as well as an appropriate control population. Individuals can be studied for conditions occurring before a specific diagnosis and treatment as well as end points such as recurrence and survival. The presence of the cancer records in both data sources allows analyses of the effect of data completeness in EDW records, the impact of records not contained in the population database, and further evaluation of the unlinked records.
Materials and methods
A number of regulatory activities were required to accomplish this project. An agreement between the University of Utah and Intermountain Healthcare was created which included the standard legal recitals. This allowed demographic information on Intermountain patients to be provided as a temporary file to the Resource for Genetic and Epidemiologic Research, which is the governance body of the UPDB.25 The project was approved by institutional review boards at the University of Utah and Intermountain Healthcare.
Our methodology allows the record linking activity to be completed using patient demographic information without exposing any medical information. After the linking is complete, a master subject index (MSI) is created, the identifying information of the EDW is deleted, and a copy of the MSI is held by each institution to facilitate future projects. Thus the linkage does not create a new combined database. The MSI allows each institution to maintain control of their information and protects the confidentiality of the individuals within each institution. When research projects request use of the new research infrastructure, the investigator will be required to obtain approval from Resource for Genetic and Epidemiologic Research and the institutional review boards from each institution, and it is only at this point that information from both institutions is accessed and combined.
For the purposes of this paper, the term ‘record’ refers to demographic information about a person that may or may not contain additional information (health information, family relationships, etc). In this way, the number of records reported is analogous to the number of distinct people those records represent. When additional information is mentioned, such as the amount of health information contained in the EDW, it is referred to as ‘information’ or ‘data’, not as a record.
Utah Population Database
The UPDB is a research resource held at the University of Utah that contains demographic and family history information linked to medical information.25 26 It was created in the mid-1970s from genealogy records and included data from the Utah Cancer Registry (UCR) and Utah death certificates. Since the mid-1990s, the UPDB has been expanded to incorporate other high-quality, statewide datasets; these include driver license, vital records from the State of Utah (birth, marriage, divorce, and fetal deaths), cancer data from the Cancer Data Registry of Idaho, and information from inpatient hospital discharges in Utah. The source data are internally linked within the UPDB and result in information for over 6 million distinct individuals. Because of its size and the varied sources of its information, most families living in Utah are represented. Use of this resource has been instrumental in the discovery of human disease genes,27–31 familial risk associated with heritable diseases,32–36 and quantification of other disease risk factors.37–39
Intermountain Healthcare EDW
Intermountain Healthcare is the largest healthcare system in Utah and operates multiple hospitals, outpatient clinics, ambulatory surgery centers, laboratories, and health insurance plans. It is a not-for-profit healthcare delivery system covering Utah and southeastern Idaho. In addition to tertiary-level teaching and research facilities, Intermountain also has several small hospitals and clinics that are the only source of care in some rural Utah communities. The Intermountain Healthcare EDW was created to bring together both health and administrative data from all facilities to allow researchers to study patient care from both an individual and a population perspective. Patients in the EDW are identified by an enterprise master patient index, which is used to link data resulting from all patient encounters. There are regular audits of enterprise master patient index numbers that look for duplicates and inaccuracy across systems. There are more than five million patients listed in the EDW connected to more than 35 billion health-related data points, such as laboratory results, discharge summaries, and diagnosis codes.
Information for linkage and evaluation
For the general linkage, demographic fields from the UPDB available for use in record linking include: full name (including maiden name), sex, birth date, multiplicity (to identify twins and other multiple births), death date, social security number, and residential history (street address, city, state, and zip codes). The familial structure in the UPDB also makes available names, social security number, and residential history of parents, siblings, and spouses. The EDW has similar demographic fields available for linkage to those listed for the UPDB. The EDW also contains phone number fields. However, there is no familial information available.
For the validation analysis, UCR data from UPDB were available. The UCR is a statewide, population-based registry containing diagnoses since 1966 and has been a member of the National Cancer Institute's Surveillance, Epidemiology, and End Results (SEER) Program since 1973. The UCR tracks incident cases of cancer diagnosed or treated in Utah. The UCR provides annual updates of invasive and in situ cancer cases to UPDB. This study used 213 828 UCR records through 2005.
