Abstract
Quantitative 1H NMR (qHNMR) provides a value-added dimension to the standard spectroscopic data set involved in structure analysis, especially when analyzing bioactive molecules and elucidating new natural products. The qHNMR method can be integrated into any routine qualitative workflow without much additional effort by simply establishing quantitative conditions for the standard solution 1H NMR experiments. Moreover, examination of different chemical lots of taxol and a Taxus brevifolia extract as working examples led to a blueprint for a generic approach to performing a routinely practiced 13C-decoupled qHNMR experiment, and for recognizing its potential and main limitations. The proposed protocol is based on a newly assembled 13C GARP broadband decoupled proton acquisition sequence that reduces spectroscopic complexity by removal of carbon satellites. The method is capable of providing qualitative and quantitative NMR data simultaneously and covers various analytes from pure compounds to complex mixtures such as metabolomes. Due to a routinely achievable dynamic range of 300:1 (0.3%) or better, qHNMR qualifies for applications ranging from reference standards to biologically active compounds to metabolome analysis. Providing a “cookbook” approach to qHNMR, acquisition conditions are described that can be adapted for contemporary NMR spectrometers of all major manufacturers.
The world’s pool of natural products plays an important role as an (in)exhaustible resource for evolutionary-shaped molecules. Natural products are valuable research tools, which in part is due to their biological potency (see comprehensive reviews 1–4). When natural products are used as biomedical agents, and in consistency with the pharmacophore model, it is the combination of their specific chemical structure and/or reactivity that forms their relationship with a biological target and ultimately defines their essential structural features. Consequently, chemical constitution plays a key role in biological activity, and, therefore, all structure related information obtainable from a biologically active agent is by default relevant. Ultimately, any variation of structural parameters has the potential to introduce variations in biological activity. This relationship holds regardless of the magnitude of the biological perturbation, i.e., whether there is slight or a substantial change in potency, or even an alteration in the type of biological response. The well-documented subtleties of mammalian hormonal steroids can serve as a distinguished example in this regard.
Typically, the relationship between chemical structure and biological activity is explored through semi-empirical systematic, structural alterations by performing (semi-)synthetic variation or investigation of families of closely related structures such as natural isolates or synthetic derivatives. In the case of natural products, and in the context of the chemistry-biology interface, it is equally important to recognize that the chemical composition of natural or synthetic bioactive agents may not necessarily be represented by a single chemical entity (SCE). Instead, the composition might be rather complex and involve a mixture of chemical entities. As a consequence, biological activity becomes closely related to the purity of isolates and/or the impurity profiles of major constituents, isolates, and reference materials.
By virtue of both its comprehensive qualitative (δ, J, nOe, T1, T2; see 5 and references therein) and quantitative capabilities,5 nuclear magnetic resonance methodology (NMR and qNMR, respectively) can contribute in multiple ways to the aforementioned biology-chemistry relationship. Moreover, the combination of NMR and qNMR can act synergistically and supply, in a concurrent manner, valuable structural and quantitative information about nature's "small" molecules that are involved in biological action by providing the following: (i) confirmation of chemical structure and structure-activity relationships; (ii) insight into structural equilibria (e.g., tautomerism or pH-dependent species formation); (iii) purity determination of bioactive agents and/or determination of impurities, which might interfere with the bioassay; (iv) exploration of the structural analogues contained in complex matrices (“biosynthetic cocktails”) and the metabolomic composition of mixtures.
Because natural products, at least initially, have to be purified from complex matrices, their chromatographic purification and structural analysis are relatively complicated. It should be kept in mind that through chemical shift (δ) dispersion, NMR also has intrinsic differentiation capability and can resolve nuclei of the same molecule from nuclei of other molecular species contained in the same sample. Significant additions to the "separation" power of NMR arise from is it capability to create multi-dimensional (nD) dispersion via spin-spin (J) or dipolar (D) coupling, heteronuclear chemical shifts (δX), and/or diffusion (D). While nD qNMR is yet to be fully established, the 1D qNMR method already allows a powerful discrimination of multi-component mixtures or impurities in isolates. In particular the 1D 1H variant of qNMR, qHNMR, adds a combined quantitative-qualitative dimension to the structural NMR portfolio of any natural product, and has the potential to make a fundamental contribution to the bigger picture of structure-activity correlations.
Considering the above, and in the context of more than 100 reports on the use of quantitative NMR in natural products and related sciences, mainly pharmaceutical, food and drug analyses,5,6 there is a significant potential for a standardized qHNMR method, i.e., a method that can be applied in routine purity analysis and quantitation of complex mixtures.7
Results and Discussion
Choice of Taxol as qHNMR Model Sample
While qNMR analyses have been routinely performed in our laboratory since 1999, using a variety of natural products and synthetic or semi-synthetic compounds, taxol was selected as a representative sample to develop the proposed qHNMR protocol for the following reasons: (i) taxol provides a rather complex 1H NMR spectrum, covering almost the entire 1H chemical shift range that is typically of interest; (ii) taxol represents a key example of a natural product with both interesting biological activity and structural novelty; (iii) the NMR spectroscopic assignments have been previously reported and its conformational dynamics in solution have been elucidated, making taxol a well documented molecule; (iv) because of the known solution dynamics associated with taxol, in which Z/E isomerism about an amide linkage are readily observable at room temperature, additional challenges were presented for implementation of the qHNMR approach.
In order to demonstrate the suitability of the proposed qHNMR method for routine analysis, the study was designed to require the least amount of structural information on the impurities. Therefore, instead of relying on involved studies required to clarify the precise nature of the most abundant taxoid analogues/impurities present in each sample, the impurities were compounded into the calculation under the assumption of having equal or at least similar molecular weights. Because a complete molar mass balance was not achievable, the 100% model was employed, representing a method typically used in chromatographic impurity profiling where response factors are also often unknown. Finally, the samples were analyzed in a blinded fashion, and without prior knowledge of LC-based impurity profiles.
