Abstract
Environmental stresses caused by either abiotic or biotic factors greatly affect agriculture. As for soybean [Glycine max (L.) Merril], one of the most important crop species in the world, the situation is not different. In order to deal with these stresses, plants have evolved a variety of sophisticated molecular mechanisms, to which the transcriptional regulation of target-genes by transcription factors is crucial. Even though the involvement of several transcription factor families has been widely reported in stress response, there still is a lot to be uncovered, especially in soybean. Therefore, the objective of this study was to investigate the role of bHLH and trihelix-GT transcription factors in soybean responses to environmental stresses. Gene annotation, data mining for stress response, and phylogenetic analysis of members from both families are presented herein. At least 45 bHLH (from subgroup 25) and 63 trihelix-GT putative genes reside in the soybean genome. Among them, at least 14 bHLH and 11 trihelix-GT seem to be involved in responses to abiotic/biotic stresses. Phylogenetic analysis successfully clustered these with members from other plant species. Nevertheless, bHLH and trihelix-GT genes encompass almost three times more members in soybean than in Arabidopsis or rice, with many of these grouping into new clades with no apparent near orthologs in the other analyzed species. Our results represent an important step towards unraveling the functional roles of plant bHLH and trihelix-GT transcription factors in response to environmental cues.
Keywords: drought, gene expression, Glycine max, phylogeny, plant-microbe interactions
Introduction
Soybean [Glycine max (L.) Merril] is one of the most important crop species in the world. It is widely used for both human and animal consumption due to the high protein and oil contents of its grains. More recently, the potential for using soybean oil in renewable fuel production has also emerged (Programa Nacional de Produção e Uso de Biodiesel). Since it belongs to the Fabaceae family, soybean also takes part in the process of organic nitrogen fertilizer production through its symbiotic association with nitrogen-fixing bacteria (Gepts et al., 2005). Currently, soybean producers are primarily concerned with losses caused by drought stress, Asian Soybean Rust (ASR, caused by the fungus Phakopsora pachyrhizi) and soybean cyst nematode (SCN, caused by Heterodera glycines) (EMBRAPA, 2007). Furthermore, the genetic variability found in soybean germplasm for those characteristics is restricted, which increases the vulnerability of this species to environmental stresses (Priolli et al., 2002; Miles et al., 2006).
As sessile organisms, higher plants are continuously exposed to a great variety of environmental stimuli. Because their survival depends on the ability to cope with those stimuli, plants have evolved a variety of sophisticated molecular mechanisms in response to environmental stresses. These generally involve alterations in gene expression, leading to changes in plant physiology, metabolism and developmental activities. Whether caused by abiotic (such as drought, salt and cold) or biotic factors (such as pathogens and insects), environmental stresses have serious adverse effects on agriculture. Therefore, a thorough understanding of the molecular mechanisms involved in plant stress tolerance has become pivotal for the development of new strategies and technologies related to the increasing demand on agricultural production (Rao et al., 2006; Yoshioda and Shinozaki, 2009).
Upon stimuli perception, responses of plants to environmental stresses comprise the activation of a multitude of interconnected signaling pathways (Singh et al., 2002). The phytohormones abscisic acid (ABA), ethylene (ET), jasmonic acid (JA) and salicylic acid (SA), aside from reactive oxygen species (ROS), are known to act as messenger molecules that trigger specific (but at times overlapping) pathways of this complex network, leading to the accumulation of stress-related gene products (Yoshioda and Shinozaki, 2009). Besides, a great number of studies have highlighted the importance of the transcriptional regulation of target-genes through transcription factors in plant responses to environmental stresses (Zhou et al., 2008; Chen et al., 2009; Zhang et al., 2009). Transcription factors act by binding to cis-elements in the promoter regions of target-genes, thereby activating or repressing their expression. Transcriptional reprogramming is known to result in both spatially and temporally altered expression patterns of stress-related genes. Thus, transcription factors are key players in fine-tuning stress responses at the molecular level (Singh et al., 2002; Eulgem, 2005).
A large part of a plant’s genome is devoted to transcription. With the recent completion of the soybean genome sequencing and assembly, a comparative analysis of putative transcription factor-encoding genes found in both soybean and the model dicot Arabidopsis thaliana can be performed. In the leguminous plant (whose genome is six times larger than that of A. thaliana), over 5,600 transcription factors were identified, these corresponding to about 12% of the predicted protein-coding loci (Schmutz et al., 2010). In contrast, in the model plant the total number of transcription factors (∼2,300) comprises only up to 7% of the predicted protein-coding loci (Singh et al., 2002). The overall distribution of these genes among known transcription-factor families is similar among the two genomes, although some families are relatively sparser or more abundant in soybean. Thus, even though the A. thaliana genome often serves general comparisons, differences in biological function between species might occur (Schmutz et al., 2010).
Basic helix-loop-helix (bHLH) proteins constitute one of the largest families of transcription factors. They are found in all three eukaryotic kingdoms and are involved in a myriad of regulatory processes. Members of this family share the bHLH signature domain, which consists of ∼60 amino acids comprising two distinct regions, a basic stretch at the N-terminus consisting of ∼15 amino-acids involved in DNA binding, and a C-terminal region of ∼40 amino-acids composed of two amphipathic α-helices, mainly consisting of hydrophobic residues linked by a variable loop (the “helix-loop-helix” region). This region is responsible for promoting protein-protein interactions through the formation of homo- and hetero-dimeric complexes (Toledo-Ortiz et al., 2003; Carretero-Paulet et al., 2010; Pires and Dolan, 2010). The Lc protein from Zea mays, reported as a transcriptional activator in the anthocyanin biosynthetic pathway (Ludwig et al., 1989), was the first plant bHLH member identified. The involvement of bHLH members in plant developmental processes (Szecsi et al., 2006; Menand et al., 2007), light perception (Liu et al., 2008), iron and phosphate homeostasis (Yi et al., 2005; Long et al., 2010; Zheng et al., 2010), and phytohormone signalling pathways (Abe et al., 1997; Friedrichsen et al., 2002; Lorenzo et al., 2004; Anderson et al., 2004; Fernandez-Calvo et al., 2011; Hiruma et al., 2011; Seo et al., 2011) has also been reported. In fact, Arabidopsis MYC2 is to date the most extensively characterized plant bHLH transcription factor, and it seems to be a global regulator of hormone signalling. MYC2 has been described as an activator of ABA-mediated drought stress-response (Abe et al., 1997, 2003). It also regulates JA/ET-induced genes, either as an activator in response to wounding, or as a suppressor in pathogen responses (Anderson et al., 2004; Lorenzo et al., 2004; Hiruma et al., 2011). In these cases, the activity of MYC2 is itself subject to regulation by JAZ proteins, in a SCFCOI1 proteosome degradation – dependent pathway (Chini et al., 2007). Additionally, MYC2 seems to form homo- and heterodimers with two other closely-related bHLH proteins (MYC3 and MYC4), and their interaction is essential for full regulation of JA responses in Arabidopsis (Fernandez-Calvo et al., 2011).
Trihelix-GT factors constitute another family of plant-specific transcription factors. They are characterized by binding specificity for GT-elements present in the promoter region of many plant genes (Hiratsuka et al., 1994; Nagano et al., 2001) and are among the first transcription factors identified in plants (McCarty and Chory, 2000). They share one or two trihelix (helix – loop – helix – loop –helix) structures, each consisting of three putative α-helices, which are responsible for binding to DNA (Zhou, 1999). Dimerization of GT factors, or interaction between trihelix-GT and other transcription factors appear to play a major role in the regulatory function of this family (Zhou, 1999). In addition, recent studies demonstrated that post-translational modifications may occur in at least some GT-factors, as shown for Arabidopsis light-responsive GT-1 (Maréchal et al., 1999; Nagata et al., 2010). Members of the trihelix-GT family were first described as being involved in the regulation of light-responsive genes (Green et al., 1987, 1988). Nevertheless, further studies in rice and Arabidopsis showed that some GT factors are not light-responsive at the transcriptional level (Dehesh et al., 1990; Kuhn et al., 1993). The involvement of this family in seed maturation (Gao et al., 2009), control of flower morphogenesis (Griffith et al., 1999; Brewer et al., 2004; Li et al., 2008), and response to environmental cues (O’Grady et al., 2001; Park et al., 2004; Wang et al., 2004; Xie et al., 2009; Fang et al., 2010) has also been reported.
