Abstract
The Genosoja consortium is an initiative to integrate different omics research approaches carried out in Brazil. Basically, the aim of the project is to improve the plant by identifying genes involved in responses against stresses that affect domestic production, like drought stress and Asian Rust fungal disease. To do so, the project generated several types of sequence data using different methodologies, most of them sequenced by next generation sequencers. The initial stage of the project is highly dependent on bioinformatics analysis, providing suitable tools and integrated databases. In this work, we describe the main features of the Genosoja web database, including the pipelines to analyze some kinds of data (ESTs, SuperSAGE, microRNAs, subtractive cDNA libraries), as well as web interfaces to access information about soybean gene annotation and expression.
Keywords: bioinformatics, database, gene expression, soybean, Genosoja
Introduction
Soybean is a legume of great economic importance in the international market, with a world production of almost 260 million tons for the 2009/2010 harvest. Brazil appears as the world’s second largest producer, with a share of about 25%, and the crop is responsible for 10% of the country’s total exports.
In recent years, the Brazilian soybean crop has been constantly threatened by climatic constraints (especially long periods of drought) and some attacks by pathogens - such as Asian Rust disease (Yorinori et al., 2005), resulting in millions in losses for producers. Such constraints increase the importance of the breeding programs, established to discover new techniques for planting and prevention, increase production and lower costs. High-throughput sequencing technologies (like 454 pyrosequencing, Illumina/Solexa and ABI/SOLiD) represent significant advantages in these areas by producing millions of reads that can be used to measure levels of gene expression, allowing the identification of new genes or novel splice variants. On the other hand, it is necessary to intensify efforts by bioinformatics groups to develop new pipelines and data integration.
To this end, the Brazilian Soybean Genome Consortium (Genosoja) was established in 2007 with the goal of integrating several institutions currently working with soybean genomics in Brazil. The project promotes the search for solutions regarding possible treats, and to improve the soybean production process, emphasizing stresses that affect domestic production, like the occurrence of droughts, and pathogen attacks such as Asian Rust disease.
Among the main objectives of the Genosoja consortium is the creation of a relational database, integrating the results achieved by different methodologies and research groups working in the project. Despite the existence of other integrated soybean databases, such as SoyXpress (Cheng and Stromvik, 2008) and the “Soybean full-length cDNA database” (Umezava et al., 2008), as well as free pipelines available for data integration, such as Distributed Annotation System (DAS) (Dowell et al., 2001; Jenkinson et al., 2008) none of them integrates data from multiple experiments or provides transcriptome data from high-throughput sequencing technologies like the database described in this work.
In light of this context, we created a soybean database connecting public soybean data (like ESTs and genomic sequences) and project data (like SuperSAGE tags, microRNAs and subtractive cDNA libraries). This database offers search tools for users, including keyword searches, statistical comparisons, automatic annotation (using some protein databases such as NR, Uniref, KEGG and Pfam), gene ontology classification and gene expression profiles under several conditions. Moreover, searches by sequence homology are possible using the local BLAST. All data are stored in a Fedora Linux machine, running the MySQL database server. The web interfaces (http://www.lge.ibi.unicamp.br/soybean) are based on a combination of CGI scripts using Perl language (including BioPerl module) and the Apache Web Server. As soon as the private data are published, the database will be freely available.
Methods, Results and Discussion
Public soybean data
In order to construct the Genosoja database we first collected all soybean data available at public biology databases. The genome of the cultivar Williams 82 and their predicted genes (66,153 sequences) were downloaded from the Phytozome (Schmutz et al., 2010). One full-length cDNA library from the Japanese cultivar Nourin2 was downloaded from the “Soybean full-length cDNA database”. From NCBI (National Center for Biotechnology Information) we obtained 1,276,813 EST sequences (sequenced by SANGER and pyrosequencing technologies) and their equivalent GenBank files. All sequences were renamed in accordance to libraries, tissues and cultivars. This information was extracted from the GenBank files using homemade PERL scripts (Supplementary Material Figure S1). The bdtrimmer software (Baudet and Dias, 2005) was used to exclude ribosomal, vector, low quality and short (less than 100 bp) sequences. The EST assembly process was divided into two steps: (1) the ESTs were mapped into the soybean genome using the BLASTn algorithm (Altschul et al., 1997) (e-value cutoff of 1e−10) and (2) all reads that aligned in same region of the reference were assembled together using the CAP3 program (Huang and Madan, 1999). The final result consists of 60,747 unigenes (30,809 contigs and 29,938 singlets). The effort to obtain the unigenes from assembled ESTs was important to increase the databases with information on untranslated regions (UTR), alternative splicing variants and gene expression profiling.