A Cookbook Approach to qHNMR
When it comes to the practical aspects, qHNMR is a rather uncomplicated 1D NMR experiment to implement, and only little additional effort is required to obtain the quantitative information through integral-based calculations as part of the routine post-acquisition processing of the spectra. There are, however, certain experimental details that need to be considered, and instrument parameters that require optimization. In general, once the acquisition and processing parameters have been established for any qHNMR experiment, either a parameter set or a set-up macro may be created on the NMR spectrometer, which can then be used to initiate and control the experiment in a much more routine manner and typically require little or no modification to the acquisition parameters (potential exceptions are the use of unusual solvents and/or relaxation behavior of the analyte). Two instrumental conditions distinguish the proposed qHNMR approach from a “normal” survey proton NMR experiment: (i) the qHNMR data are obtained non-spinning, and (ii) the qHNMR spectra are acquired with inverse gated 13C decoupling using a GARP (Globally-optimized Alternating-phase Rectangular Pulses)8 decoupling scheme. All additional key factors that have to be taken into account when establishing a routine qHNMR experimental protocol are discussed in the following. This includes a summary of key acquisition parameter settings, their effect on the qHNMR experiment, and some guidelines for their optimization. Summarized in the form of a "cookbook approach" to qHNMR (see also Table S1), the proposed experimental setup will be generally applicable and independent of the NMR equipment manufacturer. Parameter information is provided for implementation on instruments from the major manufacturers (Supplemental Information), which will hopefully serve as a set of guidelines for acquiring qHNMR data on virtually any contemporary pulsed FT-NMR spectrometer.
qHNMR Factor 1: Non-Spinning and Shimming of the Sample
The qHNMR experiment acquires data on a static sample; i.e., in non-spinning mode. This eliminates residual spinning sidebands, which represent artifacts that are frequently comparable in magnitude to low-level sample impurities and, thus, can be confusing. The presence of spinning sidebands can also lead to resonance distortion, due to signal overlap, and to errors in the accuracy of integrated intensities of the signals arising from both major component(s) of the sample as well as the minor impurities. Because of the excellent static magnetic field (Bo) homogeneity of the contemporary commercial superconducting magnets, most samples for NMR analysis can routinely be analyzed in a static non-spinning mode while maintaining the high resolution required for the detection of small coupling constants.
As required for all high-resolution NMR work, proper shimming of each sample to achieve good lineshape with good signal-to-noise (S/N) is clearly an important prerequisite. While both manual and automated shimming routines are options, gradient shimming capability will, in general, lead to the best lineshape in the shortest timeframe.
qHNMR Factor 2: Removal of 13C Satellites
In the proposed protocol, qHNMR spectra are acquired with broadband decoupling of the 13C region to remove the 13C satellites from the 1H spectrum. Therefore, a pulse sequence profile for the qHNMR experiment was assembled as illustrated in Figure 1. Because the frequency range of 13C spectra is considerably larger compared to 1H, relatively high power is required to achieve broadband decoupling, which in turn leads to sample heating. As a consequence, a composite-pulse decoupling scheme is employed for broadband decoupling, which applies efficient decoupling to the sample with minimum heat generation. The proposed method used here specifically employs the decoupling scheme known as GARP8 for the use in a qHNMR experiment, and allows coverage of the entire 13C shift range of protonated carbons, while minimizing sample heating and related effects (see also qHNMR Factor 3). GARP decoupling has the advantage over the Waltz-16 decoupling scheme, initially described as part of a qHNMR experiment,9,10 of decoupling over a broader 13C chemical shift range. However, Waltz-16 may still be employed if decoupling over a narrower 13C spectroscopic window (<100 ppm) is desired. Other composite pulse decoupling schemes such as WURST11 or STUD,12 which employ adiabatic pulses for decoupling, could be used as an alternative for further minimizing the heat produced during decoupling, especially for concentrated aqueous ionic samples.
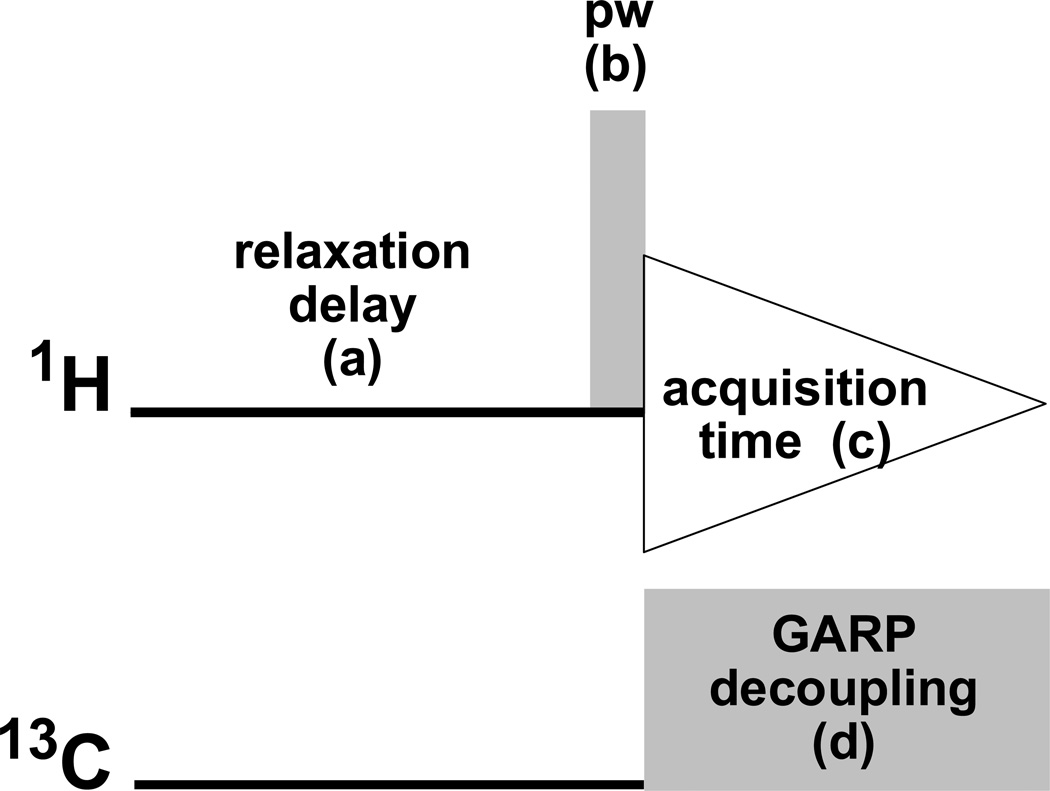
Figure 1.
