Myo1c directly interacts with G-actin and is important for its transport to the leading edge during endothelial cell migration.
Abstract
Addition of actin monomer (G-actin) to growing actin filaments (F-actin) at the leading edge generates force for cell locomotion. The polymerization reaction and its regulation have been studied in depth. However, the mechanism responsible for transport of G-actin substrate to the cell front is largely unknown; random diffusion, facilitated transport via myosin II contraction, local synthesis as a result of messenger ribonucleic acid localization, or F-actin turnover all might contribute. By tracking a photoactivatable, nonpolymerizable actin mutant, we show vectorial transport of G-actin in live migrating endothelial cells (ECs). Mass spectrometric analysis identified Myo1c, an unconventional F-actin–binding motor protein, as a major G-actin–interacting protein. The cargo-binding tail domain of Myo1c interacted with G-actin, and the motor domain was required for the transport. Local microinjection of Myo1c promoted G-actin accumulation and plasma membrane ruffling, and Myo1c knockdown confirmed its contribution to G-actin delivery to the leading edge and for cell motility. In addition, there is no obvious requirement for myosin II contractile–based transport of G-actin in ECs. Thus, Myo1c-facilitated G-actin transport might be a critical node for control of cell polarity and motility.
Introduction
Actin polymerization drives cell locomotion, proceeding by addition of monomeric actin (G-actin) to the barbed end of actin filaments (F-actin; Pollard and Borisy, 2003). Actin polymerization is highly polarized and spatially restricted in lamellipodia within a band ∼1–3 µm in width along the leading edge of a moving cell (Watanabe and Mitchison, 2002; Ponti et al., 2004; Lai et al., 2008). A high amount of lamellipodial G-actin is consumed to drive movement—for example, ∼3.6 million actin molecules per minute in a crawling breast cancer cell (Chan et al., 1998). Passive diffusion has been suggested to be the major pathway for providing G-actin to the cell leading edge (Koestler et al., 2009). However, diffusion might be insufficient for entering and traversing the viscous, dense, and highly structured lamellipodial space. Recent experimental and theoretical studies are consistent with diffusion-limited actin polymerization (Noireaux et al., 2000; Mogilner and Edelstein-Keshet, 2002; Plastino et al., 2004). Other mechanisms might contribute to delivery of G-actin to lamellipodia, including local synthesis as a result of mRNA relocalization (Lawrence and Singer, 1986; Shestakova et al., 2001), facilitated transport via myosin II contraction (Peckham et al., 2001; Zicha et al., 2003), or actin treadmilling by rapid F-actin turnover (Cramer, 1999). Forward actin flow reported in the protrusion region suggests active transport of G-actin to the leading edge (Zicha et al., 2003). However, little is known about molecular mechanisms regulating G-actin delivery to the leading edge. Here, we reveal an important contributory role of Myo1c in G-actin transport during endothelial cell (EC) migration.
Results and discussion
Vectorial transport of G-actin to the EC leading edge during migration
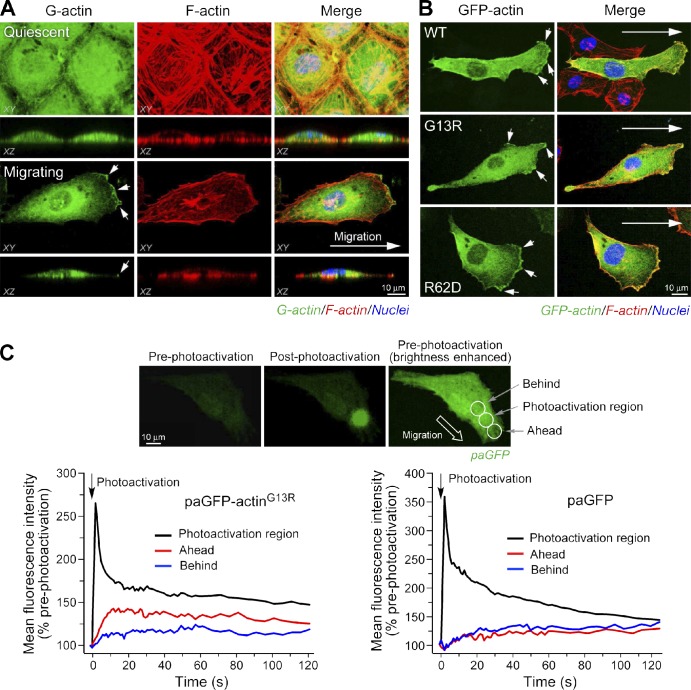
To examine G-actin localization during cell migration, bovine aortic ECs were induced to move by razor wound (Ghosh et al., 2002) and stained with fluorescence-labeled DNase I. Confocal microscopy showed uniform distribution in quiescent cells but pronounced G-actin accumulation at the leading edge of migrating cells (Fig. 1 A), consistent with a previous study in fibroblasts (Cao et al., 1993). To determine the contribution of F-actin turnover to G-actin localization, two nonpolymerizable actin mutants, G13R and R62D, mutated at the nucleotide-binding pocket and the salt bridge that joins actin subdomains, respectively (Posern et al., 2002), were expressed as GFP chimeras. Both mutant proteins accumulated at the leading edge (Figs. 1 B and S1), suggesting an F-actin turnover–independent mechanism for G-actin polarization. To determine the potential contribution of actin mRNA relocalization (Lawrence and Singer, 1986; Shestakova et al., 2001), cells were pretreated with cycloheximide to block de novo actin synthesis. The protein synthesis inhibitor did not alter G-actin accumulation in the lamellipodia (unpublished data), consistent with a previous study showing that de novo synthesis contributes only ∼7% of the G-actin required for polymerization in migrating cells (Condeelis and Singer, 2005). Fluorescent Alexa Fluor 488–labeled actin, introduced exogenously to permeabilized cells, also accumulated at the cell leading edge, directly showing mRNA-independent G-actin translocation (unpublished data). To investigate the role of vectorial transport, directed movement of G-actin was measured by photoactivation of a chimera of nonpolymerizable actinG13R and photoactivatable GFP (paGFP; Patterson and Lippincott-Schwartz, 2002). The reporter was photoactivated near the leading edge of live migrating cells, and time-lapse fluorescence intensity was measured in front of and behind the photoactivation region. The initial rate of forward movement of paGFP-actinG13R was about twice that of the rearward rate (Fig. 1 C). No difference was detected between forward and rearward rates of movement of the paGFP control protein, which is likely a result of random diffusion. Furthermore, FRAP for GFP-actinG13R at the leading edge of migrating ECs is about twice that in the cell center (Fig. S1 B). These results suggest a directed G-actin transport mechanism, consistent with previous studies in which FRAP, photoactivation, and fluorescence localization after photobleaching using GFP-tagged wild-type (WT) actin all indicated that actin movement into protrusions of transformed rat fibroblasts and mouse melanoma cells was too fast to be explained by diffusion alone (Zicha et al., 2003; Lai et al., 2008).
