Vacuole- and mitochondria-specific cargo adaptors compete for an overlapping binding site on Myo2 to determine the inheritance of these organelles during budding.
Abstract
During cell division, organelles are distributed to distinct locations at specific times. For the yeast vacuole, the myosin V motor, Myo2, and its vacuole-specific cargo adaptor, Vac17, regulate where the vacuole is deposited and the timing of vacuole movement. In this paper, we show that Mmr1 functions as a mitochondria-specific cargo adaptor early in the cell cycle and that Mmr1 binds Myo2 at the site that binds Vac17. We demonstrate that Vac17 and Mmr1 compete for binding at this site. Unexpectedly, this competition regulates the volume of vacuoles and mitochondria inherited by the daughter cell. Furthermore, eight of the nine known Myo2 cargo adaptors overlap at one of two sites. Vac17 and Mmr1 overlap at one site, whereas Ypt11 and Kar9 bind subsets of residues that also bind Ypt31/Ypt32, Sec4, and Inp2. These observations predict that competition for access to Myo2 may be a common mechanism to coordinate the inheritance of diverse cargoes.
Introduction
Multiple cellular functions are executed via cytoplasmic organelles. The volume of each organelle likely depends on several factors, including cell type, its functions, metabolic status, and stage in the cell cycle. Moreover, during cell division, organelle volume must be coordinated with transient changes in cell volume. Little is known about how organelle size is controlled.
In the budding yeast Saccharomyces cerevisiae, during the cell cycle, a portion of each cytoplasmic organelle is transported across the mother–bud neck to distinct locations in the bud. The myosin V motor, Myo2, transports most of these cytoplasmic organelles as well as other cargoes. Myo2 carries secretory vesicles (Govindan et al., 1995), vacuoles (Hill et al., 1996; Catlett and Weisman, 1998), mitochondria (Itoh et al., 2002), peroxisomes (Hoepfner et al., 2001; Fagarasanu et al., 2006), the late Golgi (Rossanese et al., 2001), and astral microtubules (Beach et al., 2000; Yin et al., 2000). This raises the question of how Myo2 carries diverse cargoes to the proper place at the right time.
Insight into how Myo2 interacts with cargoes comes from analysis of the structure of the Myo2 cargo-binding domain (CBD; Pashkova et al., 2006). The CBD is the C-terminal 487 residues of the motor heavy chain, which fold into 15 antiparallel helices, ending with a long loop containing two helices. The loop wraps back to the “beginning” of the CBD. Residues that are critical for the inheritance of the vacuole map to a single region on the surface of the CBD (Catlett et al., 2000; Pashkova et al., 2006). Mutation of these residues disrupted Myo2 interaction with Vac17, the vacuole-specific cargo adaptor protein (Ishikawa et al., 2003). Vac17 also binds Vac8, a vacuole membrane protein (Tang et al., 2003). The Myo2–Vac17–Vac8 complex attaches Myo2 to the vacuole. These observations suggested that specificity for an individual Myo2 cargo derives in part from adaptor proteins that link the cargo to the Myo2 CBD (Weisman, 2006).
Indeed several adaptor proteins have been identified. In yeast, the peroxisomal protein Inp2 binds Myo2 and is required for proper inheritance of peroxisomes (Fagarasanu et al., 2006). Kar9 links Myo2 to the microtubule plus ends via direct interaction with the capping protein Bim1 (Korinek et al., 2000; Miller et al., 2000). The Myo2–Kar9 interaction is important for proper orientation of the mitotic spindle during the cell cycle (Beach et al., 2000; Yin et al., 2000). Although the Myo2 CBD exhibits high conservation with most eukaryotic myosin V CBDs, the adaptor proteins are not uniformly conserved (Mast et al., 2012), which suggests that modes of attachment to myosin V rapidly evolve, whereas the mechanisms used by the CBD are conserved.
Mmr1 is proposed to be an adaptor protein that links Myo2 to mitochondria. The mitochondria protein Mmr1 binds directly to Myo2 (Itoh et al., 2004) and has a role in mitochondria distribution to the bud (Itoh et al., 2004; Frederick et al., 2008). Mitochondria are often missing from small buds of mmr1Δ yeast, but as bud size increases, mitochondria distribution is partially corrected, indicating that additional mechanisms of inheritance are involved. One additional mechanism involves the ER–mitochondrial encounter structure (ERMES)/mitochore complex, which forms junctions between mitochondria and the ER (Kornmann et al., 2009). The GTPase Gem1, required for distribution of mitochondria to the bud (Frederick et al., 2004), is part of the ERMES complex (Kornmann et al., 2011).
The Rab GTPase Ypt11, which has a role in mitochondrial distribution to the bud, also interacts with Myo2 (Itoh et al., 2002; Boldogh et al., 2004; Frederick et al., 2008; Förtsch et al., 2011). However, Ypt11 localizes to the ER (Buvelot Frei et al., 2006) and late Golgi vesicles (Arai et al., 2008), which raises questions about whether Ypt11 directly attaches mitochondria to Myo2. Given its localization, Ypt11 may move the ER and/or late Golgi. Moreover, Ypt11 likely functions in parallel with Mmr1; a double deletion of YPT11 and MMR1 has a synthetic effect on mitochondrial distribution (Frederick et al., 2008).
