Abstract
Plants experience various environmental stresses, but tolerance to these adverse conditions is a very complex phenomenon. The present research aimed to evaluate a set of genes involved in osmotic response, comparing soybean and medicago with the well-described Arabidopsis thaliana model plant. Based on 103 Arabidopsis proteins from 27 categories of osmotic stress response, comparative analyses against Genosoja and Medicago truncatula databases allowed the identification of 1,088 soybean and 1,210 Medicago sequences. The analysis showed a high number of sequences and high diversity, comprising genes from all categories in both organisms. Genes with unknown function were among the most representative, followed by transcription factors, ion transport proteins, water channel, plant defense, protein degradation, cellular structure, organization & biogenesis and senescence. An analysis of sequences with unknown function allowed the annotation of 174 soybean and 217 Medicago sequences, most of them concerning transcription factors. However, for about 30% of the sequences no function could be attributed using in silico procedures. The establishment of a gene set involved in osmotic stress responses in soybean and barrel medic will help to better understand the survival mechanisms for this type of stress condition in legumes.
Keywords: osmotic stress, stress-responsive genes, Glycine max, Medicago truncatula
Introduction
In the course of evolution, plants have acquired a myriad of developmental and metabolic strategies to cope with the adverse effects of environmental stresses during vegetative growth and reproduction (Parry et al., 2005), making stress tolerance a complex phenomenon.
Stress perception and the immediate induction of signals that culminate in adaptive responses are key steps leading to plant stress tolerance. Tolerance stress differences between genotypes or different developmental stages of a single genotype may arise from peculiarities in signal perception and transduction mechanisms (Chinnusamy et al., 2004). Under osmotic stress conditions diverse sets of physiological responses are activated, including metabolic and defense systems used to sustain growth and for survival. The stress-inducible genes are classified into two major groups: one of them protects the plant directly against stresses, whereas the other regulates gene expression and signal transduction (Valliyodan and Nguyen, 2006).
Because plant tolerance against osmotic stress is a complex multigenic trait, a demand exists for genome wide analysis, including ‘omics’ approaches suitable for uncovering important gene sets involved in this important process (Hirayama and Shinozaki, 2010).
After the ‘sequencing era’, genetic information was then available for several non-model plants, including some legume species, a group that exhibits unique features, such as the ability to carry the nodulation process. Nitrogen fixation mediated by nodule activities abolishes the need for external nitrogen sources from fertilizers, while providing the so-called ‘green manuring’ that enriches the soil. Moreover, some legumes, such as soybean, barrel medic and cowpea, are important economic crops that provide humans with food, livestock for feeding purposes, and industry with raw materials (Graham and Vance, 2003).
Soybean is an example of a non-model plant with plentiful transcriptome information available. Among available databases, the Genosoja platform connects public and restricted data, providing 60,747 unigenes (Nascimento et al., 2012, this issue).
The identification of candidate genes in soybean and barrel medic will provide additional evidence of the response mechanisms for osmotic stresses in Fabaceae, yielding useful information for crop improvement. As osmotic stress cannot be solved solely via remedial land management, tolerant crops - able to maintain cellular turgor and osmotic balance - may contribute significantly to reduce this economic burden. The key to plant engineering for osmotic tolerance lies in the knowledge of the underlying mechanisms of plant adaptive responses (Hariadi et al., 2011).
In the present work the main categories of osmotic stress genes known from A. thaliana were identified in the soybean (Genosoja Project) and barrel medic (M. Truncatula database) transcriptomes through an in silico approach, in order to contribute to a better understanding of the early molecular adaptation to osmotic (drought and salinity) stress in both leguminous plants.
Materials and Methods
In a previous study based on 7,000 Arabidopsis genes, Seki et al. (2002) identified 103 coding genes distributed over 27 functional categories (Table 1) whose expression increased more than five times in response to osmotic stress. The protein sequences of these stress-inducible genes were obtained at the RIKEN Arabidopsis Full-Length Clone Database, and used as query sequences.