The Autofact program (Koski et al., 2005) was used to perform an automatic annotation of the predicted genes and the assembled unigenes. The main contribution of Autofact is the capacity to resume the annotation based on sequence similarity searches in several databases. For this, we used the BLASTx procedure (e-value cutoff of 1e−5) to align the genes against certain protein databases, including: non-redundant (NR) database of NCBI, swissprot - databases containing only manually curated proteins (Suzek et al., 2007), uniref90 and uniref100 - databases containing clustered sets of proteins from UniProt, Pfam - a database of protein families (Bateman et al., 2002) and KEGG - a database of metabolic pathways (Kanehisa and Goto, 2000). The Autofact pipeline assigned function to 85% of the protein dataset. Figure 1 shows the complete pipeline of the public soybean data analysis.
Figure 1.
Complete pipeline of the public soybean data analysis. We found many occurrences of vector and poly A/T sequences in the NCBI ESTs. After trimming, a reference assembly was performed using 1,101,986 sequences. Moreover, the predicted Williams 82 genes (66,153 sequences) and the assembled unigenes (60,747 sequences) were automatically annotated using the AutoFACT pipeline based on certain BLASTx results against several protein databases.
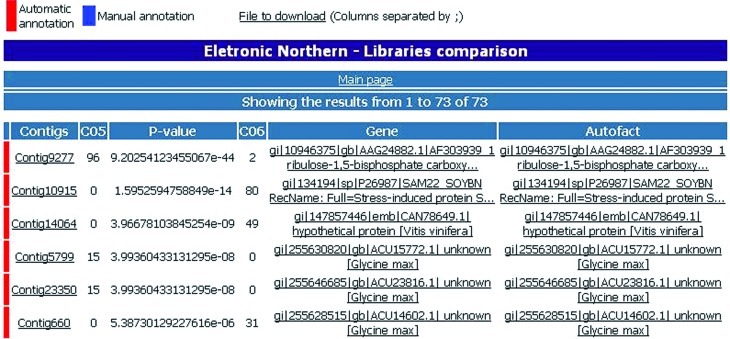
Using the description of the origin of the ESTs (tissues and conditions), normalization procedures and statistical data analysis (Audic and Claverie, 1997), it was possible to infer differential gene expression among the assembled unigenes. This approach, called Electronic Northern, allows the users to compare gene expression profiles between two or more libraries and the results are available through a web interface (Figure 2).
Figure 2.
Electronic northern interface. With this tool it is possible to infer gene expression using an assembly of ESTs. A statistical test (p-value) is performed in real time when comparing two libraries. The description of the libraries and tissue ESTs were obtained from a GenBank sequences file using a specifically made PERL script (Supplementary Material Figure S1). Furthermore, a file with the results shown in the interface is available for download.
Finally, the users can perform keyword and BLAST searches directly from the EST reads using the Gene Projects software (Carazzolle et al., 2007). This software also allows the user to perform assembly and annotation in these reads in an effort to improve unigene assembly. After generating a login/password it is possible to work on specific projects which users can develop and organize thematically by adding sequences to the assembly. After the assembly it is possible to view and to edit the results, improving the quality of the contigs.
Solexa SuperSAGE data
The Genosoja project generated three libraries using SuperSAGE methodology and these were sequenced by Illumina/Solexa technology. One library was constructed exploring gene expression in plants (Brazilian cultivar PI561356 - resistant) infected by the fungus Phakopsora pachyrhizi (Asian Rust disease) and two samples of plants (Brazilians cultivars: BR 16 - susceptible and Embrapa 48 - resistant) submitted to drought stress - for descriptions see Soares-Cavalcanti et al. (2012, this issue) and Wanderley et al. (2012, this issue). In total, the SuperSAGE approach generated 4,373,053 tags with 26 bp each.