Adaptor proteins that attach Myo2 to secretory vesicles include the Rab GTPases Ypt31/32 and Sec4, which bind a site on Myo2 that is offset 180° from the Vac17 binding site (Lipatova et al., 2008; Jin et al., 2011; Santiago-Tirado et al., 2011). Mutations of residues that disrupt Rab GTPase binding produce a severe growth defect caused by a defect in secretory vesicle transport to the plasma membrane. Sec15, a subunit of the exocyst-tethering complex, binds Myo2 on the opposite side from the Rab GTPase binding site (Jin et al., 2011). Binding of both Sec15 and the Rab GTPases is required for the normal distribution of secretory vesicles.
Here, we map the binding sites of the other known cargo adaptors for Myo2. Together with previous studies, we find that eight adaptor proteins bind to one of two overlapping binding sites (Lipatova et al., 2008; Fagarasanu et al., 2009; Jin et al., 2011). Binding sites for Mmr1 and Vac17 overlap at a simpler region. These proteins compete for access to Myo2 in vivo and in vitro. Surprisingly, mutations that affect Myo2 interaction with Mmr1 only result in an increase in the volume of vacuoles in the bud. Similarly, mutations that affect Myo2 interaction with Vac17 only result in an increase in the volume of mitochondria in the bud. Thus, overlap of the Vac17 and Mmr1 binding sites on Myo2 likely has a role in regulating organelle volume. Similarly, the other adaptor proteins bind at a site that overlaps on the opposite side of Myo2. The Ypt11 and Kar9 binding sites overlap with each other and with the secretory vesicle Rab GTPases and Inp2 binding sites. Furthermore, the Rab GTPase/Kar9/Inp2 binding region, Mmr1/Vac17 binding region, and the Sec15 binding site are potentially connected through shared helices and loops. This raises the possibility that the binding of any single cargo adaptor may enhance or inhibit binding of adaptor proteins at spatially distinct regions. Thus, the CBD of Myo2 may be a focal point to integrate the distribution of all of its cargoes.
Results
Mmr1, a cargo adaptor protein for mitochondria, binds Myo2 at a site that overlaps with the Vac17 binding site
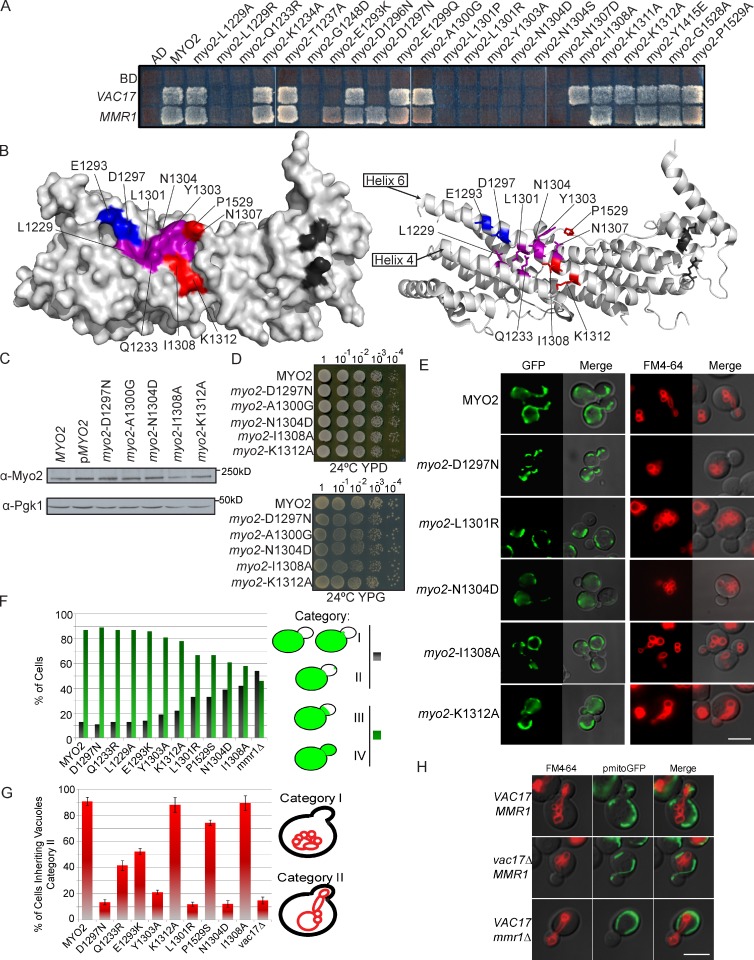
Mmr1 is required for normal distribution of mitochondria to the yeast bud. Movement of mitochondria is disrupted by mutation of specific surface residues on Myo2 (Altmann et al., 2008). These residues overlap with residues required for Myo2 interaction with Vac17, the vacuole-specific adaptor protein (Ishikawa et al., 2003). Thus, we tested whether mutations in this region disrupted the interaction between Myo2 and Mmr1 and also Myo2 and Vac17 (Fig. 1 A). In addition, we tested the six residues mutated in myo2-573, a mutant defective in binding Mmr1 and mitochondria inheritance (Itoh et al., 2004). Using a yeast two-hybrid test, we identified several mutants that disrupted interactions between both Myo2 and Mmr1 and between Myo2 and Vac17 (Fig. 1, A and B). Mutation of myo2-E1293K and myo2-D1297N affected the ability of Myo2 to interact with Vac17 and not Mmr1 (Fig. 1 B, blue). myo2-P1529S was the single residue in myo2-573 that disrupted Myo2 interaction with Mmr1 (Fig. S1 A and Fig. 1 B). It did not affect Myo2 interaction with Vac17. Two additional mutants, myo2-I1308A and myo2-K1312A, interacted with Vac17 but not Mmr1 (Fig. 1 B).
