Drosophila Evi5 is a Rab-GAP that acts on the membrane-trafficking mediator Rab11 to promote guidance receptor polarization during border cell migration.
Abstract
Membrane trafficking has well-defined roles during cell migration. However, its regulation is poorly characterized. In this paper, we describe the first screen for putative Rab–GTPase-activating proteins (GAPs) during collective cell migration of Drosophila melanogaster border cells (BCs), identify the uncharacterized Drosophila protein Evi5 as an essential membrane trafficking regulator, and describe the molecular mechanism by which Evi5 regulates BC migration. Evi5 requires its Rab-GAP activity to fulfill its functions during migration and acts as a GAP protein for Rab11. Both loss and gain of Evi5 function blocked BC migration by disrupting the Rab11-dependent polarization of active guidance receptors. Altogether, our findings deepen our understanding of the molecular machinery regulating endocytosis and subsequently cell signaling during migration.
Introduction
Small GTPases from the Rab family are important regulators of vesicular trafficking (Zerial and McBride, 2001). They control many cellular and developmental processes, such as proliferation, differentiation, and cell migration. In particular, they play a fundamental role in regulating cell signaling because they control the compartmentalization of signaling molecules (Scita and Di Fiore, 2010).
Like every GTPase, Rab proteins cycle between a GTP-bound active state and a GDP-bound inactive state. In the active state, they recruit specific effectors necessary for their function. Guanine nucleotide exchange factor (GEF) proteins are responsible for the exchange of GDP to GTP by catalyzing GDP release, whereas GTPase-activating proteins (GAPs) catalyze GTP hydrolysis. Diverse Rab-GEF domains have been identified (Barr and Lambright, 2010). In contrast, the Tre-2, Bub2, and Cdc16 (TBC) domain is the only Rab-GAP domain identified (Fukuda, 2011). The TBC domain supplies a catalytic arginine finger characterized by the consensus sequence IXXDXXR responsible for GAP activity (Pan et al., 2006).
Many questions about the regulators of Rab proteins are still unanswered. In particular, very few GAPs and GEFs have been attributed to specific Rab proteins. Furthermore, most of the work performed so far was performed in cultured cells or in vitro. Although this approach is efficient to demonstrate a GAP/GEF activity and to measure catalytic activity, the specificity of the interaction between a Rab and its regulator might be lost in vitro, as well as the compartmentalization of the interaction and possible posttranslational regulations. Hence, it is fundamental to study in vivo the regulation of Rab proteins (Frasa et al., 2012). Recent work in Drosophila melanogaster and in Caenorhabditis elegans has provided the first descriptions of in vivo regulation of Rab proteins by GAPs (Chotard et al., 2010; Houalla et al., 2010; Uytterhoeven et al., 2011). But no systematic analysis of all the GAPs has been performed so far.
In this study, we examined the implication of Rab-GAP proteins during border cell (BC) migration in the Drosophila ovary. Because membrane trafficking has been shown to be an essential regulator of BC migration (Fig. 1 A, schematic representation; Assaker et al., 2010; Janssens et al., 2010), it is a potent model to study the in vivo regulation of vesicular trafficking. BCs allow the combination of in vivo cell biology techniques, high resolution imaging, and Drosophila genetics, such as gene knockdown and overexpression. Thus, we screened for GAP proteins necessary for normal BC migration and identified Evi5 as an essential regulator of cell migration.
Figure 1.
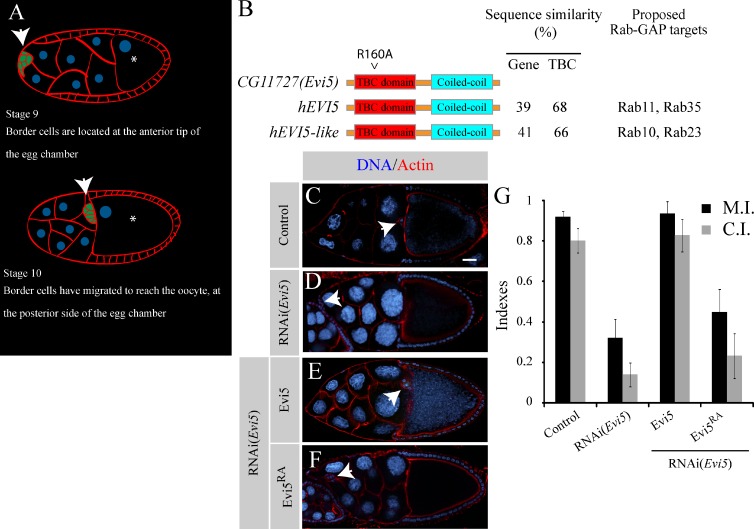
Rescue experiments demonstrate that CG11727 requires its GAP activity in vivo. (A) Schematic representation of Drosophila egg chambers of stage 9 and 10. Red represents the actin staining, blue represents the nucleus, and green represents the BCs. BCs migrate from the anterior tip toward the posterior oocyte, left to the right in all images. Arrows point the BC clusters. Asterisks mark the oocytes. (B) Schematic representation of the Evi5 gene and its human orthologues EVI5 and EVI5-like. The sequence similarity in the TBC domain between Evi5 and its human orthologue and the proposed targets of EVI5 and EVI5-like are depicted. The position of the catalytic arginine residue in Evi5 mutated for an alanine is illustrated. (C–F) Representative examples of BC migration in a stage 10 egg chamber under the indicated conditions using the C306-Gal4 driver. Control (C) or RNAi(Evi5) (D) is represented. Representative examples of BCs expressing RNAi(Evi5) and in which Evi5 was reintroduced by expression of Evi5-mcherry (E) or Evi5RA-mcherry (F). BCs are stained with phalloidin (red) and DAPI (blue). Arrows point to BC clusters. Bar, 20 µm. (G) The percentage of migration is expressed as the migration index (M.I.) and completion index (C.I.) for the indicated conditions above (35 < n < 161). Error bars are standard error of the mean.
Here, we demonstrate that Evi5 acts as a Rab11-GAP because it fulfills various predictions that can be attributed to a GAP: (a) Evi5 physically and genetically interacts with Rab11, (b) overexpressing Evi5 increases the fraction of the GDP-bound Rab11 and mimics the expression of a Rab11 dominant-negative (DN) form both in vivo and in vitro, and (c) knocking down Evi5 increases the fraction of the GTP-bound Rab11 and mimics the expression of a Rab11 constitutive active (CA) form in vivo. We found that Evi5 is the critical Rab11-GAP essential for BC migration by maintaining active receptor tyrosine kinases (RTKs) at the leading edge of the BC cluster.