In recent years, a growing number of transcription factors belonging to families, such as AP2, NAC and WRKY, have been connected to the responses of soybean against environmental stresses (Zhang et al., 2009; Pinheiro, 2009; Zhou, 2008). In addition, the involvement of two soybean trihelix-GT factors [GmGT-2A (Glyma04g39400) and GmGT-2B (Glyma10g30300)] in abiotic stress tolerance has recently been proposed, following heterologous expression in Arabidopsis (Xie, 2009). Nevertheless information regarding soybean bHLH and trihelix-GT members and their roles in this species remains scarce. In the present study we, therefore, aimed at identifying soybean bHLH-and trihelix-GT-encoding genes, as well as investigating their involvement in response to environmental stresses. Given the dimension of the bHLH family in plants (with more than 600 members in Arabidopsis divided into 32 groups), we decided to focus on a single monophyletic group (subfamily 25, Carretero-Paulet et al., 2010), once we had found some interesting soybean candidates within the LGE Soybean Genome database (Nascimento et al., 2012) that belong to this group. At least 45 bHLH (from subgroup 25) and 63 trihelix-GT putative genes reside in the soybean genome. Among these, at least 14 bHLH and 11 trihelix-GT seem to be involved in responses to abiotic/biotic stresses. A phylogenetic analysis allowed us to successfully cluster these genes with members of bHLH and trihelix-GT proteins from other plant species. All together, our results represent an important step towards understanding the molecular mechanisms by which soybean responds to environmental cues.
Material and Methods
Sequence identification and annotation
In order to identify putative soybean bHLH sequences, the TAIR (The Arabidopsis Information Resource) gene id from all 17 bHLH proteins belonging to subgroup 25 in Arabidopsis was used to search the soybean database in Phytozome and at JGI (Joint Genome Institute). Soybean peptide homologs for each A. thaliana sequence were identified from a BLASTP search with default parameters in Phytozome and redundant sequences were manually discarded. The protein sequences obtained were scanned for the existence of the bHLH domain using the SMART database. The software MEME (multiple EM for motif elicitation) version 4.4.0 was used for motif identification, using the following parameters: minimum and maximum motif width set to 6 and 50 amino acids, respectively, with any number of motif repetitions. Motif detection was restricted to a maximum of 10. Identified motifs were also compared with conserved compositions already described for bHLH sequences. In addition, the bHLH domain was manually delimited according to plant-specific boundaries, as determined by Toledo-Ortiz et al. (2003) and Carretero-Paulet et al. (2010). Classification of soybean sequences in subgroup 25 was accomplished by mismatch counting from the consensus established for A. thaliana (Carretero-Paulet et al., 2010). Sequences with more than 8 mismatches in conserved positions were discarded. Moreover, no mismatches were allowed at residues H9, E13 and R16 of the basic region, since these are crucial for DNA-binding activity, and a consensus among subgroup 25 sequences.
The identification of putative trihelix-GT protein sequences from soybean was accomplished as follows: the conserved trihelix sequence of previously reported soybean genes (O’Grady et al., 2001; Xie et al., 2009) along with motifs predicted for this family (Fang et al., 2010), were blasted (TBLASTN) against the soybean genome in Phytozome. All homologous sequences with an E-value of less than 0.0001 were scanned for the existence of the trihelix domain using SMART (domains with less significant scores than default cut-offs were also analyzed). Motif identification and comparison with conserved trihelix-GT compositions were performed using MEME. Sequences that did not fit these criteria were removed from the analysis.
To determine the intron-exon organization of all bHLH and trihelix-GT genes, the full length coding sequences were aligned with the corresponding genomic sequences available on Phytozome. Intron-exon maps of the genes were drawn using Fancy Gene v1.4 software.
Gene expression data mining
Expression profiles of the identified bHLH and trihelix-GT sequences in both biotic and abiotic situations were obtained by mining the LGE Soybean Genome database. A “gene” search was carried out using Phytozome’s gene model codes and each gene had its 5′ and 3′ untranslated regions verified in Gbrowse. Gene expression was confirmed by database searches in NCBI ESTs and LGE superSAGE stress experiments with soybean leaves infected with Asian soybean rust (accession PI 561356, resistant) vs. uninfected leaves, and soybean roots subjected to drought (cultivar BR16, susceptible / cultivar Embrapa-48, tolerant) vs. untreated roots from both cultivars.
Phylogenetic analysis
The phylogenetic analysis of plant trihelix-GT factors was performed using protein sequences from A. thaliana, G. max, Medicago truncatula and Oryza sativa. For plant bHLH transcription factors, protein sequences from A. thaliana, G. max, O. sativa and Physcomitrella patens were used. In both cases, multiple sequence alignments were conducted with full-length protein sequences using the CLUSTALW tool (Thompson et al., 1994) implemented in MEGA ver. 4.0 (Tamura et al., 2007). The phylogenetic analysis was performed by two different and independent approaches, viz. the neighbor-joining (NJ) and Bayesian methods. The NJ method was performed within MEGA v4.0. Molecular distances of the aligned sequences were calculated according to the p-distance parameter, with gaps and missing data treated as pairwise deletions. Branch points were tested for significance by bootstrapping with 1000 replications. Bayesian analysis was conducted in MrBayes 3.1.2 software (Huelsenbeck et al., 2001; Ronquist and Huelsenbeck, 2003) with the mixed amino-acid substitution model + gamma + invariant sites. Two independent runs of 5,000,000 generations each, with two Metropolis-coupled Monte Carlo Markov chains (MCMCMC) were run in parallel, each one starting from a random tree. Markov chains were sampled every 100 generations and the first 25% of the trees were discarded as burn-in. The remaining ones were used to compute the majority rule consensus tree (MrBayes command allcompat), and the posterior probability of clades and branch lengths. The unrooted phylogenetic trees of trihelix-GT and bHLH proteins were visualized and edited using the software FigTree ver. 1.3.1.
Results and Discussion
Identification and analysis of soybean bHLH-encoding genes
In the past few years several phylogenetic studies have emerged as attempts to perform the classification of bHLH proteins in plants (Heim et al., 2003; Toledo-Ortiz et al., 2003; Carretero-Paulet et al., 2010; Pires and Dolan, 2010). Nevertheless, the number of proposed subfamilies varies considerably among these studies. In the present one, the classification suggested by Carretero-Paulet et al. (2010) proposing the division of plant bHLH transcription factors into 32 subfamilies was used, since it represents the most recent and comprehensive study, so far.
From the BLASTP search at Phytozome, using all 17 Arabidopsis bHLH protein sequences from subgroup 25, 67 non-redundant homolog peptides were identified in the soybean genome. Seven of these were removed from the analysis as they did not contain any bHLH domain. Another 15 sequences were discarded after mismatch counting performed with their aligned domains. Using MEME, two other highly conserved motifs (with E-values of less than 1.7 e−851) were identified among the soybean subgroup 25 sequences. They are formed by residues right adjacent to the bHLH domain and had been previously reported (Heim et al., 2003; Li et al., 2006; Carretero-Paulet et al., 2010; Pires and Dolan, 2010). General characteristics related to the 45 remaining putative soybean bHLH genes are shown in Table 1. Remarkably, members of this subgroup were found spread throughout the 20 soybean chromosomes, with protein sequences ranging from 165 to 691 amino acids. Among the 45 annotated ORFs, 42 presented corresponding ESTs, suggesting that they are expressed genes and not pseudogenes. A complete overview of the gene expression results obtained for this group is presented in Figure 1. Differential expression in at least one of the stress situations/experiments available in LGE database was detected for 14 ORFs, four of these were differentially expressed in more than one situation and three respond to both abiotic and biotic stresses.
Table 1.