Inverse-gated decoupling scheme for eliminating the 13C satellites from 1H NMR spectra, proposed as a routine experiment for acquisition of qHNMR data sets. The main building blocks of this experiment are the relaxation delay (a), the pulse width [pw] (b), the acquisition time (c), and the composite pulse decoupling GARP (d).
qHNMR Factor 3: Relaxation Delay (d1)
The delay, in seconds, which precedes the pulsed qHNMR experiment is referred to as the relaxation delay, denoted on most commercial NMR spectrometers as d1. This delay is inserted to allow the excited nuclei to re-establish their equilibrium z-magnetization after the acquisition of the FID information and prior to the application of the next pulse or pulse train. If the pulse excitation is a 90-degree pulse, i.e., all equilibrium z-magnetization is converted into transverse (x,y) magnetization, the relaxation delay is generally set to 5 times the longest proton relaxation time (T1) in the sample (determined by considering all proton resonances in the sample) in order to avoid distortion of integrated signal intensity due to relaxation effects. If the pulse excitation is a <90-degree pulse, then a shorter relaxation delay can in principle be employed. However, the relationship between the d1 delay, the proton relaxation times of the sample, and the “flip-angle” of the pulse used must also be considered. A further aspect of setting of the relaxation delay that requires comment relates to the application of composite pulse decoupling, in this instance 13C GARP decoupling, and the heat produced from the broadband decoupling during the acquisition time. While the length of the relaxation delay can be reduced, thus avoiding relaxation time effects and obtaining good quantitative results, decoupling of the carbon frequency range occurs during the acquisition time. Therefore, it is recommended to lengthen the relaxation delay to maintain a reasonable duty cycle (relaxation delay + acquisition time = pulse repetition rate) for the composite pulse decoupling and to minimize unfavorable heating effects. Minimizing heating effects serves to reduce excessive line broadening and, if the sample is heat sensitive, degradation of the sample during the course of the NMR data acquisition. In general, a duty cycle of 10–20% is recommended.
qHNMR Factor 4: Spectral Window Selection
The spectral window (syn. acquisition window or sweep width) is the region of radio frequency excitation that is used in a qHNMR experiment and, in part, depends on where the signals of interest reside. A wide qHNMR spectral window should be predetermined empirically, optimized for each solvent, and later can be adjusted to the specific conditions of a sample. Selection of an acquisition window for qHNMR should, however, always include a broader range, having additional region (~2 ppm) added on to both the high field and low field ends of the desired spectral window of interest. This is recommended in order to compensate for the “roll off” (signal attenuation) of the analog filters (hardware) on the NMR spectrometer, which are used to restrict or eliminate aliasing (folding) of unwanted signals from outside of the desired spectral window. As a result, signal intensity appearing at the ends of the spectral window (out of the linear region of the analog filter) is severely attenuated (>70%) on either extreme end of the spectrum. This would lead to significant errors in the integrated intensities of the spectrum when attempting any kind of quantitative application. It is very important to ensure that the spectral window of interest falls in the linear region of the analog filter. This will lead to both "flatter" baselines with good integrated intensities and elimination of signal attenuation at the edges of the spectrum. On most contemporary NMR spectrometers, the use of digital filtering (in contrast to analog filtering) and over-sampling generally provides improved baseline response, and the roll off problem tends to be reduced or eliminated. For a survey spectrum, a proton spectral window of 20 ppm is recommended as a general starting point. All contemporary NMR spectrometers are equipped with digital oversampling and digital filtering capabilities as an integral component of their hardware, so this aspect of the qHNMR acquisition is transparent to the spectroscopist. Oversampling improves the effective dynamic range (detection of small in the presence of large peaks), improves S/N, and leads to flatter baselines, all important factors for quantitation.
qHNMR Factor 5: Transmitter Position
Once the desired spectral window or sweep width is set in the spectrometer software, the transmitter offset frequency for excitation of the desired spectrum is positioned in the center of the spectral window. On most spectrometers the transmitter offset frequency is automatically adjusted depending on the spectral window selected. It can also be pre-defined as a solvent-dependent parameter value in a standardized qHNMR parameter set or be derived from an appropriate setup macro.
qHNMR Factor 6: Pulse Width Selection
The pulse width (pw) represents the length of the pulse excitation (in microseconds) that converts equilibrium magnetization (z) into transverse magnetization (x, y) at a specified transmitter power level. If all equilibrium magnetization is converted into transverse magnetization by the application of a radio frequency pulse, this is defined as a 90- degree pulse. Use of a 90-degree pulse results in creation of maximum signal intensity in the resultant spectrum. In order to acquire good quantitative NMR data in a reasonable period of time, however, a trade-off in the pulse width is employed. In practice, pulse widths of less than 90 degrees are often used. However, with prior knowledge of the value of the longest T1 or 1H relaxation time in the sample and a reasonable value for the relaxation delay, an optimum pulse width or flip angle (Ernst angle) can be determined13 and employed for qHNMR measurements.