Results and discussion
Analysis of potential Rab-GAP proteins during BC migration
In Drosophila, there are ≥27 potential Rab-GAP proteins: 26 containing a TBC domain and the orthologue of human RAB3GAP2 (CG7061), a Rab-GAP devoid of a TBC domain (Nagano et al., 1998). To identify Rab-GAP proteins regulating BC migration, we depleted by RNAi each of the 27 putative Rab-GAP proteins specifically in BCs and determined the consequence on BC migration (Table 1). We then calculated the migration index (M.I.) and the completion index (C.I.; see Material and methods for details) to compare migration phenotypes. The strongest hit was CG11727 (Table 1 and Fig. 1, C and D). Also, we observed that RNAi lines against other candidates (RN-tre, CG4552, CG7061, and CG8155) induce BC migration defects (Table 1). Although we did not pursue their characterization, it is interesting to note that mammalian RN-tre acts as a GAP for Rab5 (Lanzetti et al., 2000), which is also involved in BC migration (Assaker et al., 2010).
Table 1.
M.I and C.I. resulting from knockdown of every potential Rab-GAP protein during BC migration
| Drosophila identifier | Common name | dsRNA (VDRC no.) | M.I. | C.I. | n | Human orthologue |
| CG1093 | Pollux | 27335 | N/A | N/A | N/A | TBC1D1 and TBC1D4 |
| 106969 | 0.98 | 0.95 | 20 | |||
| CG1695 | CG1695 | 20340 | 0.88 | 0.69 | 29 | SGSM1-2 |
| 48062 | 0.98 | 0.95 | 22 | |||
| 106947 | 0.875 | 0.74 | 34 | |||
| CG4041 | CG4041 | 34780 | 0.83 | 0.67 | 3 | TBCK |
| 108887 | 0.98 | 0.95 | 55 | |||
| CG4552 | CG4552 | 40538 | 0.51 | 0.30 | 41 | TBC1D23 |
| 110700 | 0.98 | 0.96 | 47 | |||
| CG5337 | CG5337 | 22070 | 0.99 | 0.94 | 17 | TBC1D16 |
| 22069 | 0.96 | 0.90 | 20 | |||
| 110067 | 1.00 | 1.00 | 20 | |||
| CG5344 | Whacked | 22081 | 0.96 | 0.92 | 24 | TBC1D10A-B-C |
| 22082 | 0.88 | 0.75 | 44 | |||
| CG5745 | CG5745 | 35034 | 0.97 | 0.93 | 45 | TBC1D22A-B |
| 35036 | 0.94 | 0.84 | 32 | |||
| 108659 | 1.00 | 1.00 | 21 | |||
| CG5916 | CG5916 | 110561 | 0.95 | 0.90 | 20 | TBC1D6 |
| CG5978 | CG5978 | 21000 | 0.98 | 0.94 | 72 | TBC1D13 |
| 21001 | 0.97 | 0.92 | 38 | |||
| 110396 | 0.97 | 0.93 | 28 | |||
| CG6182 | CG6182 | 14705 | 0.88 | 0.78 | 32 | TBC1D7 |
| 14706 | 0.97 | 0.93 | 15 | |||
| 106667 | 0.98 | 0.94 | 34 | |||
| CG7061a | CG7061 | 27823 | 0.97 | 0.94 | 17 | RAB3GAP2 |
| 27824 | 0.88 | 0.79 | 39 | |||
| 106905 | 0.54 | 0.41 | 39 | |||
| CG7112 | GapcenAb | 35174 | 0.92 | 0.82 | 91 | RAB-GAP1(GAPCENA) and TBC1D18 |
| 103588 | 0.97 | 0.93 | 61 | |||
| CG7324 | CG7324 | 32929d | 0.96 | 0.89 | 232 | TBC1D8B-9B |
| CG7742 | CG7742 | 25535 | 1.00 | 1.00 | 16 | TBC1D19 |
| 25536 | 0.99 | 0.97 | 32 | |||
| 100125 | 0.99 | 0.96 | 28 | |||
| CG8085 | RN-tre | 28192ce | 0.56 | 0.36 | 96 | RNTRE (USP6NL), TBC1D28, USP6, TBC1D3F-G-H |
| 28194c | 0.65 | 0.42 | 73 | |||
| 108670 | 0.87 | 0.75 | 60 | |||
| CG8155 | CG8155 | 24218 | 0.85 | 0.75 | 28 | TBC1D25 |
| 24221 | 0.58 | 0.42 | 31 | |||
| 108444 | 0.99 | 0.95 | 22 | |||
| CG8449 | CG8449 | 24102 | 1.00 | 1.00 | 17 | TBC1D5 |
| CG9339 | CG9339 | 44655 | 0.99 | 0.98 | 42 | TBC1D24 |
| 108736 | 0.99 | 0.96 | 23 | |||
| CG11490 | Tbc1d15-17b | 20040 | 0.95 | 0.90 | 66 | TBC1D15-17 |
| 1096668 | 0.94 | 0.84 | 51 | |||
| CG11727 | Evi5b | 17548 | 0.76 | 0.61 | 85 | EVI5 and EVI5L |
| 17549e | 0.32 | 0.14 | 161 | |||
| 105146 | 0.88 | 0.73 | 49 | |||
| CG12241 | CG12241 | 33729d | 0.95 | 0.90 | 201 | SGSM3 |
| CG16896 | CG16896 | 20315 | 0.82 | 0.76 | 17 | WDR67 |
| 20316 | 0.97 | 0.88 | 18 | |||
| 107134 | 0.91 | 0.81 | 43 | |||
| CG17883 | CG17883 | 30277 | 0.94 | 0.83 | 30 | TBC1D20 |
| CG32506 | CG32506 | 28776d | 0.96 | 0.89 | 232 | SGSM1-2 |
| CG32580 | CG32580 | 105591 | 0.99 | 0.96 | 24 | MUC16 |
| CG33715 | Msp-300 | 25906 | 0.97 | 0.92 | 83 | SYNE1-2 and CLMN |
| 40143 | 1.00 | 1.00 | 12 | |||
| 40145 | 0.98 | 0.95 | 42 | |||
| 50192 | 0.71 | 0.58 | 19 | |||
| 107183 | 0.99 | 0.98 | 56 | |||
| 109023 | 0.97 | 0.93 | 29 | |||
| CG42795 | CG42795 | 17314 | 0.95 | 0.90 | 73 | TBC1D30 |
| 108779 | 0.98 | 0.93 | 29 |
N/A, not applicable: this dsRNA line affects drastically the morphology of the egg chamber. VDRC, Vienna Drosophila RNAi Center.
This Rab-GAP protein does not have a TBC domain.
These common names have been introduced in this study.
Crosses were performed at 25°C to minimize morphological phenotypes.
Lines obtained from TRiP at Harvard Medical School.
This RNAi fly line is used subsequently is this study.
CG11727 encodes a protein with a predicted size of 89–93 kD containing a TBC and a coiled-coil domain. BLAST (Basic Local Alignment Search Tool) analysis reveals two well-conserved CG11727 orthologues in humans: EVI5 and EVI5-like (Fig. 1 B). Therefore, we refer to CG11727 as Evi5. In humans, the functions of EVI5 and EVI5-like are controversial because they were shown to target in vitro either Rab11 (Dabbeekeh et al., 2007) or Rab35 (Fuchs et al., 2007) and either Rab10 (Itoh et al., 2006) or Rab23 (Yoshimura et al., 2007), respectively (Fig. 1 B).