The EDW contains data marts where information is consolidated for specific clinical programs. The oncology data mart was used to select patients with bone marrow, breast, or prostate cancer for the validation analysis. Records were included for individuals with a matching diagnosis code from the International Classification of Diseases for Oncology (ICD-O). In addition to the demographic fields, some clinical data were available from the data mart for this study. This included cancer diagnosis as an ICD-O code, with morphology and histology, date of diagnosis, the number and date of visits, and which facilities were visited. There were 25 797 records on oncology patients with diagnosis dates from about 1993 through 2005 that were identified and used to evaluate record linking.
General linkage
The Pedigree and Population Resource at the Huntsman Cancer Institute University of Utah (University) maintains the UPDB and is responsible for linking resources to the UPDB. The University has extensive experience with linking diverse datasets25 40 and currently uses the commercial software package IBM InfoSphere QualityStage, a program based on Jaro's probabilistic AUTOMATCH algorithm.41 QualityStage allows users to input parameters for which algorithms should be used to compare different data types, such as dates, addresses, and text strings and tolerance levels for different values. In addition, the University uses a set of programs developed in-house to check for common issues known to produce false links, such as twins or siblings with similar names, described in the online appendix. Candidate links identified by the validation programs as needing special attention are reviewed. Finally, records identified as valid matches, compiled from QualityStage based on scores, validation programs, and human review, are recorded in the MSI. The MSI includes a unique UPDB ID number, UPDB pedigree quality indicator (described below), and an Intermountain Healthcare ID number.
Records are analyzed for completeness and demographic trends in an effort to understand the characteristics of unlinked records. Records are considered ‘minimally complete’ if they contain a first name, last name, birth date, and social security number. In addition, the number of fields missing values is calculated using these additional fields: a middle name, maiden name, address, and death date.
The family structure that is available in UPDB was used to calculate the depth of the pedigree for each linked EDW record. These relationships are measured in ‘pedigree quality’—an indication of how useful a record is for genetic and familial analysis. Records that linked to UPDB were assigned one of the following levels of pedigree quality:
no family relationships
parent–child set or siblings with parents who had only name information
two-generation family with four or more members
multi-generational pedigree with three or more generations. Some pedigrees have as many as 11 generations.
Linkage evaluation
To evaluate the accuracy of the UPDB–EDW linkage and test the generalizabilty of the resource to support disease-specific projects, we focused on a subset of cancer records identified in both resources. We selected three cancer types—bone marrow, breast, and prostate—in order to evaluate linkage among diagnoses that affect people across a range of demographic characteristics. Records for patients who had multiple same-site cancers or a diagnosis more recent than 2005 were excluded from the analysis. Selected ICD-O codes were used in this comparison: C42.1 for bone marrow cancer, C50.0–C50.9 for breast cancer, and C61.9 for prostate cancer. Because all cancer cases documented in the EDW should be reported to UCR, it was anticipated that target records would exist in the UPDB. With this assumption, EDW records were classified into one of the three following groups.
Group 1: records in the EDW oncology data mart that were linked to the UPDB and had a corresponding UCR record. For this analysis, our measure of accuracy is defined as the proportion of records in this group.
Group 2: records in the EDW oncology data mart that were linked to the UPDB, but did not have a corresponding UCR record or had different cancer diagnosis in UCR.
Group 3: records in the EDW oncology data mart that were not linked to the UPDB.
Linkage rate prediction
For the subset of records from the oncology data mart, a logistic regression model was used to determine characteristics in the quality of records that influenced linkage probability. The factors used to train the model were:
whether a field was missing a value (for each field);
the relative frequency of a value compared with all other values in the field (for each field);
a patient's connection to Utah (either by having Utah listed as state of residence or a social security number indicative of Utah birth);
the cancer type (bone marrow, breast, or prostate);
sex;
having a temporary social security number (usually denoting recent immigration);
being identified as Hispanic (identified from UPDB).
The resulting model was run on the full set of EDW records to estimate the expected linkage rate assuming that a matching record existed in the UPDB. The expected linkage rate was compared with the percentage of records that actually linked. In addition, the expected linkage rate prediction scores were analyzed by state of residence and age, two factors found in preliminary studies to be of interest.40
Results
General linkage
The UPDB–EDW linkage resulted in 3 429 337 (60.8%) records out of 5 636 907 EDW records linking to a UPDB record. Some EDW records mapped to the same UPDB record and were flagged as potential duplicate records. These included 445 113 EDW records mapped to 217 087 UPDB records. This duplicate record rate is consistent with previous findings.42
Records in the EDW had on average 1.9 fields with missing values. Table 1 shows a comparison of linkage rates for all EDW records, minimally complete EDW records (those with at least first name, last name, birth date, and social security number), and generally for records missing values. Minimally complete records linked at 75.4%. Virtually all records (99.3%) that had values for every field could be linked compared with 80.7% for records missing values in any two fields and 29.5% for those missing values in any four fields.