qHNMR Factor 7: Selection of Acquisition Time
As noted earlier, following radio frequency excitation, creation and measurement of the free-induction decay (FID) represents the basic acquisition of a 1D NMR spectrum. The length of time that is spent to sample (or digitize) the FID is defined as the acquisition time. The acquisition time selected, in part, is related to the spectral window (see above) and to the desired level of digitization of the resultant NMR spectrum upon Fourier Transformation (FT), and should conform to the Nyquist relationship.14,15 In general, ample digitization (low Hz/point values; typically <0.2 Hz) is advantageous for quantitative work. Acquisition times of 2–4 sec @ 400 MHz are recommended.
qHNMR Factor 8: Selection of the Number of Scans or Transients
Selection of the number of scans (ns) or transients (nt) will depend on the level of desired sensitivity, which in turn will depend on the molecular weight and/or molar concentration of the analyte(s), and thus will be sample dependent. Sensitivity is generally defined as the achieved S/N of the spectrum with respect to a particular signal in the sample, and increases as the square root of the number of scans or transients. For a qHNMR spectrum, sensitivity will not only depend on the amount of sample, but also on the complexity of or the level of impurities present in the sample. For the proposed routine qHNMR experiment, the better the S/N of the spectrum, and especially the better the S/N with respect to the lowest level component/impurity in the sample, the better the quantitative accuracy that can be achieved. Thus, selection of the number of scans or transients for a qHNMR experiment will be largely variable and sample dependent. However as a general guideline, acquisition of routine qHNMR spectra using a 400 MHz magnet and a 5 mm room temperature probe typically requires 256 transients for a 10 mg sample of a 500 amu compound (20 µmol).
qHNMR Factor 9: Receiver Gain Setting
The receiver gain (rg) is generally set automatically on most NMR spectrometers, but can be manually overridden, as it is a critical parameter to set correctly. It is set prior to initiation of any data collection on the NMR spectrometer including qHNMR experiments. If the receiver gain is set too high, saturation of the receiver can result (“FID clipping”), and attenuation or in some cases signal elimination can result. In addition, severe baseline distortions may occur, which can have significant negative impact on the accuracy of quantitation. Setting the receiver gain too low can afford a spectrum with low S/N, requiring artificially high data collection times to achieve the desired S/N. Automated receiver gain optimization procedures are an integral feature on all modern NMR spectrometers.
qHNMR Factor 10: Steady State or “Dummy” Pulses
The use of steady state pulses, or “dummy” pulses, generally permits an equilibrium condition of any NMR experiment to be established prior to actually collecting and digitizing the FID information. All NMR spectrometers have this capability within their acquisition control software and, depending on the nature of the experiment, the number of steady-state pulses or dummy pulses used will be variable. The function of these pulses is simply to improve the reproducibility and to reduce variability of the data being collected. Once the experiment is in a “steady-state condition”, collection of the FID information will begin. Typically, 2–4 steady-state pulses are sufficient for a qHNMR experiment.
qHNMR Factor 11: 13C Decoupling
With further regard to the nature of the 13C decoupling noted above, the following two parameters need to be set correctly: the 13C spectral window, and the position of the 13C decoupler. The width of the 13C window to be decoupled can be limited to cover only the region concerned with protonated carbons, typically, δ 0 – 180 ppm. However, to ensure that all protonated carbons are decoupled, and in order to standardize the acquisition parameter set, a spectral window of δ 0 – 220 ppm may be recommended. In that fashion, aldehyde carbon resonances will, for example, be included in the decoupling process. The decoupler transmitter is then centered within the carbon window (δ 110 ppm). The advantageous “baseline cleaning” (13C satellite removal) effect of, in this case, the 13C broadband GARP decoupling scheme on the acquired qHNMR spectra is illustrated for taxol in Figure 2.
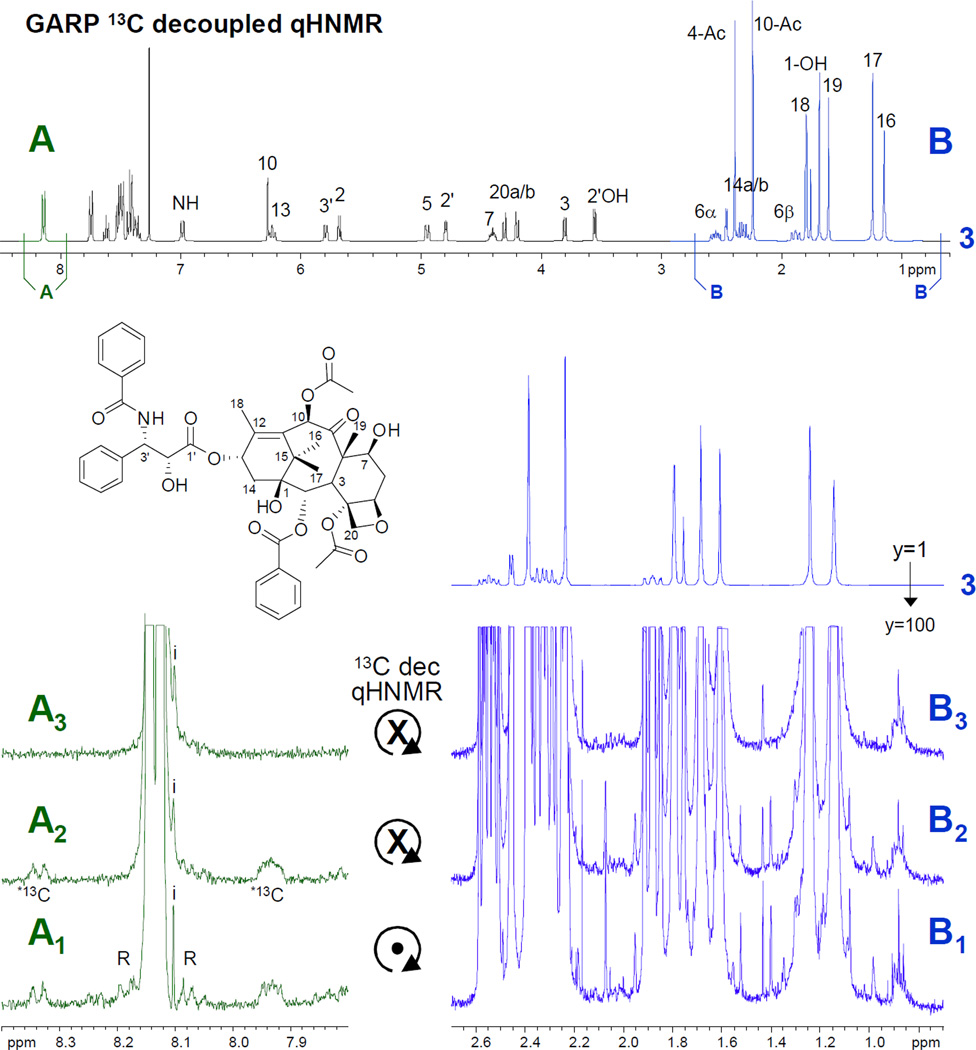
Figure 2.