To demonstrate that the observed phenotypes induced by Evi5 knockdown are not caused by artifacts such as off-target effects, we performed rescue experiments. We observed that Evi5-depleted BCs expressing an Evi5-mcherry transgene migrated in a similar fashion as the control (Fig. 1, E and G), indicating that the RNAi(Evi5) targets endogenous Evi5 and that the Evi5-tagged transgene used is functional. However, we could not rescue the Evi5 knockdown phenotype by expressing a catalytically dead form of Evi5 (Evi5RA; see Material and methods; Fig. 1, F and G), indicating that the Rab-GAP activity of Evi5 is required for its function during BC migration. We next aimed at identifying the molecular target of Evi5 during BC migration.
Evi5 is recruited to the recycling endosome by the active form of Rab11
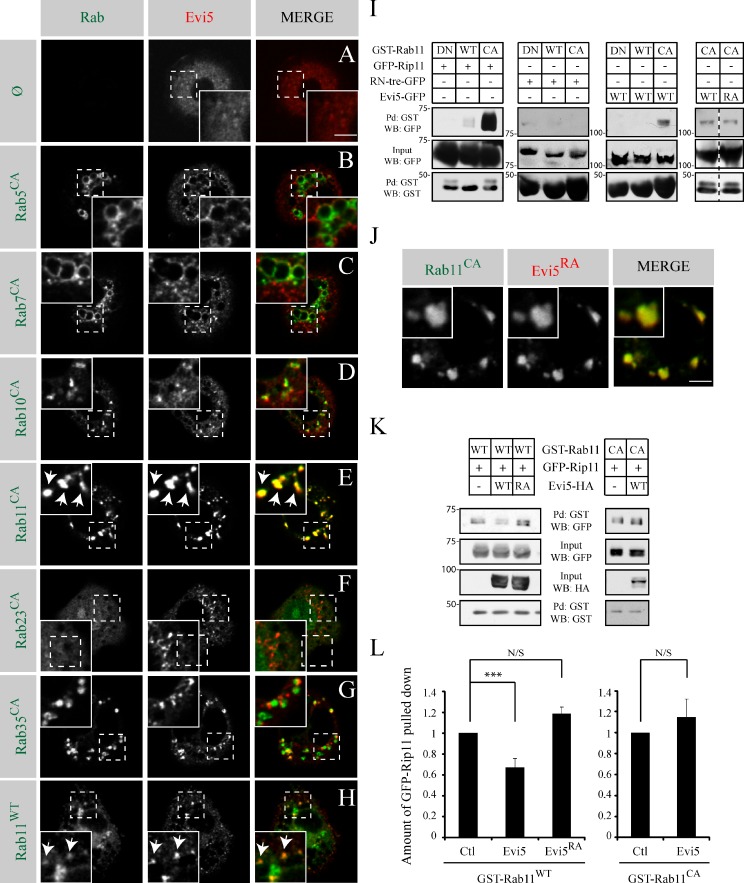
To narrow down the potential substrates of Evi5, we screened for Rab proteins, which colocalize with Evi5. We coexpressed Evi5-mCherry with candidate GFP- or YFP-tagged Rabs in Drosophila S2 cells. The distribution of Evi5-mCherry is cytoplasmic and vesicular (Fig. 2 A). We tested whether Evi5 colocalizes in these vesicular structures with endocytic Rab proteins or with Rabs previously proposed to be targets of its orthologues. We expressed Rab5, 7, 10, 11, 23, and 35 in their GTP-locked forms (CA), which should increase their interaction with their GAP. The expression of Rab11CA drastically changed Evi5 distribution, as Evi5 was recruited to Rab11CA-positive vesicles and disappeared from the cytoplasm (Fig. 2 E). Evi5 was not recruited to Rab5CA or Rab7CA-enlarged endosomes (Fig. 2, B and C). Rab10CA formed vesicles that are often closely juxtaposed to Evi5 punctae, without overlapping (Fig. 2 D). In accordance, we saw a partial colocalization between Rab11 and Rab10 in S2 cells (unpublished data). Rab23CA was mainly cytoplasmic and did not colocalize with Evi5 (Fig. 2 F). Finally, Rab35CA formed vesicles that rarely localized with Evi5 (Fig. 2 G).
Figure 2.
Evi5 acts as a Rab11-GAP in vitro. (A–H) Representative images of S2 cells transfected with Evi5-mcherry and the indicated GFP- or YFP- tagged form of Rab. Rab proteins are either expressed in their wild-type (WT) or CA form. A grayscale image of both the green and red channel is shown for every image. The insets show higher magnifications of the regions marked by dashed line squares. Arrows point to structures where Evi5 and Rab11 colocalize. (I) S2 cells were cotransfected with the wild-type, DN, or CA forms of GST-Rab11 together with the indicated GFP constructs. Pull-downs (Pd) using glutathione beads were performed on lysates. Proteins bound to GST-Rab11 were detected by Western blotting (WB). (J) YFP-Rab11CA was coexpressed with Evi5RA-mcherry in S2 cells. Insets show higher magnification. (K) Effector pull-down assays performed with lysates from S2 cells cotransfected with GST-Rab11WT (left) or GST-Rab11CA (right) together with GFP-Rip11 with or without HA-Evi5. Pull-down of GST-Rab11 and protein analysis were performed as in I. (L) Quantification of the total pulled down GFP-Rip11 normalized to the control on three (GST-Rab11CA) or four (GST-Rab11WT) independent experiments. (***, P < 0.05; t test). Ctl, control. Error bars are standard error of the mean. Bars, 5 µm.
Because Evi5 colocalizes preferentially with Rab11CA, we tested whether Evi5 colocalizes with Rab11WT and found them on the same vesicles (Fig. 2 H). Altogether, our data show that Evi5 colocalizes with Rab11 and is recruited to vesicles by active Rab11.
Evi5 interacts with and inactivates Rab11
We tested whether Evi5 binds to GTP-bound Rab11 by coexpressing GST-Rab11 with other GFP-tagged proteins in S2 cells. GST pulled down experiments were performed on lysates followed by Western blotting. We expressed wild-type, GDP-locked (DN), or CA mutant forms of Rab11. As a positive control, we expressed a GFP fusion of the Rab11 effector Rip11 known to interact preferentially with Rab11-GTP (Li et al., 2007). As expected, GFP-Rip11 can be pulled down efficiently with Rab11CA and to a lesser extent with Rab11WT (Fig. 2 I, left). As a negative control, we expressed RN-tre together with GST-Rab11 because RN-tre should not bind Rab11 (Haas et al., 2005). Indeed, RN-tre was not pulled down by any form of Rab11 (Fig. 2 I, middle left). In contrast, Evi5 was pulled down with Rab11CA (Fig. 2 I, middle right). The catalytic dead mutant form of Evi5, Evi5RA, colocalizes (Fig. 2 J) and interacts with Rab11CA to the same extent as Evi5WT (Fig. 2 I, right). The interaction between Rab11 and Evi5 is thus independent of the catalytic arginine finger.
