Abstract
Heat shock (HS) leads to the activation of molecular mechanisms, known as HS-response, that prevent damage and enhance survival under stress. Plants have a flexible and specialized network of Heat Shock Factors (HSFs), which are transcription factors that induce the expression of heat shock proteins. The present work aimed to identify and characterize the Glycine max HSF repertory in the Soybean Genome Project (GENOSOJA platform), comparing them with other legumes (Medicago truncatula and Lotus japonicus) in view of current knowledge of Arabidopsis thaliana. The HSF characterization in leguminous plants led to the identification of 25, 19 and 21 candidate ESTs in soybean, Lotus and Medicago, respectively. A search in the SuperSAGE libraries revealed 68 tags distributed in seven HSF gene types. From the total number of obtained tags, more than 70% were related to root tissues (water deficit stress libraries vs. controls), indicating their role in abiotic stress responses, since the root is the first tissue to sense and respond to abiotic stress. Moreover, as heat stress is related to the pressure of dryness, a higher HSF expression was expected at the water deficit libraries. On the other hand, expressive HSF candidates were obtained from the library inoculated with Asian Soybean Rust, inferring crosstalk among genes associated with abiotic and biotic stresses. Evolutionary relationships among sequences were consistent with different HSF classes and subclasses. Expression profiling indicated that regulation of specific genes is associated with the stage of plant development and also with stimuli from other abiotic stresses pointing to the maintenance of HSF expression at a basal level in soybean, favoring its activation under heat-stress conditions.
Keywords: HSF, Fabaceae, bioinformatics, abiotic stress, transcription factor
Introduction
Heat stress is one of the major factors limiting the productivity and adaptation of crops, especially when temperature extremes coincide with critical stages of plant development. The major developmental performance of plants occurs at a temperature regime between 10° and 40 °C. Temperatures below or above this range generally cause temperature-induced stresses (Treshow, 1970; Hsu et al., 2010). In the case of heat stress, both the rate of temperature change and the duration and degree of high temperatures contribute to the intensity of heat stress. The degree of inherent adaptedness to heat stress of a plant is an important determinant of its ability to survive a stress period (Efeoglu, 2009). However, the expression of HSF and HSP genes has been also observed under other abiotic and biotic stresses, as cited by Pirkkala et al. (2001). In response to various inducers such as elevated temperatures, salinity, drought, oxidants, heavy metals, bacterial and viral infections, most HSFs acquire DNA binding activity to the heat shock element (HSE), thereby mediating transcription of the heat shock factor genes, which results in accumulation of heat shock proteins (HSPs). Among important transcription factors, heat shock factors (HSFs) are essential for the transcription of many HSP coding genes that are active in response to sublethal heat stress leading to increased tolerance against a subsequent, otherwise lethal, heat shock (Treshow, 1970; Hsu et al., 2010).
After stress perception, intracellular changes lead to a molecular cascade of events, initiated by HSF activation and subsequent expression of HSPs limiting stress damage (Hsu et al., 2010). In general, HSF proteins have a common core structure comprising a N-terminal DNA binding domain (DBD) characterized by a central helix-turn-helix (HTH) motif, an adjacent domain with a heptad hydrophobic repeat (HR-A/B) which is involved in oligomerization, a short peptide motif essential for nuclear import [nuclear localization signal (NLS)] and export [nuclear export signal (NES)], and a C-terminal AHA type activation domain (Mittal et al., 2009; Hsu et al., 2010).
Through the DNA binding domain, activated HSFs bind to conserved cis-acting elements called heat shock elements (HSEs). HSEs are located in the promoters of HSP genes and are defined as adjacent and inverse repeats of the motif 5-nGAAn-3, for instance 5-nGAAnnTTCnnGAAn-3 (Schöffl et al., 1998).
Some HSFs have been cloned and characterized from various plant species (Nover et al., 2001; Baniwal et al., 2007) revealing that the network of HSF genes is highly flexible and specialized in this group. Details regarding the overall HS response network were initially not clear. However, studies in Arabidopsis revealed that 21 HSFs form a complex network, in which AtHSFA1a and AtHSFA1b play important roles in the induction of HSP genes in the early stage of HSR (Nover et al., 2001).
An insight into the response of HSPs and HSFs to different abiotic stresses was provided through a number of genome-wide microarray datasets. Arabidopsis HSFs and HSPs were strongly induced by heat, cold, salinity and osmotic stresses. Furthermore, overlapping responses of HSPs and HSFs to heat and other abiotic stresses was reported, indicating that these genes are important elements in the crosstalk among different response pathways (Hu et al., 2009). In rice, over-expression of OsHsp17.7 enhanced rice tolerance to heat UV-B as well as to drought (Sato and Yokoya, 2008).
Hu et al. (2009) identified rice HSF and HSP genes and analyzed their expression profiles under different abiotic stresses. A whole-genome microarray analysis was carried out to investigate expression changes of rice HSFs and HSPs genes in response to heat stress. By comparing their experimental data with other expression data under salt, cold, and drought conditions, Hu et al. (2009) found that the rice HSF and HSP families responded to different stresses in an overlapping relationship. The analysis also indicated that some HSF and HSP genes exhibited specific expression patterns in response to distinct stress types.
In Arabidopsis, for example, the major role of the representatives of the HsfA4/A5 group, which is generally not involved in the conventional heat stress response, may reside in cell type-specific functions connected with the control of cell death triggered by pathogen infection and/or reactive oxygen species (Baniwal et al., 2007).
Although the flexible network of HSF genes has been well studied in plants, there is little information available regarding the structure and function of HSF genes in legumes. Additionally, no comparison of HSF orthologs has been carried out until now among legumes. In this study, we used well-described Arabidopsis HSF proteins as seed sequences in order to identify and characterize the pool of HSF genes present in the Glycine max genome and perform a comparative analysis against Lotus japonicus and Medicago truncatula genomes, so as to trace the panorama of the HSF genes in these leguminous plants.
Material and Methods
Based on 21 well-described Arabidopsis HSF genes in the AfTDB database, BLASTp searches (Altschul et al., 1990) were carried out for similar sequences against the GENOSOJA database. GENOSOJA connects public and project soybean data (Nascimento et al., 2012). In total, the initiative provides information on 60,747 unigenes from the NBCI, Phytozome and Soybean full-length cDNA databases (Nascimento et al., 2012). Comparative searches were made in the Medicago truncatula and Lotus japonicus databases. After searching the GENOSOJA databank, only orthologs presenting the fully characteristic HSF DNA-Binding Domain (DBD) were considered for subsequent analysis. In view of the obtained soybean, Medicago and Lotus HSF candidates together with the Arabidopsis seed-sequences, a comparative analysis with 69 aligned proteins was performed, enabling the generation of a dendrogram, using the Neighbor-Joining (NJ) method with 2,000 bootstrap replications with program MEGA program v. 5.0 (Tamura et al., 2011), to infer about HSF groups and classes within the analyzed legumes. For this purpose the sequence-coding genes from Arabidopsis that did not present similarity (orthology) with the studied legumes were excluded from the phenetic analysis. To prevent the influence of different sequence sizes, the alignments were trimmed aiming to exclude unequal 5′ and 3′ extremities.