Overview and expanded (A1–3, B1–3) plots of three representative qHNMR spectra [1–3] of a high-purity sample of taxol (6.3 mg in 600 µL CDCl3[99.8%D], 5 mm probe, 400 MHz, ns=256): [1] qHNMR acquired under quantitative conditions (see text and S1) with spinning sample; [2] same as spectrum 1, but no spin; [3] same as spectrum 2, but with 13C broadband carbon decoupling using the GARP composite pulse scheme. The expansions are plotted at 100-fold vertical scale in order to visualize the rotation artifacts (R; rotation side bands) in spectrum 1, and the carbon satellites (*13C) in spectra 1 and 2. The latter are both absent in spectrum 3 (only the signal of the very minor impurity (i) is visible), which represents the proposed method for routine qHNMR. While region B is dominated by the large methyl resonances, example B3 demonstrates how GARP decoupling improves the impurity detection by eliminating satellites, e.g. in the δ 1.35–1.55 region and from the minor impurity signals left isolated in B3 at δ 2.05.
The qHNMR evaluation of taxol reference materials and related samples
In order to demonstrate the suitability of the proposed qHNMR method, taxol was chosen as a model analyte in the form of three different samples (Table 1): reference materials of varying purity of taxol (taxol A–C), a structurally related compound (taxoid D), and a crude extract of Taxus brevifolia bark. All five samples were subject to qHNMR analysis and their (im)purity profiles were quantitatively evaluated. Quantitative calculations were performed under the qualitative assumption that structurally related analogues, as evident from marker signals similar to those of taxol, were present as impurities.16 Due to the close structural similarities of the taxoids, the assumption was made that the molecular weight of the taxoid impurities is similar to taxol, and the identical mass (854 amu) was taken into account as a ficticious weight. This approach has empirically been proven to provide very reasonable quantitative results in purity analyses that are based on the 100% method5,17 (and unpublished data using absolute quantitation; see also error discussion below).
Table 1.
Quantitative impurity profiles of three reference materials (taxol A–C, taxoid D) and a crude T. brevifolia extracta investigated by qHNMR.
| Sample | Number of impurities |
Impurity in % |
Taxol purity or content in % |
Nature of impurity |
LC-based content in % |
|---|---|---|---|---|---|
|
Reference Materials |
|||||
| taxol A | imp1 imp2 imp3 |
4.6(9) 5.1(3) 0.3(1) |
89.8(7) (w aliph. material) 94.7(3) (w/o aliph. material) |
taxoid taxoid aliphatic material |
n.a. 96.9 |
| taxol B | imp1 imp2 imp3 imp4 |
8.3(7) 5.6(8) 0.9(1) 0.2(5) |
84.7(3) | taxoid taxoid taxoid taxoid |
n.a. |
| taxol C | imp1 imp2 |
4.9(2) 9.5(1) |
85.5(7) | taxoid taxoid |
85.6 |
| taxoid Db | – | – | 1.4(4) | taxol | |
| Crude Extract |
% taxol content |
||||
|
T. brevifolia bark extracta |
3.1 | 4.0 |
The taxol content of the extract was determined as 4.0% by HPLC
10-Deacetylbaccatin
Post-acquisition data processing was performed according to a spectroscopic processing concept that had been specifically optimized for qHNMR spectra.18 Given the choice of acquisition conditions and parameters (GARP 13C decoupled qHNMR sequence as in Figure 1, acquisition parameters according to S1), the optimum choice of processing parameters was as follows. A Lorentzian-Gaussian resolution enhancement (LG) with a Gaussian factor of 0.05 (5% of AQ) and a line broadening factor of (−)0.3 Hz was used. The digital resolution of the 64k-sized frequency domain spectra was maintained by adding an equal number of zeros to the end of the original FID data, i.e., by single zero filling. Additional zero filling to 256k data points was used to increase the overall digital resolution for the purpose of integration/quantitation. In order to improve the precision of the integration, further steps were taken for each individual qHNMR spectrum: (i) the baseline of the FID was corrected (DC correction); (ii) broad resonances, such as those of water, other –OH and exchangeable protons, were eliminated by repeated line fitting and subtraction; and (iii) the overall baseline of the spectra was flattened by applying an nth order polynomial correction (n<10).