Annotation of soybean bHLH (subgroup 25) encoding-genes.
| Accession number in Phytozome | Chromosome | ORF (bp) | Expression confirmed by EST (GenBank Accession) |
|---|---|---|---|
| Glyma01g04610 | 1 | 795 | BE021678.1 |
| Glyma01g09400 | 1 | 1587 | BU765737.1 |
| Glyma01g39450 | 1 | 667 | AW782148.1 |
| Glyma02g13860 | 2 | 1539 | BI786324.1 |
| Glyma02g16110 | 2 | 861 | AW460021.1 |
| Glyma03g21770 | 3 | 1575 | FK005566.1 |
| Glyma03g29710 | 3 | 1203 | BI427219.1 |
| Glyma03g31510 | 3 | 879 | BW666688.1 |
| Glyma03g32740 | 3 | 1446 | BM732402.1 |
| Glyma04g01400 | 4 | 1293 | CA853113.1 |
| Glyma04g05090 | 4 | 855 | FK457664.1 |
| Glyma04g34660 | 4 | 732 | FG990727.1 |
| Glyma04g37690 | 4 | 1041 | CA937888.1 |
| Glyma05g01590 | 5 | 675 | EV276804.1 |
| Glyma05g35060 | 5 | 741 | BE473364.1 |
| Glyma05g38450 | 5 | 1029 | BF325330.1 |
| Glyma06g01430 | 6 | 1173 | BU551063.1 |
| Glyma06g17420 | 6 | 1050 | FG995242.1 |
| Glyma06g20000 | 6 | 810 | CO978579.1 |
| Glyma07g10310 | 7 | 498 | BE347561.1 |
| Glyma08g01210 | 8 | 942 | FG994001.1 |
| Glyma08g04660 | 8 | 528 | - |
| Glyma08g46040 | 8 | 1761 | BM885094.1 |
| Glyma09g14380 | 9 | 1473 | CA936197.1 |
| Glyma09g31580 | 9 | 906 | - |
| Glyma10g03690 | 10 | 852 | BW657011.1 |
| Glyma10g04890 | 10 | 1302 | BI785116.1 |
| Glyma10g12210 | 10 | 1074 | CO978592.1 |
| Glyma10g28290 | 10 | 2076 | BW675573.1 |
| Glyma10g30430 | 10 | 987 | FG999826.1 |
| Glyma11g05810 | 11 | 1146 | GR843316.1 |
| Glyma11g12450 | 11 | 1263 | BU082612.1 |
| Glyma12g04670 | 12 | 1215 | BE661807.1 |
| Glyma13g19250 | 13 | 1437 | BQ741548.1 |
| Glyma14g10180 | 14 | 1269 | EV269688.1 |
| Glyma15g33020 | 15 | 1428 | BI699764.1 |
| Glyma16g10620 | 16 | 1788 | FK024158.1 |
| Glyma17g08300 | 17 | 1098 | CX708610.1 |
| Glyma17g10290 | 17 | 690 | FG993937.1 |
| Glyma17g34010 | 17 | 807 | - |
| Glyma18g32560 | 18 | 1743 | BI317112.1 |
| Glyma19g32570 | 19 | 1101 | FG996268.1 |
| Glyma19g34360 | 19 | 879 | GR826097.1 |
| Glyma20g22280 | 20 | 1281 | BE658194.1 |
| Glyma20g36770 | 20 | 999 | BE474708.1 |
Figure 1.
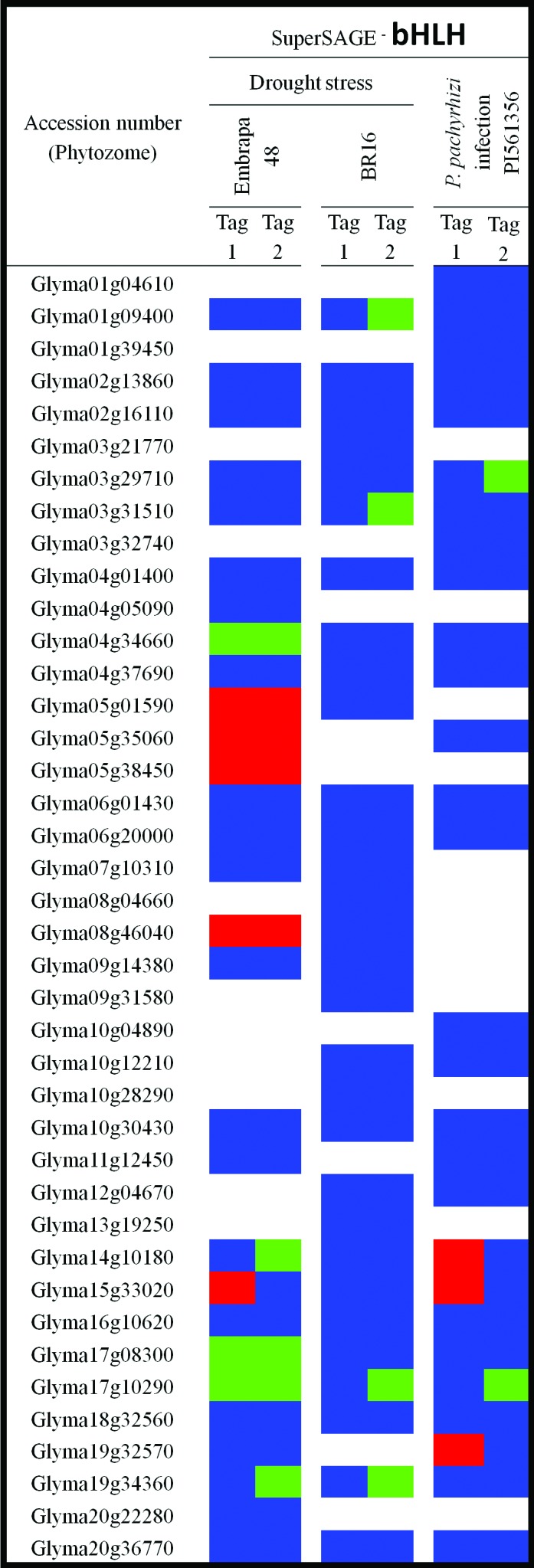
Expression pattern of bHLH encoding-genes under drought stress and P. pachyrhizi infection. The expression data were obtained from superSAGE experiments available at www.lge.ibi.unicamp.br/soja/. Blocks indicate up-regulation (red), down-regulation (green), non-significant differences (p > 0.05) but expression detected (blue), and expression not detected (white). Contrasting expression might reflect detection of a single gene by different tags. Drought stress was carried out in roots from Embrapa-48 (tolerant cultivar) and BR 16 (susceptible cultivar). Soybean leaves from PI561356 (resistant genotype) were infected with P. pachyrhizi.
Lately, a growing number of studies accessing the functional role of specific plant bHLH transcription factors have been reported (Friedrichsen et al., 2002; Szécsi et al., 2006; Liu et al., 2008; Chandler et al., 2009; Todd et al., 2010; Zheng et al., 2010). Nevertheless, a deeper (and broader) functional characterization of this family, focusing on the connection of members/subgroups to the biological processes they control, remains to be done. A first step in this direction has been recently taken by Carretero-Paulet et al. (2010) and Pires and Dolan (2010), where comprehensive information relating both classification and function of previously characterized plant bHLH transcription factors was assembled. More specifically, information regarding the function of subgroup 25 members is still scarce and concerns Arabidopsis members only. An alternative transcript of At1g59640 (ZCW32/BPE) seems to be involved in the control of petal size, whereas its counterpart is expressed ubiquitously (Szécsi et al., 2006). Furthermore, At4g34530 (CIB1) and At1g26260 (CIB5) were shown to interact with blue-light receptor CRY2 and promote floral initiation (Liu et al., 2008). Of most interest for this study, is the redundant role of At1g18400 (BEE1, Brassinosteroid Enhanced Expression1), At4g36540 (BEE2) and At1g73830 (BEE3) in brassinosteroids (BRs)/ABA antagonistic cross-talk during cell elongation (Friedrichsen et al., 2002). According to these authors, BEE1, 2 and 3 are early-response genes induced by BRs through the BRI1 receptor complex, and their expression is repressed by ABA through a yet unknown ABA receptor. Whether this pathway is also related to the ABA-dependent stress-responsive network, still requires further study. Moreover, Poppenberger et al. (2011) have demonstrated that At1g25330 (CESTA), a close homolog of BEE1 and BEE3 (Figure 2), is also involved in BR signaling, possibly by heterodimerization with its closest homologs. Remarkably, it has also been shown that lack of CESTA activity results in the misregulation of genes that are not only BR-responsive but also stress-responsive, such as Arabidopsis ERD5 (Early Responsive to Dehydration 5), TTL4 (Tetratricopetide-Repeat Thioredoxin-Like 4), WRKY18 and a putative LRR-disease resistance protein (Poppenberger et al., 2011), further suggesting that these pathways might indeed share common features.