To evaluate the HSF-related tags represented in the SuperSAGE libraries generated by the GENOSOJA project, a comparative analysis using the same seed sequences and the MegaBLAST algorithm was carried out according to Altschul et al. (1990). For this purpose the parameters were adjusted to an e-value equal to or less than 0.1 and word size equal to 7. The low complexity filter was deactivated. Results considered only tags with identity equal to or larger than 23 bp.
The GENOSOJA databank is comprised of six SuperSAGE libraries and allowed the generation of three comparisons, including two from root tissues subjected to water deficit stress and one inoculated with Asian Soybean Rust fungus (Phakopsora pachyrhizi). For the water deficit libraries, seeds of a drought tolerant cultivar (Embrapa 48) and a drought susceptible cultivar (BR 16) were germinated on filter paper for four days in a growth chamber at 25 ± 1 °C and 100% relative humidity (RH). Seedlings were placed in 36 L boxes containing 50% Hoagland’s solution (Hoagland and Arnon, 1950) continuously aerated and replaced on a weekly basis. These boxes were then transferred to a greenhouse under natural photoperiod of approximately 12/12 h light/dark cycle, temperature of 30 ± 5 °C and 60 ± 10% RH. The plants were allowed to grow until the V4 stage (Fehr et al., 1971). The experimental plan was a randomized complete block 2x7 factorial design with three repetitions. The treatments included two cultivars (BR 16 and Embrapa 48) and seven water deficit periods (0, 25, 50, 75, 100, 125 and 150 min). Water stress was applied by removing the plants from the hydroponic solution and leaving them in boxes without nutrient solution for up to 150 min under ambient-air exposure. For each stress exposure time, roots from 10 plants were collected, pooled and frozen in liquid nitrogen before storage at −80 °C. The above mentioned exposure times were bulked together generating a library from drought tolerant genotype Embrapa 48 after stress as compared with the negative control (T0); the same procedure was also applied to the drought sensitive genotype (BR16 cultivar). The comparison regarding Asian Soybean Rust infection was generated from leaves of the resistant cultivar PI561356 collected at different times (12, 24 and 48 h) after spraying with a P. pachyrhizi spore suspension (6 x 105 uredospores.mL−1). The urediniospores were collected from Phakopsora pachyrhizi infected soybean fields in the state of Mato Grosso, Brazil, and maintained for over 10 generations on the susceptible cv. BRSMS-Bacuri. The suspension of spores was sprayed onto three plants per pot at the V2 to V3 growth stage (Fehr and Caviness, 1977). The same solution without the spores was used for the false inoculations (Mock). The different times were bulked together to form a single resistant library, which was compared with the false inoculated negative control collected at the same time points.
Considering the identified G. max EST transcripts, standard statistical methods (see Eisen et al., 1998) were used to arrange the HSF genes according to their gene expression pattern, generating a graphic with colors (green, red and black) indicating their quantitative and qualitative expression (down-, up- and unregulated genes, respectively), while gray stood for absence of information. The gene expression data analyzed were collected from soybean during a variety of challenging and control conditions available at the GENOSOJA database. So as to obtain a picture of how HSFs contribute to sensing the environmental up-shifts in temperature, we applied Self-Organizing Maps followed by pairwise average-linkage cluster analysis to normalized gene expression data (Eisen et al., 1998). Relationships among genes and libraries were represented by dendrograms in which branch lengths reflect the degree of gene co-expression.
An available genome browser for soybean (Phytozome database) was used to anchor identified EST candidate sequences on G. max virtual chromosomes, aiming to identify their distribution, relative position, and abundance. For this purpose the MegaBLAST tool was used to identify the exact location of the HSF genes in the genome, using at least 80% identity as a parameter. For the construction of a virtual karyotype representation, a CorelDRAW12 graphic application was used. The soybean chromosome information for the schematic representation was obtained from the SOYBASE site. For the design of chromosomes, considering the need for high-resolution bands (data anchored in the genome), a proportion of 1:1 (cm:Mb) was adopted for all chromosomes; thus, for the sequence positioning, each millimeter corresponded to 100,000 bp. On the representation each transversal black line corresponded to an HSF gene.
Results and Discussion
Heat and cold can have damaging consequences on both vegetative and reproductive tissues. Temperature changes can also regulate plant movements, resetting internal clocks and diurnal synchronization, flowering and germination in some species (Ruelland and Zachowski, 2010). Moreover, temperature changes can induce metabolic changes so that plants adapt and tolerate moderate cold, freezing and heat stresses (Ruelland and Zachowski, 2010). HSFs are important components of the heat shock regulatory network, with a single gene identified for yeast and drosophila, while vertebrates accounted with only four genes of this category (Swindell et al., 2007). Nevertheless, unlike other organisms, plant genomes encode extraordinarily complex HSF families, both in terms of the total number of genes (usually more than 20), as well as in terms of their structural and functional diversification (Nover et al., 2001). This abundance and diversity can be also seen in legumes. An extensive BLAST search of Arabidopsis HSF orthologs in soybean, Lotus and Medicago EST databases led to the identification of a total of 25, 19 and 21 expressed sequences, respectively (Table 1).
Table 1.
Arabidopsis thaliana Heat Shock Factors used as seed sequences and respective matches from Glycine max, Lotus japonica and Medicago truncatula, with corresponding domain information. Abbreviations: HSF = Heat Shock Factor; ID, identification; Gm, Glycine max; Lj, Lotus japonicus; Mt, Medicago truncatula.