For the reference materials, quantitation was based on the proportionality of the integrals of all detected resonances, and by assigning arbitrary values to presumably non-overlapped (“most pure”) reference resonances of the major taxoid in samples A–D, respectively (Table 1). Due to the conformational molecular dynamics of taxol, integration of the apparently non-overlapped signals at lower field still lead to slightly disproportionate integrals. Taking into account the underlying dynamic peak broadening/splitting, this deviation could be eliminated and almost the nominal integrals (value 100) be obtained by using relatively wide integration limits, which were necessary to cover the entire resonance signal. The proportionality was then calculated on the basis of all detected impurities, which were normalized to 100% of the total sample (“100 % method” or “100%-minus-impurity approach”).5,17,19,20 Specifically, the signal groups of the H-10 and H-13 resonances, which appear in the range 6.18–6.27 ppm (sample taxol A and C), as well as the resonances assigned to H-5 and H-2’, appearing at 4.72–4.97 ppm for taxol B and taxoid D, were assigned integration values of 200 and, thus, served as internal reference signals with an arbitrary integral value of 100 per proton. The assignment of these two groups of non-overlapped reference signals was based on their maximum signal purity, as indicated by their minimum integral per proton compared to a the other resonances of the spectra. In order to aid in the assignment and detection of overlapping and non-overlapping impurity signals, 2D COSY spectra were employed.21,22
Three of the four reference materials A–D were found to contain 2–4 different taxol analogues as the only impurities, while one of the samples contained a significant amount of aliphatic material (Table 1). The purities of the reference materials ranged from 84.8 to 94.7%. One of the samples (sample D), due to the blinded study, was initially analyzed as taxol, but turned out to be 10-deacetylbaccatin containing taxol as a minor impurity (1.44%). In general, the qHNMR based quantitative impurity profiles and sample purity values determined in this study were congruent with the LC-based purity evaluation (Table 1). A noteworthy detail is that the qHNMR purities tend to be somewhat lower than LC-based values, which matches previously made observations (unpublished data). There are three obvious factors that might explain differences between NMR and chromatographic results. (i) NMR does not require calibration by a response factor, which is required in chromatographic quantifications, but often is unknown when doing chromatographic impurity profiling using the 100% method; also, chromatographic response factors can exhibit large differences. (ii) Chromatographic co-elution is more difficult to detect than NMR peak overlap: in chromatography only a single peak is obtained per analyte, whereas in NMR multiple signals arise from one analyte. (iii) Ubiquitously occurring natural "matrix substances" are likely to be omitted in chromatographic impurity profiling due to the lack of corresponding peaks (e.g., they may only give rise to an elevated baseline), whereas their detection in qHNMR will more likely occur as long as they contain protons. Systematic studies will be needed comparing LC with qHNMR purity of natural products at different levels of purification in order to fully establish correlations between LC and qHNMR purities of natural products.
In one of the taxol samples (sample C) a minor impurity of an unassigned taxoid could be detected and was quantified to 0.25% using the proposed qHNMR method. This demonstrates that routine qHNMR is capable of quantifying minor components in complex spectra with a dynamic range of 300:1 or better, as illustrated in Figure 3. As this study was performed with blinded samples and structural information about the main analytes and the impurities was lacking, a systematic error related to the abovementioned molecular weight assumptions remains. However, proving the identity of all taxoid impurities in samples A–D was clearly beyond the scope of this study. Moreover, it is noteworthy that errors from molecular weight assumptions are diluted in proportion to the relative content of the minor component in the whole mixture. For example, for an impurity at the 1% level, even a 20 % assumption error translates into a mere 0.2% deviation in the final 100% qHNMR mass balance.
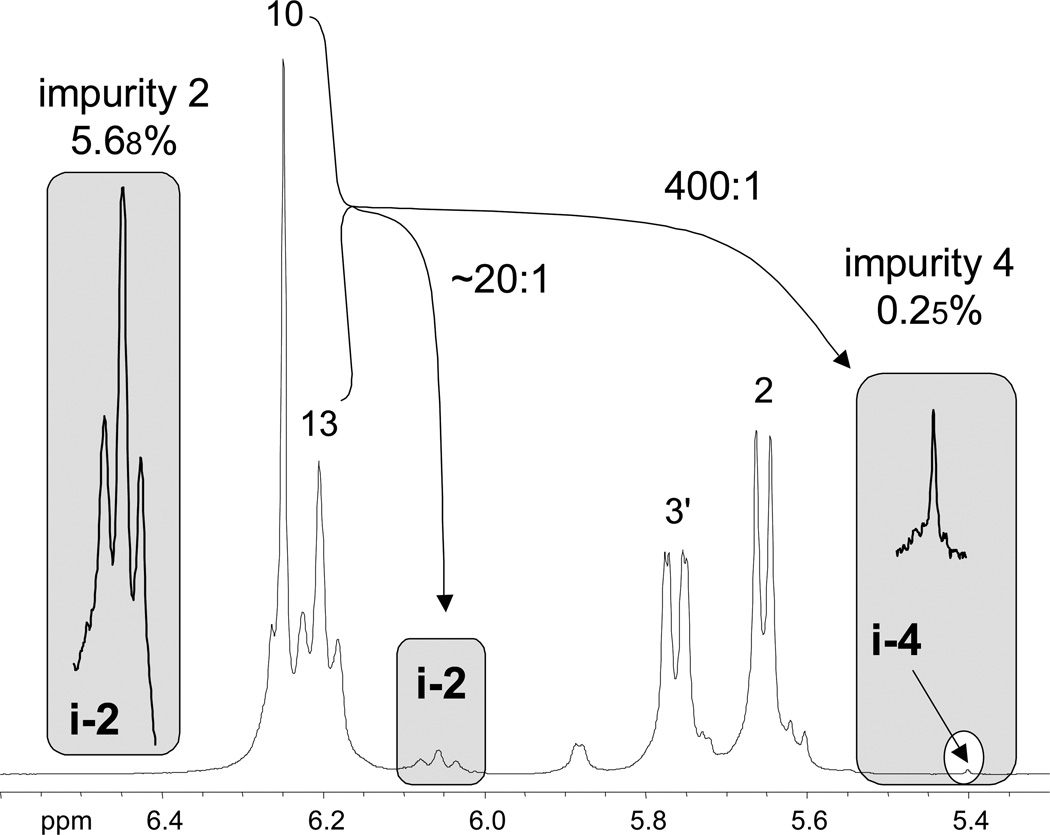
Figure 3.