Figure 1.
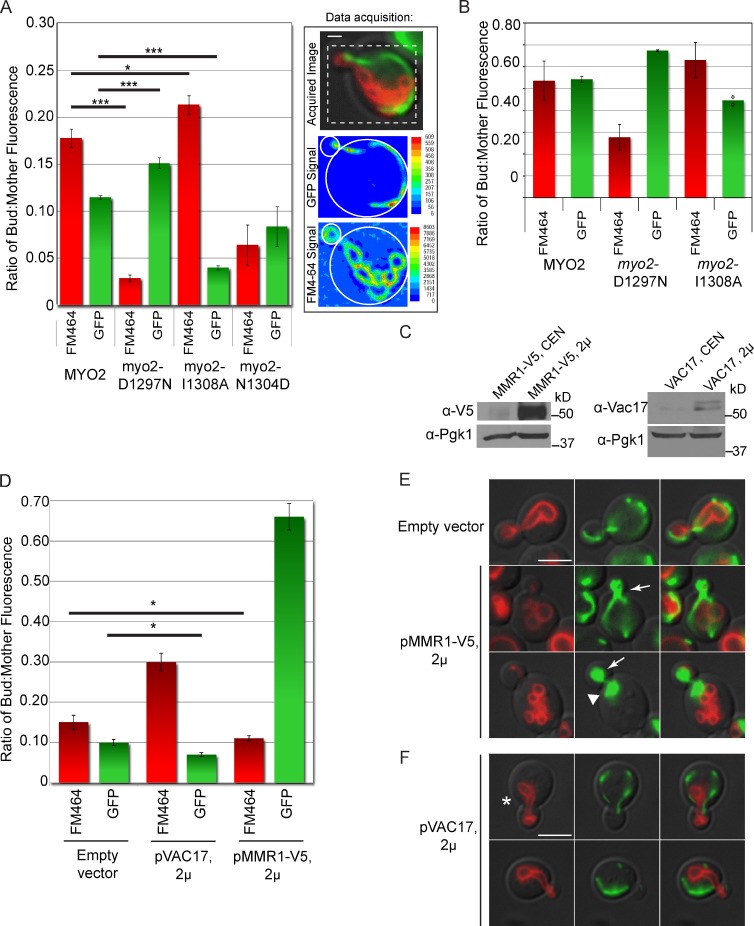
Mmr1 and Vac17 binding sites overlap on Myo2. (A) Mutations on the Myo2 CBD surface reveal overlapping binding regions essential for interaction with Vac17 and Mmr1. Top left squares show empty vector controls. (B) Surface (left) and ribbon (right) view of Myo2 CBD. Blue: Vac17 only; red: Mmr1 only; purple: Vac17 and Mmr1; dark gray: Sec15 binding site. (C) Mutations did not affect the levels of Myo2. (D) Myo2 mutants affecting Mmr1 and/or Vac17 interactions grow on glucose (YPD [yeast extract, peptone, dextrose]) and glycerol (YPG [yeast extract, peptone, glucose]). (E) myo2-D1297N is defective in vacuole inheritance, myo2-I1308A and -K1312A are defective in mitochondria inheritance, and myo2-L1301R and -N1304D are defective in mitochondria and vacuole inheritance. For mitochondria, strains with pmitoGFP (green) were grown in SC minus leucine media for at least six doubling times; density of <5 × 106 cells/ml. For vacuoles, cells were pulsed with FM4-64 for 1 h and chased for one doubling. (F) Mitochondria inheritance was scored in small and medium budded cells according to position of mitochondria in the bud. Categories I and II were mutant and grouped together (dark bars). Categories III and IV were wild type–like and grouped together (green bars). n = 2; ≥200 cells per strain. (G) Cells scored for the presence or absence of vacuoles in small and medium budded cells. n = 3; ≥200 cells per strain. Error bars represent SDs. (H) Vacuoles (FM6-46) and mitochondria (mitoGFP) in wild-type, vac17Δ, and mmr1Δ yeast. AD, activation domain; BD, binding domain. Bars, 5 µm.
Additional phenotypes were not observed in these mutants. When expressed from plasmids, the mutant and wild-type proteins were present at the same steady-state levels (Fig. 1 C) and had normal actin cable organization (Fig. S1 B), cell viability, and mitochondria function as measured by growth of the mutant strains on a nonfermentable carbon source, glycerol (Fig. 1 D). Surface mutations near this region did not disrupt binding to other cargo adaptor proteins (Fig. S1 C and Table S3).