Table 1.
Functional categories procured and respective seed-sequence number. Abbreviation: TF = Transcription Factor.
| Functional category | # Seed sequence | Functional category | # Seed sequence |
|---|---|---|---|
| bZIP TF | 1 | WRKY TF | 2 |
| Photosynthesis | 1 | Osmoprotectant | 3 |
| Signaling | 1 | ZincFinger TF | 3 |
| Reproductive development | 1 | Detoxification enzyme | 2 |
| Respiration | 1 | Cellular metabolism | 3 |
| DNA nucleus | 1 | DREB and ERF TF | 2 |
| Ferritin | 1 | Ethylene biosynthesis | 2 |
| LEA protein | 1 | Cytochrome P450 | 2 |
| MYB TF | 1 | Fatty Acid metabolism | 4 |
| Homeodomain TF | 1 | Heat Shock protein | 2 |
| Membrane protein | 2 | Kinase protein | 2 |
| Senescence-related | 1 | Carbohydrate metabolism | 6 |
| Degradation protein | 1 | Plant defense | 4 |
| Secondary metabolism | 1 | Transport protein ion channel carrier | 4 |
| Water channel protein | 1 | Cellular struct. organiz. and biogenesis | 5 |
| NAC TF | 2 | Unknown protein | 37 |
| Protein phosphatase | 2 | ||
| Total | 103 |
After this step, a local bank with the retrieved sequences was generated in order to make searches for similar sequences against the Genosoja platform (Nascimento et al., 2012) and the M. truncatula database (Quackenbush et al., 2000) using the tBLASTn algorithm (Altschul et al., 1990) with a cut-off of 1e−05. The results were annotated in other local databank for further analyses and for comparisons among studied organisms and literature information. In view of the different number of seed sequences per category, the results obtained from each category and organism were normalized. The soybean and Medicago genes with unknown function were submitted to the AutoFACT program (Koski et al., 2005), and annotated according to the data available in the largest functional annotation databanks (KEGG, COG, PFAM, SMART, nr). This step was performed in order to categorize these sequences and assign function to them, based on a comparative analysis.
Results and Discussion
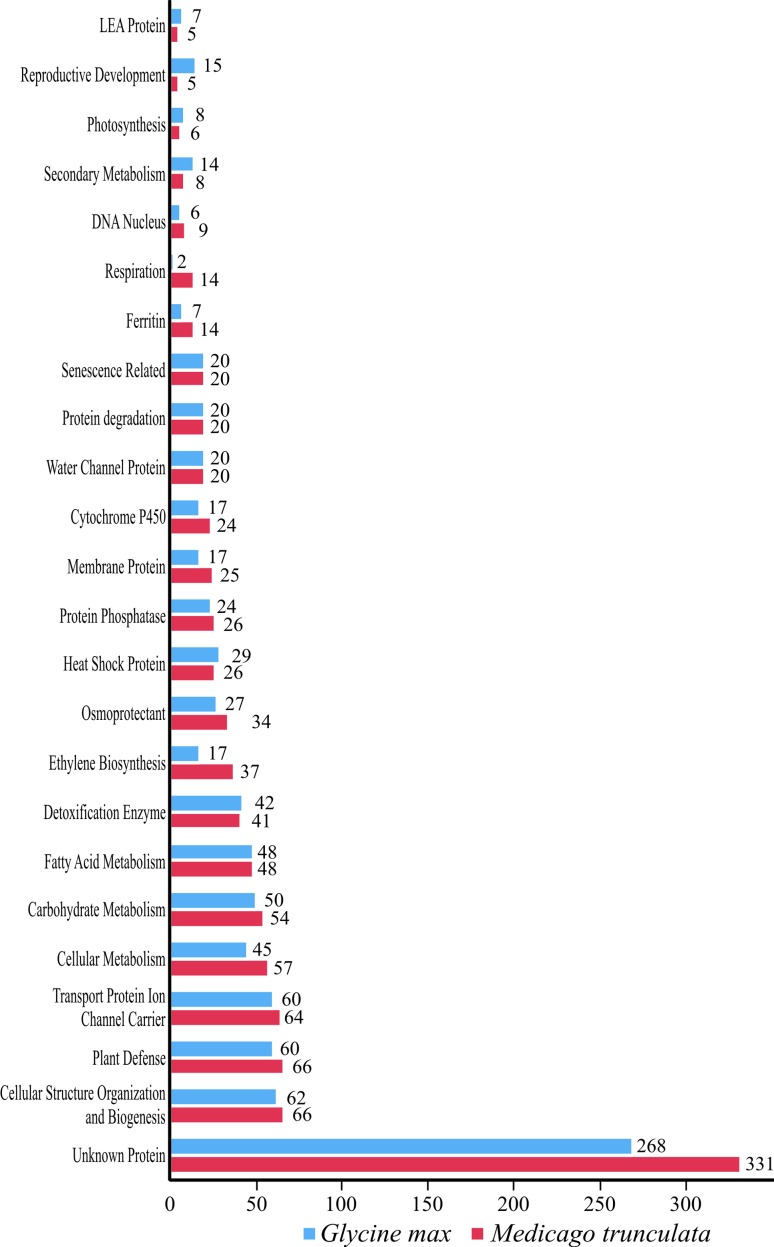
The stress-inducible gene products were classified into two main groups: (I) those that are at the front line of defense, protecting the plant against adverse conditions and (II) those that regulate genic expression and signal transduction in the stress response (Seki et al., 2003). The first group included proteins that probably act in the protection of plant cells from dehydration, such as the enzymes required for the biosynthesis of various osmoprotectants, LEA