Provided that the chemical shift dispersion is sufficient, the proposed qHNMR experiment can routinely detect and quantitate minor impurities with a dynamic range of 300:1 or better. This example demonstrates the relative intensities of the signals of the impurities i-2 (5.68%) and i-4 (0.25%) and their relationship with the main resonances of the 84.8% pure taxol sample B. The remarkable 400:1 dynamic range between taxol and its impurity i-4 also visualizes how readily the signals of minor impurities can be overlooked and/or buried under noise unless NMR spectra are acquired with adequate S/N.
In order to mimic a situation typically found in metabolome analysis, the lower quantitation level of a structurally complex phytochemical contained in a chemically diverse mixture was assessed as model system. Accordingly, a crude extract of Taxus brevifolia bark was analyzed by qHNMR as described, and taxol as a minor constituent in this crude metabolome mixture was determined to be present in the amount of 3.1(2)%. Due to severe signal overlap in this very complex T. brevifolia bark extract, additional processing of the qHNMR spectrum was necessary. The only taxol signal that was sufficiently isolated and amenable for quantitation was the signal of H-10 at 6.250 ppm. Prior to integration, interfering signals resulting from the numerous other components contained in the extract, which were convoluted to an underlying hump, were line-fitted and subtracted from the spectrum. In addition, it became evident from the analysis of the taxol reference materials A–C that the integral for H-10 had to be set to an arbitrary value of 125 instead of 100, since the H-10 resonance demonstrated overlap (25% by integral) with the resonance of H-13. The final calculation of the taxol content in the extract was based on the relative integrals in the region between 7.29 and 8.20 ppm as follows. In pure taxol, the signal of 15 protons resonated in this chemical shift range. Therefore, their arbitrary integral value was set to 1500 for “pure” taxol, and was subsequently related to the measured total integral (48144) of the signals of the extract sample resonating in the same chemical shift range. This led to the calculation of a content of 3.1(2)% of taxol in the investigated T. brevifolia bark extract (see Figure 3), which correlated reasonably well with a value of 4.0% as determined by HPLC, taking into account the likelihood of both chromatographic peak and NMR signal overlap. While this observed congruence generally supports the potential role of qHNMR in metabolomic analyses, qHNMR assay specificity will have to be demonstrated for each analyte of interest and within the margins of error to be achieved.
Routine qHNMR of natural products and biological reference materials
The results of this study clearly illustrate the principle feasibility of routine GARP 13C-decopled qHNMR analysis of rather complex molecules. The qHNMR concept performs even when working with deliberately flawed assumptions, e.g., when exact structures and MWs of impurities are unavailable (see above); when using imperfect molar masses in calculations that apply the 100% normalization/mass balance method; and when dealing with relatively demanding analytes such as taxol that exhibit a highly complex 1H resonance pattern that are complicated by the, presence of various conformers at RT.
The key advantages of qHNMR besides being a relatively inexpensive and fast method can be summarized as follows: (i) qHNMR can reliably distinguish “highly pure” from “less pure” reference material with precisions in the range of 0.1%; (ii) qHNMR allows detection of even very low proportions of a target analyte (e.g., 3.1% of taxol in T. brevifolia extract) or impurities (< 1%) in complex matrices by detailed targeted processing of single signals, e.g., in crude extract and metabolome analysis; (iii) the qHNMR concept comes included with a tool for the verification of the authenticity of the (target) analyte through the direct structural evidence contained in individual 1H resonances (marker signals); (iv) qHNMR has the potential to become a routine technique of almost universal applicability, even in research environments with entry-level instrumentation (300–400 MHz) and/or when using relatively small amounts (< 10 mg) of high molecular weight samples (taxol: 854 Daltons).
Because of the potential biological implication of the various constituents and impurities that in a given sample can be present at widely varying abundances (~10 to 0.1%), qHNMR can provide a value-added dimension to the standard set of spectroscopic data. Consequently, it is reasonable to suggest that qHNMR spectra be routinely obtained for bioactive molecules and as part of the structure elucidation process for new natural products. Because the qHNMR method realistically requires almost no additional effort, except for the simple task of establishing the quantitative conditions for the standard 1H NMR experiments, it can be easily integrated into the workflow of any routine qualitative NMR protocol. Considering that qHNMR can routinely cover a dynamic range of 300:1 or better, its suitability for quantitative metabolome analysis is evident. At the same time, the method is capable of positive identification of metabolites through marker signals, which adds to the portfolio of qHNMR in metabolome and natural products research.
Experimental Section
NMR Spectroscopy
All of the 1H NMR data (400 MHz) described here were obtained on either a Bruker DPX-400 NMR spectrometer using a 5-mm QNP probe or a Bruker AVANCE-400 NMR spectrometer using a 5-mm broadband ATM probe. The taxol samples (about 10 mg) were weighed to precision, dissolved in CDCl3 (Aldrich Chemical/Isotech), and then transferred, with filtering through a “plug” of cotton wool, into a 5-mm NMR tube with a total volume of solution of ~ 700 uL. A standard 1H NMR experiment (without 13C decoupling) was quickly obtained to evaluate the sample shimming. A 13C GARP decoupled qHNMR spectrum was then obtained using the pulse profile described in Figure 1 and in the Supplementary Information (S1). The latter also provides parameter sets that encode the acquisition conditions for modern spectrometers from all major NMR instrument manufacturers (Bruker, Jeol, and Varian). An alternative implementation of GARP-1 decoupling using a scan-dependant decoupler offset of a limited 13C spectral region has recently been reported.23
Qualitative NMR of taxol
The NMR data of taxol shown here generally refer to assignments that are well documented,16, 24–27 and the observed spectra (Figure 2) were found to be qualitatively identical with the published data.