| Arabidopsis seed-sequence (Loci) | Orthologous information
|
BLAST results
|
|||
|---|---|---|---|---|---|
| Fabaceae species | Sequence ID | E-value | Score | HSF domain | |
| ATHSFA1A (AT4G17750.1) | Glycine max | Gm_HSFA1A.1 | 1.00 e−77 | 166 | Complete |
| Lotus japonicus | Lj_HSFA1A.1 | 3.00 e−30 | 88 | Incomplete | |
| Medicago truncatula | Mt_HSFA1A.1 | 1.00 e−106 | 381 | Complete | |
|
| |||||
| ATHSFA1B (AT5G16820.1) | - | - | - | - | - |
|
| |||||
| ATHSFA1D (AT1G32330.1) | Lotus japonicus | Lj_HSFA1D.1 | 4.00 e−67 | 406 | Incomplete |
|
| |||||
| ATHSFA1E (AT3G02990.1) | Glycine max | Gm_HSFA1E.1 | 2.00 e−32 | 159 | Complete |
| Gm_HSFA1E.2 | 5.00 e−23 | 124 | Complete | ||
| Lotus japonicus | Lj_HSFA1E.1 | 4.00 e−91 | 487 | Complete | |
| Medicago truncatula | Mt_HSFA1E.1 | 6.00 e−98 | 354 | Complete | |
| Mt_HSFA1E.2 | 6.00 e−98 | 354 | Complete | ||
|
| |||||
| ATHSFA2 (AT2G26150.1) | Glycine max | Gm_HSFA2.1 | 6.00 e−30 | 68.9 | Complete |
| Lotus japonicus | Lj_HSFA2.1 | 4.00 e−91 | 387 | Complete | |
| Lj_HSFA2.2 | 1.00 e−64 | 373 | Incomplete | ||
| Medicago truncatula | Mt_HSFA2.1 | 4.00 e−92 | 334 | Complete | |
|
| |||||
| ATHSFA3 (AT5G03720.1) | Lotus japonicus | Lj_HSFA3.1 | 1.00 e−74 | 324 | Complete |
|
| |||||
| ATHSFA4A (AT4G18880.1) | Glycine max | Gm_HSFA4A.1 | 1.00 e−82 | 143 | Complete |
| Gm_HSFA4A.2 | 7.00 e−12 | 121 | Complete | ||
| Lotus japonicus | Lj_HSFA4A.1 | 3.00 e−90 | 401 | Complete | |
| Lj_HSFA4A.2 | 6.00 e−89 | 393 | Complete | ||
| Medicago truncatula | Mt_HSFA4A.1 | 1.00 e−84 | 309 | Complete | |
|
| |||||
| ATHSFA4C (AT5G45710.1) | Medicago truncatula | Mt_HSFA4C.1 | 9.00 e−47 | 183 | Complete |
|
| |||||
| ATHSFA5 (AT4G13980.1) | Glycine max | Gm_HSFA5.1 | 2.00 e−58 | 117 | Complete |
| Gm_HSFA5.2 | 1.00 e−109 | 392 | Complete | ||
| Medicago truncatula | Mt_HSFA5.1 | 1.00 e−104 | 374 | Complete | |
| Mt_HSFA5.2 | 2.00 e−72 | 269 | Complete | ||
| Mt_HSFA5.3 | 2.00 e−61 | 232 | Complete | ||
| Mt_HSFA5.4 | 1.00 e−21 | 100 | Incomplete | ||
| Mt_HSFA5.5 | 2.00 e−15 | 79.7 | Incomplete | ||
|
| |||||
| ATHSFA6A (AT5G43840.1) | - | - | - | - | - |
|
| |||||
| ATHSFA6B (AT3G22830.1) | Glycine max | Gm_HSFA6B.1 | 1.00 e−49 | 153 | Complete |
| Lotus japonicus | Lj_HSFA6B.1 | 9.00 e−82 | 291 | Complete | |
| Lj_HSFA6B.2 | 7.00 e−81 | 311 | Complete | ||
| Lj_HSFA6B.3 | 7.00 e−97 | 347 | Complete | ||
| Medicago truncatula | Mt_HSFA6B.1 | 1.00 e−66 | 249 | Complete | |
|
| |||||
| ATHSFA7A (AT3G51910.1) | Glycine max | Gm_HSFA7B.1 | 5.00 e−21 | 117 | Incomplete |
|
| |||||
| ATHSFA7B (AT3G63350.1) | - | - | - | - | - |
|
| |||||
| ATHSFA8 (AT1G67970.1) | Glycine max | Gm_HSFA8.1 | 3.00 e−63 | 131 | Complete |
| Gm_HSFA8.2 | 0.0 | 678 | Complete | ||
| Lotus japonicus | Lj_HSFA8.1 | 9.00 e−65 | 318 | Complete | |
| Medicago truncatula | Mt_HSFA8.1 | 1.00 e−61 | 233 | Complete | |
|
| |||||
| ATHSFA9 (AT5G54070.1) | - | - | - | - | - |
|
| |||||
| ATHSFB1 (AT4G36990.1) | Glycine max | Gm_HSFB1.1 | e−144 | 508 | Complete |
| Gm_HSFB1.2 | e−127 | 450 | Complete | ||
| Lotus japonicus | Lj_HSFB1.1 | 3.00 e−18 | 187 | Incomplete | |
| Lj_HSFB1.2 | 1.00 e−26 | 68 | Incomplete | ||
| Medicago truncatula | Mt_HSFB1.1 | 4.00 e−51 | 197 | Complete | |
| Mt_HSFB1.2 | 3.00 e−36 | 148 | Complete | ||
| Mt_HSFAB1.3 | 7.00 e−19 | 90.9 | Incomplete | ||
|
| |||||
| ATHSFB2A (AT5G62020.1) | Glycine max | Gm_HSFB2A.1 | 3.00 e−48 | 80.5 | Complete |
| Gm_HSFB2A.2 | 5.00 e−73 | 270 | Complete | ||
| Lotus japonicus | Lj_HSFB2A.1 | 9.00 e−30 | 197 | Complete | |
| Medicago truncatula | Mt_HSFB2B.1 | 3.00 e−06 | 48.9 | Incomplete | |
|
| |||||
| ATHSFB2B (AT4G11660.1) | Glycine max | Gm_HSFB2B.1 | 2.00 e−44 | 222 | Complete |
| Gm_HSFB2B.2 | 5.00 e−81 | 296 | Complete | ||
| Lotus japonicus | Lj_HSFB2B.1 | 8.00 e−68 | 280 | Complete | |
| Medicago truncatula | Mt_HSFB2B.1 | 6.00 e−67 | 251 | Complete | |
|
| |||||
| ATHSFB3 (AT2G41690.1) | Glycine max | Gm_HSFB3.1 | 1.00 e−47 | 160 | Complete |
| Gm_HSFB3.2 | 6.00 e−32 | 97.4 | Complete | ||
| Lotus japonicus | Lj_HSFB3.1 | 5.00 e−56 | 194 | Complete | |
| Medicago truncatula | Mt_HSFB3.1 | 1.00 e−51 | 199 | Complete | |
|
| |||||
| ATHSFB4 (AT1G46264.1) | Glycine max | Gm_HSFB4.1 | 3.00 e−61 | 96.3 | Complete |
| Gm_HSFB4.2 | 1.00 e−44 | 86.3 | Complete | ||
| Gm_HSFB4.3 | 1.00 e−126 | 448 | Complete | ||
| Gm_HSFB4.4 | 1.00 e−120 | 427 | Complete | ||
| Lotus japonicus | Lj_HSFB4.1 | 1.00 e−66 | 202 | Incomplete | |
| Lj_HSFB4.2 | 1.00 e−19 | 52 | Complete | ||
| Medicago truncatula | Mt_HSFB4.1 | 2.00 e−89 | 325 | complete | |
| Mt_HSFB4.2 | 8.00 e−65 | 243 | Complete | ||
|
| |||||
| ATHSFC1 (AT3G24520.1) | Glycine max | Gm_HSFC1.1 | 8.00 e−60 | 73.6 | Complete |
HSF expressed sequence tags
The characteristic HSF domains were complete in 24, 13 and 17 orthologous candidates identified among the three species, respectively (Table 1). From the 21 types of Arabidopsis HSF genes only 13 types were identified in soybean and Lotus and 14 in Medicago (Table 1). In our evaluation, HSFA1B, HSFA6A, HSFA7B and HSFA9 were absent in all species analyzed (Table 1). HSFA1A and HSFA1B interact as regulators responsible for immediate-early transcription of a subset of HS genes in Arabidopsis, and are independently important for the initial phase of HS-responsive gene expression, while their interaction enhances the expression of their target genes (Li et al., 2010). The absence of HSFA1B may render soybean more sensitive to heat stress but another class A HSF may alternately play this role (Sung et al., 2003; Kotak et al., 2004; Li et al., 2004). Whether another gene substitutes the role of HSFA1B in soybean could be tested by heterologous expression of HSFA1B; in the case of the existence of different pathways, the over-expression of HSFA1B might change the performance of soybean plants, especially under heat stress.