Table 1.
Rate of linkage of EDW records to Utah Population Database by completeness of record
| Number of records | Linkage rate (%) | |
| All EDW records | 5 636 907 | 60.8 |
| Minimally complete EDW records | 3 385 716 | 75.4 |
| Records without any missing values | 13 489 | 99.3 |
| Records missing 1 value | 375 915 | 95.6 |
| Records missing 2 values | 2 017 797 | 80.7 |
| Records missing 3 values | 1 929 683 | 57.2 |
| Records missing 4 values | 1 062 850 | 29.5 |
| Records missing 5 values | 166 115 | 3.6 |
| Records missing 6 or more values | 71 058 | 0.0 |
| Average number of missing values per record | 1.9 |
EDW, enterprise data warehouse.
A comparison was made between the percentage of records missing values in particular fields in linked and unlinked records. All records in the EDW had values for sex and birth date. Only 5705 records were missing values for first name, and only 150 records were missing values for last name. A total of 36.3% of records were missing social security number; for linked records, 24.5% were missing this value, and 60.8% for unlinked records. Street address was more complete, with 8.7% missing; for linked records, it was 2.0%, and 10.7% for unlinked.
EDW records of Utah residents were more likely to link to UPDB than non-residents, with 75.6% of 4 099 565 records of Utah residents linking compared with 21.6% of 1 182 671 records of non-Utah residents. Records where the current state of residence was not known linked at 19.7%. Records of persons with Utah connections (as defined above) were also much more likely to link to the UPDB, with 75.2% out of 4 226 734 records linking.
Linkage evaluation
The evaluation analysis focused on the subset of cancer records in both sources. The mean age for these patients with bone marrow cancer was 49.1 compared with 60.2 and 68.6 for patients with breast and prostate cancer, respectively. In addition, almost 25% of patients with bone marrow records received a diagnosis before the age of 18, a patient population with virtually no cases of breast or prostate cancer.
The average linkage rate for the cancer records was 99.0%. A comparison of linkage rates between the cancer types by completeness of records is provided in table 2. The number of missing fields between linked and unlinked records for the cancer records and which fields were missing values followed the same trend as the overall linkage. These records had on average 0.9 fields missing values compared with 1.9 for all EDW records. The low number of fields missing values could be attributed to the high number of visits (34.7 average) and facilities visited (2.7 average) per cancer patient. Similarly, records for persons with Utah connections linked at a higher rate (99.2%) than persons without Utah connections (96.1%) for the cancer records.
Table 2.
Linkage rate of EDW oncology data mart records by completeness
| Bone marrow (%) | Breast (%) | Prostate (%) | |
| Number | 2632 | 11 334 | 11 831 |
| % Linked records | 97.2 | 99.3 | 99.1 |
| Minimally complete records | 98.8 | 99.5 | 99.2 |
| Records without any missing values | 100.0 | 99.9 | 100.0 |
| Records missing 1 value | 99.8 | 99.8 | 99.6 |
| Records missing 2 values | 97.8 | 99.3 | 99.5 |
| Records missing 3 values | 93.8 | 96.0 | 96.0 |
| Records missing 4 values | 70.7 | 82.1 | 57.9 |
| Records missing 5 values | 0.0 | 0.0 | N/A |
| Records missing 6 or more values | N/A | N/A | N/A |
| Average number of missing fields | 1.2 | 0.6 | 1.2 |
EDW, enterprise data warehouse; N/A, no records.
Prostate and breast cancer records linked at a higher rate than bone marrow cancer records. Although there are a number of possible reasons for the lack of UCR records, the higher rate of failed links for bone marrow cases may be attributable to the known under-reporting of bone marrow cancers, as not all cases undergo pathologic testing (via communication with UCR).