Next, to test whether Evi5 provides GAP activity to Rab11, we developed an effector pull-down assay based on the preferential binding of Rip11 to active Rab11 (Fig. 2 I). Here, GST-Rab11 is coexpressed in S2 cells together with GFP-Rip11, in the presence or in the absence of HA-Evi5. The activity of Rab11 is determined by measuring the amount of Rip11 that can be coprecipitated with GST-Rab11 from S2 cell lysates. If Evi5 is a Rab11-GAP, overexpressing Evi5WT should decrease the amount of Rip11 coprecipitated with Rab11, as the Rab11-GTP pool will decrease. Accordingly, the amount of GFP-Rip11 bound to Rab11 dropped when Evi5WT was expressed compared with the situation in which Evi5 was not expressed (Fig. 2, K [left] and L). As expected, expressing Evi5RA did not inactivate Rab11 (Fig. 2, K [left] and L), as the amount of Rip11 coprecipitated was similar to the control. Moreover, expressing Evi5WT did not affect the general binding profile of Rip11 bound to Rab11CA (Fig. 2, K [right] and L). This last experiment demonstrates that the decrease of Rip11 coprecipitated with Rab11WT in the presence of Evi5 is not caused by a steric effect. Overall, the data presented thus far demonstrate that Evi5 is a GAP for Rab11.
Evi5 genetically interacts with Rab11 during BC migration
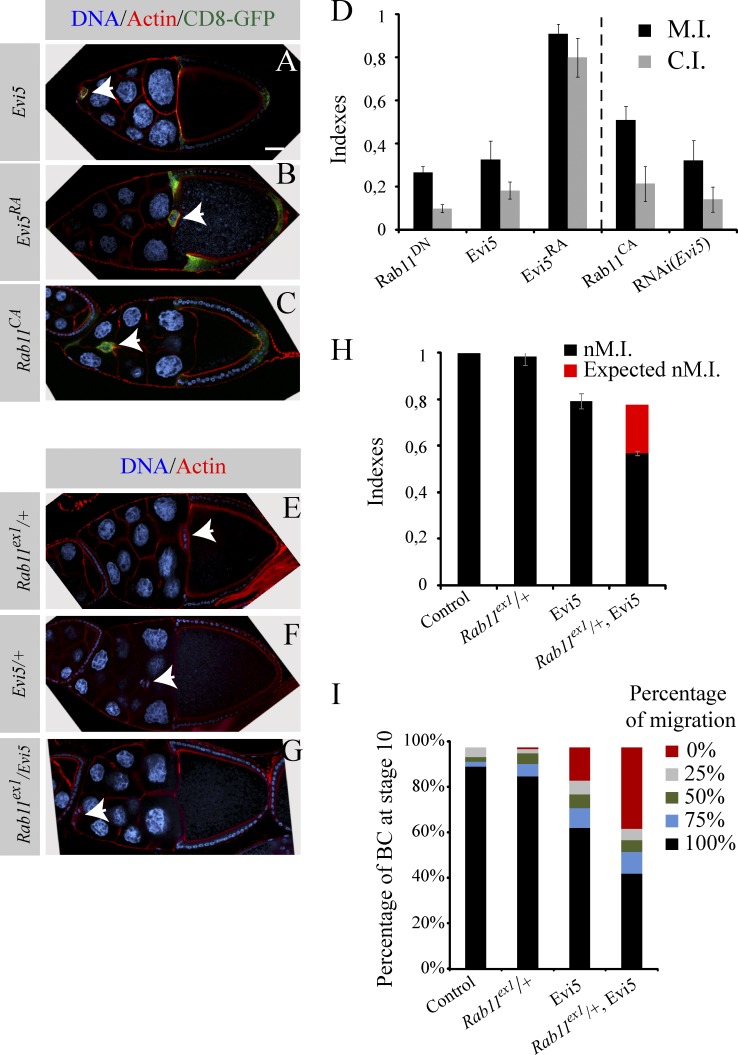
Overexpression of a specific Rab-GAP protein is expected to inhibit a particular membrane trafficking step because it increases the rate of inactivation of its Rab target, thus mimicking the expression of a DN form of the Rab (Fukuda, 2011). As the expression of Rab11DN leads to a strong migration block (Fig. 3 D; Assaker et al., 2010), we asked whether overexpressing Evi5 phenocopies Rab11DN. We observed a strong migration block in BCs expressing Evi5 (Fig. 3, A and D). Expression of Evi5RA did not affect BC migration, confirming that Evi5 requires its Rab-GAP activity to fulfill its function (Fig. 3, B and D). Then, we performed the converse experiment and showed that knocking down Evi5 mimics the phenotype induced by expressing Rab11CA (Fig. 3, C and D).
Figure 3.
Evi5 genetically interacts with Rab11. (A–C) Representative examples of egg chambers expressing Evi5-mcherry (A), Evi5RA-mcherry (B), and Rab11CA using the slbo-Gal4 driver (the arrowheads indicate BCs). (D) M.I. and C.I. for the indicated conditions (51 < n < 161). The dotted line serves as a visual separator between conditions in which Rab11 should be preferential in an inactive (left) and active (right) conformation. (E–G) Representative images of c306-Gal4, Rab11ex1/+ (E), c306-Gal4, Evi5-mcherry/+ (F), and c306-Gal4, Rab11ex1/ +, Evi5-mcherry (G) egg chambers. (H) M.I. for the indicated conditions. The red bar represents the difference between the expected indexes if the phenotype were only additive (46 < n < 188). (I) Percentage of BCs presented in H having completed 0, 25, 50, 75, and 100% (standard errors of the mean ≤ 4.3). Error bars are standard error of the mean.
To further explore the relationship between Evi5 and Rab11, we tested for a potential genetic interaction between the two genes by measuring normalized M.I. (nM.I.) as in Assaker et al. (2010). Flies heterozygous for the null allele Rab11ex1 (Dollar et al., 2002) show no significant BC migration defect (nM.I. of 0.98; Fig. 3, E and H). Although expression of Evi5 using c306-Gal4 slightly affected migration in a control background (nM.I. of 0.79; Fig. 3, F and H), the Rab11ex1 heterozygous background enhanced the phenotype (nM.I. of 0.57; Fig. 3, G and H). Also, the number of clusters unable to initiate migration is strongly increased when Evi5 is expressed and a copy of Rab11 is removed (Fig. 3 I). Collectively, these results show a synergistic interaction between Rab11 and Evi5, suggesting that Evi5 collaborates with Rab11 during BC migration.