Supplementary Material
Acknowledgment
We are indebted to Dr. James McChesney, NaproBio, Boulder (CA), and to Dr. Nick Oberlies, RTI (NC), for generously providing the taxol and Taxus brevifolia extract samples, respectively. Furthermore, the authors are grateful to their colleagues at UIC, Chicago, for fostering a stimulating academic environment. We would also like to acknowledge the assistance of Dr. George Gray (Varian) for checking the accuracy of the Varian macro, as well as Dr. Ashok Krishnaswami (JEOL) for providing the description and code that allows implementation of the qHNMR experiment on JEOL NMR spectrometers. Finally, the authors wish to acknowledge partial support of this work through the following grants: P50-AT00155 from the Office of Dietary Supplements (ODS) and the National Center for Complementary and Alternative Medicine (NCCAM), and R21-AI052847-01 from NIAID/NIH.
Footnotes
Dedicated to Dr. Norman R. Farnsworth on the occasion of his 77th birthday.
Supporting Information Available. This material is available free of charge via the Internet at http://pubs.acs.org. Further information on qNMR methodology, instrument parameter files for the NMR major manufacturer’s spectrometers, and original FIDs of the taxol qHNMR experiments in JCAMP-DX format can be found at http://qnmr.org.
References and Notes
- 1.Newman DJ, Cragg GM, Snader KM. J. Nat. Prod. 2003;66:1022. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- 2.Newman DJ, Cragg GM, Snader KM. Nat. Prod. Rep. 2000;17:215. doi: 10.1039/a902202c. [DOI] [PubMed] [Google Scholar]
- 3.Cragg GM, Newman DJ, Snader KM. J. Nat. Prod. 1997;60:52. doi: 10.1021/np9604893. [DOI] [PubMed] [Google Scholar]
- 4.Eldridge GR, Vervoort HC, Lee CM, Cremin PA, Williams CT, Hart SM, Goering MG, O'Neil-Johnson M, Zeng L. Anal. Chem. 2002;74:3963. doi: 10.1021/ac025534s. [DOI] [PubMed] [Google Scholar]
- 5.Pauli GF, Jaki B, Lankin D. J. Nat. Prod. 2005;68:133. doi: 10.1021/np0497301. [DOI] [PubMed] [Google Scholar]
- 6.Pauli GF, Jaki B, Lankin D, Walter JA, Burton I. Quantitative NMR of Bioactive Natural Products. In: Colegate S, Molyneux R, editors. Bioactive Natural Products. 2nd ed. New York: Taylor & Francis; 2007. in press. [Google Scholar]
- 7.Szantay C, Beni Z, Balogh G, Gati T. TrAC, Trends in Analytical Chemistry. 2006;25:806. [Google Scholar]
- 8.Shaka AJ, Barker PB, Freeman R. J. Magn. Reson. 1985;64:547. [Google Scholar]
- 9.Simeral LS. Today's Chemist at Work. 1993;11:52. [Google Scholar]
- 10.Simeral LS. Appl. Spectrosc. 1995;49:400. [Google Scholar]
- 11.Kupce E, Freeman R. J. Magn. Res. Series A. 1995;115:273. [Google Scholar]
- 12.Bendall MR, Skinner TE. J. Magn. Reson. 1998;134:331. doi: 10.1006/jmre.1998.1522. [DOI] [PubMed] [Google Scholar]
- 13.Ernst RR, Anderson WA. Rev. Sci. Instru. 1966;37:93. [Google Scholar]
- 14.Claridge TDW. High-resolution NMR Techniques in Organic Chemistry. New York: Pergamon; 1999. [Google Scholar]
- 15.Derome AE. Modern NMR Techniques for Chemistry Research. New York: Pergamon Press; 1987. [Google Scholar]
- 16.Chmurny GN, Paukstelis JV, Alvarado AB, McGuire MT, Snader KM, Muschik GM, Hilton BD. Phytochemistry. 1993;34:477. [Google Scholar]
- 17.Pauli GF. Phytochem. Anal. 2001;12:28. doi: 10.1002/1099-1565(200101/02)12:1<28::AID-PCA549>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 18.Jaki B, Chadwick L, Chen S-N, Pauli GF. The Pivotal Role of Spectral Processing in the Quantitative 1H NMR Analysis of Natural Products. Annual Meeting of the American Society of Pharmacognosy: Corvallis (OR); 2005, July 23–27; (#O34). [Google Scholar]
- 19.Pinciroli V, Biancardi R, Visentin G, Rizzo V. Org. Proc. Res. Devel. 2004;8:381. [Google Scholar]
- 20.Rizzo V, Pinciroli V. J. Pharm. Biomed. Anal. 2005;38:851. doi: 10.1016/j.jpba.2005.01.045. [DOI] [PubMed] [Google Scholar]
- 21.Malz F. Ph.D. Dissertation. Berlin: Humboldt-Universität Berlin; 2003. Quantitative NMR-Spektroskopie als Referenzverfahren in der Analytischen Chemie; p. 147. [Google Scholar]
- 22.Malz F, Jancke H. J. Pharm. Biomed. Anal. 2005;38:813. doi: 10.1016/j.jpba.2005.01.043. [DOI] [PubMed] [Google Scholar]
- 23.Soininen P, Haarala J, Vepsäläinen J, Niemitz M, Laatikainen R. Anal. Chim. Acta. 2005;542:178–185. [Google Scholar]
- 24.Hilton BD, Chmurny GN, Muschik GM. J. Nat. Prod. 1992;55:1157. doi: 10.1021/np50086a023. [DOI] [PubMed] [Google Scholar]
- 25.Chmurny GN, Hilton BD, Brobst S, Look SA, Witherup KM, Beutler JA. J. Nat. Prod. 1992;55:414. doi: 10.1021/np50082a002. [DOI] [PubMed] [Google Scholar]
- 26.Barboni L, Lambertucci C, Appendino G, Vander Velde DG, Himes RH, Bombardelli E, Wang M, Snyder JP. J. Med. Chem. 2001;44:1576. doi: 10.1021/jm001103v. [DOI] [PubMed] [Google Scholar]
- 27.Snyder JP, Nevins N, Cicero DO, Jansen J. J. Am. Chem. Soc. 2000;122:724. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.