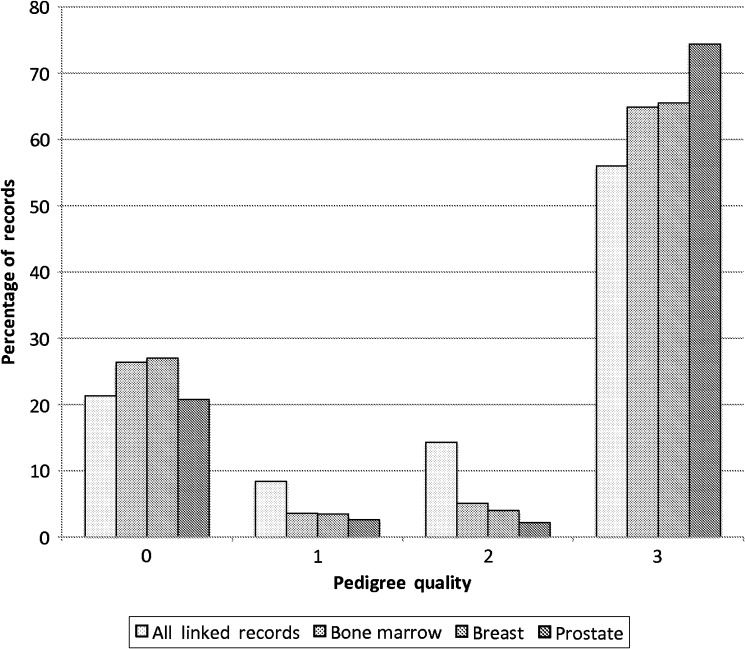
The pedigree quality for all linked records and for each cancer type is shown in figure 1. Of the records that linked to UPDB, over half linked to a multi-generational family (pedigree quality 3). While cancer groups have a relatively high level of familial information compared with all linked patient records, patients with prostate cancer have the highest proportion with deep pedigrees and are more likely to represent earlier birth cohorts.
Figure 1.
Proportion of all linked records and cancer type by pedigree quality.
The percentage of cancer records in each of the linking groups is shown in table 3. Of the cancer records, 96.3% had a corresponding UCR record and were assigned to group 1. The records in group 1 are considered true positives and provide the lower-bound accuracy of the linkage method. The positive predictive value was 97.2%, and the sensitivity was 99.0%. The majority of patients in group 2 linked to UPDB, but did not link to a cancer record in UCR. False positives (incorrect links) would appear in group 2, and false negatives (missed linked) would appear in group 3. Groups 2 and 3 could include a variety of possibilities: (a) patients who are non-residents of Utah and were diagnosed as having cancer outside of Utah; (b) diagnoses in the data mart that do not meet SEER reporting guidelines; (c) UCR policies that exclude some data from routine UPDB linkage. These possibilities suggest that some records in groups 2 and 3 are not mistakes in the linkage and that the number of records in group 1 provides the minimum accuracy of the linkage.
Table 3.
Assigned linkage group for oncology data mart records
| Bone marrow (%) | Breast (%) | Prostate (%) | Combined (%) | |
| Group 1 | 89.7 | 96.9 | 97.1 | 96.3 |
| Group 2 | 7.4 | 2.4 | 2.0 | 2.7 |
| Group 3 | 2.8 | 0.7 | 0.9 | 1.0 |
Linkage rate prediction
Although the oncology data mart analysis provided an estimate of accuracy, it was based on accuracy given that the target records existed in the UPDB. A regression model was fitted on this subset and, when run on all EDW records, provided an estimate of the coverage of the UPDB. The regression model identified the significant factors influencing linkage as having a social security number, a middle name, a maiden name, a death date, and a connection to Utah. Odds ratio (OR), 95% confidence interval (CI), and p value are shown in table 4. The regression model predicted that 80.7% of all non-cancer records in the EDW and 80.8% of all EDW records would link when the target records existed in the UPDB. The predicted linkage rate was 91.1% for Utah residents and 50.1% for non-Utah residents.
Table 4.