Figure 1.
Vectorial transport of G-actin to the EC leading edge during migration. (A) Accumulation of G-actin at the leading edge. EC migration was induced by razor wound, and quiescent or migrating ECs were fixed; G-actin was visualized with Alexa Fluor 488–DNase I and F-actin with Alexa Fluor 568–phalloidin followed by confocal microscopy. (B) Localization of nonpolymerizable actin mutants in migrating ECs. Cells were transfected with pEGFP-actin (WT, G13R, or R62D mutant). (A and B) Arrows indicate the direction of cell migration. (C) Directed movement of paGFP-actinG13R. Cells transfected with paGFP or paGFP-actinG13R were photoactivated in the lamellipodium, and fluorescence intensity was monitored in three regions. (top) Representative fluorescent images of paGFP before (left) and after (center) photoactivation; brightness of preactivated paGFP was enhanced to show cell and regions of photoactivation and signal acquisition (right). (bottom) Fluorescence intensity in each of three regions was measured for 120 s. The data shown are representative from multiple experiments (n = 5–7 cells).
Cargo-binding tail domain of Myo1c interacts with G-actin
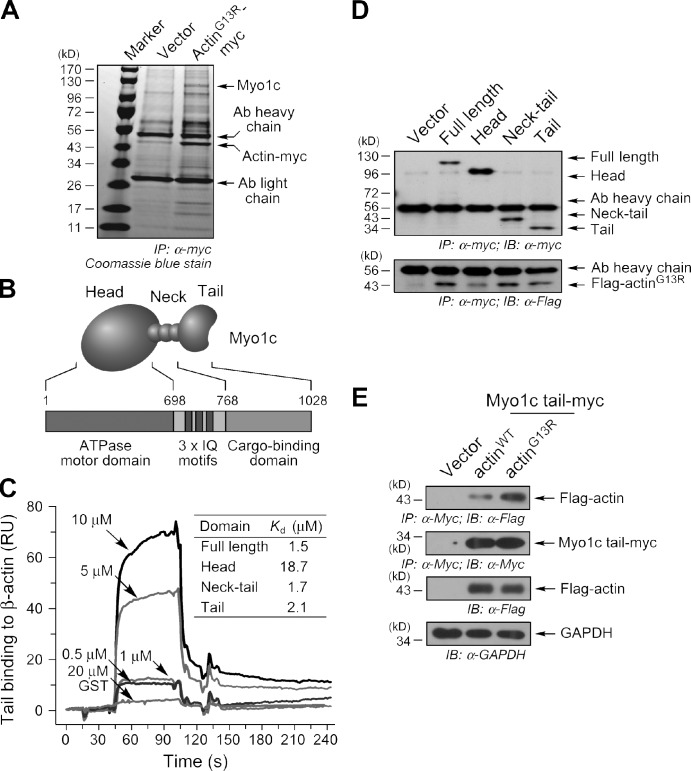
To investigate the possible role of carrier proteins in G-actin transport, myc-tagged actinG13R was immunoprecipitated from a lysate made from migrating ECs and endogenous interacting proteins determined by mass spectrometry (MS). The nonpolymerizable actin mutant was used to enhance pull-down of specific G-actin–binding proteins in the background of abundant F-actin–binding proteins. Multiple peptides consistent with the sequence of bovine Myo1c (previously called myosin-Ic, myosin-Iβ, or Myr 2) were identified (16 peptides spanning 25.2% of coding sequence; Fig. 2 A). Myo1c is a single-headed, nonfilamentous F-actin–binding motor protein consisting of head, neck containing three IQ motifs, and tail domain, responsible for ATP hydrolysis and motor function, CaM and light chain binding, and cargo-binding activities, respectively (Fig. 2 B; Reizes et al., 1994). To gain insight into the functional consequences of this interaction, the G-actin–interacting domain of Myo1c was investigated. Interaction of G-actin with the cargo-binding tail domain would suggest a transport function. Alternatively, interaction with the Myo1c head domain could promote actin polymerization, as shown for the interaction of G-actin with the head domain of myosin II, a domain conserved in all myosins (Miller et al., 1988; DasGupta et al., 1990). Full-length Myo1c and deletion domains corresponding to the head, tail, and neck plus tail were expressed in Escherichia coli as GST-tagged chimeras. Binding of Myo1c domains to immobilized G-actin was determined by surface plasmon resonance (SPR). The full-length protein and deletion fragments containing the tail domain all bound G-actin with moderately high affinity (i.e., ∼2 µM or less), but the head domain exhibited an order-of-magnitude lower affinity (Fig. 2 C). To investigate the interaction in vivo, myc-tagged Myo1c and Flag-tagged actinG13R were coexpressed in subconfluent, migrating ECs. Full-length Myo1c and tail-containing fragments were efficiently coprecipitated with Flag-actinG13R, but weaker interaction with Myo1c head domain was observed (Fig. 2 D). To determine binding of Myo1c tail domain to both G- and F-actin, myc-tagged tail domain and Flag-actinWT or Flag-actinG13R were coexpressed in ECs and cells treated with jasplakinolide to stabilize polymerized actin. Coimmunoprecipitation showed preferential binding of the tail domain to Flag-actinG13R, suggesting the domain is more likely to influence G-actin dynamics rather than interactions between nearby F-actin filaments (Fig. 2 E). Together, these results support the in vivo interaction of G-actin with the tail domain and suggest that Myo1c might function as a G-actin transport protein during cell migration.
Figure 2.
Cargo-binding tail domain of Myo1c interacts with G-actin. (A) Identification of Myo1c as a G-actin–interacting protein. Lysates from migrating cells transfected with pcDNA-actinG13R-myc were immunoprecipitated (IP) with anti-myc antibody (Ab) and resolved by SDS-PAGE. Proteins in gel-eluted bands were identified by MS. (B) Domain structure of Myo1c including head, IQ-containing neck, and tail domains. (C) Binding of Myo1c tail domain to G-actin. G-actin was immobilized on a sensor chip, purified Myo1c was injected, and the interaction was measured by SPR. (inset) Kd calculated from three experiments. RU, relative unit. (D) Tail domain interacts with G-actin in migrating cells. ECs were cotransfected with pDream-Flag-actinG13R and pcDNA-Myo1c-myc. Cell lysates were immunoprecipitated with anti-myc antibody and immunoblotted (IB) with anti-myc, -Flag, and -actin antibodies. (E) Myo1c tail domain preferentially binds G-actin. ECs were cotransfected with pcDNA–Myo1c tail–myc and pDream-Flag-actinG13R or pDream-Flag-actinWT and treated with 1 µM jasplakinolide for 1 h. Cell lysates were immunoprecipitated with anti-myc antibody and immunoblotted with anti-myc and -Flag antibodies. Lysates were immunoblotted with anti-Flag and anti–glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibodies.