Evi5 acts as a Rab11-GAP during BC migration
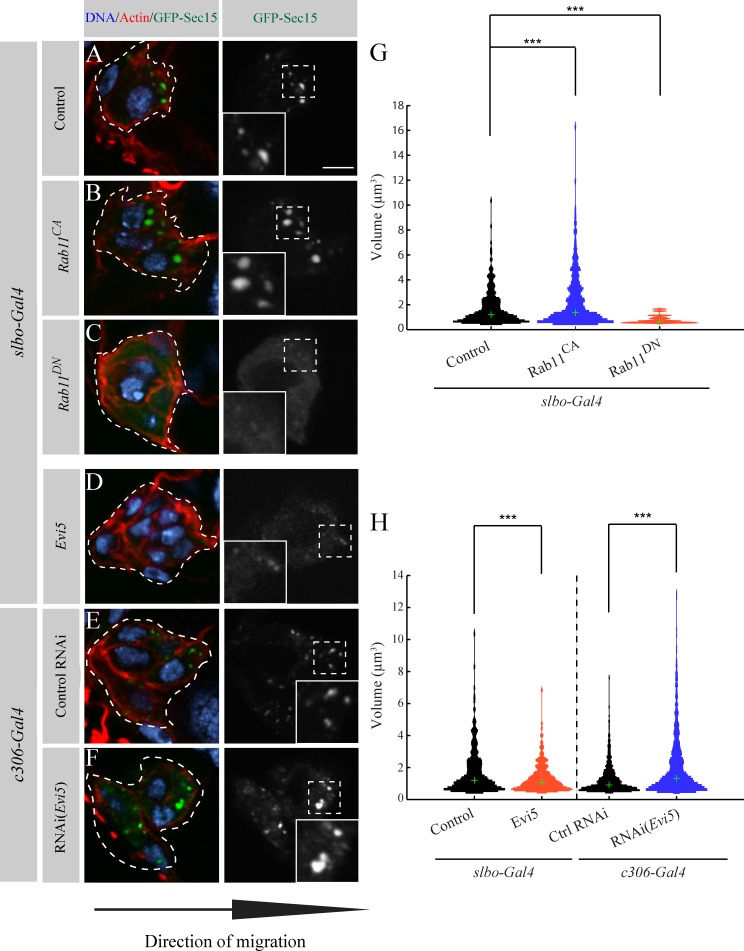
We next wanted to demonstrate that Evi5 acts as a Rab11-GAP during BC migration. Sec15 is an effector of Rab11 in Drosophila (Wu et al., 2005), which forms vesicular-like structures at the leading edge of BCs (Figs. S1 and 4 A). We previously observed that expression of Rab11DN affects these structures (Assaker et al., 2010). We rationalize that quantifying the Sec15 structure volume could serve as a readout for Rab11 activity.
Figure 4.
Evi5 regulates Rab11 during BC migration. (A–F) Representative images showing the distribution of GFP-Sec15 at the onset of migration (stage 9) for the indicated conditions. The dotted lines outline BC clusters as determined by the GFP signal. The insets show a higher magnification of the regions marked by dashed line squares. Bar, 5 µm. (G and H) Computational analysis of the conditions (A–F) as in Fig. S1 F (504 < n vesicles < 1,682; ***, P < 0.05; KS test and rank sum test; Fig. S1 G). Green crosses indicate the medians of the thresholded distribution.
To determine the volume of GFP-Sec15 structures, we used high resolution imaging combined with computational image analysis. To this end, we developed an algorithm that segmented GFP-Sec15 structures from three-dimensionally reconstituted BC cluster images and quantified their volume. To confirm that our method could be used to monitor Rab11 activity, we expressed DN forms of various endocytic Rabs and calculated the volume of GFP-Sec15 structures. Expression of Rab4DN and Rab5DN did not affect significantly the volume distribution of Sec15 structures, whereas Rab7DN slightly reduces their volume but to a lesser extent than Rab11DN (Fig. S1). Rab11DN reduced GFP-Sec15 structures to a volume <2 µm3 and causes a cytoplasmic distribution of GFP-Sec15 (Fig. 4, C and G). To demonstrate that we can also detect gain of Rab11 activity, we expressed Rab11CA and found that it increases the size of Sec15 structures (Figs. 4, B and G; and S1 G). These differences were significant both when comparing the volume distribution (Kolmogorov–Smirnov [KS] test) or the median volume (rank sum test). Thus, our method is highly sensitive to changes of Rab11 activity.
We next modulated Evi5 expression and tested its effects on endogenous Rab11 activity in BCs. We expected that overexpressing Evi5 would mimic a Rab11 loss-of-function phenotype and that knocking down Evi5 would mimic a Rab11 gain of function. Accordingly, we observed that overexpressing Evi5 increases the cytoplasmic distribution of GFP-Sec15, whereas depleting Evi5 increases the size of GFP-Sec15 structures (Fig. 4, D–F). Measured Sec15 structure volumes changed accordingly after modulating Evi5 expression (Figs. 4 H and S1 G).
To confirm the data obtained with Sec15, we performed a similar analysis with Rip11, a specific Rab11 effector (Horgan and McCaffrey, 2009). We expressed the Rab11-binding domain of Rip11 tagged to GFP (Li et al., 2007) in BC clusters and performed the same analysis as for Sec15. Similarly, we found that expressing Evi5 reduces the size of Rip11-GFP structures, whereas depleting Evi5 had the converse effect (Fig. S2). Overall, these results demonstrate that Evi5 acts as a Rab11-GAP during BC migration.
Orthologues of other Rab11-GAPs do not regulate Rab11 during BC migration
Two other Rab-GAPs have been demonstrated to accelerate the GTP hydrolysis of human Rab11 in vitro: GAPCENA (Fuchs et al., 2007) and TBC1D15 (Zhang et al., 2005). Accordingly, both CG7112 and CG11490, the orthologues of mammalian GAPCENA and TBC1D15, respectively (thereafter called GapcenA and Tbc1d15-17) colocalize with Rab11CA in S2 cells, although Tbc1d15-17 colocalizes to a lesser degree (Fig. S3 A). However, the RNAi screen results indicate that both GapcenA and Tbc1d15-17 knockdown did not affect BC migration (Table 1). To further confirm that these two GAPs do not act on Rab11 during BC migration, we expressed GapcenA and Tbc1d15-17 in BCs and did not observe any migration block (Fig. S3, B and C). These results suggest that Evi5 is the critical Rab11-GAP involved in BC migration.