Inheritance of vacuoles and mitochondria was assayed in selected mutants that blocked Myo2 interaction with Vac17 or Mmr1 (Fig. 1 E). Mutations that disrupted interaction with both Vac17 and Mmr1 also disrupted inheritance of both mitochondria (Fig. 1 F) and vacuoles (Fig. 1 G). Thus, a central region on helix 6 and part of helix 4 contain residues important for inheritance of both mitochondria and vacuoles.
myo2-E1293K and myo2-D1297N mutants that affected Myo2 interaction with Vac17, but not Mmr1, disrupted the inheritance of vacuoles only. Conversely, myo2-I1308A, myo2-K1312A, and myo2-P1529S mutants only disrupted mitochondria inheritance. Thus, Vac17 and Mmr1 interact with Myo2 at overlapping, but not identical, sites.
myo2-P1529S disrupted the interaction of Myo2 with Mmr1 (Fig. S1 D) and caused a mitochondria inheritance defect. However, because substitution of a proline may disrupt tertiary structure, it is unclear whether P1529 contributes directly to the Mmr1 binding site.
We tested whether Vac17 and Mmr1 are each solely dedicated to moving vacuoles or mitochondria, respectively. The mmr1Δ mutant has a mitochondria inheritance defect (Fig. 1 H), but vacuole inheritance was normal. Conversely, in vac17Δ cells, mitochondria were inherited normally, but vacuole inheritance was disrupted (Fig. 1 H). Thus, inheritance of mitochondria requires Mmr1, whereas inheritance of vacuoles requires Vac17. Furthermore, Vac17 binds Myo2 in the absence of Mmr1, and conversely, Mmr1 binds Myo2 in the absence of Vac17.
We also tested the relative importance of Mmr1 and Ypt11 for inheritance of mitochondria. Ypt11 binds directly to the Myo2 CBD at a distinct site from Mmr1/Vac17. The mmr1Δ mutant had a greater mitochondria inheritance defect than the ypt11Δ mutant (49 vs. 21% buds had no mitochondria; Fig. S1 E). Moreover, Mmr1 and Ypt11 do not interact with each other (Fig. S1 F). Thus, we chose to study Vac17 and Mmr1 independent of potential contributions from Ypt11.
Vac17 and Mmr1 localize to the leading edge of vacuoles and mitochondria in the bud and mother cell
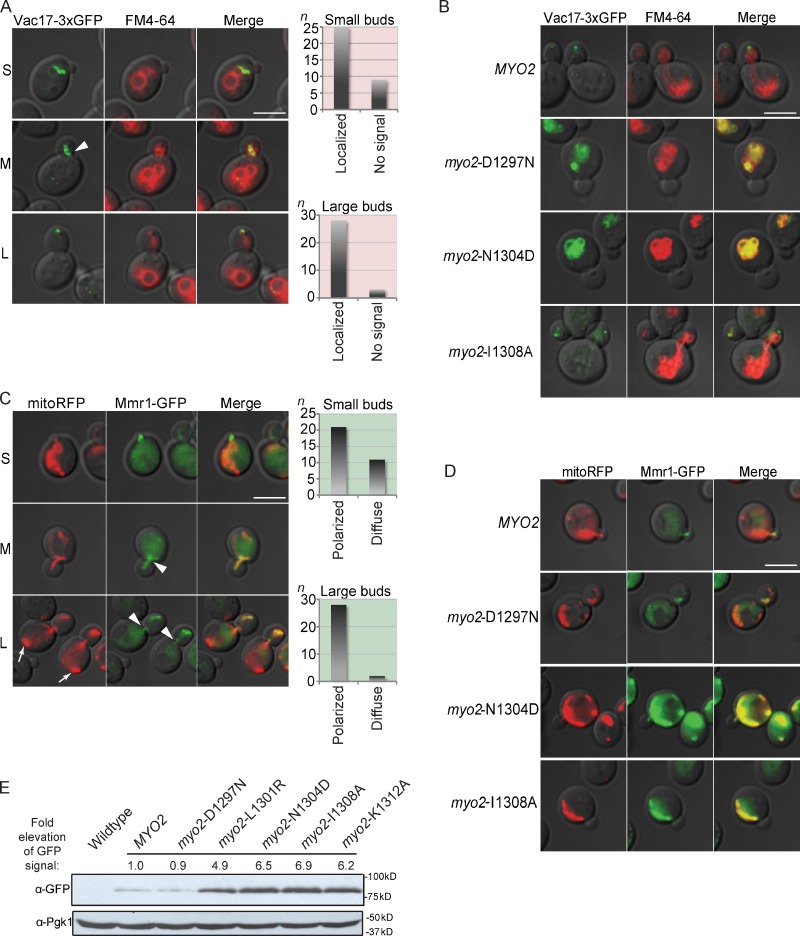
To determine functions of Vac17 and Mmr1, we studied the localization of these proteins in wild-type cells and in myo2 mutants. Vac17-3×GFP localized on the leading edge of vacuoles in the bud as well as to portions of vacuoles in the mother cell near the mother–bud neck (Fig. 2 A; Jin et al., 2009). In wild-type cells, the Vac17 protein is degraded after vacuole inheritance (Tang et al., 2003). In the myo2-D1297N mutant, which is defective in vacuole inheritance, there is unpolarized accumulation of Vac17-3×GFP protein on the vacuole membrane in the mother cell (Fig. 2 B). Similarly, Vac17 protein levels are elevated on the mother cell vacuole in the myo2-N1304S mutant (Tang et al., 2003). These findings support the hypothesis that Myo2 and Vac17 bind vacuoles in the mother and move a portion of the vacuole to the bud. Vac17-3×GFP did not accumulate in myo2-I1308A, which does not have a vacuole inheritance defect.