Localization of G-actin in lamellipodia by motor activity of Myo1c
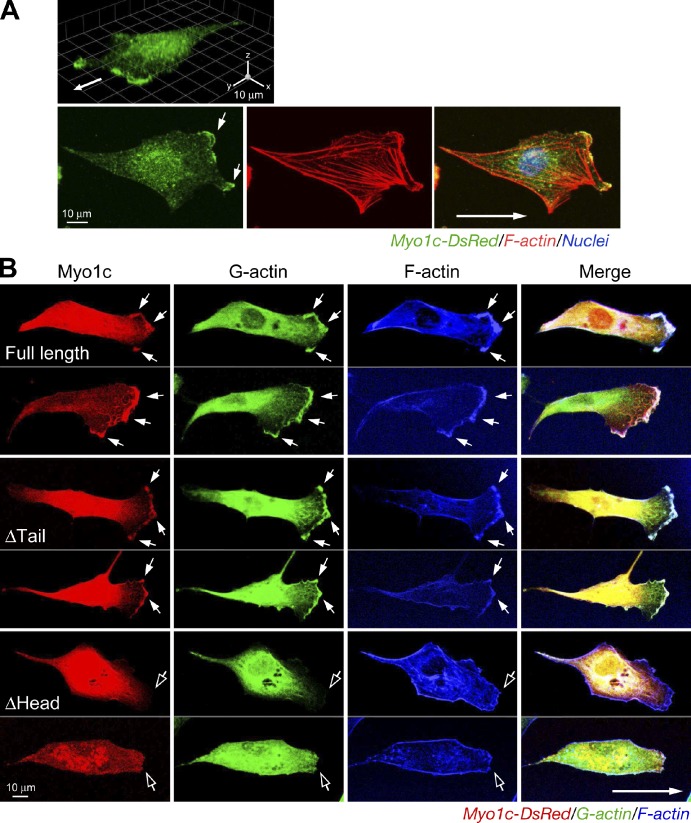
To determine Myo1c subcellular localization during EC movement, migrating cells were visualized with anti-Myo1c antibody. Myo1c was enriched and colocalized with F-actin at the leading edge of migrating ECs (Fig. 3 A). To determine whether Myo1c specifically colocalizes with G-actin, monomeric DsRed fusion protein of full-length Myo1c or proteins lacking the head or tail domains were expressed in migrating ECs. Full-length Myo1c colocalized with G-actin at the protrusive leading edge (Fig. 3 B). Myo1c lacking the tail region also accumulated in the leading edge. However, deletion of the head markedly blocked polarization of Myo1c localization, consistent with a previous study showing that Myo1b localization in the protrusive edge of spreading epithelial cells requires the motor domain (Tang and Ostap, 2001). Remarkably, overexpression of headless Myo1c prevented G-actin accumulation at the cell leading edge, possibly by competing with endogenous Myo1c for G-actin binding and transport. The interaction of Myo1c with G-actin and its spatial distribution during cell migration were investigated by Förster resonance energy transfer (FRET) coupled to confocal microscopy. An elevated, but nonuniform, interaction was observed in the lamellipodial region (Fig. S2 A), suggesting that a principal function of Myo1c might be local G-actin transport in lamellipodia to the leading edge. Interestingly, a weak interaction was observed in a protruding region of the leading edge where G-actin was most highly concentrated, suggestive of local release of G-actin in protrusive structures. Together, these results indicate that Myo1c mediates short-range G-actin trafficking in lamellipodia, compatible with structural and biochemical studies identifying myosin I isoforms as low–duty ratio motor proteins (De La Cruz and Ostap, 2004; O’Connell et al., 2007).
Figure 3.
Motor domain–dependent localization of Myo1c at the cell leading edge. (A) Myo1c localization at the cell leading edge. ECs were stained with anti-Myo1c antibody and visualized with Alexa Fluor 488–IgG and Alexa Fluor 568–phalloidin. 3D reconvolution (top) and single-layer confocal scanning images are shown. Arrows indicate the direction of cell migration. (B) Localization of Myo1c at the leading edge requires the motor domain. ECs were transfected with plasmids encoding full-length or domain-deleted Myo1c fused with DsRed. Migrating cells were stained with Alexa Fluor 488–DNase I. Filled arrows indicate colocalized Myo1c and G-actin at the leading edge, and open arrows show the leading edge with minimal colocalization.
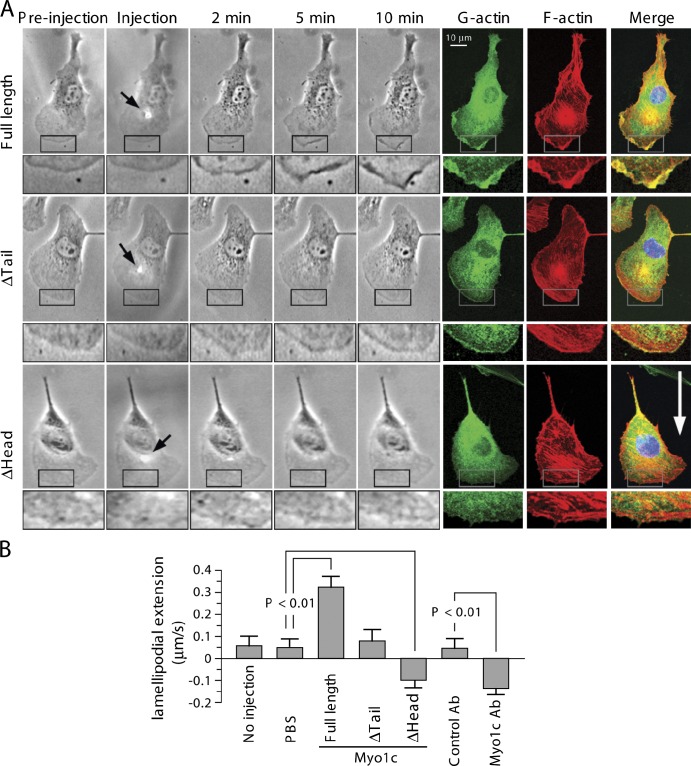
We determined the effect of Myo1c on leading edge dynamics by microinjecting purified Myo1c into lamellipodia. Local injection of full-length Myo1c protein, but not motor domain, increased G-actin localization and F-actin content at the leading edge, rapidly inducing plasma membrane ruffling and lamellipodia extension (Fig. 4). Microinjection of Myo1c tail domain reduced G-actin accumulation and induced retraction of lamellipodia. Microinjection of anti-Myo1c antibody targeting the head domain responsible for motor activity rapidly induced G-actin delocalization at the leading edge and lamellipodial retraction (unpublished data). Collectively, these results indicate that Myo1c dynamically traffics to the leading edge and mediates G-actin localization to the leading edge, membrane ruffling, and lamellipodial extension.