Initially the tags of each sample were grouped in unique sequences. The unique sequences that presented low read counts (read count < 2) were discarded from the list. The Audic-Claverie statistic (Audic and Claverie, 1997) with a 95% confidence level (cutoff of 0.05) was used to identify tags as up-regulated (more expressed in the treated library) or down-regulated (more expressed in the control library).
In order to connect the unique tag with a gene sequence, the SOAP2 aligner program (Li et al., 2009) was used to align the unique tags with three databases (shown previously): (i) assembled unigenes (60,747 sequences), (ii) predicted genes (66,153 sequences) and (iii) the soybean genome. The program has been configured to allow for up to 2 mismatches in the alignments (SNPs can generate mismatches in the alignment, especially in this case because the assembled unigenes are generated by ESTs from different cultivars) and return all optimal alignments. In cases where more than one optimal alignment exists we decided to use the results from assembled unigenes, because they contain the UTR regions (a large part of the Super-SAGE tags are in the 3′ UTR region), followed by the alignment with the predicted genes and, in the last case, the genome alignment was considered. Figure 3 shows the pipeline used in the SuperSAGE analysis.
Figure 3.
Complete pipeline of the SuperSAGE data analysis. All tags (control and treated libraries) of each sample were grouped in unique tags. The unique tags were aligned against the public soybean databases using the SOAP2 aligner. Most of the tags have one alignment (83%) and up to one mismatch (85%). A summary of the total, unique, mapped and differential tags is shown in Table 1.
For the sample submitted under drought stress (BR 16 and Embrapa 48 cultivars) we mapped 84.3% of the unique tags (Table 1), whereby the remaining 15.7% could represent new soybean genes specific to Brazilian cultivars. Similar results were obtained from a sample infected by Asian Rust, which yielded a total of 21,338 unique tags (20.42%) (Table 1) that did not align with any soybean database. In this case the unique tags from fungi genes may have contributed to increase this value. We tried to map these tags against Phakopsora pachyrhizi databases, but we did not find any fungus genes, probably due to the limited amount of fungus data available in the literature. The genome of this fungus, for example, has an estimated size of 500 Mb, but there is only 50 Mb available at NCBI.
Table 1.
Summary of Solexa SuperSAGE data deposited in the Genosoja databank.
| Total tags (control) | Total tags (treated) | Unique tags | Mapped tags | Differential tags | |
|---|---|---|---|---|---|
| Asian Rust | 813,205 | 885,439 | 104,725 | 83,337 (79.58%) | 15,761 (15.05%) |
| Drought BR 16 | 1,092,374 | 509,465 | 89,205 | 75,233 (84.34%) | 14,450 (16.20%) |
| Drought Embrapa 48 | 653,352 | 419,218 | 74,833 | 63,083 (84.30%) | 25,364 (33.89%) |
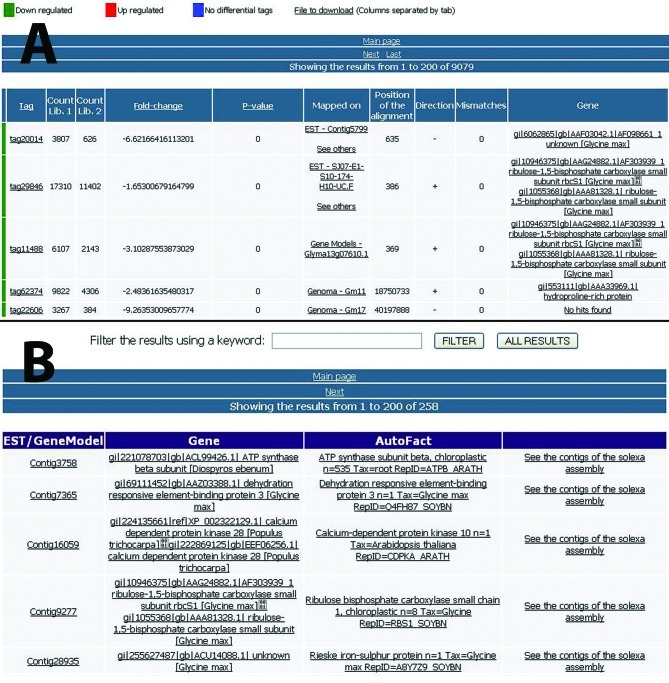
We constructed a web interface for SuperSAGE analysis (Figure 4A). This interface shows, for each tag, the count number in both libraries (control and treated), the correspondent gene and its annotation (NR and Autofact result), as well as the position and the number of mismatches in the alignment. The user can filter the results using a keyword or gene name.