Figure 2.
Mmr1 and Vac17 concentrate at the leading edge of inherited mitochondria and vacuoles, respectively. (A–E) Vac17-3×GFP (A and B) and Mmr1-GFP (C–E) expressed from their endogenous loci. (A) Vac17 (green) concentrated at the leading edge of the vacuole (red). Small (S), medium (M), and large (L) budded cells. Small budded (top graph; n = 34) and large budded (bottom graph; n = 31) cells were scored. When GFP was detected, it was exclusively on the vacuole membrane. Arrowhead indicates an example of Vac17 accumulation at the mother–bud neck. (B) Vac17-3×GFP is elevated and on the mother vacuole membrane in myo2-D1297N and myo2-N1304D mutants. myo2Δ cells with the indicated myo2 mutant were expressed from a plasmid (CEN and HIS3) as the sole copy of MYO2. (A and B) Before FM4-64 labeling, cells were grown in rich media for five doubling times at a density of <5 × 106 cells/ml. Cells were chased for one doubling. (C) Mmr1 (green) is concentrated on the leading edge of mitochondria (red; mitoRFP expressed from a plasmid). Small budded (top graph; n = 32) and large budded (bottom graph; n = 30) cells were scored for polarized GFP signal or a diffuse GFP signal on the mitochondria membrane. Arrowheads indicate examples of Mmr1 signal at the mother–bud neck region. Arrows show mitochondria at the rear of the mother cell. (A and C) Three independent experiments. (D) Mmr1-GFP is elevated and localized throughout the mitochondria membrane in myo2-N1304D and myo2-I1308A mutants. myo2Δ cells with the indicated myo2 mutant or wild type expressed from a plasmid (CEN and HIS3) as the sole copy of MYO2 were transformed with pmitoRFP. (E) Western analysis of Myo2 in Mmr1-GFP cells containing MYO2 or myo2 mutant alleles. Anti-Pgk1 is the loading control. Bars, 5 µm.
Two postulated roles for Mmr1 function are consistent with the finding that Mmr1 directly contacts Myo2. Mmr1 localizes to mitochondria at bud tips in wild-type cells (Itoh et al., 2004) and may connect Myo2 to mitochondria for their transport (Itoh et al., 2004; Frederick et al., 2008). Alternatively, Mmr1 protein may be transported to the bud tip via Myo2, where it functions to tether mitochondria (Shepard et al., 2003; Peraza-Reyes et al., 2010; Swayne et al., 2011). These hypotheses are not mutually exclusive. We focused on whether the Mmr1–Myo2 complex moves mitochondria across the mother–bud neck.
Mmr1-GFP localized to portions of mitochondria in the bud as well as mitochondria in the mother near the mother–bud neck (Fig. 2 C). Notably, mitochondria at the rear of the mother lack Mmr1-GFP. Thus, localization of both Vac17 and Mmr1 is consistent with their roles as adaptor proteins, which link Myo2 to portions of vacuoles and mitochondria that move into the bud. In further support, in the myo2-I1308A mutant, which cannot bind Mmr1, Mmr1 was distributed throughout the mitochondrial surface, with a concomitant increase in Mmr1-GFP (Fig. 2 D). Elevation of Mmr1-GFP did not occur in cells containing myo2-D1297N, which disrupts Vac17, but not Mmr1, interaction with Myo2 (Fig. 2 D). Mmr1 protein was elevated five- to sevenfold in myo2 mutants defective in binding Mmr1 (Fig. 2 E). Vac17 and Mmr1 are elevated in myo2-N1304D, which has both a vacuole and mitochondria inheritance defect (Fig. 2, B and D). That Mmr1 and Vac17 accumulate in specific myo2 mutants in the mother cell on mitochondria and vacuoles, respectively, strongly suggests that Mmr1 and Vac17 interact with the organelles in the mother and attach to Myo2 to transport mitochondria and vacuoles to the bud.