Figure 4.
Myo1c-induced actin dynamics in lamellipodia. (A) Representative images. Microinjection of Myo1c induces rapid G-actin accumulation and plasma membrane ruffling at the leading edge, increasing lamellipodial extension. Cells were microinjected with purified full-length or truncated Myo1c, and cell morphology was monitored for 10 min followed by staining for visualization of G- or F-actin. Arrows indicate the microinjection spots. The boxed areas are magnified below each image. The white arrow indicates the direction of cell migration. (B) Quantification of lamellipodial extension/retraction speed (mean ± SEM; n = 5–9 cells). Ab, antibody.
Yeast Myo1 directly stimulates F-actin polymerization (Lee et al., 2000; Lechler et al., 2001; Sirotkin et al., 2005), and mammalian Myo1b promotes assembly of F-actin foci (Almeida et al., 2011), suggesting that Myo1c might increase G-actin at the cell leading edge by facilitating F-actin polymerization. To test this possibility, the effect of purified Myo1c on in vitro actin polymerization was investigated and found to be without effect (Fig. S2 B). Furthermore, inhibition of actin turnover with jasplakinolide did not influence G-actin localization at the cell leading edge following overexpression of Myo1c, suggesting an F-actin turnover–independent mechanism for Myo1c-induced G-actin localization (Fig. S3 A).
Myo1c contributes to G-actin delivery to leading edge and optimal cell migration
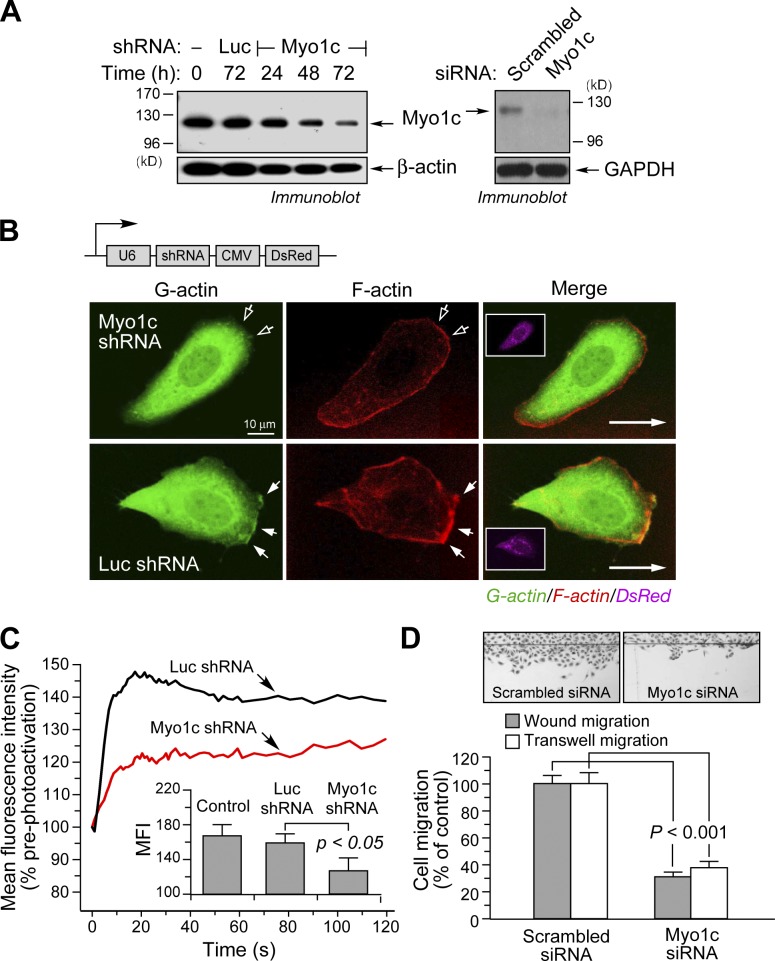
The role of Myo1c in G-actin transport during cell migration was investigated by knockdown experiments. Transfected, U6 promoter–driven short hairpin RNA (shRNA) and siRNA targeting identical Myo1c sequences reduced Myo1c expression by ∼70 and 90%, respectively, after 72 h (Fig. 5 A). Myo1c knockdown markedly reduced G-actin localization at the leading edge compared with cells transfected with control shRNA–targeting luciferase (Luc) mRNA (Fig. 5 B). Lamellipodial F-actin was also reduced. To investigate the role of Myo1c in G-actin translocation, cells were cotransfected with plasmids expressing Myo1c shRNA and the photoactivatable, nonpolymerizable G-actin chimera paGFP-actinG13R. Myo1c knockdown markedly reduced the rate of forward G-actin transport (Fig. 5 C). Finally, we examined the role of Myo1c in EC movement. Transfection of ECs with siRNA-targeting Myo1c reduced wound-induced planar migration and VEGF-A–induced chemotaxis by ∼70% and 60%, respectively, compared with scrambled siRNA (Fig. 5 D). Transient overexpression of the Myo1c tail domain reduced planar EC migration by ∼40% (P < 0.01), but overexpression of the head domain was ineffective (unpublished data). Together, these experiments establish the requirement for Myo1c in vectorial G-actin trafficking and EC motility.
Figure 5.
Myo1c is required for G-actin delivery to the leading edge and optimal cell migration. (A) Knockdown of Myo1c by shRNA and siRNA. Cells were transfected with shRNA targeted against Myo1c or Luc or Myo1c or scrambled siRNA, and Myo1c protein was detected by immunoblotting. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. (B) Cells were transfected with a plasmid coexpressing DsRed and shRNA targeted against Luc or Myo1c and stained to visualize G- or F-actin. The insets show coexpressed DsRed. Long arrows indicate the direction of cell migration. Filled arrows indicate regions enriched in G- or F-actin, and open arrows indicate the cell leading edge in regions of reduced G- or F-actin. CMV, cytomegalovirus. (C) Cells were cotransfected with vectors expressing paGFP-actinG13R and shRNA targeted against Luc or Myo1c, and paGFP-actinG13R movement was detected by photoactivation. Dynamics in the forward region near the leading edge fluorescence intensity after photoactivation in the lamellipodia are shown. (inset) Mean fluorescence intensity (MFI) 15 s after photoactivation (mean fluorescence intensity ± SEM; n = 8–12 cells). (D) Cells were transfected with Myo1c or scrambled siRNA and subjected to wound-induced migration and VEGF-A–induced chemotaxis. The number of migrating cells was determined (mean ± SEM; three independent experiments).