Figure 2.
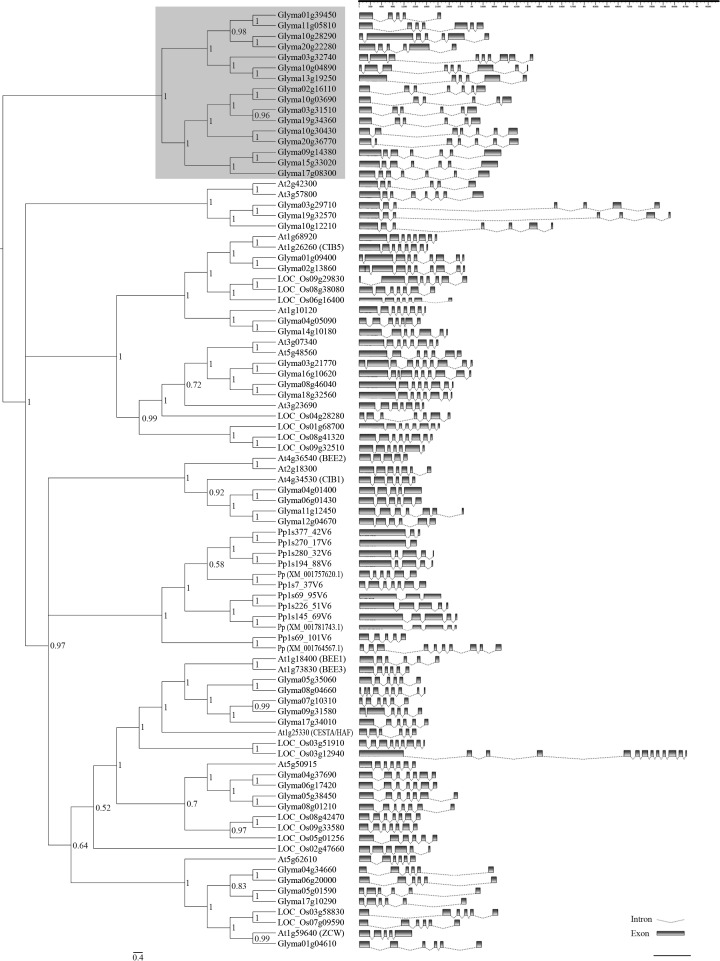
Phylogenetic relationships among bHLH subgroup 25 members. The phylogenetic tree shown on the left comprises 89 plant bHLH protein sequences. The Bayesian analysis was conducted using Mr.Bayes v3.1.2, after alignment of full-length bHLH proteins from selected plant species by means of ClustalW. The unrooted cladogram was edited using Fig Tree v1.3.1 software. Nodal support is given by posteriori probability values shown next to the corresponding nodes. The scale bar indicates the estimated number of amino acid substitutions per site. The gray area denotes a specific soybean cluster. Previously reported bHLH genes were identified according to their accession/locus numbers, the other genes were designated according to their locus ID in Phytozome. A. thaliana (At); G. max (Glyma); O. sativa (LOC_Os) and P. patens (Pp). The graph on the right shows gene organization of full-length coding sequences from 89 plant bHLHs. Intron-exon maps were drawn using Fancy Gene v1.4 software, according to sequence data available in Phytozome.
As an attempt to predict gene function of the annotated genes, a comparison of their amino-acid sequences with subgroup 25 bHLH protein sequences from three other model plant species was carried out. Indeed, representative members from diverse taxonomic groups (P. patens, bryophytes; O. sativa, monocotyledonous; and A. thaliana, dicotyledonous) were included in the phylogenetic analysis in order to access the evolutionary features of this subgroup. The results obtained from the phylogenetic analysis proved to be consistent, since the clades formed were highly supported by posteriori probabilities (Figure 2, on left) and bootstrap (data not shown) analyses. Unlike previous phylogenetic reconstructions of the bHLH family that used the bHLH domain only, this study presents a tree reconstructed from full-length protein sequences. This adds accuracy and reliability to the tree resolution, since the short length of the bHLH domain (∼60 amino-acids), along with its extremely high conservation within subgroups may compromise the reliability of the analysis (Amoutzias et al., 2004).
Patterns of intron distribution among bHLH-encoding genes from diverse species were shown to be conserved within subgroups and provide another criterion in phylogenetic analysis (Li et al., 2006; Carretero-Paulet et al., 2010). In this study, the overall intron-exon organization of bHLH subfamily 25-encoding genes from soybean and other three species was established (Figure 2, on right). Among 89 sequences, the number of introns ranged from 1 (Pp1s270_17v6) to up to 12 (LOC_Os03g12940), and in many cases, phylogenetically related proteins exhibited a closely related gene structure, corroborating the clustering results.
Since it is a basal species among land plants, the moss P. patens was added to this classification in order to help infer about this group’s ancestral state (Rensing et al., 2008). Notably, all 12 members from P. patens grouped together into a clade, instead of grouping with the other plant species, indicating that the radiation within this subgroup has occurred independently in mosses and vascular plants, after the divergence of these taxonomic groups. The same result was obtained by Carretero-Paulet et al. (2010), even when a different method was applied [maximum likelihood (ML) analysis from bHLH-domain alignments]. Nevertheless, the chance that genes belonging to this subgroup might have independently evolved similar functions in both mosses and vascular plants should not be discarded, as suggested by Menand et al. (2007). In fact, while studying plant bHLH ancestry, Pires and Dolan (2010) concluded that the complex regulatory machinery that may be observed in modern plant lineages actually arose early in plant evolution.
The most striking feature that can be inferred from our phylogenetic analysis, which is in accordance with other previously published plant bHLH phylogenies (mentioned above), is the importance of gene duplication during the evolution of this family as a whole. Recurring events of single-gene duplications (“birth-and-death evolution”), combined with domain shuffling seem to rule bHLH evolution and diversification (Morgenstern and Atchley, 1999; Amoutzias et al., 2004; Nei and Rooney, 2005). Furthermore, whole genome duplication (WGD) events also seem to have had an active effect (as seen in the outer clades in Figure 2, on the left), and this seems to be even more intense in the soybean genome. According to our results, the subgroup in question encompasses almost three times more members in soybean than in Arabidopsis or rice (Table 1), with many of these grouping into new clades with no apparent near orthologs in the other analyzed species (Figure 2, in gray on the left side). Indeed, soybean suffered from two WGD events with an impressive retention of homologous blocks (Schmutz et al., 2010). Furthermore, specifically in the case of transcription factors (and other genes working in complex networks), duplications resulting from WGD events are vastly overretained, simply because they may be too costly to be removed, thus making functional redundancy a common feature among transcription factors, especially in plant species. Once retained, homologous duplicates might diverge in function or even subfunctionalize (Freeling, 2009), thus providing a source of evolutionary novelty in the form of new regulatory networks (Carretero-Paulet et al., 2010).
With all that in mind, an integrated analysis of both the expression profile (Figure 1) and the phylogeny (Figure 2) presented herein provides a hint at the roles of subgroup 25 bHLH soybean genes. By focusing on soybean-near homologs shown in the tree (Figure 2 on left) we could see that for most of the paralogs whose expression has been detected, a divergent profile seems to prevail. An exception would be the cases of Glyma03g31510 and Glyma19g34360, which were both repressed during drought stress, with a broadly negative response in the latter, as its mRNA levels were down-regulated in both the susceptible and the tolerant cultivars analyzed. Moreover, the transcripts from Glyma19g32570 were up-regulated during ASR infection in the resistant genotype, whereas its counterpart Glyma03g29710 exhibited opposite differential expression. The near paralogs Glyma05g01590 and Glyma17g10290 also seem to be moving in different directions. Whereas the first seems to be up-regulated in response to fungal stress, the latter seems to be broadly down-regulated, in both susceptible and tolerant cultivars submitted to drought, as well as in P. pachyrhizi’s infection. Furthermore, while Glyma15g33020 seems to be positively involved in soybean defense against ASR and during drought stress in tolerant Embrapa-48 cultivar, its nearest paralog (Glyma09g14380) was not differentially expressed in any of the situations assessed, and their near homolog Glyma17g08300 seems to be negatively involved in drought stress responses, since it was down-regulated in the same cultivar. Whether the examples mentioned above reflect functional divergence or subfunctionalization among duplicate homologs still requires further analysis.