proteins, antifreeze proteins, chaperones and detoxification enzymes. The second group included signaling molecules such as transcription factors and protein kinases, among others (Seki et al., 2003). Twenty-seven categories of these two groups classified according to Seki et al. (2002) were analyzed, resulting in 1,088 (soybean) and 1,210 (Medicago) sequences (Table S1, supplementary material). In both genomes the ‘unknown protein’ category was the most representative (Figure 1), with 268 candidates for soybean and 331 for Medicago, followed by ‘cellular structure organization and biogenesis’, ‘plant defense’ and ‘transport protein ion channel carrier’ categories (Figure 1).
Figure 1.
Main categories of Group I stress-inducible genes (protective molecules), indicating the number of orthologs identified in Glycine max and Medicago truncatula.
The highest number of sequences for genes with ‘unknown function’ - a very common category in expression essays regarding osmotic stress response in plants – attracting great interest from researchers, since those genes represent a clear source of new candidates for breeding purposes. Previous studies highlighted the importance of analyzing the role of stress-induced genes, not only for a further understanding of the molecular mechanisms of stress tolerance in higher plants, but also for improving crop performance using gene manipulation (Seki et al., 2002).
Osmotic stress greatly affects cells both at the micro (i.e., membrane structure), and at the macro level (i.e. the physiology of the whole plant), with results that reflect the variety of responses involved in the acquisition of tolerance. At the microcellular level, the activation of genes in the categories ‘cellular structure, organization and biogenesis’ (soybean: 62; Medicago: 66) and ‘transport protein ion channel carrier’ (soybean: 64; Medicago: 60) was observed, showing the importance of the maintenance of cellular structures and of the control of ion exchange with the environment.
Furthermore, we observed the activation of genes in the category ‘plant defense’ (soybean: 66; Medicago: 60), indicating the presence of a cross-talk process between pathways, a common mechanism in plants under stressful conditions. In addition to stress-specific adaptive responses, plants also share responses that protect them from more than one type of stress (Seki et al., 2002; DeFalco et al., 2010; Nuruzzaman et al., 2010), a response also observed in cowpea, another Fabaceae member (Kido et al., 2011).