Figure 4.
Web interfaces. (A) Results for the SuperSAGE analysis. For each unique tag are available: tag count in control and treated libraries (columns 3 and 4), fold-change (column 5), p-value (column 6), the correspondent gene (column 7), alignment information (columns 8, 9 and 10) and gene annotation (column 11). (B) Results for one subtractive library of the project, showing all genes found in the library and their respective annotation. The interface allows the user to search using a keyword in the annotation results.
Solexa cDNA subtractive libraries data
Twenty-two cDNA subtractive libraries from different cultivars were sequenced in the Genosoja context, using many treatments with different time courses (Table 2) (Rodrigues et al., 2012, this issue). The reads were generated by Illumina/Solexa technology with read lengths of 45 or 76 bp, depending on the library.
Table 2.
Summary of Solexa cDNA data from subtractive libraries deposited in the Genosoja databank.
| Genotype | Time course | Read length | Reads | Aligned reads (%) | Genes | |
|---|---|---|---|---|---|---|
| Asian Rust | PI1356 - resistant | 12, 24 and 48 h | 76 bp | 5,185,015 | 82.65 | 3,103 |
| Asian Rust | PI1356 - resistant | 72 and 96 h | 76 bp | 5,000,616 | 81.43 | 1,303 |
| Asian Rust | PI1356 - resistant | 192 h | 76 bp | 4,700,869 | 71.32 | 1,318 |
| Asian Rust | PI230970 - resistant | 1 and 6 h | 76 bp | 4,679,963 | 79.87 | 948 |
| Asian Rust | PI230970 - resistant | 12 and 24 h | 76 bp | 4,878,530 | 79.44 | 950 |
| Asian Rust | PI230970 - resistant | 48 and 72 h | 76 bp | 4,335,862 | 78.87 | 3,309 |
| Virus | CD206 - resistant | 5 and 13 days | 76 bp | 5,963,145 | 31.67 | 1,855 |
| Virus | BRSGO - susceptible | 6 and 13 days | 76 bp | 5,345,985 | 81.42 | 1,541 |
| Nitrogen* | MG/BR 46 | - | 76 bp | 4,621,072 | 75.11 | 6,815 |
| Nitrogen* | MG/BR 46 | - | 76 bp | 5,343,969 | 77.02 | 18,921 |
| Drought - leaf | BR 16 - sensitive | 25–50 min | 45 bp | 1,854,641 | 81.13 | 1,560 |
| Drought - leaf | BR 16 - sensitive | 75–100 min | 45 bp | 519,031 | 80.09 | 2,009 |
| Drought - leaf | BR 16 - sensitive | 125–150 min | 45 bp | 2,035,320 | 81.01 | 3,124 |
| Drought - root | BR 16 - sensitive | 25–50 min | 45 bp | 2,486,569 | 65.71 | 258 |
| Drought - root | BR 16 - sensitive | 75–100 min | 45 bp | 2,458,847 | 76.83 | 600 |
| Drought - root | BR 16 - sensitive | 125–150 min | 45 bp | 2,428,923 | 74.57 | 657 |
| Drought - leaf | Embrapa 48 - tolerant | 25–50 min | 76 bp | 5,144,645 | 79.66 | 10,495 |
| Drought - leaf | Embrapa 48 - tolerant | 75–100 min | 76 bp | 5,644,473 | 81,57 | 17,810 |
| Drought - leaf | Embrapa 48 - tolerant | 125–150 min | 76 bp | 5,359,395 | 80.53 | 8,970 |
| Drought - root | Embrapa 48 - tolerant | 25–50 min | 76 bp | 3,095,694 | 82.34 | 3,187 |
| Drought - root | Embrapa 48 - tolerant | 75–100 min | 76 bp | 5,731,156 | 74.72 | 17,218 |
| Drought - root | Embrapa 48 - tolerant | 125–150 min | 76 bp | 5,545,375 | 78.63 | 17,520 |
Inoculated with B. japonicum.