Vacuole and mitochondria inheritance occurs early in the cell cycle, at similar but not identical times
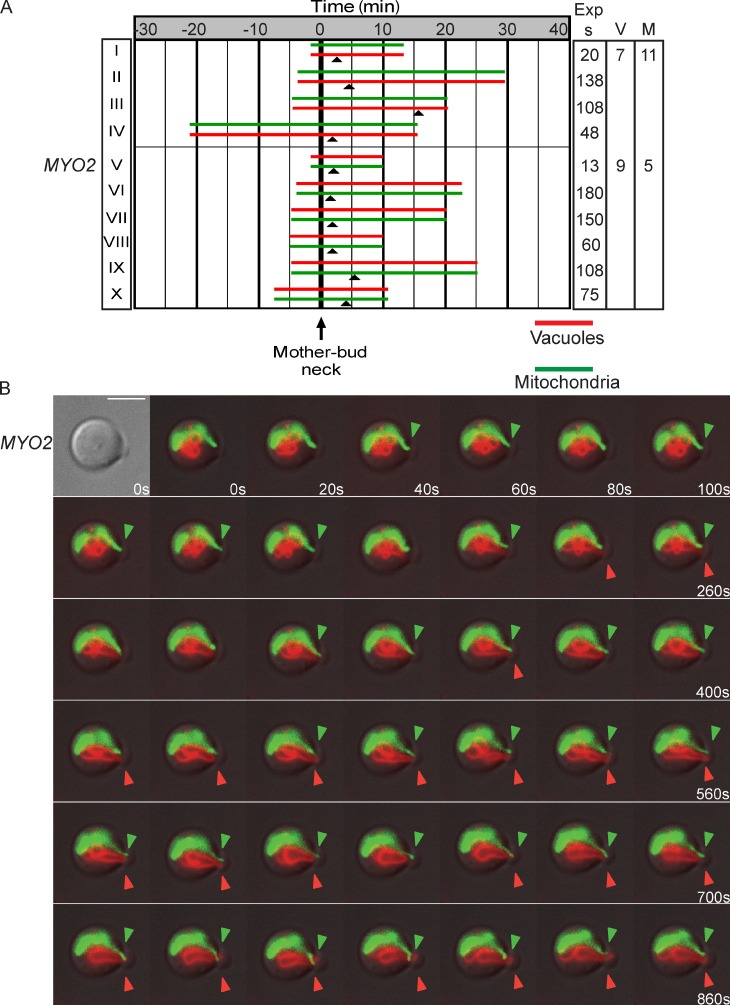
To further test the role of Mmr1 and Vac17 interactions with Myo2, we performed in vivo time-lapse imaging. Cells with buds initially devoid of mitochondria and vacuoles were visualized until both mitochondria (mitoGFP) and vacuole (FM4-64) tubules moved into the bud (Fig. 3 A). In Fig. 3, each red and green pair represents one cell. The time the vacuole or mitochondria crossed the mother–bud neck occurred independently; time 0 indicates the time that the first organelle crossed the mother–bud neck; closed arrowheads indicate when the second organelle crossed the mother–bud neck. For example, in Fig. 3 A (I), the top green bar indicates that mitochondria moved into the bud first and crossed the mother–bud neck ∼1 min from the start of imaging. After ∼4 min, the vacuole also crossed the mother–bud neck.
Figure 3.
Vacuole and mitochondria inheritance occurs early in the cell cycle at similar but not identical times. (A) Vacuole and mitochondria inheritance occurs at similar times. A pair of bars represents one cell. Bar length indicates total time of the time lapse. t = 0, time the first organelle (top bar) entered the bud. Closed arrowheads show time the second organelle entered the bud. Right columns indicate time between images. V or M indicates the number of times a vacuole (V) or mitochondria (M) crossed the mother–bud neck for the two time-lapse series with <60 s between images. Wild-type cells were transformed with pmitoGFP, grown in log phase for a minimum of six doublings, labeled with FM4-64, and chased for one doubling. Small buds initially devoid of mitochondria and vacuoles, with both mitochondria and vacuoles visible in the region around the mother–bud neck, were analyzed. Images for all acquired time-lapse series are presented in the JCB DataViewer. (B) Images of A (I; top red–green pair) as mitochondria and vacuoles crossed the mother–bud neck. Images were taken every 20 s for 13 min. Closed arrowheads show organelle in the bud. Exp s, seconds of exposure. Bar, 5 µm.
To maximize the number of time courses analyzed, we used time-lapse series ranging from 13 to 180 s between images. For most cells, both vacuoles and mitochondria crossed into the bud within 7 min from the start of imaging. Organelles frequently retracted back into the mother cell and then recrossed from the mother to the bud (Fig. 3 B, arrowheads), which was observed in 6 of the 10 time-lapse series (Fig. 3 A). Moreover, for the two cells in which images were acquired <60 s apart (Fig. 3 B, I and V), the vacuole tubule crossed the mother–bud neck seven and nine times, respectively, and the mitochondrial tubule crossed the mother–bud neck 11 and five times, respectively.
The time-lapse series revealed several aspects regarding the inheritance of vacuoles and mitochondria: (a) There is not a designated order for whether vacuoles or mitochondria enter the bud first. Either vacuoles or mitochondria enter the bud first. Mitochondria moved into the bud before vacuoles in 4 of 10 cells analyzed, whereas vacuoles moved first in the other six. (b) Although the time each organelle crossed the mother–bud neck was independent of the other, mitochondria and vacuoles moved into the bud at similar times in the cell cycle. Thus, there is not a chronological order to binding of Vac17 versus Mmr1 to Myo2, yet binding of each is regulated by the cell cycle. (c) The movement of vacuoles and mitochondria is a combination of anterograde and retrograde events (rocking) across the mother–bud neck. The timing of rocking of each organelle was independent of the other.