Other members of class A, such as HSFA9, are less active or may be active only under certain conditions. The reason seems to be the presence of interesting regulators (HSFs or other transcription factors) with specialized functions. In fact, HSFA9 was found to be specific to seed development in sunflower and was exclusively detected in yellow siliques of Arabidopsis mRNA (Kotak et al., 2004). Hence the lack of identification of some HSF classes may correlate with specialized functions other than those represented among the conditions analyzed herein.
A similar result was reported by Nover et al. (2001) after carrying out an analysis of HSFs in A. thaliana. Among the 21 described genes, HSFs A3, A6A, A6B, A7B, B2A and B3 could not be detected in any of the tissues analyzed (etiolated seedlings, roots, leaves from vegetative plants stems, flowers, siliques, and developing seeds) or conditions (heat stressed leaves and cell cultures vs. control). According to the authors it was not surprising that no matching EST was found in libraries created exclusively from RNA isolated from control tissues; a serious limitation of the data from EST libraries for these studies is the lack of samples from heat stressed tissues.
Comparing the obtained results with the data available in the Legume Transcription Factor database (Legume TFDB, Mochida et al., 2009a) an increased number of Lotus and Medicago HSF representatives were observed, since the LegumeTFDB includes 18 and 16 genes, respectively, and our searches identified 19 and 21, respectively, revealing that both organisms presented a similar number of HSFs as Arabidopsis. However, the results considering the ESTs deposited at the GENOSOJA platform revealed a surprisingly low number of HSFs (25 sequences) as compared to the LegumeTFDB information (65 sequences). This may be due to the type of databases (LegumeTFDB is sourced from large-scale shotgun sequencing whereas GENOSOJA is sourced from transcriptomic approaches), besides the fact that the LegumeTFDB bank considers both HSF and HSF-like sequences with data annotation based on different databanks (NCBI nr, A. thaliana, TIGR rice, L. japonicus, M. truncatula, Populus trichocarpa and UniProt). On the other hand, Kotak et al. (2004) listed 34 soybean sequences, a higher number of HSF representatives than those in GENOSOJA, but these authors did not indicate the methods and procedures used in the acquisition of these HSFs. Finally, the soybean candidates identified herein represent the active (expressed) HSFs bearing the complete DBD-domain. This set size was similar to that described for Arabidopsis and also for the Lotus and Medicago orthologs identified in this study; both being evolutionarily closely related species when compared to soybean (Fabaceae family, Papilionoideae subfamily).
Notwithstanding, it is important to highlight that evolutionary studies and haploid genome analysis suggested that the soybean genome experienced a tetraploidization event approximately 10–15 million years ago. Since then, the soybean genome has gone through gene rearrangements and deletions, reverting to diploid state. Therefore, soybean multigene families, including the heat shock factor family, may contain highly related but diversified genes (Mochida et al., 2009b).
HSF matching to SuperSAGE tags
Regarding SuperSAGE, 68 different tags could be identified, including 26 tags unique to water deficit experiments with the tolerant comparison (water deficit stressed Embrapa 48 vs. control), 28 tags unique to water deficit experiments with the susceptible comparison (water deficit stressed BR16 vs. control) and 14 regarding Asian Soybean Rust (PI561356 inoculated vs. control) (Table 2; Figure 1). No common tags were identified. It is important to note that among 25 HSF EST clusters, 18 had no representative in the tags database, while five clusters were represented in all libraries. The sequence Gmax_HSFB1_SJ09-E1-R06-064-B09-UC.F was not identified in ‘Embrapa 48’ and ‘PI561356’ libraries, and Gmax_HSFB3_Contig20961 was present in the water deficit stressed libraries only. When looked at from a different point of view, from the 14 HSF types compared, only six HSF types (HSFB1, HSFA1E, HSFB2A, HSFB3, HSFA8 and HSFA4A) were identified (Table 2; Figure 1), indicating their induction during the stress response.
Table 2.
Distribution of unique SuperSAGE tags in the three comparisons considered, as compared with the identified soybean EST contigs.
| Soybean Contig | Drought Embrapa 48 (tolerant) vs. Control | Drought BR16 (susceptible) vs. Control | Asian Rust PI561356 vs. Control | Total |
|---|---|---|---|---|
| Gmax_HSFB1_Contig12262 | 16 | 17 | 3 | 36 |
| Gmax_HSFA1E_Contig12828 | 2 | 3 | 4 | 9 |
| Gmax_HSFB2A_Contig14439 | 3 | 2 | 3 | 8 |
| Gmax_HSFB3_Contig20961 | 2 | 2 | 0 | 4 |
| Gmax_HSFA8_Contig22657 | 1 | 1 | 1 | 3 |
| Gmax_HSFA4A_Contig4226 | 2 | 2 | 3 | 7 |
| Gmax_HSFB1_SJ09-E1-R06-064-B09-UC.F | 0 | 1 | 0 | 1 |
| Total | 26 | 28 | 14 | 68 |
Figure 1.
Graphic representation of SuperSAGE tag distribution per cluster and compared libraries. A) Quantitative analysis (number of unitags per category). B) Qualitative analysis of tag prevalence (in %).
Despite the small number of identified sequences in the Asian rust ‘PI561356’ stress analysis, when compared to water deficit experiments, the presence of HSF representatives indicates the involvement of HS-response also during biotic stresses. The stress condition by itself can activate non-specific stress-responsive-pathways, due to the debility caused to plants by biotic stressful conditions, which can activate a crosstalk among different stress related pathways, as observed in other plants (Glombitza et al., 2004; Kido et al., 2011). Moreover, it is important to consider the tissue from which the library was generated, since leaves are among the first organs to present stress symptoms (especially to abiotic ones). These are necessary for the maintenance of photosynthesis and evapotranspiration processes to ensure plant survival. Moreover, the Gmax_HSFB3_Contig20961 gene seems to be expressed specifically under abiotic stress, such as water deficit.
The analysis of SuperSAGE transcript abundance revealed a higher number of orthologous tags for the Gmax_HSFB1_Contig12262 cluster (more than 50% of the identified SuperSAGE tags), followed by Gmax_HSFA1E_Contig12828 (Table 2; Figure 1A). There is evidence suggesting that HSFB1 plays a special role in gene activation as a cooperative partner of HSFA1 and that coexpression of low levels of HSFB1 with HSFA1 can result in strong synergistic effects in reporter gene activation. Experiments in tomato showed that HSFB1 acts as a novel type of coactivator and may be able to cooperate with HSFA1a or other activators to control expression of certain housekeeping genes (Bharti et al., 2004).
Evaluating the results for the comparisons among water deficit libraries (susceptible X tolerant), a similar proportion of HSF genes was observed, with the exception of the Gmax_HSFB1_SJ09-E1-R06-064-B09-UC.F transcript, which was recorded exclusively in the susceptible genotype. In both libraries, Gmax_HSFB1_Contig12262 (Figure 1B) was more represented, indicating that HSF genes are expressed under water stress conditions in a similar way in both susceptible and tolerant cultivars.
As expected, most SuperSAGE tags were identified from water deficit libraries. However, it is worth noting that more than 60% of the HSF gene types obtained from soybean ESTs were not identified in the SuperSAGE comparisons, suggesting that the seed EST sequences used were not complete, lacking the necessary 3′ extremity for anchoring of SuperSAGE tags. This opens the possibility of identifying additional candidates upon using other annotation approaches. A role of these factors in water deficit response may exist, since their expression was reported also in association with other abiotic stresses (Kotak et al., 2007). Moreover, the 68 identified tags could be potentially useful for 3′ RACE (3′-rapid amplification of cDNA ends) experiments to identify the complete transcript, besides expression validation using RT-qPCR with the same mRNA samples.