Significant factors affecting linkage based on the regression model
| Factor | OR (95% CI) | p Value |
| Presence of social security number | 7.7423 (5.9528 to 10.0698) | <2e–16 |
| Presence of middle name | 3.1528 (2.6075 to 3.8123) | <2e–16 |
| Presence of maiden name | 1.5606 (1.2094 to 2.0139) | 0.000625 |
| Presence of death date | 3.3029 (2.3758 to 4.5918) | 1.17e–12 |
| Having a Utah connection | 6.3815 (5.2210 to 7.7998) | <2e–16 |
Since the general linkage only produced links with 60.8% of the EDW records (table 1), an estimated 24.8% (1 – (60.8/80.8)) of the individuals in the EDW do not have records in the UPDB. For Utah residents, 17.0% (1 – (75.6/91.1)) of individuals and for non-Utah residents about 56.9% (1 – (21.6/50.1)) of individuals in the EDW do not exist in the UPDB. This is consistent with the statement that the majority of Utah residents have records in the UPDB. The high number of non-Utah residents with records in the UPDB (43.1%) suggests that many persons treated at Intermountain Healthcare facilities have Utah connections.
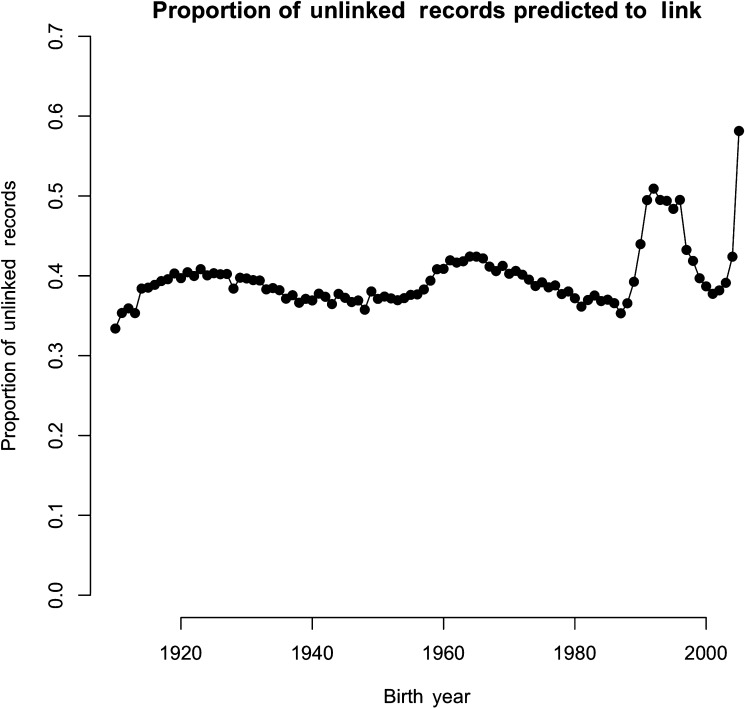
Figure 2 shows the proportion of all records for each birth year that did not link, but had a very high predicted probability of linkage (>90%). These records represent persons most likely to be missing a target record in the UPDB. Because the UPDB relies on source datasets for information, a person may not have a record in the UPDB until they have an event in Utah that triggers the creation of a record. This includes being born, obtaining a driver license, getting married or divorced, being hospitalized, having children, having a cancer diagnosis, or dying. Thus the UPDB will not include records of children born outside Utah and persons who have recently moved to Utah until they get a driver license or have some other event that creates a record. A higher proportion of these records are for persons born from 1989 to 1998, ages 7–16 as calculated from the cancer record update as of 2005. This also explains the higher proportion of patients with bone marrow cancer classified in group 3 (table 3).
Figure 2.
Proportion of unlinked records with predicted probability of linkage greater than 90% by birth year.
Discussion
The diversity of information available through the UPDB from its various source records adds considerable value to the more than 3.4 million records in the EDW that linked. As more than half of the linked records have multi-generational family information, the linked records provide the power to detect and localize genetic traits.43 Since 78.7% of all linked records have at least some family information, parent–child and sibling pairs can be analyzed when varying amounts of pedigree information is available.44 45 The use of this data infrastructure and the MSI was recently demonstrated in a study of inflammatory bowel disease in Utah kindreds.46
The question remains why did the remaining 2.2 million not link? This can be answered in two parts. First, the analysis of the general linkage indicates that the completeness of EDW records affected the ability to match to records in the UPDB. This reinforces the importance of providers reviewing patient demographic information for changes and additions at each encounter. The more information a record had, the more likely it was to link; however, which fields were missing values also affected linkage. The significant factors identified in the regression model also support the idea that certain demographic fields are more valuable than others.