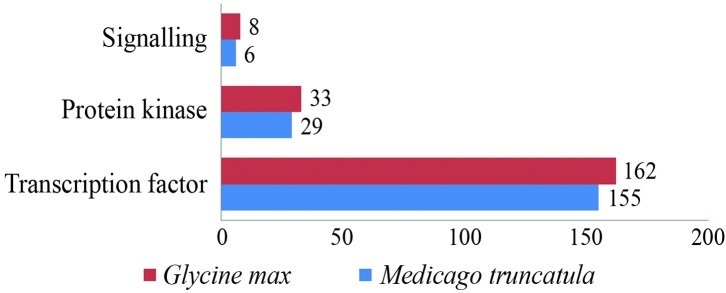
Amongst the candidates of the second group of responses, composed of genes involved in signal transduction and regulation of expression (203 in soybean and 190 in Medicago; Figure 2), the category transcription factor (TF) was the most prevalent, representing up to 80% in soybean and 82% in Medicago (Figure 2). The high number of transcription factors suggests that transcriptional regulation is an important mechanism in the signal transduction triggered by osmotic stresses in both legumes.
Figure 2.
Percentage of stress-inducible genes (Group II), including cell signaling factors identified in Glycine max and Medicago truncatula.
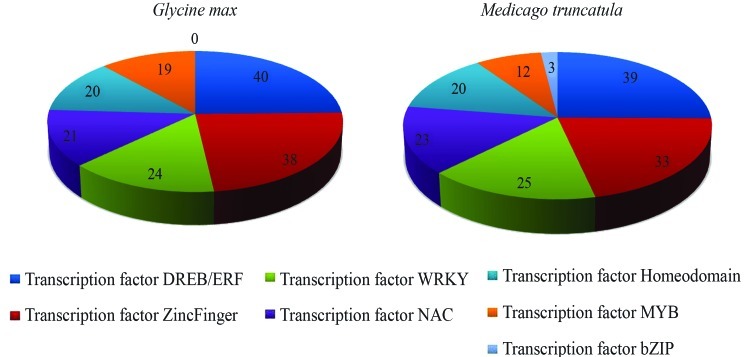
A surprising result was the absence of a bZIP representative in the soybean database, while in Medicago this category was represented by three candidates (Figure 3). This transcription factor has been identified in many plants and is known to participate in various responsive pathways, including abiotic stress response.
Figure 3.
Graphic representation of transcription factors identified in Glycine max and Medicago truncatula.
Among the transcription factors, the DREB/ERF and Zinc-finger families had the highest number of sequences (Figure 3). This result was expected, since from more than 1,600 transcription factors encoded by A. thaliana, 9% are members of the DREB/ERF-like family (Dietz et al., 2010). Due to the versatility of functions that the zinc finger family may have, as well as the variety of their structural proteins, the obtained result was expected. According to Takatsuji (1998), plants seem to have adopted preexisting prototype zinc-finger motifs, generating new zinc-finger domains to adapt them to various regulatory processes. The zinc finger domain can be present in a number of transcription factors and play critical roles in interactions with other molecules. Mutations in some of the genes coding for zinc-finger proteins have been found to cause profound developmental aberrations or defective responses to environmental cues (Takatsuji, 1998). Zinc finger proteins are required for key cellular processes including transcriptional regulation, development, pathogen defense, and stress responses (Ciftci-Yilmaz and Mittler, 2008). A recent study of rice showed that the C2H2-type zinc finger family alone was represented by 189 members and demonstrated that at least 26 of them respond to different environmental stresses (Agarwal et al., 2007). Moreover, Gong et al. (2010), in a study on transcriptional regulation in drought-tolerant tomato genotypes, also identified and characterized the zinc-finger family as the main activated group during the drought response.
It is important to note that the number of seed-sequences used in the search was different for each category; the ‘unknown protein’ category, for example, was represented by 37 sequences, while the ‘bZIP transcription factor’ category comprised a single sequence. Thus, it was expected that the more abundant orthologous categories would be those obtained through comparative searches with the categories composed of more query sequences.