Structure and evolution of HSF candidates in soybean, Medicago, Lotus and Arabidopsis
The functional properties of HSFs are attributed to conserved structural domains, with the highest degree of conservation being observed for the DNA-binding domain (DBD) composed of helix-turn-helix (HTH) structures, and an adjacent domain with a heptad hydrophobic repeat (HR-A/B) which is involved in oligomerization. In addition, there are two further characteristic components: (i) the short peptide motif essential for nuclear import (NLS: nuclear localization signal) and export (NES: nuclear export signal), and (ii) a C-terminal AHA type activation domain (Li et al., 2010). Primarily based on the structural features of the oligomerization domain, plant HSFs are classified into three evolutionarily-conserved classes, namely A, B and C, bearing 14 sub-classes (Nover et al., 2001). The high degree of conservation within the HSF family is corroborated by our in silico analysis, as in the generated dendrogram it was possible to observe the differentiation of sequences according to their classes, and within each class there was a grouping of sequences according to their subclasses (Figure 2). A clear differentiation among the HSF classes A and B classes from a basal ancestral sequence has been established, as expected, since class B- and non-plant-HSFs differ from class A- and C-HSFs by an additional 21 or 7 amino acids, respectively, which separate the two subdomains HR-A and HR-B located in the hydrophobic regions (Nover et al., 2001). Furthermore, the AHA type acidic activation domain is exclusively represented by class A members (Mittal et al., 2009).
Figure 2.
Dendrogram showing relationships among HSF proteins of A. thaliana, G. max, L. japonicus and M. truncatula. Numbers on the base of the nodes represent bootstrap values. Dotted line divides Type A and C from type B. Gray circles indicate each node shared by HSF groups: ‘Group I (Class A and C HSFs)’ = HSFA4 + HSFA5, ‘Group II (Class A and C HSFs)’ = HSFA1 + HSFA3 + HSFA6 + HSFA8 + HSFC, ‘Group I (Class B HSF)’ = HSFB1 + HSFB2 + HSFB4 and ‘Group II (Class B HSF)’ = HSFB3 + L. japonicus HSFB2A.1. Bar represents similarity coefficient.
With respect to class A, two main groups emerged in the present evaluation: one (I) with HSFA4 and HSFA5 representatives and the other (II) with the remaining HSFA and HSFC members (Figure 2). This is a predictable result, since HSFs A4 and A5 form a group distinct from the remaining HSFs by structural features of their oligomerization domains and by a number of conserved signatures. This is also consistent with their role, since A4 HSFs are potent activators of heat stress-related gene expression, whereas A5 HFSs act as a specific repressor of HSFA4 activity, while other members of class A are not affected due to the high specificity of their oligomerization domains (Baniwal et al., 2007).
The second group included three branches, with a basal one including HSFA8 and HSFC1 (Figure 2). Although class C is more similar to class A than to B, it was expected that this class would behave as a separate group. Nevertheless, the high diversity in the response of different HSF genes to different stresses suggests that there is a high degree of specialization regarding the response of specific HSFs to a particular stress condition. This is consistent with the fact that both HSFA8 and HSFC presented increased expression under cold stress (Miller and Mittler, 2006), indicating that this adaptive response to tolerate cold conditions may be responsible for characteristics shared by these two genes. In fact, in the multiple alignment analysis, two regions comprising 15 residues each (amino acid positions 125 to 139 and 154 to 168) were shared by both HSFA8 and HSFC protein sequences, though absent in other class A HSF members. Furthermore, peculiarities shared by HSFCs, such as deletions of six amino acids at position 106-111 and probable mutations in two segments (intervals: 161–168 and 195–220) may justify the differentiation of class C proteins from class A ones, as evidenced in the dendrogram.
Regarding the specific function of class C, remarkable little information is currently available. According to Nover et al. (2001), HSFCs were well represented in expressed sequence tags (ESTs) from libraries of tomato, soybean, potato, barley and Arabidopsis. The HSFC type is clearly separated from all others by sequence details of the DBDs and by the characteristics of the HR-A/B region. However the significance of these extended oligomerization domains in class A and C HSFs for the coiled-coil structure and oligomerization behavior is not yet clear (Nover et al., 2001).
We denoted a conservation in the position and function of AHA motifs and NES in the C-terminal regions of class A. These regions, in addition to the flanking amino acid residues, were sufficient to identify the HSFs without prior knowledge about the respective DBDs or HR-A/B regions (Kotak et al., 2004). Furthermore, the results were positive for ESTs encoding representatives of HSF groups A1, A2 and A6 (Kotak et al., 2004). Thus, it can be inferred that the observed grouping formed by HSFA1, HSFA2 and HSFA6B in the dendrogram (Figure 2) was based on the similarity of AHA motifs and NES in the C-terminal regions.
It is noteworthy that the C-terminal domains (CTDs) of class B HSFs are completely different (Nover et al., 2001), justifying their isolation in a separate branch, composed of two main groups. The first one includes the B3 sub-class members together with a single member of the B2 sub-class from L. japonicus. This unexpected grouping of the Lotus B2 sub-class member seems to result from a deletion in a region rich in alanine, valine, isoleucine and methionine. Apparently, this deletion was responsible for the exclusion of this sequence from the branch including the remaining class B members. The second group includes B1, B2 and B4 sub-classes, these being separated in different branches according to their sub-classes (Figure 2). This grouping may be explained by differences observed in a cluster containing arginine and lysine residues close to the C-terminus of HSFB1, probably responsible for permanent nuclear localization (Heerklotz et al., 2001) and also by the fact that similar motifs were found in other representatives of this group and also in groups B2 and B4 (with the exception of the HSFB3 sub-class) which is the smallest of all HSFs identified so far.
Although our knowledge is still limited, functional diversification seems to be the main reason for the coexistence of more than 20 HSF types in plants (Baniwal et al., 2007). A systems analysis of tomato HSFs revealed two interesting peculiarities: (i) there are at least four different HSF groups (Scharf et al., 1990, 1993; Treuter et al., 1993; Bharti et al., 2000) belonging to two classes (i.e., class A with HSFs A1, A2, and A3 and class B with HSFB1), and (ii) two of the four HSFs (HSFA2 and B1) are heat stress-inducible proteins (Nover et al., 2001; Kotak et al., 2004). In most cases, all identified gene classes and sub-classes were expressed and identified in the four evaluated legumes, suggesting that the family members diverged before the species differentiated. Alternatively, such gene classes and sub-classes may have already functioned as independent genes in the common ancestor, thus favoring divergent evolution.
HSF expression in soybean
Plant cells constitutively express a pool of HSF proteins that are maintained in an inactive state. Certain results suggest that heat-induced protein denaturation participates in the activation of these HSFs (Yamada et al., 2007). This molecular device is normally based on changes in protein conformation and can respond very quickly, playing therefore a central role in transcriptomic remodeling induced upon heat exposure. Accordingly, all HSFs expressed in soybean identified in this study were derived from experiments in the absence of heat stress.