Second, the linked repositories cover slightly different geographic areas. In this study having a Utah connection affected which records linked. The UPDB primarily represents Utah residents, and persons without a Utah connection have a much smaller chance of having a record in the UPDB. In addition to Utah, Intermountain Healthcare provides coverage to southeastern Idaho and serves as a referral center for the neighboring states. Thus not all patients would have a Utah connection. A person who temporarily resides in Utah or who recently moved to the state will not be present in the UPDB.
The fact that certain demographic values are more informative in linkage can help guide policy decisions. For example, privacy concerns have fostered a trend for healthcare institutions to not require social security number. This study suggests that social security number is one of the most informative fields and that the decision not to collect it reduces the ability to correctly identify which records belong to which patient. Where privacy concerns or other policy decisions drive which information is collected, it is important to ensure that other fields always have correct, up-to-date values. In addition, collecting other demographic, but possibly less sensitive, information may mitigate the inability of records to link.
The subset of cancer records allowed the accuracy of the linkage to be evaluated. Cancer records had fewer missing values than the general linkage on average, and the number of missing fields in those records correlated positively with the number of visits and the number of facilities visited. This underlines the advantage of using well-defined clinical information that includes carefully collected data, such as data marts.
Conclusion
This project provides a blueprint for the linkage of records from an EDW with a population database to create a large general-purpose resource useful for clinical research, epidemiological risk studies, familial investigation, short or long term follow-up, and case–control studies. The creation of a MSI is beyond the scope of a single investigator or research team. It would take a skilled researcher months or years to acquire, link, and extract meaningful information from a myriad of secondary datasets.21 Such an effort requires collaboration across institutions. The usefulness of creating a MSI between institutions should be weighed along with concerns of confidentiality.
Acknowledgments
We wish to acknowledge Solange Gomes and Anne Zeller for their valuable record-linking skills. David E Avrin, Matthew H Samore, and Richard A Kerber provided graduate committee oversight. Timothy N Trautman reviewed and edited the manuscript.
Footnotes
Funding: Funds for this work were provided by training grant No LM007124-11 from the National Library of Medicine and Robert Wood Johnson Foundation. This project was sponsored by the Huntsman Intermountain Cancer Control Program. Partial support for all datasets within the Utah Population Database (UPDB) was provided by the University of Utah Huntsman Cancer Institute and the Huntsman Cancer Institute Cancer Center Support grant, P30 CA42014 from National Cancer Institute. Support for the Utah Cancer Registry is provided by Contract No HHSN 261201000026C from the National Cancer Institute with additional support from the Utah Department of Health and the University of Utah. Support for this project was also provided by the Division of Genetic Epidemiology in the Department of Biomedical Informatics University of Utah. This work was supported using resources and facilities at the VA Salt Lake City Health Care System with funding support from the VA Informatics and Computing Infrastructure (VINCI), VA HSR HIR 08-204, and the Consortium for Healthcare Informatics Research (CHIR), VA HSR HIR 08-374.
Competing interests: None.
Ethics approval: This study was approved by University of Utah and Intermountain Healthcare.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Goldacre M. Benefits of Linking Data: An International Perspective. Data Symposium, Link and Multiply: the Benefits of Data Linkage. The Sax Institute and the Centre of Health Record Linkage Management, Sydney, Australia, 27 July 2006 [Google Scholar]
- 2.Brooks JM, Chrischilles E, Scott S, et al. Information gained from linking SEER Cancer Registry Data to state-level hospital discharge abstracts. Surveillance, Epidemiology, and End Results. Med Care 2000;38:1131–40 [DOI] [PubMed] [Google Scholar]