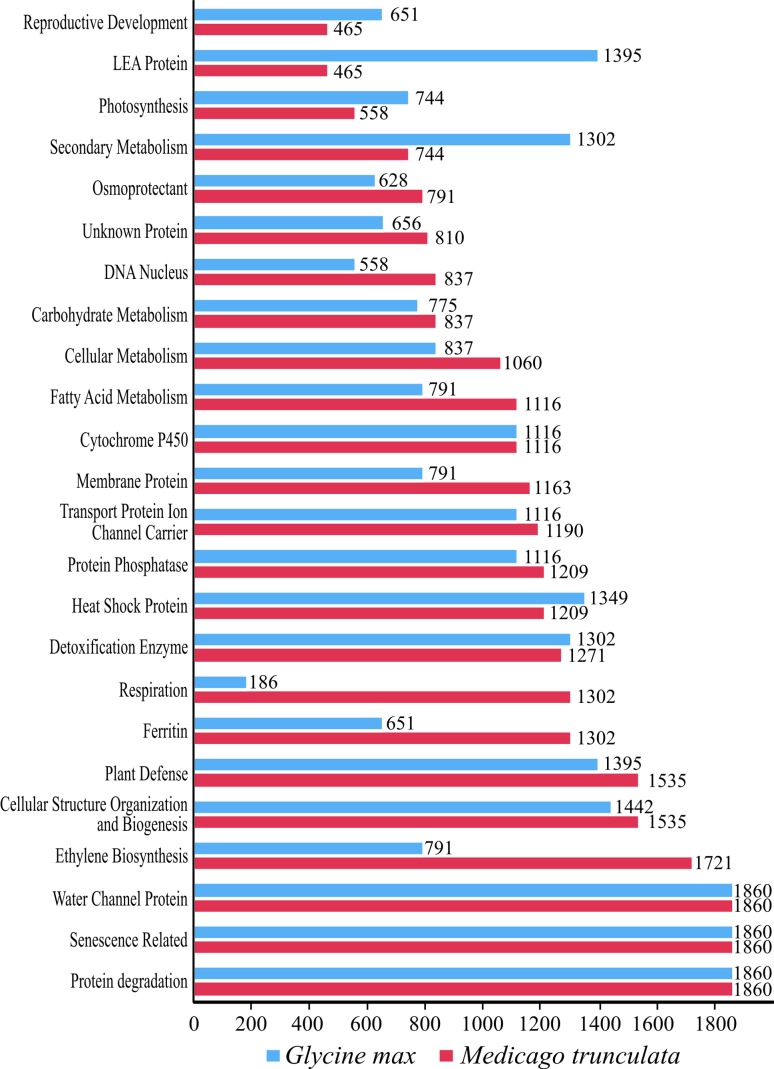
As for the remainder, after normalizing the results, proportionally the most representative categories (7% each) were: ‘water channel proteins’, ‘protein degradation’ and ‘senescence-related’ (Figure 4). Without doubt, all categories analyzed may contribute to an improvement in osmotic tolerance, although some functions are more relevant than others. Proteins associated with ion channels and water channels are essential in the acquisition of resistance in the presence of soluble salts and water shortages, the former controlling the entry and exit of ions such as Na+, which are toxic in high concentrations, and the latter controlling water loss to the environment. Besides these proteins, those falling into the category ‘protein degradation’ are required for protein turnover and recycling of essential amino acids, while ‘senescence-related’ genes are key components in the abiotic stress response, with genes controlling subcellular changes that lead to tolerance (Seki et al., 2002).
Figure 4.
Number of gene candidates from Group I for Medicago truncatula and Glycine max, after data normalization.