In order to identify the genes in these libraries, the reads were mapped into soybean genes. First, we aligned the sequences against the unigenes using the SOAP2 aligner configured to allow up to two mismatches, discarding fragments with “Ns”, and returning all optimal alignments. The sequences that did not align with unigenes were aligned against the predicted genes with the same parameters. A web interface (Figure 4B) provides users with all genes identified in each library and enables searches by gene name and keywords (in annotation results).
Solexa microRNA data
The Genosoja project generated eight small RNA libraries from soybean - four of the plants with Asian Rust disease (Brazilians cultivar PI561356 - resistant and Embrapa 48 - susceptible) and four under drought stress (Brazilians cultivars BR 16 - susceptible and Embrapa 48 -resistant) (Molina et al., 2012, this issue). These libraries were sequenced using Illumina/Solexa technology and for each library the reads size range from 19 to 24 bp (Table 3).
Table 3.
Data from Solexa MicroRNA libraries deposited in the Genosoja databank.
| Sequence sizes
|
||||||||
|---|---|---|---|---|---|---|---|---|
| 19 bp | 20 bp | 21 bp | 22 bp | 23 bp | 24 bp | |||
| Drought stress | Resistant | Control | 327,448 | 271,772 | 531,595 | 357,980 | 203,722 | 208,377 |
| Treated | 71,011 | 72,628 | 154,808 | 87,326 | 77,045 | 177,626 | ||
| Susceptible | Control | 89,040 | 91,816 | 215,419 | 128,524 | 142,446 | 200,087 | |
| Treated | 266,165 | 220,714 | 353,003 | 244,641 | 138,051 | 250,213 | ||
|
| ||||||||
| Asian Rust | Resistant | Control | 91,908 | 205,404 | 1,177,303 | 394,378 | 175,063 | 285,064 |
| Treated | 100,045 | 155,849 | 779,788 | 426,383 | 187,926 | 859,624 | ||
| Susceptible | Control | 115,824 | 236,750 | 921,964 | 340,129 | 167,306 | 540,949 | |
| Treated | 123,423 | 190,799 | 962,676 | 363,983 | 86,230 | 176,753 | ||
Initially, the reads were grouped into unique sequences and read frequencies computed. The unique sequences that presented low read counts (read count = 2) were discarded from the list, as they were possibly caused by sequencing errors. In order to perform differential expression analysis between libraries, both a normalization and statistical significance analysis were applied using DEGseq software (Wang et al., 2009) considering a confidence level of 95% (cutoff of 0.05). Table 4 presents the number of unique and differential sequences in each library. For the statistical significance analysis, the treated over control libraries were considered.
Table 4.
Unique and differential sequences in microRNA libraries generated by the Genosoja consortium.