Moreover, it is well known that heat often occurs in combination with drought or other stresses that cause extensive agricultural losses worldwide. HSFs serve as the terminal components of signal transduction, mediating the expression of HSPs and other HS-induced transcripts, but their diverse temporal and spatial expression has also been demonstrated under the influence of other abiotic stresses (Kotak et al., 2007).
HSFs are involved in stress sensing and signaling but can also be part in the regulation of other cellular processes, including development, where a role is strongly suggested by expression profiles in libraries of tissues from young stages. The only exceptions seen herein were mature adult and drought-stressed leaves where the expression of HSFB1 and HSFB2A1 was diametrically and remarkably down- and up-regulated, respectively (Figure 3).
Figure 3.

Hierarchical clustering (Cluster3.0) of up-regulated (red), down-regulated (green) and non-regulated (black) soybean EST clusters (p < 0.05) related to HS response; gray stands for absence of information. Dendrograms above and to the left of the graph show the relationships among libraries and expressed genes, respectively. Library codes: C04, immature cotyledons of greenhouse grown plants; C05, 8-day-old cotyledons; C08, 3- and 7-day-old cotyledons; F03, mature flowers of field grown plants; F04, floral meristem; H03, hypocotyl and plumule, germinating seeds; L05, unexpanded leaves and shoot tips of 2-week-old seedlings; L06, drought stressed leaf tissue; L07, leaf, 3-week-old, greenhouse grown; L08, leaf; R02, roots of 7-day-old plants; R04, roots of bulked times; R05, roots of 8-day-old seedlings; R06, root; S07, seed coats of greenhouse grown plants; S08, 11-day-old seedlings; S09, whole seedlings of greenhouse grown plants; S10, seedlings; S11, seedlings, minus the cotyledons; S12, Seeds containing globular-stage embryos; SH2, germinating shoots; SO1, in vitro cultivated somatic embryos; ST2, stem tissue of greenhouse grown plants.
Plant HSFs may also function as H2O2 sensors, as is also the case in humans and Drosophila, where HSFs directly sense H2O2 and assemble into homotrimers in a redox-regulated manner. HSFA2 controls expression under prolonged HS and recovery conditions. Interestingly, its expression is induced by high luminosity and exposition to H2O2, emphasizing its importance under various stress conditions (Miller and Mittler, 2006). HSFA4A and HSFA8 are likely to act as sensors of reactive oxygen species (ROS), with HSFA5 acting as a repressor of HSFA4. Indeed, in soybean the profiles of HSFA4A and HSFA8 were quite similar, considering the number of libraries where they were detected. On the other hand, and considering the same libraries, HSFA5 was absent, except in immature seeds containing globular embryo stages where none of the three genes were detectably expressed (Figure 3). It is also interesting to note that HSFB1.1 was up-regulated in seven-day-old root libraries (R02) and in seedlings (without cotyledons) (S11), situations in which HSFB2A.2 was down-regulated, indicating that these genes may act as antagonists during the initial phases of plant development. This assumption is corroborated by the fact that HSFB1.1 was down-regulated, while HSFB2A.2 was up-regulated in the mature root library (L08).
The similarity in expression patterns of HSF genes in specific libraries (in specific developmental stages or conditions) indicates that the activation of these genes might be evoked by the same cis-regulatory elements in their promoters. Such co-expression was observed for HSFA2.1, HSFA2.2, HSFA6B.1 and HSFA4A.1 in the library S07 from ‘seed coats of greenhouse grown plants’. Co-expression could indicate that these genes play the same role or are co-participants in the same pathway.
The induction of transcriptomic remodeling through the HSF network is very important but complex, as it involves several HSFs. This network is only a part of the orchestration that contributes to survival under high temperature stress. The panel exposed by our work suggests that HSFs also mediate cross-talk between signaling cascades in soybean for HS and other abiotic stresses, with possible roles in soybean development. Nevertheless, the questions raised here may have to be addressed in subsequent experiments in which the tissues and conditions should be pooled for different and sequential time points.
Distribution of HSF genes in the soybean genome
The comparative analysis of G. max EST sequences (25 in total) and genomic sequences enabled the identification of 62 loci bearing HSF genes (Table 3; Figure 4) from 65 HSFs previously described for soybean (Mochida et al., 2009a), a crop with a supposed polyploid recent past (McClean et al., 2010). From the 25 obtained candidates, two did not align significantly with the characterized heat shock factor genes, which can be justified by differences in the cultivars used in genomic and expression sequencing projects. In addition, three described genes for soybean were not identified among the EST sequences, indicating a lack of expression of these genes in the libraries of the GENOSOJA database. Differences among the analyzed cultivars may also explain this lack of similarity.