While the normalized results evidenced similar amounts of data in the most representative categories for both organisms, in some categories there were significant variations in the number of sequences between both leguminous species (Figure 4); this difference was even greater than 50% for the categories ‘Reproductive development’ (soybean: 1,395; Medicago: 465), ‘Ferritin’ (soybean: 651; Medicago: 1,392), ‘Respiration’ (soybean: 186; Medicago: 1,302) and ‘Ethylene biosynthesis’ (soybean: 791; Medicago: 1.721). Nevertheless, this variation may be related to the conditions under which the data were generated and deposited, as well as to the number of sequences available in the respective databases. Additionally, species-specific features could be responsible for these variations, to a lesser extent.
Regarding the category ‘Unknown Protein’, screened candidates from soybean (268) and Medicago (331) were subjected to the AutoFACT program in order to assign function to these sequences, allowing the recognition of the function of 174 and 217 sequences, respectively.
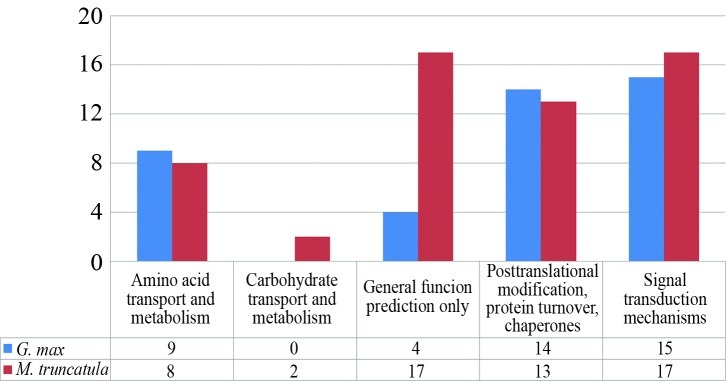
As a result, 42 and 57 G. max and M. truncatula were categorized according to the COG (Cluster of Orthologous Groups) functional database in five categories (Table 2; Figure 5). Within each category, the annotation revealed that they present the same description as the matched sequences deposited in the databank. For example, the ‘Amino acid transport and metabolism’ functional category was represented just by ‘Amino Acid Permease’ sequences (Table 2). Two candidates of Medicago, which were functionally classified into the ‘Carbohydrate transport and metabolism’ category, were also annotated on the KEGG database as involved in the beta-galactosidase pathway (Galactose Metabolism Glycan Structure – degradation), (Table 2).
Table 2.
Sequence description annotated according to the COG (Cluster of Orthologous Groups) functional category in Glycine max and Medicago truncatula.
| COG functional category | Sequence description | Sequence amount
|
|
|---|---|---|---|
| G. max | M. truncatula | ||
| Amino acid transport and metabolism | Amino acid permease | 9 | 8 |
| Carbohydrate transport and metabolism | Beta-galactosidase | 0 | 2 |
| General function prediction only | Patatin | 4 | 17 |
| Posttranslational modification, protein turnover, chaperones | DnaJ-like protein | 14 | 13 |
| Signal transduction mechanisms | Universal Stress Protein (USP) family protein | 15 | 17 |
| Total | 42 | 57 | |
Figure 5.
Categorization of soybean and Medicago ‘unknown category’ candidates based on COG (Cluster of Orthologous Groups) functional database.
The remaining previously ‘unknown’ sequences were annotated as shown in Table 3. The analysis through AutoFACT allowed a function assignment to 132 and 160 soybean and Medicago sequences, respectively. In general, the highest number of sequences was categorized as transcription factors, essential genes participating in the transcriptional regulation of plants. Although it was possible to record more than 65% of the sequences, 35% of ‘unknown’ soybean and 34% of ‘unknown’ Medicago sequences remained without their putative function identified. These are relevant data to be worked out in future functional studies, since they may represent new genes not yet described and unique to legumes.
Table 3.