Evi5 is essential to restrict locally the RTK activity at the leading edge of BCs
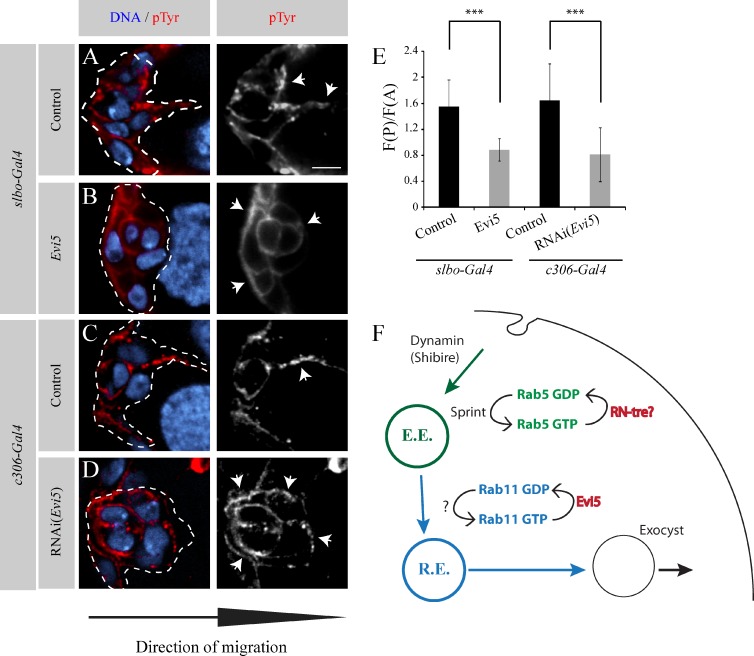
To achieve their directed collective cell migration toward the oocyte, BCs need to polarize their guidance receptors, which consist of RTKs, at their leading edge (Jékely et al., 2005). We have recently demonstrated that Rab11 is involved in restricting the activation of the RTKs at the front of the cell cluster (Assaker et al., 2010). Thus, we hypothesize that Evi5 might also be important for the polarization of active RTKs. To test this, we assessed the distribution of pTyr (phosphorylated tyrosine), which was previously used as a marker of RTK activity in BCs (Jékely et al., 2005; Assaker et al., 2010). As previously published, we observed a strong polarization of the pTyr signal at the leading edge of control BCs, particularly within membrane protrusions (Fig. 5, A and C). Either overexpression or depletion of Evi5 abolishes the enrichment of the pTyr signal at the leading edge (Fig. 5, B and D), as shown by the ratio of posterior over anterior pTyr signal in BCs (Fig. 5 E). These data demonstrate that a strict regulation of the recycling endosome is necessary for the proper spatial restriction of RTK activity.
Figure 5.
Evi5 is necessary to properly localize active RTKs at the leading edge of BCs. (A–D) Representative images showing the distribution of pTyr at the onset of the migration process in stage 9 egg chambers in the indicated conditions. Nuclei are stained with DAPI (blue). A grayscale image of the red channel is shown for every image. Dashed lines outline the BC cluster in the colored image. The arrows point to regions with a high pTyr signal. Bar, 5 µm. (E) Quantification of pTyr fluorescence ratio in the posterior half of the cluster (F(P)) to the anterior half (F(A)) at the onset of migration for the indicated conditions. (10 < n < 17; ***, P < 0.005; t test). Error bars are standard deviations. (F) Schematic representation of the endocytic cycle regulating the polarization of RTKs during BC migration. Vesicle trafficking steps illustrated in green and in blue are regulated by Rab5 and Rab11, respectively. Moreover, Evi5, the Rab11-GAP protein identified in this study as a novel regulator of BC migration, is shown in red. RN-tre is shown as the potential Rab5-GAP involved in BC, as it was found in RNAi screen as necessary for BC migration, and its mammalian orthologue is known to act on Rab5. E.E., early endosome; R.E., recycling endosome.
Most of the previous studies focusing on GAP proteins were performed in cell culture or in vitro (Fukuda, 2011). Furthermore, only fragments of the proteins, such as the TBC domain, were used in most cases because GAP proteins are frequently large and insoluble. To circumvent these limitations, we have combined in vitro and in vivo approaches to demonstrate that the Rab-GAP Evi5 regulates Rab11 in Drosophila. Furthermore, our work identifies Evi5 as a new regulator of collective cell migration, necessary for the maintenance of active RTK at the leading edge.
Materials and methods
Plasmid constructs
Copper-inducible pMET vectors were used for the GST pull-down assay in S2 cells. GST-Rab11WT, GST-Rab11DN, and GST-Rab11CA were cloned in pMet-picoblast-GFP (gift from V. Archambault laboratory, Université de Montréal, Montréal, Québec, Canada), and S2 stable cell lines expressing these constructs were generated.
The different Rab-GAP proteins were cloned in pUASt-Dest29 (Wirtz-Peitz et al., 2008) to be expressed in flies and in S2 cells. Evi5(CG11727) (EST: GH14362), GapcenA(CG7112) (EST: RE63030), and Tbc1d15-17(CG11490) (EST: LD27216) were first cloned in pDonor221 and transferred in pUASt-Dest29 using the Gateway system (Invitrogen).
To generate the constitutively active Rab proteins the following mutations were generated by mutagenesis (QuikChange; Agilent Technologies; Zhang et al., 2007): of Rab5 (EST: GH24702) Gln88 to Leu; Rab7 (EST: GH03685) Gln67 to Leu; Rab10 (EST: LD39986) Gln68 to Leu; Rab11 (EST: LD14551) Gln70 to Leu; Rab23 (RH23273) Gln96 to Leu; and Rab35 (LD21953) Gln67 to Leu. Using the Gateway system, Rab proteins were cloned in pAGW (EGFP in N terminal), except Rab11, which was cloned in pAVW (Venus in N terminal). Evi5 Arg160 to Ala (Evi5RA) was generated the same way. Evi5WT and Evi5RA were cloned in pDest29, pAWG (EGFP in C terminal) and pAWH (HA tag in C terminal). Rip11 (EST: GH05001) was cloned in pAGW. ESTs were obtained from the Drosophila Genomics Resource Center (Bloomington, IN).
Fly genetics
Expression of proteins was driven specifically in the BCs using slbo-Gal4, UAS-CD8-GFP (Assaker et al., 2010) at 29°C. Double-stranded RNA (dsRNA) fly lines were obtained from the Vienna Drosophila RNAi Center (Dietzl et al., 2007) or, when none were available for a certain gene, from the TRiP (Transgenic RNAi Project) consortium at Harvard Medical School (obtained from the Bloomington Stock Collection) and were expressed using c306-Gal4, UAS-GFP-Sec15 (Assaker et al., 2010) at the indicated temperature. Both CD8-GFP and GFP-Sec15 were expressed to facilitate the identification of the BCs inside the egg chamber. Evi5 rescue experiments were performed using c306-Gal4. Also, slbo-Gal4, UAS-GFP-Sec15 was used for GFP-Sec15 analysis upon changes in Rab11 activity. Other stocks used include UAS-Rab11DN (Assaker et al., 2010), UAS-Rab11CA (Emery et al., 2005), UAS-Rip11-GFP (gift from D.F. Ready, Purdue University, West Lafayette, IN; Li et al., 2007), and Rab11ex1 (Dollar et al., 2002). Evi5, GapcenA, and Tbc1d15-17 in the pDest29 vector were injected into Drosophila, and insertion strains containing a single copy of each transgene were generated using standard methods. Transgenic flies expressing Evi5RA were generated by BestGene Inc.