Table 3.
Correspondence among identified GENOSOJA expressed sequence tags (ESTs) and characterized genes of Glycine max. Abbreviations: ID = identification; HSF, Heat Shock Factor; Gm, Glycine max.
| Genosoja ID | Gene ID | Genosoja ID | Gene ID |
|---|---|---|---|
| Gm_HSFA1A.1 | Glyma01g01990 | Gm_HSFA1A.4 | Glyma10g07620 |
| Gm_HSFB1.1 | Glyma01g22910 | Gm_HSFA2.3 | Glyma10g09460 |
| Gm_HSFB3.1 | Glyma01g34490 | Gm_HSFA5.2 | Glyma10g36910 |
| Gm_HSFB4.1 | Glyma01g34490 | Gm_HSFB2B.1 | Glyma10g38240 |
| Gm_HSFB1.2 | Glyma01g39260 | Gm_HSFA1E.2 | Glyma10g38930 |
| Gm_HSFB2A.1 | Glyma01g42640 | Gm_HSFA1E.3 | Glyma11g01190 |
| Gm_HSFA1E.1 | Glyma01g44330 | Gm_HSFB2A.2 | Glyma11g02800 |
| Gm_HSFB4.2 | Glyma02g44670 | Gm_HSFB1.5 | Glyma11g06010 |
| Gm_HSFB3.2 | Glyma03g29190 | Gm_HSFA2.4 | Glyma11g33630 |
| Gm_HSFA1A.2 | Glyma03g31380 | Gm_HSFA1A.5 | Glyma13g16510 |
| Gm_HSFA6B.1 | Glyma03g31380 | Gm_HSFA1A.6 | Glyma13g21490 |
| Gm_HSFA7A.1 | Glyma03g31380 | Gm_HSFB1.6 | Glyma13g24860 |
| Gm_HSFA1A.3 | Glyma03g34900 | Gm_HSFA4A.3 | Glyma13g29760 |
| Gm_HSFB4.3 | Glyma04g04200 | Gm_HSFB4.7 | Glyma14g04070 |
| Gm_HSFB4.4 | Glyma04g04200 | Gm_HSFB4.8 | Glyma14g09190 |
| Gm_HSFA2.1 | Glyma04g05500 | Gm_HSFA1A.7 | Glyma14g11030 |
| Gm_HSFA2.2 | Glyma04g05500 | Gm_HSFA4A.4 | Glyma15g09280 |
| Gm_HSFB1.3 | Glyma05g20460 | Gm_HSFA1A.8 | Glyma16g13400 |
| Gm_HSFB1.4 | Glyma05g20460 | Gm_HSFA1E.4 | Glyma16g19500 |
| Gm_HSFA5.1 | Glyma05g28460 | Gm_HSFB3.3 | Glyma16g29750 |
| Gm_HSFA4A.1 | Glyma05g28460 | Gm_HSFB2A.3 | Glyma16g32070 |
| Gm_HSFA4A.2 | Glyma05g29470 | Gm_HSFA1A.9 | Glyma17g06160 |
| Gm_HSFA8.1 | Glyma05g34450 | Gm_HSFB1.7 | Glyma17g20070 |
| Gm_HSFB4.5 | Glyma06g04390 | Gm_HSFA1A.10 | Glyma17g34540 |
| Gm_HSFC1.1 | Glyma07g09510 | Gm_HSFB4.9 | Glyma17g35980 |
| Gm_HSFC1.2 | Glyma07g09520 | Gm_HSFB2A.4 | Glyma18g14700 |
| Gm_HSFB4.6 | Glyma07g36370 | Gm_HSFA1E.5 | Glyma19g26460 |
| Gm_HSFA8.2 | Glyma08g05220 | Gm_HSFA5.4 | Glyma19g26750 |
| Gm_HSFA5.3 | Glyma08g11460 | Gm_HSFB3.4 | Glyma19g31940 |
| Gm_HSFA4A.5 | Glyma08g12630 | Gm_HSFB3.5 | Glyma19g31940 |
| Gm_HSFB2A.5 | Glyma09g26510 | Gm_HSFA6B.3 | Glyma19g34210 |
| Gm_HSFB2A.6 | Glyma09g26510 | Gm_HSFA8.3 | Glyma19g37580 |
| Gm_HSFC1.3 | Glyma09g32300 | Gm_HSFB4.10 | Glyma20g08250 |
| Gm_HSFA1A.11 | Glyma09g33920 | Gm_HSFA1A.13 | Glyma20g28870 |
| Gm_HSFA1A.12 | Glyma10g00560 | Gm_HSFB2B.2 | Glyma20g29610 |
| Gm_HSFA6B.2 | Glyma10g03530 | - | - |
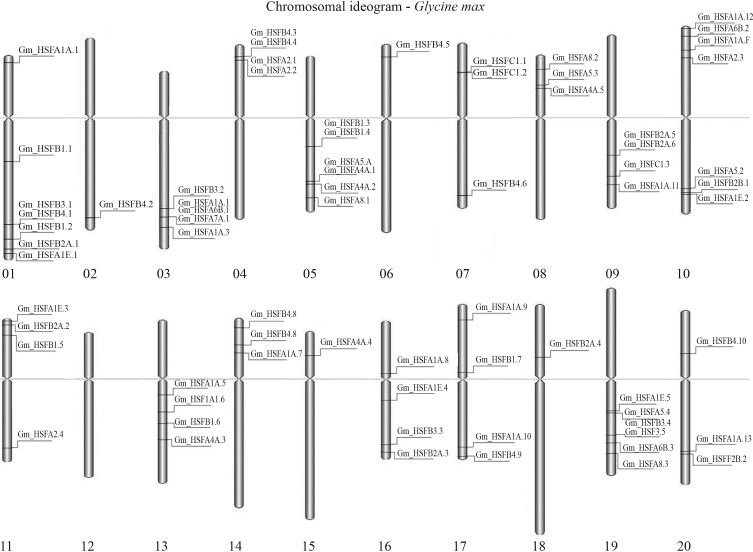
Figure 4.
In silico hybridization of HSF sequences against the SOYBASE database. Schematic representation of clusters that were anchored in soybean based on BLAST similarity results (see Table 3 for correspondence between EST cluster identification and HSF described genes).
With respect to the genomic distribution of the HSF family, nine gene clusters could be identified in chromosomes 01, 03, 04, 05, 10, 11 and 19 (Figure 4). According to Mochida et al. (2009a) these clusters may consist of paralogous genes. In soybean, the relative physical distribution of transcription factor genes is of interest, and two types of clusters can be distinguished based on their evolutionary history. The first type consists of a series of genes that arose through repeated tandem duplications (originated from a founding locus). The second type, which is not considered as consisting of paralogous genes, probably arose independently and then relocated to form these duplications and clusters (Mochida et al., 2009b). Pairs of duplicated genes on different chromosomes are common and gene clusters of three or more highly related genes are also widely found (Mochida et al., 2009a). Considering the distance of their occurrence, a few of the duplicated genes could be classified arbitrarily as either genes that were not duplicated in tandem on the same chromosome, or genes that were so (Mochida et al., 2009a).
Moreover, none of the EST clusters aligned on chromosome 12. This was expected, since in this chromosome there is no description of HSF family members (Mochida et al., 2009b), while other chromosomes (02, 06, 15 and 18) presented a single representative of the group.
Concluding Remarks
Results from the present investigation indicate that gene duplication and diversification occurred during plant evolution, whilst differences in their expression patterns caused species-specific variability in the composition of the HSF family members, which can be divided into three different classes and several sub-classes according to their particular motifs and residue-specific rich regions. Although not all of the previously described genes could be found for the three species studied when using a transcriptomic approach, we expect that experiments directed at heat-stress conditions may provide additional sequences related to the HS response, including other HSF genes. Furthermore, the absence of soybean ESTs for some HSF members did not impair the evaluation of the distribution of the HSF family in the soybean genome. The family is present in 19 of the 20 chromosomes, including clustered distribution in some.