Description of sequences with unknown function after AutoFACT analysis.
| Description | G. max | M. truncatula |
|---|---|---|
| Amino acid permease | 7 | 4 |
| ATP binding / kinase / protein serine/threonine kinase | 0 | 3 |
| Auxin-responsive GH3 product [Glycine max] | 2 | 8 |
| BTB/POZ domain-containing protein | 0 | 2 |
| Calcium ion binding | 2 | 4 |
| Calmodulin binding | 10 | 14 |
| CCT_2 domain containing protein | 4 | 5 |
| Copper ion binding / electron transporter | 4 | 1 |
| Cu-binding-like domain containing protein | 4 | 10 |
| Dev_Cell_Death domain containing protein | 9 | 17 |
| DFL1 (DWARF IN LIGHT 1) | 1 | 0 |
| DnaJ-like protein [Phaseolus vulgaris] | 3 | 2 |
| F-box family protein | 5 | 4 |
| Heat shock protein binding | 3 | 4 |
| Herpes_BLLF1 domain containing protein | 1 | 0 |
| Hydroxyproline-rich glycoprotein family protein | 0 | 1 |
| IFRD1; interferon-related developmental regulator 1 | 8 | 0 |
| Indole-3-acetic acid-amido synthetase GH3.17, putative | 3 | 5 |
| NAC Transcription Factor | 4 | 10 |
| Nucleic acid binding / transcription factor | 18 | 14 |
| Patatin B2 precursor, putative | 1 | 0 |
| PHI-1 (PHOSPHATE-INDUCED 1) | 19 | 20 |
| Plastocyanin-like domain-containing protein | 0 | 1 |
| RCI2A (RARE-COLD-INDUCIBLE 2A) | 0 | 2 |
| SMC_N multi-domain protein | 1 | 3 |
| SPX domain-containing protein | 2 | 0 |
| Stress-inducible protein | 0 | 2 |
| Tify domain containing protein | 8 | 12 |
| Triacylglycerol lipase | 5 | 5 |
| Uncharacterized protein family/Unassigned protein/Protein of unknown function | 94 | 114 |
| Universal stress protein (USP) family protein | 1 | 3 |
| Zinc finger family protein | 7 | 4 |
In conclusion, even in the absence of libraries restricted to osmotic stress in the Genosoja databank, this study indicated that most of the genes involved in the osmotic stress pathways were expressed by the non-stressed soybean and Medicago libraries at least in a baseline way. The data also revealed that soybean and Medicago are a rich source of stress-responsive candidates, which can be also applied to improve soybean and other legumes. It also highlights the existence of significant diversity for most genes, useful for comparative physiological essays. The obtained data are available for gene-targeted functional evaluation using qRT-PCR, as well as other biotechnological approaches. The molecular differences detected between the compared libraries will permit the identification of important candidates by additional approaches including PCR walking, as previously done for other crops (e.g. Coemans et al., 2005).
The identified candidates are also being monitored in further expression assays carried out in the Genosoja project (considering contrasting combinations of tolerant and susceptible plants under drought stress as compared with their negative control in a time frame) providing a more complete picture of genes involved in osmotic stress response and useful for breeding and biotechnological purposes.
Acknowledgments
The authors would like to thank CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), FACEPE (Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco), and CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) for their financial support.
Supplementary Material
The following online material is available for this article:
Table S1 - Identified candidates among abiotic stress responsive gene categories in soybean and Medicago genomes.
This material is available as part of the online article from http://www.scielo.br/gmb.