Even though comparison of soybean genes with their orthologs in other species (such as Arabidopsis) is a tentative approach, and as such needs to be performed carefully. In this context it would be interesting to address the function of BEE orthologs in soybean, so as to determine whether they are similar to their Arabidopsis counterparts, and whether they somehow connected to stress responses. In this respect, special attention should be given to Glyma05g35060, which clustered together with the Arabidopsis BR-responsive genes, and whose transcripts turned out to be up-regulated in Embrapa-48 tolerant cultivar in response to drought.
Identification and analysis of soybean trihelix-GT encoding genes
The first isolated and described soybean GT-factor was GmGT-2 (Glyma02g09060), which binds to an element within the Aux28 promoter, and whose mRNA levels were down-regulated by light in a phytochrome-dependent manner (O’Grady et al., 2001). In a global approach using massive EST analysis, Tian et al. (2004) identified 13 putative trihelix genes in the soybean genome. Two of these [GmGT-2A (Glyma04g39400) and GmGT-2B (Glyma10g30300)] were cloned and had their roles in abiotic stress tolerance described using transgenic Arabidopsis plants (Xie et al., 2009). The current annotation analysis indicates the occurrence of at least 63 GT-like genes in the soybean genome. 56 of these had their expression confirmed in the NCBI databases (Table 2). Unfortunately, since information available in Phytozome is not yet definitive, full-length cDNAs were not obtained for most sequences, so only gene-models were considered for this analysis. The 63 soybean trihelix-GT genes encode proteins with lengths ranging from 201 to 885 amino acids, distributed across most of the soybean chromosomes, except for chromosomes 5 and 14. There is an average of 3.5 GT-factor-encoding genes per chromosome, with the highest number of 9 genes found in chromosome 10, whereas a single member was detected in chromosomes 12 and 17, respectively. Three genes (Glyma09g19750, Glyma10g34610 and Glyma20g30630) with incorrect gene model predictions were manually curated.
Table 2.
Annotation of soybean trihelix-GT encoding-genes.
| Accession number in Phytozome (gene) | Chromosome | ORF (bp) | Expression confirmed by EST (GenBank Accession) |
|---|---|---|---|
| Glyma01g29760 | 1 | 819 | BW682708.1 |
| Glyma01g35370 | 1 | 834 | GR826253.1 |
| Glyma02g09050 | 2 | 1653 | FG988995.1 |
| Glyma02g09060 (GmGT-2) | 2 | 1896 | AF372498.1 |
| Glyma03g18750 | 3 | 765 | DB957166.1 |
| Glyma03g34730 | 3 | 1368 | FK016354.1 |
| Glyma03g07590 | 3 | 822 | - |
| Glyma03g34960 | 3 | 1617 | BE555145.1 |
| Glyma03g40610 | 3 | 1626 | - |
| Glyma04g37020 | 4 | 2217 | CO982525.1 |
| Glyma04g39400 (GmGT-2A) | 4 | 1335 | AI900211.1 |
| Glyma06g15500 | 6 | 1494 | BW678214.1 |
| Glyma06g17980 | 6 | 2655 | EH258249.1 |
| Glyma07g04790 | 7 | 1107 | CO981809.1 |
| Glyma07g09690 | 7 | 1083 | BM731493.1 |
| Glyma07g18320 | 7 | 876 | - |
| Glyma08g05630 | 8 | 942 | AW351117.1 |
| Glyma08g28880 | 8 | 981 | CO979268.1 |
| Glyma09g01670 | 9 | 918 | FK019218.1 |
| Glyma09g19750 | 9 | 1155 | BE659959.1 |
| Glyma09g32130 | 9 | 1014 | GR829369.1 |
| Glyma09g38050 | 9 | 969 | AI460860.1 |
| Glyma10g36980 | 10 | 1335 | BU765094.1 |
| Glyma10g07490 | 10 | 1494 | GD961953.1 |
| Glyma10g34520 | 10 | 1374 | BE820805.1 |
| Glyma10g36950 | 10 | 1350 | BU549085.1 |
| Glyma10g36960 | 10 | 2004 | BW666798.1 |
| Glyma10g07730 | 10 | 1785 | FG992486.1 |
| Glyma10g30300 (GmGT-2B) | 10 | 1746 | CA953306.1 |
| Glyma10g34610 | 10 | 1017 | - |
| Glyma10g44620 | 10 | 978 | GR827102.1 |
| Glyma11g25570 | 11 | 1026 | CO979922.1 |
| Glyma11g37390 | 11 | 1125 | BI317190.1 |
| Glyma12g33850 | 12 | 924 | CD415252.1 |
| Glyma13g21350 | 13 | 1410 | CX708572.1 |
| Glyma13g26550 | 13 | 957 | BI702330.1 |
| Glyma13g30280 | 13 | 939 | DB955747.1 |
| Glyma13g21370 | 13 | 1464 | CO981764.1 |
| Glyma13g36650 | 13 | 921 | CA800657.1 |
| Glyma13g41550 | 13 | 1221 | GD834531.1 |
| Glyma13g43650 | 13 | 1014 | EV282528.1 |
| Glyma15g03850 | 15 | 1233 | BF068981.1 |
| Glyma15g08890 | 15 | 603 | BM085616.1 |
| Glyma15g12590 | 15 | 696 | - |
| Glyma15g01730 | 15 | 1113 | GD914877.1 |
| Glyma16g01370 | 16 | 1113 | CA801229.1 |
| Glyma16g14040 | 16 | 801 | CO980073.1 |
| Glyma16g28240 | 16 | 1785 | FK012336.1 |
| Glyma16g28250 | 16 | 1395 | BQ296282.1 |
| Glyma16g28270 | 16 | 1332 | - |
| Glyma17g13780 | 17 | 2433 | BQ273464.1 |
| Glyma18g01360 (GmGT-1) | 18 | 1131 | BG406222.1 |
| Glyma18g43190 | 18 | 879 | - |
| Glyma18g51790 | 18 | 990 | BQ786728.1 |
| Glyma19g37410 | 19 | 1359 | GR845650.1 |
| Glyma19g37660 | 19 | 1641 | BF066376.1 |
| Glyma19g43280 | 19 | 1803 | FK019637.1 |
| Glyma20g30630 | 20 | 1338 | BG726775.1 |
| Glyma20g30640 | 20 | 1935 | BW679178.1 |
| Glyma20g30650 | 20 | 1893 | EH261764.1 |
| Glyma20g32940 | 20 | 1572 | FG988154.1 |
| Glyma20g36680 | 20 | 1773 | BE607585.1 |
| Glyma20g39410 | 20 | 960 | BI699475.1 |
Mining the LGE gene expression superSAGE experiments revealed that 11 soybean trihelix-GT genes were differentially expressed in the abiotic/biotic conditions tested (Figure 3). In accordance with our analyses, five trihelix-GT genes were up-regulated under drought in the tolerant cultivar (Embrapa-48), whereas only two genes were down-regulated in this genotype. In the susceptible cultivar (BR16), Glyma10g34520 had its transcript levels increased in response to water deficit and the opposite situation occurred with Glyma10g36950. When plants were infected with P. pachyrhizi, only two genes displayed up-regulation of mRNA levels in response to biotic stress whereas two others seemed to be down-regulated. Interestingly, none of the soybean trihelix-GT previously reported as responsive to stress conditions and particularly to abiotic stress [GmGT-2A (Glyma04g39400) and GmGT-2B (Glyma10g30300)] were detected in the superSAGE experiments herein assessed. Divergence in experimental parameters and genotypes used might explain this unexpected result.
Figure 3.