Indexes
The M.I. represents the mean distance migrated by the BCs, whereas the C.I. represents the percentage of clusters having completed migration, and both are calculated as previously described (Assaker et al., 2010). In brief, the M.I. was calculated with the formula M.I. = (1 × n(100%) + 0.75 × n(75%) + 0.5 × n(50%) + 0.25 × n(25%) + 0 × n(0%) + 0.5 × n(split clusters))/n(total), in which n(100%) corresponds to the number of BCs that reached the oocyte at stage 10, and n(75%) corresponds to the number of BCs that reached 75% of the total distance, etc. The C.I. was calculated as follows: C.I. = n(100%)/n(total).
Rab-GAP screen
Identification of potential Rab-GAP proteins was performed based on the presence of a TBC domain: FlyMine Software (Cambridge Systems Biology Centre, Cambridge University) was used to identify genes containing the TBC domain and orthologues of previously described Rab-GAP proteins, revealing 27 proteins. Females were kept at 29°C for 2 d before ovaries were dissected. Variation in the penetrance of the phenotype between RNAi fly lines targeting the same gene can be explained by the strength of the dsRNA expression, the sequence of the dsRNA used, and possible off-target effects.
Details of image acquisition and quantitative analysis
Images were acquired using an inverted confocal microscope (LSM 510 META; Carl Zeiss), using either a Plan Apochromat 63×, NA 1.4 differential interference contrast objective or an EC Plan Neofluar 40×, NA 1.3. Images used for subsequent Sec15 or Rip11 vesicle analysis were acquired using the 63× objective by sequential scans of 0.5 µm in multiple channels. For figure assembly, images were processed with Photoshop (Adobe) by using the Gaussian blur and the level functions. For a better rendering of the blue channel, a posttreatment with the selective color function was performed.
The enrichment of pTyr intensity at the leading edge (F(P)/F(A)) was determined as follows: mean pTyr fluorescence intensities were quantified in an area following the leading edge (posterior fluorescence or F(P)) and in an area at the trailing edge (anterior fluorescence or F(A)) of BC clusters in three consecutive frames from a z scan separated by 1 µm using original images and the ImageJ software (National Institutes of Health). Only images with a signal in the linear range were considered for quantification. We used as a central plan the one containing the two polar cells, which do not express upstream activation sequence constructs with the same intensity as the other cells of the cluster. Furthermore, only clusters detached from the epithelium but still at the onset of migration were considered. GFP was used to determine the periphery of the cluster. The background, determined outside the egg chamber, was systematically subtracted to the fluorescence intensities determined. The ratio between the posterior and the anterior mean fluorescence (F(P)/F(A)) is shown in Fig. 5 E.
Reagents
Glutathione–Sepharose beads were obtained from GE Healthcare. The transfection reagent used in S2 cells is TransIT-L1 obtained from Mirus Bio LLC. Antibodies used for immunostaining or immunoblotting were supplied from Cell Signaling Technology (GST), USBiological (GFP), the laboratory of G. Rubin (Howard Hughes Medical Institute, Ashburn, VA; HA), and mouse monoclonal anti-pTyr (4G10). Secondary antibodies were purchased from Invitrogen and coupled to Alexa Fluor 555 or 647 dyes. Alexa Fluor 555– and 647–labeled phalloidin were used to visualize F-actin. DAPI (Sigma-Aldrich) was used to stain nuclei. Egg chambers were mounted in Mowiol 4-88 (Sigma-Aldrich). Concanavalin A (Sigma-Aldrich) and 96 wells were purchased from PerkinElmer.
GST pull-down analysis
S2 cells were cultured in Schneider’s medium supplemented with 10% FBS and transfected with TransIT-LT1 on day 1. Protein expression was induced with 0.8 mM CuSO4 on day 2. On day 4, cells were lysed in Nonidet P-40 lysis buffer (20 mM Tris, pH 8.0, 137 mM NaCl, 1% Nonidet P-40, 10% glycerol, and 1 mM EDTA) with protease inhibitors. For GST pull-down assays, 50 µl of 50% slurry of glutathione–Sepharose beads equilibrated in lysis buffer was added to protein lysates and rocked for 4 h at 4°C. Beads were then washed three times with 1 ml lysis buffer. Total protein lysates or eluted proteins were resolved on an 8–10% SDS-PAGE, transferred to nitrocellulose membranes, and immune detected using specific antibodies.
Imaging of S2 cells
For fluorescence analysis, Rab and Rab-GAP proteins were cotransfected for 3 d, transferred for 1 h on concanavalin A–coated 96-well plates, and fixed in 4% formaldehyde for 30 min.
Vesicle analysis
Fluorescent images were analyzed using semiautomated software custom written in MATLAB (MathWorks). Image analysis was performed in two steps: (1) BCs were identified by automated thresholding of the GFP-Sec15 channel. (2) Within the region of the BCs, fluorescent signals were detected in 3D by identifying connected groups of voxels that were significantly brighter than the local noise. Where nearby vesicles were close enough to one another so that their signals overlapped, the voxel groups were split into as many parts, as they contained local maxima of intensity.
Online supplemental material
Fig. S1 shows that expression of other DN Rabs did not affect Sec15-GFP distribution to the same extent as Rab11DN, demonstrating that monitoring the distribution of Sec15-GFP can be used to estimate the activity of Rab11 in vivo. Fig. S2 confirms that Evi5 is a Rab11-GAP during BC migration by monitoring the distribution of a second effector of Rab11, Rip11. Fig. S3 shows that orthologues of mammalian GAPs proposed to target Rab11 do not affect BC migration, suggesting that Evi5 is the critical Rab11-GAP in this process. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201112114/DC1.
Supplementary Material
Acknowledgments
We thank Donald F. Ready, Vincent Archambault, the Bloomington Stock Collection, the Vienna Drosophila RNAi Center, TRiP at Harvard Medical School (National Institutes of Health/National Institute of General Medical Sciences R01-GM084947), and BestGene Inc. for fly stocks and plasmids. We are grateful to Peter S. McPherson for critical reading of the manuscript and to Francis Barr for advice. We also thank Carole Iampietro for technical assistance.
This work was supported by a grant from the Canadian Institute for Health Research to G. Emery (MOP 114899). G. Emery holds a Canada Research Chair (Tier II) in Vesicular Trafficking and Cell Signaling. P.S. Maddox is supported by grants from the Canadian Institute for Health Research (MOP 106548 and MOP 115171), the National Sciences and Engineering Research Council (355641), and the Canadian Cancer Society Research Institute (700824). P.S. Maddox is the Canada Research Chair in Cell Division and Chromosomal Organization. C. Laflamme holds a Canadian Institute for Health Research Doctoral award.