- 3.Coté TR, Manns A, Hardy CR, et al. Epidemiology of brain lymphoma among people with or without acquired immunodeficiency syndrome. AIDS/Cancer Study Group. J Natl Cancer Inst 1996;88:675–9 [DOI] [PubMed] [Google Scholar]
- 4.Melnikow J, McGahan C, Sawaya GF, et al. Cervical intraepithelial neoplasia outcomes after treatment: long-term follow-up from the British Columbia Cohort Study. J Natl Cancer Inst 2009;101:721–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Travis LB, Fosså SD, Schonfeld SJ, et al. Second cancers among 40,576 testicular cancer patients: focus on long-term survivors. J Natl Cancer Inst 2005;97:1354–65 [DOI] [PubMed] [Google Scholar]
- 6.Ejlertsen B, Jensen MB, Rank F, et al. ; Danish Breast Cancer Cooperative Group Population-based study of peritumoral lymphovascular invasion and outcome among patients with operable breast cancer. J Natl Cancer Inst 2009;101:729–35 [DOI] [PubMed] [Google Scholar]
- 7.Hisada M, Chen BE, Jaffe ES, et al. Second cancer incidence and cause-specific mortality among 3104 patients with hairy cell leukemia: a population-based study. J Natl Cancer Inst 2007;99:215–22 [DOI] [PubMed] [Google Scholar]
- 8.Steyerberg EW, Neville BA, Koppert LB, et al. Surgical mortality in patients with esophageal cancer: development and validation of a simple risk score. J Clin Oncol 2006;24:4277–84 [DOI] [PubMed] [Google Scholar]
- 9.Yabroff KR, Davis WW, Lamont EB, et al. Patient time costs associated with cancer care. J Natl Cancer Inst 2007;99:14–23 [DOI] [PubMed] [Google Scholar]
- 10.Edwards BK, Brown ML, Wingo PA, et al. Annual report to the nation on the status of cancer, 1975-2002, featuring population-based trends in cancer treatment. J Natl Cancer Inst 2005;97:1407–27 [DOI] [PubMed] [Google Scholar]
- 11.Chang MH, You SL, Chen CJ, et al. ; the Taiwan Hepatoma Study Group Decreased incidence of hepatocellular carcinoma in hepatitis B vaccinees: a 20-year follow-up study. J Natl Cancer Inst 2009;101:1348–55 [DOI] [PubMed] [Google Scholar]
- 12.Ambs A, Warren JL, Bellizzi KM, et al. Overview of the SEER-Medicare Health Outcomes Survey linked dataset. Health Care Financ Rev 2008;29:5–21 [PMC free article] [PubMed] [Google Scholar]
- 13.Earle CC, Nattinger AB, Potosky AL, et al. Identifying cancer relapse using SEER-Medicare data. Med Care 2002;40:IV-75–81 [DOI] [PubMed] [Google Scholar]
- 14.Bluhm E, McNeil DE, Cnattingius S, et al. Prenatal and perinatal risk factors for neuroblastoma. Int J Cancer 2008;123:2885–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winther JF, Boice JD, Christensen J, et al. Hopsitalizations among children of survivors of childhood and adolescent cancer: a population-based cohort study. Int J Cancer 2010;127:2879–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clegg LX, Reichman ME, Miller BA, et al. Impact of socioeconomic status on cancer incidence and stage at diagnosis: selected findings from the surveillance, epidemiology, and end results: National Longitudinal Mortality Study. Cancer Causes Control 2009;20:417–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reeve BB, Potosky AL, Smith AW, et al. Impact of cancer on health-related quality of life of older Americans. J Natl Cancer Inst 2009;101:860–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doebbeling BN, Wyant DK, McCoy KD, et al. Linked insurance-tumor registry database for health services research. Med Care 1999;37:1105–15 [DOI] [PubMed] [Google Scholar]
- 19.Beelen R, Hoek G, van den Brandt PA, et al. Long-term exposure to traffic-related air pollution and lung cancer risk. Epidemiology 2008;19:702–10 [DOI] [PubMed] [Google Scholar]
- 20.Rankin J, Silf KA, Pearce MS, et al. Congenital anomaly and childhood cancer: a population-based, record linkage study. Pediatr Blood Cancer 2008;51:608–12 [DOI] [PubMed] [Google Scholar]
- 21.Bradley CJ, Penberthy L, Devers KJ, et al. Health services research and data linkages: issues, methods, and directions for the future. Heath Serv Res 2010;45:1468–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyons RA, Jones KH, John G, et al. The SAIL databank: linking multiple health and social care datasets. BMC Med Inform Decis Mak 2009;9:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cameron CM, Purdie DM, Kliewer EV, et al. Population health and clinical data linkage: the importance of a population registry. Aust N Z J Public Health 2007;31:459–63 [DOI] [PubMed] [Google Scholar]