References
- Agarwal P, Arora R, Ray S, Singh AK, Singh VP, Takatsuji H, Kapoor S, Tyagi AK. Genome-wide identification of C(2)H(2) zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol Biol. 2007;65:467–485. doi: 10.1007/s11103-007-9199-y. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Schumaker K, Zhu J-K. Molecular genetics perspectives on cross-talk and specificity in abiotic stress signaling in plants. J Exp Bot. 2004;55:225–236. doi: 10.1093/jxb/erh005. [DOI] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S, Mittler R. The zinc finger network of plants. Cell Mol Life Sci. 2008;65:1150–1160. doi: 10.1007/s00018-007-7473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coemans B, Matsumura H, Terauchi R, Remy S, Swennen R, Sági L. SuperSAGE combined with PCR walking allows global gene expression profiling of banana (Musa acuminata), a non-model organism. Theor Appl Genet. 2005;111:1118–1126. doi: 10.1007/s00122-005-0039-7. [DOI] [PubMed] [Google Scholar]
- DeFalco TA, Bender KW, Snedden WA. Breaking the code: Ca2+ 27 sensors in plant signalling. Biochem J. 2010;425:27–40. doi: 10.1042/BJ20091147. [DOI] [PubMed] [Google Scholar]
- Dietz K-J, Vogel MO, Viehhauser A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma. 2010;245:3–14. doi: 10.1007/s00709-010-0142-8. [DOI] [PubMed] [Google Scholar]
- Gong P, Zhang J, Li H, Yang C, Zhang C, Zhang X, Khurram Z, Zhang Y, Wang T, Fei Z, et al. Transcriptional profiles of drought-responsive genes in modulating transcription signal transduction, and biochemical pathways in tomato. J Exp Bot. 2010;61:3563–3575. doi: 10.1093/jxb/erq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PH, Vance CP. Legumes: Importance and constraints to greater use. Plant Physiol. 2003;131:872–877. doi: 10.1104/pp.017004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariadi Y, Marandon K, Tian Y, Jacobsen S-E, Shabala S. Ionic and osmotic relations in quinoa (Chenopodium quinoa Willd.) plants grown at various salinity levels. J Exp Bot. 2011;62:185–193. doi: 10.1093/jxb/erq257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010;61:1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- Kido EA, Barbosa PK, Ferreira Neto JCR, Pandolfi V, Houllou-Kido LM, Crovella S, Benko-Iseppon AM. Identification of plant protein kinases in response to abiotic and biotic stresses using SuperSAGE. Curr Prot Pept Sci. 2011;12:643–656. doi: 10.2174/1389203711109070643. [DOI] [PubMed] [Google Scholar]
- Koski LB, Gray MW, Lang BF, Burger G. AutoFACT: An Automatic Functional Annotation and Classification Tool. BMC Bioinformatics. 2005;16:151–161. doi: 10.1186/1471-2105-6-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento LC, Costa GGL, Binneck E, Pereira GAG, Carazzolle MF. A web-based bioinformatics interface applied to Genosoja Project: Databases and pipelines. Genet Mol Biol. 2012;35(suppl 1):203–211. doi: 10.1590/S1415-47572012000200002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465:30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Parry MAJ, Flexas J, Medrano H. Prospects for crop production under drought: Research priorities and future directions. Ann Appl Biol. 2005;147:211–226. [Google Scholar]
- Quackenbush J, Liang F, Holt I, Pertea G, Upton J. The TIGR Gene Indices: Reconstruction and representation of expressed gene sequences. Nucleic Acids Res. 2000;28:141–145. doi: 10.1093/nar/28.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Seki M, Kamei A, Yamaguchi-Shinozaki K, Shinozaki K. Molecular responses to drought, salinity and frost: Common and different paths for plant protection. Curr Opin Plant Biol. 2003;14:194–199. doi: 10.1016/s0958-1669(03)00030-2. [DOI] [PubMed] [Google Scholar]
- Takatsuji H. Zinc-finger transcription factors in plants. Cell Mol Life Sci. 1998;54:582–596. doi: 10.1007/s000180050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valliyodan B, Nguyen HT. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr Opin Plant Biol. 2006;9:189–195. doi: 10.1016/j.pbi.2006.01.019. [DOI] [PubMed] [Google Scholar]
Internet Resources
- RIKEN Arabidopsis Full-Length Clone Database, http://www.brc.riken.go.jp/lab/epd/catalog/cdnaclone.html (May, 2011)
- Genosoja platform, http://bioinfo03.ibi.unicamp.br/soja/ (May, 2011)
- Medicago truncatula database, http://www.medicago.org/ (May, 2011)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 - Identified candidates among abiotic stress responsive gene categories in soybean and Medicago genomes.