Footnotes
Abbreviations used in this paper:
- BC
- border cell
- CA
- constitutive active
- C.I.
- completion index
- DN
- dominant negative
- dsRNA
- double-stranded RNA
- GAP
- GTPase-activating protein
- GEF
- guanine nucleotide exchange factor
- KS
- Kolmogorov–Smirnov
- M.I.
- migration index
- nM.I.
- normalized M.I.
- RTK
- receptor tyrosine kinase
- TBC
- Tre-2, Bub2, and Cdc16
References
- Assaker G., Ramel D., Wculek S.K., González-Gaitán M., Emery G. 2010. Spatial restriction of receptor tyrosine kinase activity through a polarized endocytic cycle controls border cell migration. Proc. Natl. Acad. Sci. USA. 107:22558–22563 10.1073/pnas.1010795108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F., Lambright D.G. 2010. Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22:461–470 10.1016/j.ceb.2010.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotard L., Mishra A.K., Sylvain M.A., Tuck S., Lambright D.G., Rocheleau C.E. 2010. TBC-2 regulates RAB-5/RAB-7-mediated endosomal trafficking in Caenorhabditis elegans. Mol. Biol. Cell. 21:2285–2296 10.1091/mbc.E09-11-0947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabbeekeh J.T., Faitar S.L., Dufresne C.P., Cowell J.K. 2007. The EVI5 TBC domain provides the GTPase-activating protein motif for RAB11. Oncogene. 26:2804–2808 10.1038/sj.onc.1210081 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 448:151–156 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Dollar G., Struckhoff E., Michaud J., Cohen R.S. 2002. Rab11 polarization of the Drosophila oocyte: a novel link between membrane trafficking, microtubule organization, and oskar mRNA localization and translation. Development. 129:517–526 [DOI] [PubMed] [Google Scholar]
- Emery G., Hutterer A., Berdnik D., Mayer B., Wirtz-Peitz F., Gaitan M.G., Knoblich J.A. 2005. Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell. 122:763–773 10.1016/j.cell.2005.08.017 [DOI] [PubMed] [Google Scholar]
- Frasa M.A., Koessmeier K.T., Ahmadian M.R., Braga V.M. 2012. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat. Rev. Mol. Cell Biol. 13:67–73 [DOI] [PubMed] [Google Scholar]
- Fuchs E., Haas A.K., Spooner R.A., Yoshimura S., Lord J.M., Barr F.A. 2007. Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J. Cell Biol. 177:1133–1143 10.1083/jcb.200612068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M. 2011. TBC proteins: GAPs for mammalian small GTPase Rab? Biosci. Rep. 31:159–168 10.1042/BSR20100112 [DOI] [PubMed] [Google Scholar]
- Haas A.K., Fuchs E., Kopajtich R., Barr F.A. 2005. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat. Cell Biol. 7:887–893 10.1038/ncb1290 [DOI] [PubMed] [Google Scholar]
- Horgan C.P., McCaffrey M.W. 2009. The dynamic Rab11-FIPs. Biochem. Soc. Trans. 37:1032–1036 10.1042/BST0371032 [DOI] [PubMed] [Google Scholar]
- Houalla T., Shi L., van Meyel D.J., Rao Y. 2010. Rab-mediated vesicular transport is required for neuronal positioning in the developing Drosophila visual system. Mol. Brain. 3:19 10.1186/1756-6606-3-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Satoh M., Kanno E., Fukuda M. 2006. Screening for target Rabs of TBC (Tre-2/Bub2/Cdc16) domain-containing proteins based on their Rab-binding activity. Genes Cells. 11:1023–1037 10.1111/j.1365-2443.2006.00997.x [DOI] [PubMed] [Google Scholar]
- Janssens K., Sung H.H., Rørth P. 2010. Direct detection of guidance receptor activity during border cell migration. Proc. Natl. Acad. Sci. USA. 107:7323–7328 10.1073/pnas.0915075107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jékely G., Sung H.H., Luque C.M., Rørth P. 2005. Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Dev. Cell. 9:197–207 10.1016/j.devcel.2005.06.004 [DOI] [PubMed] [Google Scholar]
- Lanzetti L., Rybin V., Malabarba M.G., Christoforidis S., Scita G., Zerial M., Di Fiore P.P. 2000. The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature. 408:374–377 10.1038/35042605 [DOI] [PubMed] [Google Scholar]
- Li B.X., Satoh A.K., Ready D.F. 2007. Myosin V, Rab11, and dRip11 direct apical secretion and cellular morphogenesis in developing Drosophila photoreceptors. J. Cell Biol. 177:659–669 10.1083/jcb.200610157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano F., Sasaki T., Fukui K., Asakura T., Imazumi K., Takai Y. 1998. Molecular cloning and characterization of the noncatalytic subunit of the Rab3 subfamily-specific GTPase-activating protein. J. Biol. Chem. 273:24781–24785 10.1074/jbc.273.38.24781 [DOI] [PubMed] [Google Scholar]
- Pan X., Eathiraj S., Munson M., Lambright D.G. 2006. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 442:303–306 10.1038/nature04847 [DOI] [PubMed] [Google Scholar]
- Scita G., Di Fiore P.P. 2010. The endocytic matrix. Nature. 463:464–473 10.1038/nature08910 [DOI] [PubMed] [Google Scholar]
- Uytterhoeven V., Kuenen S., Kasprowicz J., Miskiewicz K., Verstreken P. 2011. Loss of skywalker reveals synaptic endosomes as sorting stations for synaptic vesicle proteins. Cell. 145:117–132 10.1016/j.cell.2011.02.039 [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F., Nishimura T., Knoblich J.A. 2008. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 135:161–173 10.1016/j.cell.2008.07.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Mehta S.Q., Pichaud F., Bellen H.J., Quiocho F.A. 2005. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat. Struct. Mol. Biol. 12:879–885 10.1038/nsmb987 [DOI] [PubMed] [Google Scholar]
- Yoshimura S., Egerer J., Fuchs E., Haas A.K., Barr F.A. 2007. Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 178:363–369 10.1083/jcb.200703047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M., McBride H. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2:107–117 10.1038/35052055 [DOI] [PubMed] [Google Scholar]
- Zhang J., Schulze K.L., Hiesinger P.R., Suyama K., Wang S., Fish M., Acar M., Hoskins R.A., Bellen H.J., Scott M.P. 2007. Thirty-one flavors of Drosophila rab proteins. Genetics. 176:1307–1322 10.1534/genetics.106.066761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.M., Walsh B., Mitchell C.A., Rowe T. 2005. TBC domain family, member 15 is a novel mammalian Rab GTPase-activating protein with substrate preference for Rab7. Biochem. Biophys. Res. Commun. 335:154–161 10.1016/j.bbrc.2005.07.070 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.