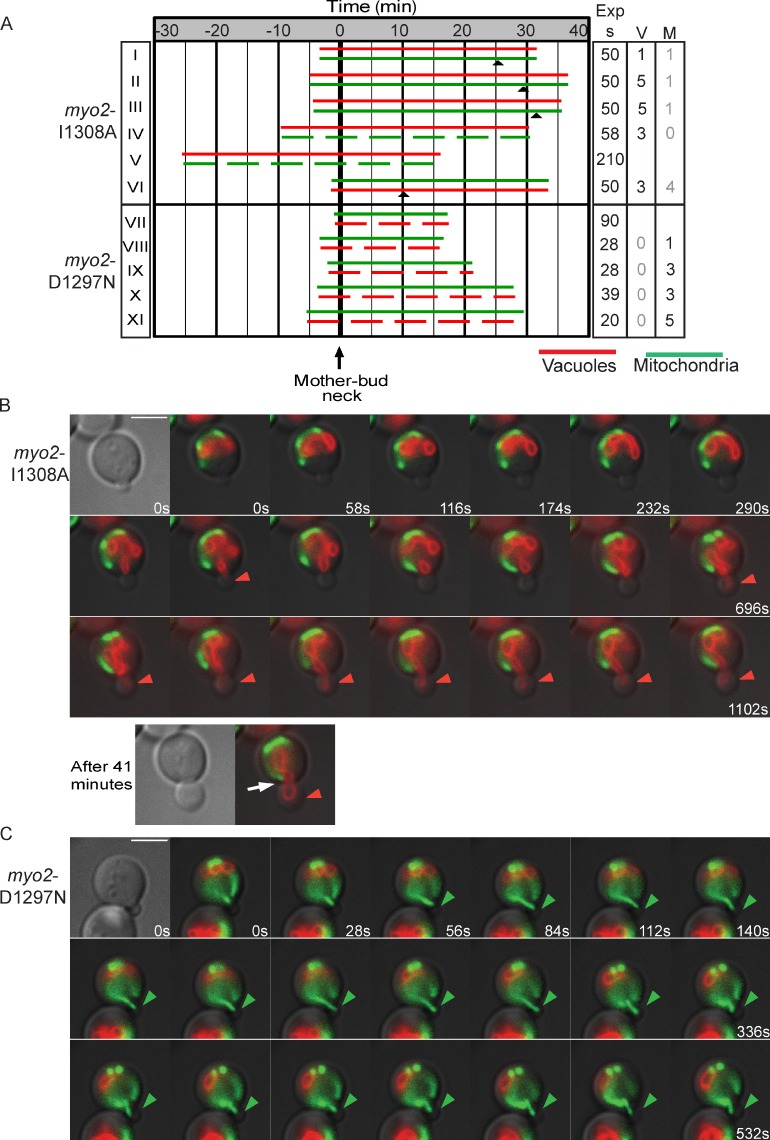
We acquired time-lapse images for myo2-I1308A, which is defective in binding Mmr1 (Fig. 4 A). In six examples, five cells had either a delay (∼32 min; Fig. 4 A, I–III) or complete defect (observed for >40 min each; Fig. 4 A, IV and V) in movement of mitochondria into the bud. Although mitochondria in myo2-I1308A mutants frequently moved to a position near the mother–bud neck, they did not move into the bud (Fig. 4 B, white arrow). That mitochondria eventually crossed the mother–bud neck in three of the six myo2-I1308A mutants fits the postulate that there are redundant mechanisms to move mitochondria across the mother–bud neck (Boldogh et al., 2001; Frederick et al., 2008; Kornmann and Walter, 2010). Alternatively, myo2-I1308A may be only partially defective in binding Mmr1. The time required for buds to inherit the vacuole in six of the myo2-I1308A mutants was similar to wild-type cells: ∼7 min. The defects observed in myo2-I1308A cells support the hypothesis that Mmr1 links the mitochondrial membrane to Myo2 for transport across the mother–bud neck.
Figure 4.
Myo2 mutations that disrupt only Vac17 or only Mmr1 binding exhibit a delay in vacuole or mitochondria inheritance, respectively. Vacuoles (FM4-64) and mitochondria (mitoGFP) in cells with myo2 mutants expressed from plasmids (CEN and HIS3) as the sole copy of MYO2. (A) t = 0, time that the first organelle (top bar) entered the bud. Arrowheads show the time that the lagging organelle entered the bud. (top) Five of six cells containing myo2-I1308A have a delay (I–III) or absence (IV and V) of mitochondria inheritance. VI was wild type–like. (bottom) Cells containing myo2-D1297N had no vacuole inheritance (VII–XI). (right) Time images per series. V or M indicates the number of times a vacuole tubule (V) or mitochondria tubule (M) crossed the mother–bud neck in series with <60 s between images. For myo2-I1308A, black numbers indicate the vacuole is inherited, whereas gray indicates a defect in mitochondria inheritance. For myo2-D1297N, black numbers indicate that mitochondria are inherited, whereas gray indicates a defect in vacuole inheritance. Dashed lines show no mitochondrial or vacuole tubule detected in the bud during the time course. Images for all acquired time-lapse series are presented in the JCB DataViewer. (B) Time-lapse images of A (IV). Arrow shows a mitochondrial tubule remaining in the mother cell, near the mother–bud neck. (C) Time-lapse images of A (VIII). (B and C) Closed arrowheads show vacuole or mitochondria tubule in bud. Exp s, seconds of exposure. Bars, 5 µm.
One myo2-I1308A mutant had normal inheritance of mitochondria (Fig. 4 A, VI). In this example, the vacuole was inherited late, at 12.3 min. This raises the possibility that Vac17 may also interact with Myo2 residue I1308, although this residue is not a major contributor to Vac17 binding.