| Sequence sizes
|
||||||||
|---|---|---|---|---|---|---|---|---|
| 19 bp | 20 bp | 21 bp | 22 bp | 23 bp | 24 bp | |||
| Drought stress | Resistant | Unique | 725 | 665 | 448 | 522 | 448 | 231 |
| Differential | 79.30% | 77.30% | 78.10% | 76.60% | 76.80% | 89.20% | ||
| Susceptible | Unique | 719 | 652 | 448 | 516 | 442 | 170 | |
| Differential | 75.50% | 75.60% | 88.00% | 81.80% | 82.10% | 95.30% | ||
|
| ||||||||
| Asian Rust | Resistant | Unique | 588 | 524 | 427 | 456 | 386 | 208 |
| Differential | 54.30% | 60.90% | 72.10% | 55.50% | 46.40% | 57.70% | ||
| Susceptible | Unique | 590 | 537 | 435 | 461 | 373 | 220 | |
| Differential | 63.40% | 57.90% | 76.30% | 69.00% | 67.80% | 73.20% | ||
To identify microRNAs from the small RNAs dataset it is necessary to identify the pre-microRNA by alignment of small RNAs (unique sequences) into the soybean genome assembly, followed by secondary structure identification. This alignment was performed using SOAP2 configured to allow for exact alignments only. The upstream and downstream genomic sequences of the read alignment position, 300 bp each in size, were extracted from the genome using homemade PERL scripts (Supplementary Material Figure S2). These genomic regions were aligned against the reverse complement of its respective tag (rc-tag) using the Smith-Waterman (Smith and Waterman, 1981) algorithm with two gaps and four mismatches allowed. The resulting sequences were considered pre-microRNA candidates, and the secondary structure was manually curated, resulting in 256 microRNAs (Figure 5) (Kulcheski et al., 2011).
Figure 5.
Pipeline used for microRNA prediction. The microRNA candidates (unique tags, Table 3) were mapped against the soybean genome with the SOAP2 aligner (no mismatches allowed). The genomic region (300 bp upstream and downstream of the alignment position) of the SOAP alignment was mapped with the reverse complement of the original microRNA using the Smith-Waterman algorithm. After manual analysis of the secondary structure, 256 microRNAs were identified. Finally, the Smith-Waterman algorithm (3 mismatches allowed) was used to identify targets in the assembled unigenes (it was possible to identify targets for 169 microRNAs).
Finally, the microRNA target prediction was performed using the Smith-Waterman algorithm (3 mismatches allowed) to align the 256 microRNAs against the assembled unigenes (shown previously). We considered only alignments in the 5′-3′ direction obtained by comparison of the unigenes with the NR database using BLASTx. This methodology was able to identify targets for 169 microRNAs, most of which (39%) presented one or two targets (Figure 5).
Conclusions
In this work we presented all the bioinformatics analysis and pipelines used in the Genosoja project. The web-based interface constructed and described herein represents an important tool to help in the discovery of genes and new drugs that will enable increased soybean productivity. This system’s use of common references (genome, assembled unigenes and predicted genes) facilitates the incorporation of new data from other sequencing methodologies or experimental conditions. Moreover, the bioinformatic pipeline discussed herein can also be applied to any genomic project, regardless of the organism.
Acknowledgments
The authors would like to acknowledge all research by the Brazilian Soybean Genome Consortium (Genosoja) involved in the generation of data used to construct the database presented in this paper. Moreover, we thank CNPQ (Conselho Nacional de Desenvolvimento Científico e Tecnológico - Brazil) for financial support to this work.
Supplementary Material
The following online material is available for this article:
Figure S1 - Perl script to extract information about sequences from GenBank files.
Figure S2 - Perl script to extract the upstream and downstream genomic sequences of the read alignment position.