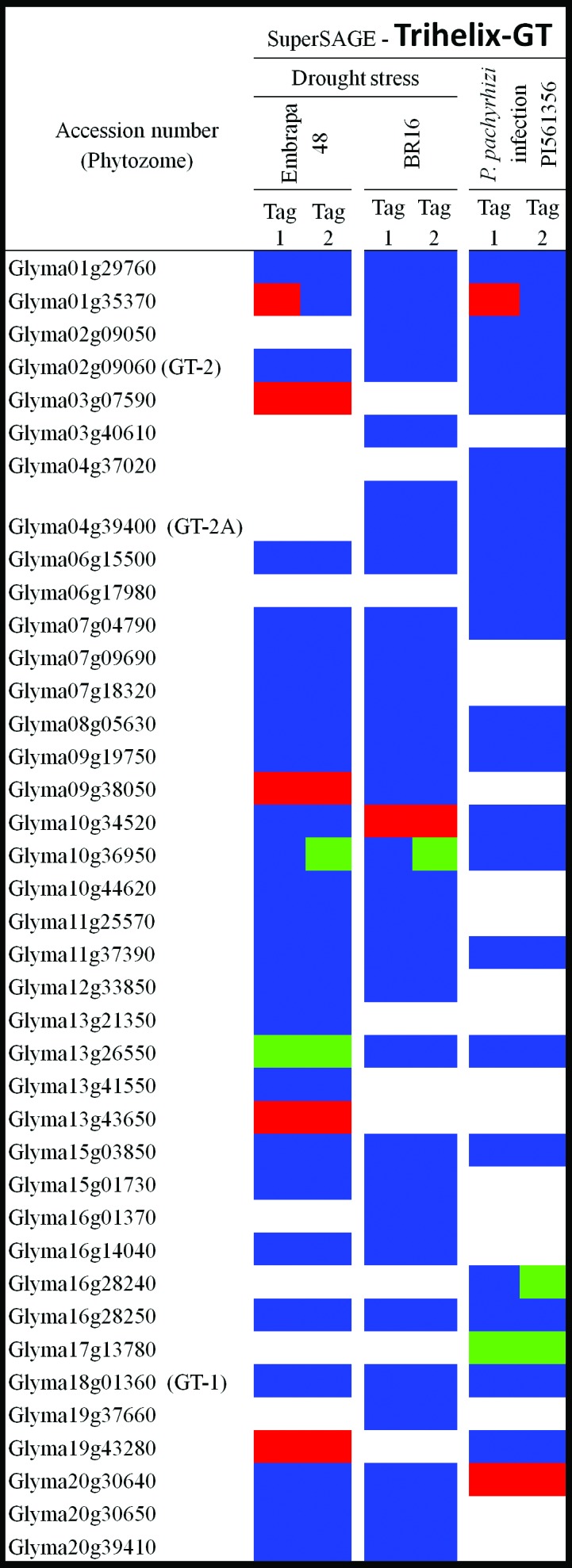
Expression pattern of trihelix-GT encoding-genes under drought stress and P. pachyrhizi infection. The expression data were obtained from superSAGE experiments available at www.lge.ibi.unicamp.br/soja/. Blocks indicate up-regulation (red), down-regulation (green), non-significant differences (p > 0.05) but expression detected (blue), and expression not detected (white). Contrasting expression might reflect detection of a single gene by different tags. Drought stress was carried out in roots from Embrapa-48 (tolerant cultivar) and BR 16 (susceptible cultivar). Soybean leaves from PI561356 (resistant genotype) were infected with P. pachyrhizi.
Transcript levels from Glyma01g35370 and Glyma20g30640 increased when plants were infected with ASR, while the opposite situation occurred with Glyma16g28240 and Glyma17g13780 mRNA levels. A rice GT-factor (OsRML1) was already reported to be upregulated in response to Magnaporthe grisea (Wang et al., 2004), which corroborates a connection between pathogen attack and trihelix-GT gene regulation. It is also possible that Glyma01g35370 may be involved in plant responses to both abiotic and biotic stresses, since the gene expression profile was modulated during water deficit and P. pachyrhizi infection.
The superSAGE experiments suggested that, at least in some cases, the same gene has variable transcript levels in different cultivars and/or in response to different stresses or agents. For example, when water deficit was imposed on soybean plants, Glyma10g36950 was down-regulated in the susceptible (BR16) and the tolerant (Embrapa-48) cultivars, whereas its transcript levels did not change in response to ASR. In another case, Glyma09g38050 was up-regulated in response to drought stress in Embrapa-48, but no differences were detected in BR16. Furthermore, Glyma13g26550 was down-regulated in response to drought stress in the tolerant cultivar, whereas its expression in cultivar BR16 did not exhibit any alterations. In these cases, in addition to differential gene regulation, there may be other factors contributing to distinct regulatory function, such as post-translational modifications or variation in dimerization partners (Zhou, 1999).
Modifications in individual cis-regulatory elements on trihelix-GT promoter regions of duplicated genes might lead to the processes of transcriptional neofunctionalization or subfunctionalization (Haberer et al., 2004), which may explain gene induction or repression without any counterpart response during the same stimuli. This seems to be the case for Glyma03g07590 and its nearest paralog Glyma01g29760, or for Glyma16g28240 and the phylogenetically related Glyma02g09050. Further studies focusing on identifying cis-elements, as well as performing promoter analyses to verify inducible expression patterns may clarify the involvement of duplicated genes in stress-related responses.
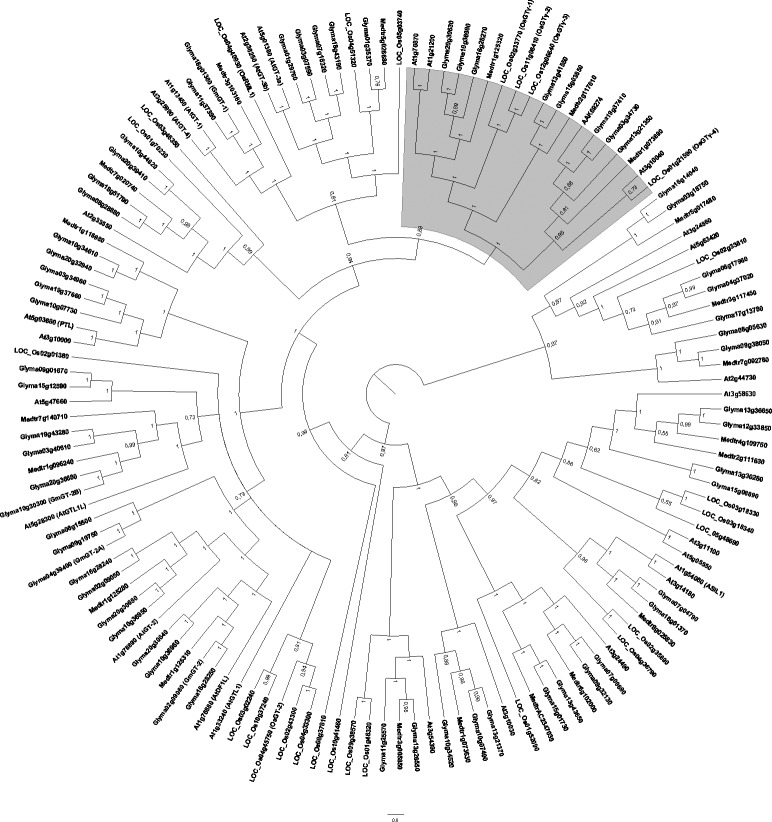
A previous study regarding the phylogenetic analysis encompassing Arabidopsis and rice GT factors (Fang et al., 2010) showed that this family could be classified into three subfamilies (α, β and γ), with unique composition of predicted motifs. Unfortunately, these results were not reproduced in our analysis, even when full-length protein sequences (Figure 4) or the trihelix domains alone were aligned (data not shown). An exception occurred with subfamily γ, which had already been described as having low sequence similarity with the other reported GT factors. The introduction of soybean and M. truncatula sequences in the phylogeny might have affected the expected distribution within those subgroups. Besides, we also inserted into our tree the soybean gene AAK69274 described by Fang et al. (2010), which could neither be identified in the soybean genome nor detected in the expression database. According to our analysis, this unexpected result seems to indicate the occurrence of an alternative splicing in Glyma19g37410 or Glyma03g34730, both considered to be phylogenetically closest to the unidentified gene locus.
Figure 4.