Cytoplasmic G-actin concentration is an important determinant of actin polymerization rate and eukaryotic cell migration (Kiuchi et al., 2011). In addition to passive diffusion, local synthesis, and F-actin turnover, active transport of G-actin can contribute to accumulation of G-actin at the cell leading edge, where it elongates F-actin and can influence cell migration speed and direction. A facilitated transport mechanism has been proposed in which myosin II–mediated contraction of the cell rear generates forward cytoplasmic fluid flow to drive actin transport (Peckham et al., 2001; Zicha et al., 2003; Keren et al., 2009). Interestingly, the role of myosin family members in actin transport is suggested by the abrogation of actin flow and cell motility by the myosin inhibitor 2,3-butanedione monoxime and by the myosin light chain kinase inhibitor ML-7 (Peckham et al., 2001; Zicha et al., 2003). However, we find that the myosin II–specific inhibitor blebbistatin does not reduce GFP-actinG13R localization at the EC leading edge (Fig. S3, B and C), consistent with a study showing that blebbistatin does not affect transport of photoactivated GFP-actin in B16-F1 melanoma cells (Koestler et al., 2009). Thus, myosin-mediated actin transport might depend on cell type, migration mode, and myosin type.
Myo1c is single headed and does not form dimers in vitro and thus is unlikely to move processively by itself along F-actin filaments. However, single-headed Myo9b is a processive motor (Inoue et al., 2002; Post et al., 2002), and a cryptic insertion in the motor domain constitutes a second F-actin–interacting site in addition to the classical ATP-sensitive actin-binding domain and is likely responsible for processive movement. It is unclear whether Myo1c has a similar domain or adaptor proteins that facilitate pairwise interaction with F-actin in vivo or whether there is cargo-mediated dimerization, by which processive activity is generated. We suggest two possible molecular mechanisms underlying Myo1c-mediated G-actin transport: processive movement of the Myo1c–G-actin complex and a ballista-in-relay–like mechanism in which Myo1c does not translocate but rather drives G-actin forward by repeated cycle of conformational switch-driven forward passes of G-actin along F-actin, involving binding and release during each cycle.
Previous biophysical studies with recombinant Myo1c suggest that it is an inefficient motor for movement, as indicated by an ATP hydrolysis–induced force of 2 pN acting against thermodynamic fluctuations (kT = ∼4 pN/nm at 37°C) and a velocity of ∼80 nm/s in vitro (Lin et al., 2011), which is much lower than the speed of forward actin flow at the cell front, ∼5 µm/s (Zicha et al., 2003). In contrast, Myo1c efficiently transports nuclear factor κB essential modulator, Neph1, and Glut4-containing vesicles in cells (Bose et al., 2002; Nakamori et al., 2006), suggesting that unknown mechanisms might facilitate Myo1c movement in vivo. Given the G-actin diffusion rate of 2–6 µm2/s in lamellipodia measured in ECs and other cells (McGrath et al., 1998; Zicha et al., 2003; Plastino et al., 2004), active transport by Myo1c and passive diffusion may both contribute to the forward actin flow.
Myo1c transport of G-actin is restricted primarily to the lamellipodia, where it can supplement diffusion in translocating G-actin from the region of treadmilling to the cell front (i.e., from ∼1 µm behind to the leading edge). Myo1c has a work stroke of ∼5 nm (Laakso et al., 2008), and, thus, it might require ∼200 strokes to move G-actin the required distance, consuming ∼200 ATP molecules. A typical mammalian cancer cell polymerizes ∼6 × 105 actin monomers per second during cell migration (Chan et al., 1998). Assuming all actin polymerization is derived from Myo1c delivery, then ∼1 × 108 ATPs per second are consumed. The mean cellular ATP concentration is about 5 mM (range is 2–10 mM), and our confocal microscopy measurement showed that migrating ECs have a mean volume of ∼2.4 × 10−14 m3 (Fig. S3 D); thus, each cell has ∼7 × 1010 ATP molecules. Finally, a typical cell has an ATP turnover rate of 1–2 min, i.e., ∼1% of total ATP is consumed and produced per second (Alberts et al., 2004), or ∼7 × 108 ATP/s. Based on these estimates, translocation of G-actin by Myo1c utilizes ∼15% of total ATP consumption of the cell. Given that ATP-independent diffusion is likely to account for a significant fraction of G-actin translocation to the cell and because ECs are relatively slow-moving cells (0.05–0.1 µM/min vs. ∼0.3 µm/min for breast cancer cells; Yuasa-Kawada et al., 2009), it is likely that ATP utilization by Myo1c-driven G-actin mobilization is considerably less than 15% of total ATP consumption.
An important role for Myo1c in cell movement is suggested by its abundance in actin-rich ruffles and by the induced ruffling activity after overexpression in 3T3-L1 adipocytes (Bose et al., 2002; Nakamori et al., 2006). Our analysis indicates Myo1c concentrations up to 10 µM in patches near the leading edge in motile ECs (Fig. S3 D). Reports of free G-actin in lamellipodia have been highly variable, ranging from 0.2 to 150 µM, depending on method, cell type, and G-actin status, i.e., interacting partners (Huber et al., 2008; Koestler et al., 2009; Kiuchi et al., 2011). At the low end of the range, there is sufficient Myo1c to transport a substantial fraction of the G-actin. At the high end, Myo1c is not sufficient to transport all G-actin at a 1:1 ratio; however, a cyclic throwing mechanism would allow each Myo1c molecule to facilitate forward movement of multiple G-actin molecules. In addition to delivery of G-actin to the cell leading edge by binding and transport, Myo1c may regulate actin-based cell motility through other mechanisms. Myo1c transports multiple signaling molecules to the plasma membrane, including rapamycin-insensitive companion of mammalian target of rapamycin (Hagan et al., 2008), nuclear factor κB essential modulator (Nakamori et al., 2006), Neph1 (Arif et al., 2011), and phosphatidylinositol 4,5-bisphosphate (Hokanson et al., 2006), which might indirectly contribute to actin dynamics. Alternatively, Myo1c-driven vesicle transport to the plasma membrane could influence actin polymerization. For example, interaction of Myo1c with RalA, a small GTPase residing in Glut4-containing vesicles, induces translocation of the glucose transporter to the cell surface, a process that may alter local bioenergetic processes (Bose et al., 2002; Chen et al., 2007; Yip et al., 2008). In summary, our results reveal an important role of Myo1c in G-actin transport to the leading edge of moving ECs and might present a critical node for control of cell polarity and motility.