This material is available as part of the online article from http://www.scielo.br/gmb.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer ELL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudet C, Dias Z. New EST trimming strategy in: Brazilian Symposium on Bioinformatics, 2005. Lect Notes Bioinf. 2005;3594:206–209. [Google Scholar]
- Carazzolle MF, Formighieri EF, Digiampietri LA, Araujo MRR, Costa GLL, Pereira GAG. Gene projects: A genome web tool for ongoing mining and annotation applied to CitEST. Genet Mol Biol. 2007;30(suppl):1030–1036. [Google Scholar]
- Cheng KCK, Stromvik MV. SoyXpress: A database for exploring the soybean transcriptome. BMC Genomics. 2008;9:e368. doi: 10.1186/1471-2164-9-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell RD, Jorkest RM, Day A, Eddy SR, Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:e7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson AM, Albrecht M, Birney E, Blankenburg H, Down T, Finn RD, Hermjakib H, Hubbard TJP, Jimenez RC, Jones P, et al. Integrating biological data - The Distributed Annotation System. BMC Bioinformatics. 2008;9(Suppl 8):S3. doi: 10.1186/1471-2105-9-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski LB, Gray LW, Lang BF, Burger G. AutoFACT: An automatic functional annotation and classification tool. BMC Bioinformatics. 2005;6:e151. doi: 10.1186/1471-2105-6-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulcheski FR, Oliveira LFV, Molina LG, Almerao MP, Rodrigues FA, Marcolino J, Barbosa JF, Stolf-Moreira R, Nepomuceno AL, Marcelino-Guimaraes FC, et al. Identification of novel soybean microRNAs involved in abiotic and biotic stress. BMC Genomics. 2011;12:e307. doi: 10.1186/1471-2164-12-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam T-W, Yiu S-M, Kristiansen K, Wang J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Molina L, Cordenonsi G, Loss G, Oliveira LFV, Carvalho K, Kulcheski F, Margis R. Metatranscriptomic analysis of small RNAs present in soybean deep sequencing libraries. Genet Mol Biol. 2012;35(suppl 1):292–303. doi: 10.1590/S1415-47572012000200010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues FA, Marcolino J, Carvalho JFC, Nascimento LC, Neumaier N, Farias JRB, Carazzolle MF, Marcelino FC, Nepomuceno AL. Using subtractive libraries to prospect differentially expressed genes in soybean plants submitted to water deficit. Genet Mol Biol. 2012;35(suppl 1):304–314. doi: 10.1590/S1415-47572012000200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the paleopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Soares-Cavalcanti NM, Belarmino LC, Kido EA, Pandolfi V, Marcelino-Guimaraes FC, Rodrigues F, Pereira GAG, Benko-Iseppon AM. Overall picture of expressed Heat Shock Factors in Glycine max, Lotus japonicus and Medicago truncatula. Genet Mol Biol. 2012;35(suppl 1):247–259. doi: 10.1590/S1415-47572012000200006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzek BE, Huang H, McGarvey P, Mazumber R, Wu CH. Uniref: Comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sakurai T, Totoki Y, Toyoda A, Seki M, Ishiwata A, Akiyama K, Kurotani A, Yoshida T, Mochida K, et al. Sequencing and analysis of approximately 40,000 soybean cDNA clones from a full-length-enriched cDNA library. DNA Res. 2008;15:333–346. doi: 10.1093/dnares/dsn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2009;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Wanderley-Nogueira AC, Belarmino LC, Soares-Cavalcanti NM, Bezerra-Neto JP, Kido EA, Pandolfi V, Abdelnoor RV, Binneck E, Carazzole MF, Benko-Iseppon AM. An overall evaluation of the Resistance (R) and Pathogenesis Related (PR) superfamilies in soybean, as compared with Medicago and Arabidopsis. Genet Mol Biol. 2012;35(suppl 1):260–271. doi: 10.1590/S1415-47572012000200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorinori JT, Paiva WM, Frederick RD, Costamilan LM, Bertagnolli PF. Epidemics of soybean rust (Phakopsora pachyrhizi) in Brazil and Paraguay from 2001 to 2003. Plant Disease. 2005;89:675–677. doi: 10.1094/PD-89-0675. [DOI] [PubMed] [Google Scholar]
Internet Resources
- NCBI, http://www.ncbi.nlm.nih.gov/ (October 10, 2011).
- Phytozome, http://www.phytozome.net/soybean.php (October 10, 2011).
- Soybean full-length cDNA database, http://rsoy.psc.riken.jp/ (October 10, 2011).
- SOAP2 aligner, http://soap.genomics.org.cn/ (October 10, 2011).
- Genosoja database, http://www.lge.ibi.unicamp.br/soybean/ (October 10, 2011).
- SoyXpress database, http://soyxpress.agrenv.mcgill.ca (October 10, 2011).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 - Perl script to extract information about sequences from GenBank files.
Figure S2 - Perl script to extract the upstream and downstream genomic sequences of the read alignment position.