Bayesian phylogenetic tree of 137 plant trihelix-GT proteins. The Bayesian analysis was conducted using Mr.Bayes v3.1.2 software after alignment of full-length trihelix-GT proteins from selected plant species using ClustalW. The unrooted cladogram was edited using Fig Tree ver. 1.3.1 software. Nodal support is given by posteriori probability values shown next to the corresponding nodes. The scale bar indicates the estimated number of amino acid substitutions per site. The gray area denotes GTγ subfamily described by Fang et al. (2010). Previously reported GT factors were identified according to their accession/locus numbers, the other genes were designated according to their locus ID at Phytozome. A. thaliana (At); G. max (Glyma); Medicago truncatula (Medtr) and O. sativa (LOC_Os).
Hence, when taking into account the full-length protein sequence, the GT-factor family might be divided into two subgroups, in one of these subgroups a branch corresponded to the already described subfamily γ (Figure 4, in gray). Despite the fact that subfamilies α and β were not distinguished, other probabilities supported our tree, especially when inner nodes were observed.
When gene organization among Arabidopsis and soybean sequences was compared (Figure 5), the number of introns ranged from zero (twenty three genes) up to 16 (At5g63420 and Glyma06g17980), and some phylogenetically close sequences showed the same gene structure. For example, the Arabidopsis At3g10040 and its soybean ortholog do not have intron, whereas At2g33550 and related members have two introns, with remarkable differences in intron size.
Figure 5.
Gene organization of phylogenetically related full-length coding sequences from Arabidopsis and soybean trihelix-GT transcription factors. Intron-exon maps were drawn using Fancy Gene ver. 1.4 software.
As observed for bHLH transcription factors, the soybean GT factor family encompasses almost three times more members than Arabidopsis or rice, a consequence of the WGD events that took place during plant evolution. In several cases, soybean paralogs clustered with one M. truncatula gene, indicating that these paralogs probably derived from a WGD event that occurred after the divergence of the two legume species. Similarly, Schmutz et al. (2010) refer to a Glycine-specific WGD event, estimated to have occurred about 13 million years ago. However, the possibility that extra M. truncatula orthologs might arise upon the completion of its genome sequencing should not be discarded.
Recently, the OsGTγ subfamily was proposed to participate in the regulation of stress tolerance in rice (Fang et al., 2010). OsGTγ -1 showed more specific expression pattern than their counterparts OsGTγ-2 and OsGTγ-3, which are supposedly redundant. None of them was responsive to light, but their transcript levels increased in response to salt and cold stresses, whereas OsGTγ-1 was upregulated by ABA and SA stimulus. It is possible that some soybean members of this subfamily may act in response to stressor agents, but more studies are required in order to understand whether the pattern seen in rice GTγ factors also occurs in soybean and M. truncatula. Our analysis, so far, does not indicate their involvement in an abiotic and/or biotic stress response. Moreover, soybean genes previously reported as involved in stress responses (Xie et al., 2009) together with other genes herein identified are dispersed in different tree branches, indicating that this family is in fact evolutionarily diversified.
Conclusion
The present study identified new members of soybean bHLH and trihelix-GT transcription factor families, some of which seem to be involved in responses to environmental stresses. It also emphasizes the role of duplication events in the expansion and evolution of soybean transcription factor families, indicating that exciting new layers of complexity might exist in this species’ regulatory mechanisms, including biotic and abiotic stress responses.
Acknowledgments
This research project was supported by CNPq, CAPES, FAPERGS-PRONEX and GENOSOJA/CNPq.
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Urao T, Ito T, Seki M, Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell. 2004;16:3460–3479. doi: 10.1105/tpc.104.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoutzias GD, Robertson DL, Oliver SG, Bornberg-Bauer E. Convergent evolution of gene networks by single-gene duplications in higher eukaryotes. EMBO Rep. 2004;5:274–279. doi: 10.1038/sj.embor.7400096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Howles PA, Dorian K, Griffith ME, Ishida T, Kaplan-Levy RN, Kilinc A, Smyth DR. PETAL LOSS, a trihelix transcription factor gene, regulates perianth architecture in the Arabidopsis flower. Development. 2004;131:4035–4045. doi: 10.1242/dev.01279. [DOI] [PubMed] [Google Scholar]
- Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martinez-Garcia JF, Bilbao-Castro JR, Robertson DL. Genome wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss and algae. Plant Physiol. 2010;153:1398–1412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JW, Cole M, Flier A, Werr W. BIM1, a bHLH protein involved in brassinosteroid signalling, controls Arabidopsis embryonic patterning via interaction with DORNROSCHEN and DORNROSCHEN-LIKE. Plant Mol Biol. 2009;69:57–68. doi: 10.1007/s11103-008-9405-6. [DOI] [PubMed] [Google Scholar]
- Chen M, Xu Z, Xia L, Li L, Cheng X, Dong J, Wang Q, Ma Y. Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean (Glycine max L.) J Exp Bot. 2009;60:121–135. doi: 10.1093/jxb/ern269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- Dehesh K, Bruce WB, Quail PH. A trans-acting factor that binds to a GT-motif in a phytochrome gene promoter. Science. 1990;250:1397–1399. doi: 10.1126/science.2255908. [DOI] [PubMed] [Google Scholar]
- EMBRAPA. 10th edition. Embrapa Soja, Londrina: 2007. Tecnologias de Produção de Soja; p. 225. [Google Scholar]
- Eulgem T. Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 2005;10:71–78. doi: 10.1016/j.tplants.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Fang Y, Xie K, Hou X, Hu H, Xiong L. Systematic analysis of GT factor family of rice reveals a novel subfamily involved in stress responses. Mol Genet Genomics. 2010;283:157–169. doi: 10.1007/s00438-009-0507-x. [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico J-M, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23:701–15. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics. 2002;162:1445–1456. doi: 10.1093/genetics/162.3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M-J, Lydiate DJ, Li X, Lui H, Gjetvaj B, Hegedus DD, Rozwadowski K. Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell. 2009;21:54–71. doi: 10.1105/tpc.108.061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepts P, Beavis WD, Brummer EC, Shoemaker RC, Stalker HT, Weeden NF, Young ND. Legumes as a model plant family: Genomics for food and feed report of the cross-legume advances through genomics conference. Plant Physiol. 2005;137:1228–1235. doi: 10.1104/pp.105.060871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PJ, Kay SA, Chua N-H. Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J. 1987;6:2543–2549. doi: 10.1002/j.1460-2075.1987.tb02542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PJ, Yong M-H, Cuozzo M, Kano-Murakami Y, Silverstein P, Chua N-H. Binding site requirements for pea nuclear protein factor GT-1 correlate with sequences required for light-dependent transcriptional activation of the rbcS-3A gene. EMBO J. 1988;7:4035–4044. doi: 10.1002/j.1460-2075.1988.tb03297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith ME, Conceição AS, Smyth DR. PETAL LOSS gene regulates initiation and orientation of second whorl organs in the Arabidopsis flower. Development. 1999;126:5635–5644. doi: 10.1242/dev.126.24.5635. [DOI] [PubMed] [Google Scholar]
- Haberer G, Hindemitt T, Meyers BC, Mayer KFX. Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol. 2004;136:3009–3022. doi: 10.1104/pp.104.046466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20:735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- Hiratsuka K, Wu X, Fukuzawa H, Chua NH. Molecular dissection of GT-1 from Arabidopsis. Plant Cell. 1994;6:1805–1813. doi: 10.1105/tpc.6.12.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma K, Nishiuchi T, Kato T, Bednarek P, Okuno T, Schulze-Lefert P, Takano Y. Arabidopsis ENHANCED DISEASE RESISTANCE 1 is required for pathogen-induced expression of plant defensins in nonhost resistance and acts through interference of MYC2-mediated repressor function. Plant J. 2011;67:980–992. doi: 10.1111/j.1365-313X.2011.04651.x. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Caspar T, Dehesh K, Quail PH. DNA binding factor GT-2 from Arabidopsis. Plant Mol Biol. 1993;23:337–348. doi: 10.1007/BF00029009. [DOI] [PubMed] [Google Scholar]
- Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Qin G, Chen Z, Gu H, Qu L-J. A gain-of-function mutation of transcriptional factor PTL results in curly leaves, dwarfism and male sterility by affecting auxin homeostasis. Plant Mol Biol. 2008;66:315–327. doi: 10.1007/s11103-007-9272-6. [DOI] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell. 2010;22:2219–2236. doi: 10.1105/tpc.110.074096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sa JJ. JASMONATE-INSEN-SITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig SR, Habera LF, Dellaporta SL, Wessler SR. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci USA. 1989;86:7092–7096. doi: 10.1073/pnas.86.18.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal E, Hiratsuka K, Delgado J, Nairn A, Qin J, Chait BT, Chua NH. Modulation of GT-1 DNA-binding activity by calcium-dependent phosphorylation. Plant Mol Biol. 1999;40:373–386. doi: 10.1023/a:1006131330930. [DOI] [PubMed] [Google Scholar]
- McCarty DR, Chory J. Conservation and innovation in plant signaling pathways. Cell. 2000;103:201–209. doi: 10.1016/s0092-8674(00)00113-6. [DOI] [PubMed] [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer DG, Dolanet L. An ancient mechanism controls the development of cells with a rooting function in land plants. Science. 2007;316:1477–1480. doi: 10.1126/science.1142618. [DOI] [PubMed] [Google Scholar]
- Miles MR, Frederick RD, Hartman GL. Evaluation of soybean germplasm for resistance to Phakopsora pachyrhizi. Plant Health Progr. 2006. [DOI]
- Morgenstern B, Atchley WR. Evolution of bHLH transcription factors: Modular evolution by domain shuffling? Mol Biol Evol. 1999;16:1654–1663. doi: 10.1093/oxfordjournals.molbev.a026079. [DOI] [PubMed] [Google Scholar]
- Nagano Y, Inaba T, Furuhashi H, Sasaki Y. Trihelix DNA-binding protein with specificities for two distinct cis-elements: Both important for light down-regulated and dark-inducible gene expression in higher plants. J Biol Chem. 2001;276:22238–22243. doi: 10.1074/jbc.M102474200. [DOI] [PubMed] [Google Scholar]
- Nagata T, Niyada E, Fujimoto N, Nagasaki Y, Noto K, Miyanoiri Y, Murata J, Hiratsuka K, Katahira M. Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: Mechanism for an increase in DNA-binding affinity through phosphorylation. Proteins. 2010;78:3033–3047. doi: 10.1002/prot.22827. [DOI] [PubMed] [Google Scholar]
- Nascimento LC, Costa GGL, Binneck E, Pereira GAG, Carazzolle MF. Databases and bioinformatics pipelines of the Genosoja project. Genet Mol Biol. 2012;35(suppl 1):203–211. doi: 10.1590/S1415-47572012000200002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Grady K, Goekjian VH, Naim CJ, Nagao RT, Key JL. The transcript abundance of GmGT-2, a new member of the GT-2 family of transcription factors from soybean, is down-regulated by light in a phytochrome-dependent manner. Plant Mol Biol. 2001;47:367–378. doi: 10.1023/a:1011629307051. [DOI] [PubMed] [Google Scholar]
- Park HC, Kim ML, Kang YH, Jeon JM, Yoo JH, Kim MC, Park CY, Jeong JC, Moon BC, Lee JH, et al. Pathogen-and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 2004;135:2150–2161. doi: 10.1104/pp.104.041442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro GL, Marques CS, Costa MDBL, Reis PAB, Alves MS, Carvalho CM, Fietto LG, Fontes EPB. Complete inventory of soybean NAC transcription factors: Sequence conservation and expression analysis uncover their distinct roles in stress response. Gene. 2009;444:10–23. doi: 10.1016/j.gene.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol. 2010;27:862–874. doi: 10.1093/molbev/msp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B, Rozhon W, Khan M, Husar S, Adam G, Luschnig C, Fujioka S, Sieberer T. CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J. 2011;30:1149–1161. doi: 10.1038/emboj.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priolli RHG, Mendes-Junior CT, Arantes NE, Contel EPB. Characterization of Brazilian soybean cultivars using microsatellite markers. Genet Mol Biol. 2002;25:185–193. [Google Scholar]
- Rao KVM, Raghavendra AS, Reddy KJ. Physiology and Molecular Biology of Stress Tolerance in Plants. Springer; Amsterdam: 2006. p. 345. [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud P-F, Lindquist EA, Kamisugi Y, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Seo J-S, Joo J, Kim M-J, Kim Y-K, Nahm BH, Song SI, Cheong J-J, Lee JS, Kim J-K, Choi YD. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011;65:907–921. doi: 10.1111/j.1365-313X.2010.04477.x. [DOI] [PubMed] [Google Scholar]
- Singh KB, Foley RC, Sánchez LO. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol. 2002;5:430–436. doi: 10.1016/s1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- Szécsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, Bendahmane M. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J. 2006;25:3912–3920. doi: 10.1038/sj.emboj.7601270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software ver. 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: Improving the sensitivy of progressive multiple alignment through sequences weighting, position-specific gap penalties and wight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian A-G, Wang J, Cui P, Han Y-J, Xu H, Cong L-J, Huang X-G, Wang X-L, Jiao Y-Z, Wang B-J, et al. Characterization of soybean genomic features by analysis of its expressed sequence tags. Theor Appl Genet. 2004;108:903–913. doi: 10.1007/s00122-003-1499-2. [DOI] [PubMed] [Google Scholar]
- Todd AT, Liu E, Polvi SL, Pammett RT, Page JE. A functional genomics screen identifies diverse transcription factors that regulate alkaloid biosynthesis in Nicotiana benthamiana. Plant J. 2010;62:589–600. doi: 10.1111/j.1365-313X.2010.04186.x. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Hong G, Han B. Transcript abundance of rml1, encoding a putative GT1-like factor in rice, is up-regulated by Magnaporthe grisea and down-regulated by light. Gene. 2004;324:105–115. doi: 10.1016/j.gene.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Xie Z-M, Zou H-F, Lei G, Wei W, Zhou Q-Y, Niu C-F, Liao Y, Tian A-G, Ma B, Zhang W-K, et al. Soybean trihelix transcription factors GmGT-2A and GmGT-2B improve plant tolerance to abiotic stresses in transgenic Arabidopsis. PloS One. 2009;4:e6898. doi: 10.1371/journal.pone.0006898. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P. OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol. 2005;138:2087–2096. doi: 10.1104/pp.105.063115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioda K, Shinozaki K. Signal Crosstalk in Plant Stress Responses. Wiley-Blackwell; Ames: 2009. p. 210. [Google Scholar]
- Zhang G, Chen M, Li L, Xu Z, Chen X, Guo J, Ma Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot. 2009;60:3781–3796. doi: 10.1093/jxb/erp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Ying Y, Wang L, Wang F, Whelan J, Shou H. Identification of a novel iron regulated basic helix-loop-helix protein involved in Fe homeostasis in Oryza sativa. BMC Plant Biol. 2010;10:e166. doi: 10.1186/1471-2229-10-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D-X. Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci. 1999;4:210–214. doi: 10.1016/s1360-1385(99)01418-1. [DOI] [PubMed] [Google Scholar]
- Zhou Q-Y, Tian A-G, Zou H-F, Xie Z-M, Lei G, Huang J, Wang C-M, Wang H-W, Zhang J-S, Chen S-Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J. 2008;6:486–503. doi: 10.1111/j.1467-7652.2008.00336.x. [DOI] [PubMed] [Google Scholar]
Internet Resources
- LGE Soybean Genome database, http://www.lge.ibi.unicamp.br/soja/ (October 21, 2011).
- Fancy Gene v1.4, http://host13.bioinfo3.ifom-ieocampus.it/fancygene/ (October 21, 2011).
- FigTree software, http://tree.bio.ed.ac.uk/software/figtree/ (October 21, 2011).
- JGI - Joint Genome Institute, http://www.jgi.doe.gov (October 21, 2011).
- LGE Soybean Genome database, http://bioinfo03.ibi.unicamp.br/soja/ (October 21, 2011).
- MEME (multiple EM for motif elicitation) software, http://meme.sdsc.edu/meme4_4_0/cgi-bin/meme.cgi (October 21, 2011).
- Programa Nacional de Produção e Uso de Biodiesel, http://www.biodiesel.gov.br/ (October 21, 2011).
- Phytozome database, http://www.phytozome.org (October 21, 2011).
- SMART database, http://smart.embl-heidelberg.de/ (October 21, 2011).