Materials and methods
Cells and reagents
ECs were isolated from adult bovine aortas and cultured in DME/Ham’s F-12 medium (Invitrogen) containing 5% FBS. Cells were induced to migrate by a razor wound method, and the number of migrating cells was determined with ImageJ software (National Institutes of Health; Ghosh et al., 2002). Rabbit polyclonal anti–bovine Myo1c antibody was raised against a synthetic peptide (REASELLRELCRKNMVWKY) and purified by peptide affinity chromatography (Thermo Fisher Scientific).
Plasmid construction and siRNA
Full-length Myo1c cDNA (NCBI Protein database accession no. NP_776821) was amplified from a bovine cDNA pool (BioChain) by PCR. Full-length and truncated fragments were subcloned into pcDNA 3.1-myc/His (Invitrogen) or pDsRed-N1 (Takara Bio Inc.). pGFP-actin (Takara Bio Inc.) was used as a template to generate G13R and R62D mutations by PCR using GeneTailor Mutagenesis System (Invitrogen). Actin cDNA was subcloned into pEGFP-C1 (Takara Bio Inc.), pPA-GFP-C1 (provided by J. Lippincott-Schwartz, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD), or pDream with a Flag tag (GenScript). siRNA duplexes (targeting sequences of bovine Myo1c, nucleotides 1,071–1,095 or 1,213–1,237, or scrambled sequence) were synthesized (Invitrogen). Duplex oligonucleotides encoding Myo1c shRNA targeting the same Myo1c sequence or control Luc shRNA (Invitrogen) were annealed and cloned into pRNAT-U6-GFP/Neo (GenScript). Plasmids and siRNA were transfected into cells with Lipofectin and Lipofectimine 2000 (Invitrogen), respectively.
Immunoprecipitation and MS
Lysates from migrating cells were subjected to immunoprecipitation using anti-myc antibody–immobilized agarose beads, and the precipitated proteins were detected by SDS-PAGE and immunoblot analysis. To identify actinG13R-myc–interacting proteins, gels were stained with Coomassie blue, and bound proteins were identified by HPLC-MS. In brief, proteins were alkylated with iodoacetamide before digestion with trypsin, and the peptides were analyzed by collisionally induced dissociation spectra in an LTQ ion trap MS system.
Protein expression and purification
Full-length or truncated Myo1c cDNA was subcloned into pET41-GST, and plasmids were transformed into Rosetta-gami 2 bacteria (EMD Millipore). Protein expression was induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside at 30°C for 5 h with chloramphenicol, streptomycin, tetracycline, and kanamycin. Soluble protein was extracted with CelLytic B lysis reagent (Sigma-Aldrich) and purified with B-PER GST purification kit (Thermo Fisher Scientific). For protein expression in insect cells, Myo1c was in vitro synthesized from pcDNA-Myo1c-myc/His vector with insect EasyXpress kit (QIAGEN) and purified with MagneHis system (Promega).
Protein–protein interaction
Purified β-actin (Cytoskeleton) was centrifuged through a 50-kD filter (EMD Millipore) to remove any polymerized actin and immobilized on a CM5 sensor chip. Binding of purified bacterial Myo1c to G-actin was determined by SPR (BiaCore 3000). Dissociation constants were calculated for a range of analyte concentrations using BIAevaluation software (BiaCore).
Confocal imaging
Migrating cells were fixed with 3.7% PFA and imaged with a 63× objective lens on an upright microscope (DMRXE) equipped with a confocal scanning system (TCS SP2; Leica). Live cells were imaged in phenol red–free DME/Ham’s F-12 medium on an inverted microscope (DMIRBE) with a confocal system (TCS SP2) in a heated chamber at 37°C. Photoactivation of paGFP-actinG13R fluorescence in lamellipodia was performed using a single full-power pulse of a 405-nm laser followed by time series imaging, and relative fluorescence intensity was corrected by background subtraction. For FRAP experiments, cells were transfected with pEGFP-actinG13R and induced to migrate. Live cells were photobleached using a 488-nm argon laser at maximal power. Recovery was monitored by repetitive scanning of bleached areas. Fluorescence recovery curves were fit by nonlinear regression with SigmaPlot software (Systat Software) and expressed as apparent lateral diffusion coefficient D (Vasanji et al., 2004). For FRET experiments, cells were cotransfected with pEGFP-actinG13R and pDsRed-Myo1c. Confocal images were acquired in three channels corresponding to GFP, DsRed, and FRET. Corrections to remove spectral bleed-through, variations in fluorophore expression, and background subtraction were performed with PFRET software (CircuSoft; Fan et al., 2009).
Cell microinjection
Purified insect full-length or truncated Myo1c protein, Myo1c antibody, or control rabbit IgG (5 µg/ml) was microinjected into lamellipodia of migrating cells seeded on glass coverslips. Morphological changes were monitored in a microscope (Leica) equipped with charge-coupled device camera (Sony). 10 min after injection, the cells were fixed and stained with Alexa Fluor 488–DNase I and Alexa Fluor 568–phalloidin, and G- and F-actin were visualized on an upright confocal microscope (TCS SP2).
In vitro actin polymerization
GST-tagged Myo1c was expressed in bacteria, purified, and dialyzed in G-buffer (5 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP, and 0.5 mM DTT) at 4°C for 2 h. 10 µM β-actin (10% labeled with pyrene; Cytoskeleton) was induced to polymerize by addition of 2 mM MgCl2 in G-buffer with or without 1 µM GST-Myo1c. Fluorescence intensity was measured with an excitation wavelength at 365 nm and an emission wavelength at 407 nm.
Online supplemental material
Fig. S1 shows GFP-actinG13R accumulation at the cell leading edge with volume controls of GFP and RFP and rapid movement of GFP-actinG13R at the cell leading edge, as determined by FRAP. Fig. S2 shows the spatially restricted Myo1c interaction with G-actin in lamellipodia, as visualized by FRET, and shows that purified Myo1c does not affect in vitro actin polymerization. Fig. S3 shows that inhibition of actin turnover by jasplakinolide does not affect G-actin localization at the cell leading edge, that the myosin II inhibitor blebbistatin does not alter localization of G-actin at the cell leading edge, and that blebbistatin dismantles stress fibers and shows the calculation of intracellular Myo1c concentration. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201111088/DC1.