To understand the complexity of a plant’s HSF family and stress response systems in general, it is important to consider that when plants became adapted to terrestrial habitats they evidently had to face and become specialized to rapidly changing and extreme environmental conditions. The present approach represents the first evaluation considering only expressed HSF genes, revealing 25 expressed ESTs and 68 SuperSAGE tags, with emphasis on root tissue (water deficit) libraries. Some HSF candidates present in Arabidopsis, that are apparently missing in the transcriptome of the evaluated legumes (for example HSFA1B), may be important candidates for biotechnological approaches in soybean and other legumes directed towards increasing their performance under temperature stress conditions. Moreover, some genes found to be induced under water deficit may constitute interesting target genes for inferences regarding the association of heat and cold stresses, especially considering current climate change scenarios.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baniwal SK, Chan KY, Scharf K-D, Nover L. Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4*. J Biol Chem. 2007;282:3605–3613. doi: 10.1074/jbc.M609545200. [DOI] [PubMed] [Google Scholar]
- Bharti K, Schimidt E, Lyck R, Bublak D, Scharf K-D. Isolation and characterization of HsfA3, a new heat stress transcription factor of Lycopersicon peruvianum. Plant J. 2000;22:355–365. doi: 10.1046/j.1365-313x.2000.00746.x. [DOI] [PubMed] [Google Scholar]
- Bharti K, von Koskull-Döring P, Bharti S, Kumar P, Tintschl-Körbitzer A, Treuter E, Nover L. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with HAC1/CBP. Plant Cell. 2004;16:1521–1535. doi: 10.1105/tpc.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeoglu B. Heat shock proteins and heat shock response in plants. G U J Sci. 2009;22:67–75. [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr WR, Caviness CE, Burmood DT, Pennington IS. Stage of development descriptions for soybeans, Glycine max (L.) Merrill. Crop Sci. 1971;11:929–931. [Google Scholar]
- Fehr WR, Caviness CE. Ames, Iowa State University of Science and Technology; Iowa: 1977. Stage of Soybean Development. Special Report n. 80; p. 12. [Google Scholar]
- Glombitza S, Dubuis P-H, Thulke O, Welzl G, Bovet L, Götz M, Affenzeller M, Geist B, Hehn A, Asnaghi C, et al. Crosstalk and differential response to abiotic and biotic stressors reflected at the transcriptional level of effector genes from secondary metabolism. Plant Mol Biol. 2004;54:817–835. doi: 10.1007/s11103-004-0274-3. [DOI] [PubMed] [Google Scholar]
- Heerklotz D, Doring P, Bonzelius F, Winkelhaus S, Nover L. The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol Cell Biol. 2001;21:1759–1768. doi: 10.1128/MCB.21.5.1759-1768.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland D, Arnon DI. The water culture method for growing plants without soil. Calif Agric Exp Stn Circ. 1950;347:1–32. [Google Scholar]
- Hsu S-F, Lai H-C, Jinn T-L. Cytosol-localized heat shock factor-binding protein, AtHSBP, functions as a negative regulator of heat shock response by translocation to the nucleus and is required for seed development in Arabidopsis. Plant Physiol. 2010;153:773–784. doi: 10.1104/pp.109.151225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Hu G, Han B. Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci. 2009;176:583–590. doi: 10.1016/j.plantsci.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Kido EA, Barbosa PK, Ferreira Neto JCR, Pandolfi V, Houllou-Kido LM, Crovella S, Benko-Iseppon AM. Identification of plant protein kinases in response to abiotic and biotic stresses using SuperSAGE. Curr Prot Pept Sci. 2011;12:643–656. doi: 10.2174/1389203711109070643. [DOI] [PubMed] [Google Scholar]
- Kotak S, Port M, Ganguli A, Bicker F, von Koskull-Doring P. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class a Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 2004;39:98–112. doi: 10.1111/j.1365-313X.2004.02111.x. [DOI] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Doring P, Vierling E, Scharf KD. Complexity of the heat stress response in plants. Curr Opin Plant Biol. 2007;10:310–316. doi: 10.1016/j.pbi.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Li H-Y, Chang C-S, Lu L-S, Liu C-A, Chan M-T, Charng Y-Y. Over-expression of Arabidopsis thaliana heat shock factor gene (AtHsfA1b) enhances chilling tolerance in transgenic tomato. Bot Bull Acad Sin. 2004;44:129–140. [Google Scholar]
- Li M, Berendzen KW, Schoffl F. Promoter specificity and interactions between early and late Arabidopsis heat shock factors. Plant Mol Biol. 2010;73:559–567. doi: 10.1007/s11103-010-9643-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClean PE, Mamidi S, McConnell M, Chikara S, Lee R. Synteny mapping between common bean and soybean reveals extensive blocks of shared loci. BMC Genomics. 2010;11:e184. doi: 10.1186/1471-2164-11-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Mittler R. Could heat shock transcription factors function as hydrogen peroxide sensors in plant? Ann Bot. 2006;98:279–288. doi: 10.1093/aob/mcl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal D, Chakrabarti S, Sarkar A, Singh A, Grover A. Heat shock factor gene family in rice: Genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol Biochem. 2009;47:785–95. doi: 10.1016/j.plaphy.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Mochida K, Yoshida T, Sakurai T, Yamaguchi-Shinozaki K, Shinozaki K, Tran L-SP. In silico analysis of transcription factor repertoire and prediction of stress responsive transcription factors in soybean. DNA Res. 2009a;16:353–369. doi: 10.1093/dnares/dsp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Yoshida T, Sakurai T, Yamaguchi-Shinozaki K, Shinozaki K, Tran L-SP. LegumeTFDB: An integrative database of Glycine max, Lotus japonicus and Medicago truncatula transcription factors. Bioinformatics. 2009b;26:290–291. doi: 10.1093/bioinformatics/btp645. [DOI] [PubMed] [Google Scholar]
- Nascimento LC, Costa GGL, Binneck E, Pereira GAG, Carazzolle MF. A web-based bioinformatics interface applied to Genosoja Project: Databases and pipelines. Genet Mol Biol. 2012;35(suppl 1):203–211. doi: 10.1590/S1415-47572012000200002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Bharti K, Doring P, Mishra SK, Ganguli A, Scharf K-D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chap. 2001;6:177–189. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirkkala L, Nykanen I, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- Ruelland E, Zachowski A. How plants sense temperature. Environ Exp Bot. 2010;69:225–232. [Google Scholar]
- Sato Y, Yokoya S. Enhanced tolerance to drought stress in transgenic rice plants overexpressing a small heat-shock protein, sHSP17.7. Plant Cell Rep. 2008;27:329–334. doi: 10.1007/s00299-007-0470-0. [DOI] [PubMed] [Google Scholar]
- Scharf K-D, Rose S, Thierfelder J, Nover L. Two cDNAs for tomato heat stress transcription factors. Plant Physiol. 1993;102:1355–1356. doi: 10.1104/pp.102.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf K-D, Rose S, Zott W, Schoffl F, Nover L. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J. 1990;9:4495–4501. doi: 10.1002/j.1460-2075.1990.tb07900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöff F, Prändl R, Reindl A. Regulation of the heat-shock response. Plant Physiol. 1998;117:1135–1141. doi: 10.1104/pp.117.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung D-Y, Kaplan F, Lee K-J, Guy CL. Acquired tolerance to temperature extremes. Trends Plant Sci. 2003;8:179–187. doi: 10.1016/S1360-1385(03)00047-5. [DOI] [PubMed] [Google Scholar]
- Swindell WR, Huebner M, Weber AP. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics. 2007;8:e125. doi: 10.1186/1471-2164-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treshow M. Environment and Plant Response. McGraw-Hill, Company; New York: 1970. p. 421. [Google Scholar]
- Treuter E, Nover L, Ohme K, Scharf K-D. Promoter specificity and deletion analysis of three tomato heat stress transcription factors. Mol Gen Genet. 1993;240:113–125. doi: 10.1007/BF00276890. [DOI] [PubMed] [Google Scholar]
- Yamada K, Fukao Y, Hayashi M, Fukazawa M, Suzuki I, Nishimura M. Cytosolic HSP90 regulated the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J Biol Chem. 2007;282:37794–37804. doi: 10.1074/jbc.M707168200. [DOI] [PubMed] [Google Scholar]
Internet Resources
- AtTFDB - Arabidopsis transcription factor database at Agris, http://arabidopsis.med.ohio-state.edu/AtTFDB/ (Jun/2011).
- GENOSOJA database, Brazilian Soybean Genome Consortium, http://bioinfo03.ibi.unicamp.br/soja/ (Jun/2011).
- Lotus japonicas database, http://www.kazusa.or.jp/lotus (Jun/2011).
- Medicago truncatula database, http://www.medicago.org/ (Jun/2011).
- Phytosome Soybean genome browser, http://www.phytozome.net/cgi-bin/gbrowse/soybean/ (Jun/2011).
- SOYBASE site, http://soybase.org/gbrowse/cgi-bin/gbrowse/gmax1.01/#se arch (Jun/2011).
- Legume Transcription Factor Database, LegumeTFDB, http://legumetfdb.psc.riken.jp/ (Jun/2011).