- 24.McDonald CJ, Overhage JM, Barnes M, et al. ; INPC Management Committee The Indiana network for patient care: a working local health information infrastructure. An example of a working infrastructure collaboration that links data from five health systems and hundreds of millions of entries. Health Aff (Millwood) 2005;24:1214–20 [DOI] [PubMed] [Google Scholar]
- 25.Wylie JE, Mineau GP. Biomedical databases: protecting privacy and promoting research. Trends Biotechnol 2003;21:113–16 [DOI] [PubMed] [Google Scholar]
- 26.Skolnick M, Bean L, Dintelman S, et al. A computerized family history database system. Sociol Soc Res 1979;63:506–23 [Google Scholar]
- 27.Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994;266:66–71 [DOI] [PubMed] [Google Scholar]
- 28.Wooster R, Neuhausen SL, Mangion J, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science 1994;265:2088–90 [DOI] [PubMed] [Google Scholar]
- 29.Cannon-Albright LA, Goldgar DE, Meyer LJ, et al. Assignment of a locus for familial melanoma, MLM, to chromosome 9p13-p22. Science 1992;258:1148–52 [DOI] [PubMed] [Google Scholar]
- 30.Groden J, Thliveris A, Samowitz W, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell 1991;66:589–600 [DOI] [PubMed] [Google Scholar]
- 31.Camp NJ, Farnham JM, Allen-Brady K, et al. Statistical recombinant mapping in extended high-risk Utah pedigrees narrows the 8q24 prostate cancer locus to 2.0 Mb. Prostate 2007;67:1456–64 [DOI] [PubMed] [Google Scholar]
- 32.Allen-Brady K, Camp NJ, Ward JH, et al. Lobular breast cancer: excess familiality observed in the Utah Population Database. Int J Cancer 2005;117:665–1 [DOI] [PubMed] [Google Scholar]
- 33.Weires MB, Tausch B, Haug PJ, et al. Familiality of diabetes mellitus. Exp Clin Endocrinol Diabetes 2007;115:634–40 [DOI] [PubMed] [Google Scholar]
- 34.Neklason DW, Thorpe BL, Ferrandez A, et al. Colonic adenoma risk in familial colorectal cancer–a study of six extended kindreds. Am J Gastroenterol 2008;103:2577–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerber RA, O'Brien E. A cohort study of cancer risk in relation to family histories of cancer in the Utah population database. Cancer 2005;103:1906–15 [DOI] [PubMed] [Google Scholar]
- 36.Luo L, Harmon J, Yang X, et al. Familial aggregation of age-related macular degeneration in the Utah population. Vision Res 2008;48:494–500 [DOI] [PubMed] [Google Scholar]
- 37.Smith KS, Mineau GP, Garibotti G, et al. Effects of childhood and middle-adulthood family conditions on later-life mortality: evidence from the Utah Population Database, 1850–2002. Soc Sci Med 2009;68:1649–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith KR, Brown BB, Yamada I, et al. Walkability and body mass index density, design, and new diversity measures. Am J Prev Med 2008;35:237–44 [DOI] [PubMed] [Google Scholar]
- 39.Esplin MS, O'Brien E, Fraser A, et al. Estimating recurrence of spontaneous preterm delivery. Obstet Gynecol 2008;112:516–23 [DOI] [PubMed] [Google Scholar]
- 40.DuVall SL, Fraser AM, Kerber RA, et al. The impact of a growing minority population on identification of duplicate records in an enterprise data warehouse. Stud Health Technol Inform 2010;160:1122–6 [PubMed] [Google Scholar]
- 41.Jaro MA. Probabilistic linkage of large public health data files. Stat Med 1995;14:491–8 [DOI] [PubMed] [Google Scholar]
- 42.Thornton SN, Hood SK. Reducing duplicate patient creation using a probabilistic matching algorithm in an open-access community data sharing environment. AMIA Annu Symp Proc 2005:1135. [PMC free article] [PubMed] [Google Scholar]
- 43.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 1998;62:1198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Badner JA, Gershon ES, Goldin LR. Optimal ascertainment strategies to detect linkage to common disease alleles. Am J Hum Genet 1998;63:880–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abecasis GR, Cardo LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet 2000;66:279–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guthery SL, Mineau G, Pimentel RP, et al. Inflammatory bowel disease aggregation in Utah kindreds. Inflamm Bowel Dis 2011;17:823–30 [DOI] [PubMed] [Google Scholar]