Supplementary Material
Acknowledgments
We are grateful to Tom Egelhoff, Alan Levine, and Sanjay Pimplikar for helpful suggestions, Arie Horowitz for critically reading the manuscript, Michael Kinter for mass spectrometric analysis, and Judith Drazba and Patricia Conrad for image analysis.
This work was supported by National Institutes of Health grants P01 HL29582, P01 HL76491, R01 HL75255, and R21 HL094841 (to P.L. Fox).
Footnotes
Abbreviations used in this paper:
- EC
- endothelial cell
- FRET
- Förster resonance energy transfer
- MS
- mass spectrometry
- paGFP
- photoactivatable GFP
- shRNA
- short hairpin RNA
- SPR
- surface plasmon resonance
- WT
- wild type
References
- Alberts B., Bray D., Lewis J., Raff M., Roberts K., Watson J.. 2004. Molecular Biology of the Cell. Fourth edition. Taylor and Francis, New York. 93 pp. [Google Scholar]
- Almeida C.G., Yamada A., Tenza D., Louvard D., Raposo G., Coudrier E.. 2011. Myosin 1b promotes the formation of post-Golgi carriers by regulating actin assembly and membrane remodelling at the trans-Golgi network. Nat. Cell Biol. 13:779–789. 10.1038/ncb2262 [DOI] [PubMed] [Google Scholar]
- Arif E., Wagner M.C., Johnstone D.B., Wong H.N., George B., Pruthi P.A., Lazzara M.J., Nihalani D.. 2011. Motor protein Myo1c is a podocyte protein that facilitates the transport of slit diaphragm protein Neph1 to the podocyte membrane. Mol. Cell. Biol. 31:2134–2150. 10.1128/MCB.05051-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose A., Guilherme A., Robida S.I., Nicoloro S.M., Zhou Q.L., Jiang Z.Y., Pomerleau D.P., Czech M.P.. 2002. Glucose transporter recycling in response to insulin is facilitated by myosin Myo1c. Nature. 420:821–824. 10.1038/nature01246 [DOI] [PubMed] [Google Scholar]
- Cao L.G., Fishkind D.J., Wang Y.L.. 1993. Localization and dynamics of nonfilamentous actin in cultured cells. J. Cell Biol. 123:173–181. 10.1083/jcb.123.1.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.Y., Raft S., Bailly M., Wyckoff J.B., Segall J.E., Condeelis J.S.. 1998. EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J. Cell Sci. 111:199–211. [DOI] [PubMed] [Google Scholar]
- Chen X.W., Leto D., Chiang S.H., Wang Q., Saltiel A.R.. 2007. Activation of RalA is required for insulin-stimulated Glut4 trafficking to the plasma membrane via the exocyst and the motor protein Myo1c. Dev. Cell. 13:391–404. 10.1016/j.devcel.2007.07.007 [DOI] [PubMed] [Google Scholar]
- Condeelis J., Singer R.H.. 2005. How and why does β-actin mRNA target? Biol. Cell. 97:97–110. 10.1042/BC20040063 [DOI] [PubMed] [Google Scholar]
- Cramer L.P. 1999. Role of actin-filament disassembly in lamellipodium protrusion in motile cells revealed using the drug jasplakinolide. Curr. Biol. 9:1095–1105. 10.1016/S0960-9822(99)80478-3 [DOI] [PubMed] [Google Scholar]
- DasGupta G., White J., Cheung P., Reisler E.. 1990. Interactions between G-actin and myosin subfragment 1: Immunochemical probing of the NH2-terminal segment on actin. Biochemistry. 29:8503–8508. 10.1021/bi00488a043 [DOI] [PubMed] [Google Scholar]
- De La Cruz E.M., Ostap E.M.. 2004. Relating biochemistry and function in the myosin superfamily. Curr. Opin. Cell Biol. 16:61–67. 10.1016/j.ceb.2003.11.011 [DOI] [PubMed] [Google Scholar]
- Fan Y., Gong Y., Ghosh P.K., Graham L.M., Fox P.L.. 2009. Spatial coordination of actin polymerization and ILK-Akt2 activity during endothelial cell migration. Dev. Cell. 16:661–674. 10.1016/j.devcel.2009.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P.K., Vasanji A., Murugesan G., Eppell S.J., Graham L.M., Fox P.L.. 2002. Membrane microviscosity regulates endothelial cell motility. Nat. Cell Biol. 4:894–900. 10.1038/ncb873 [DOI] [PubMed] [Google Scholar]
- Hagan G.N., Lin Y., Magnuson M.A., Avruch J., Czech M.P.. 2008. A Rictor-Myo1c complex participates in dynamic cortical actin events in 3T3-L1 adipocytes. Mol. Cell. Biol. 28:4215–4226. 10.1128/MCB.00867-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokanson D.E., Laakso J.M., Lin T., Sept D., Ostap E.M.. 2006. Myo1c binds phosphoinositides through a putative pleckstrin homology domain. Mol. Biol. Cell. 17:4856–4865. 10.1091/mbc.E06-05-0449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber F., Käs J., Stuhrmann B.. 2008. Growing actin networks form lamellipodium and lamellum by self-assembly. Biophys. J. 95:5508–5523. 10.1529/biophysj.108.134817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A., Saito J., Ikebe R., Ikebe M.. 2002. Myosin IXb is a single-headed minus-end-directed processive motor. Nat. Cell Biol. 4:302–306. 10.1038/ncb774 [DOI] [PubMed] [Google Scholar]
- Keren K., Yam P.T., Kinkhabwala A., Mogilner A., Theriot J.A.. 2009. Intracellular fluid flow in rapidly moving cells. Nat. Cell Biol. 11:1219–1224. 10.1038/ncb1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiuchi T., Nagai T., Ohashi K., Mizuno K.. 2011. Measurements of spatiotemporal changes in G-actin concentration reveal its effect on stimulus-induced actin assembly and lamellipodium extension. J. Cell Biol. 193:365–380. 10.1083/jcb.201101035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler S.A., Rottner K., Lai F., Block J., Vinzenz M., Small J.V.. 2009. F- and G-actin concentrations in lamellipodia of moving cells. PLoS ONE. 4:e4810. 10.1371/journal.pone.0004810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laakso J.M., Lewis J.H., Shuman H., Ostap E.M.. 2008. Myosin I can act as a molecular force sensor. Science. 321:133–136. 10.1126/science.1159419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F.P., Szczodrak M., Block J., Faix J., Breitsprecher D., Mannherz H.G., Stradal T.E., Dunn G.A., Small J.V., Rottner K.. 2008. Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 27:982–992. 10.1038/emboj.2008.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J.B., Singer R.H.. 1986. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 45:407–415. 10.1016/0092-8674(86)90326-0 [DOI] [PubMed] [Google Scholar]
- Lechler T., Jonsdottir G.A., Klee S.K., Pellman D., Li R.. 2001. A two-tiered mechanism by which Cdc42 controls the localization and activation of an Arp2/3-activating motor complex in yeast. J. Cell Biol. 155:261–270. 10.1083/jcb.200104094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.L., Bezanilla M., Pollard T.D.. 2000. Fission yeast myosin-I, Myo1p, stimulates actin assembly by Arp2/3 complex and shares functions with WASp. J. Cell Biol. 151:789–800. 10.1083/jcb.151.4.789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T., Greenberg M.J., Moore J.R., Ostap E.M.. 2011. A hearing loss-associated myo1c mutation (R156W) decreases the myosin duty ratio and force sensitivity. Biochemistry. 50:1831–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J.L., Tardy Y., Dewey C.F., Jr, Meister J.J., Hartwig J.H.. 1998. Simultaneous measurements of actin filament turnover, filament fraction, and monomer diffusion in endothelial cells. Biophys. J. 75:2070–2078. 10.1016/S0006-3495(98)77649-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L., Phillips M., Reisler E.. 1988. Polymerization of G-actin by myosin subfragment 1. J. Biol. Chem. 263:1996–2002. [PubMed] [Google Scholar]
- Mogilner A., Edelstein-Keshet L.. 2002. Regulation of actin dynamics in rapidly moving cells: A quantitative analysis. Biophys. J. 83:1237–1258. 10.1016/S0006-3495(02)73897-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamori Y., Emoto M., Fukuda N., Taguchi A., Okuya S., Tajiri M., Miyagishi M., Taira K., Wada Y., Tanizawa Y.. 2006. Myosin motor Myo1c and its receptor NEMO/IKK-γ promote TNF–α-induced serine307 phosphorylation of IRS-1. J. Cell Biol. 173:665–671. 10.1083/jcb.200601065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noireaux V., Golsteyn R.M., Friederich E., Prost J., Antony C., Louvard D., Sykes C.. 2000. Growing an actin gel on spherical surfaces. Biophys. J. 78:1643–1654. 10.1016/S0006-3495(00)76716-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell C.B., Tyska M.J., Mooseker M.S.. 2007. Myosin at work: Motor adaptations for a variety of cellular functions. Biochim. Biophys. Acta. 1773:615–630. 10.1016/j.bbamcr.2006.06.012 [DOI] [PubMed] [Google Scholar]
- Patterson G.H., Lippincott-Schwartz J.. 2002. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 297:1873–1877. 10.1126/science.1074952 [DOI] [PubMed] [Google Scholar]
- Peckham M., Miller G., Wells C., Zicha D., Dunn G.A.. 2001. Specific changes to the mechanism of cell locomotion induced by overexpression of β-actin. J. Cell Sci. 114:1367–1377. [DOI] [PubMed] [Google Scholar]
- Plastino J., Lelidis I., Prost J., Sykes C.. 2004. The effect of diffusion, depolymerization and nucleation promoting factors on actin gel growth. Eur. Biophys. J. 33:310–320. 10.1007/s00249-003-0370-3 [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Borisy G.G.. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Ponti A., Machacek M., Gupton S.L., Waterman-Storer C.M., Danuser G.. 2004. Two distinct actin networks drive the protrusion of migrating cells. Science. 305:1782–1786. 10.1126/science.1100533 [DOI] [PubMed] [Google Scholar]
- Posern G., Sotiropoulos A., Treisman R.. 2002. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol. Biol. Cell. 13:4167–4178. 10.1091/mbc.02-05-0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post P.L., Tyska M.J., O’Connell C.B., Johung K., Hayward A., Mooseker M.S.. 2002. Myosin-IXb is a single-headed and processive motor. J. Biol. Chem. 277:11679–11683. 10.1074/jbc.M111173200 [DOI] [PubMed] [Google Scholar]
- Reizes O., Barylko B., Li C., Südhof T.C., Albanesi J.P.. 1994. Domain structure of a mammalian myosin I β. Proc. Natl. Acad. Sci. USA. 91:6349–6353. 10.1073/pnas.91.14.6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova E.A., Singer R.H., Condeelis J.. 2001. The physiological significance of β-actin mRNA localization in determining cell polarity and directional motility. Proc. Natl. Acad. Sci. USA. 98:7045–7050. 10.1073/pnas.121146098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirotkin V., Beltzner C.C., Marchand J.B., Pollard T.D.. 2005. Interactions of WASp, myosin-I, and verprolin with Arp2/3 complex during actin patch assembly in fission yeast. J. Cell Biol. 170:637–648. 10.1083/jcb.200502053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N., Ostap E.M.. 2001. Motor domain-dependent localization of myo1b (myr-1). Curr. Biol. 11:1131–1135. 10.1016/S0960-9822(01)00320-7 [DOI] [PubMed] [Google Scholar]
- Vasanji A., Ghosh P.K., Graham L.M., Eppell S.J., Fox P.L.. 2004. Polarization of plasma membrane microviscosity during endothelial cell migration. Dev. Cell. 6:29–41. 10.1016/S1534-5807(03)00397-6 [DOI] [PubMed] [Google Scholar]
- Watanabe N., Mitchison T.J.. 2002. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 295:1083–1086. 10.1126/science.1067470 [DOI] [PubMed] [Google Scholar]
- Yip M.F., Ramm G., Larance M., Hoehn K.L., Wagner M.C., Guilhaus M., James D.E.. 2008. CaMKII-mediated phosphorylation of the myosin motor Myo1c is required for insulin-stimulated GLUT4 translocation in adipocytes. Cell Metab. 8:384–398. 10.1016/j.cmet.2008.09.011 [DOI] [PubMed] [Google Scholar]
- Yuasa-Kawada J., Kinoshita-Kawada M., Rao Y., Wu J.Y.. 2009. Deubiquitinating enzyme USP33/VDU1 is required for Slit signaling in inhibiting breast cancer cell migration. Proc. Natl. Acad. Sci. USA. 106:14530–14535. 10.1073/pnas.0801262106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zicha D., Dobbie I.M., Holt M.R., Monypenny J., Soong D.Y., Gray C., Dunn G.A.. 2003. Rapid actin transport during cell protrusion. Science. 300:142–145. 10.1126/science.1082026 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.