In the five time-lapse series of the myo2-D1297N mutant, vacuoles were not inherited during imaging (observation time 20–35 min; Fig. 4 A, VII–XI). Thus, Myo2 interaction with Vac17 is essential for vacuole movement across the mother–bud neck.
In the myo2-D1297N mutant, in which vacuole inheritance is defective, mitochondrial tubules moved into the bud earlier. On average, mitochondria moved into the bud within 2–3 min from the start of imaging (Fig. 4, A and C) compared with 5–6 min for wild-type cells (Fig. 3 A). In addition, in time-lapse images acquired <60 s apart, there was less rocking, mitochondria crossed the mother–bud neck, a mean of three times in the myo2-D1297N mutant. Conversely, vacuoles crossed the mother–bud neck a mean of 3.4 times in the myo2-I1308A mutant. These findings strongly suggest that in Myo2 mutants in which Vac17 binding is compromised, Mmr1 has greater access to Myo2. Conversely, in mutants in which Mmr1 binding is compromised, Vac17 has greater access to Myo2. Thus, Vac17 and Mmr1 may compete for binding to Myo2.
Mmr1 and Vac17 compete for access to Myo2 in vivo
We tested whether Mmr1 and Vac17 compete for binding to Myo2 in vivo. We examined a large population of cells to determine whether myo2-D1297N, which has a defect in binding Vac17, has increased amounts of mitochondria in the bud. Conversely, we tested whether myo2-I1308A, which has a defect in binding Mmr1, has increased amounts of vacuoles in the bud. We measured the ratio of bud fluorescence to mother fluorescence for individual cells for both mitochondria (mitoGFP) and vacuoles (FM4-64) to compare relative volumes of each organelle. A minimum of one hundred small budded cells for each strain was analyzed (Fig. 5 A). Compared with vacuoles in wild-type cells, the ratio of fluorescence in the bud versus the mother in myo2-D1297N cells was significantly lower (0.029 ± 0.004 vs. wild type, 0.178 ± 0.010; P < 0.001), consistent with observations that myo2-D1297N has a vacuole inheritance defect. Interestingly, myo2-D1297N had increased mitochondria fluorescence in the bud compared with wild-type cells (0.151 ± 0.006 vs. 0.115 ± 0.002; P < 0.001). Thus, when Vac17 cannot bind Myo2, transport of mitochondria into buds is increased, which suggests that Mmr1 has greater access to Myo2.
Figure 5.
Vac17 competes with Mmr1 for binding Myo2 in vivo. (A) Mitochondria and vacuole inheritance measured with FM4-64 and mitoGFP, respectively. Small budded cells (bud diameter ≤1/3 mother diameter) scored for relative levels of fluorescence in the bud versus the mother. (right) Example of linear intensity scales for mitochondria and vacuoles in a wild-type small budded cell. For each cell, background was subtracted, and the bud–mother ratio of fluorescence at 528 nm (GFP) and 617 nm (FM4-64) was determined. Criteria for cells analyzed were as follows: (a) Bud in focus. (b) Both vacuoles and mitochondria visible in the mother cell. (c) Cell free of surrounding cells on at least two sides. Error bars represent SEMs; Student’s t test analysis. *, P < 0.05; ***, P < 0.001. n = 3; a minimum of 33 small budded cells per strain, per experiment; 100 total cells per strain. Dotted box of the acquired image corresponds to the same area as shown in the GFP signal box and FM4-64 signal box. Bar, 1 µm. (B) Quantitative fluorescence analysis of large budded/unbudded cells; 33 cells per strain, per experiment. Error bars show SEMs of three independent experiments, with the exception of myo2-I1308A (mitoGFP) in which two experiments are indicated (diamonds). (C) Western blot of cells transformed with empty vector, pMMR1-V5, or pVAC17 expressed from multicopy plasmids grown in log phase. (D–F) Overexpression of Vac17 or Mmr1 leads to reduced inheritance of mitochondria or vacuoles. (D) Mitochondria and vacuole inheritance assessed via mitoGFP and FM4-64. Error bars indicate SDs. (E and F) Fluorescence microscopy of wild-type cells transformed with pmitoGFP (LEU2) plus either pRS416 (empty vector), pMMR1-V5 (ADH1 promoter and URA3), or pVAC17 (ADH1 promoter and URA3). Cells were grown at least six doubling times before FM4-64 labeling. Arrows show mitochondria accumulation in the bud. Arrowhead shows accumulation of mitochondria in the mother cell at the mother–bud neck. Asterisk shows accumulation of vacuoles in the mother cell at the mother–bud neck. Bars, 5 µm.
In myo2-I1308A mutant cells, which have a defect in Mmr1 binding to Myo2, there was a lower ratio of mitochondria fluorescence in buds compared with wild-type cells (0.040 ± 0.002 vs. 0.115 ± 0.002; P < 0.001). This supports the hypothesis that Mmr1 interaction with Myo2 is important for mitochondria inheritance. Interestingly, buds in the myo2-I1308A mutant had an increase in vacuole fluorescence compared with wild-type cells (0.213 ± 0.010 vs. 0.178 ± 0.010; P < 0.05). These data suggest that when Myo2 cannot bind to Mmr1, a larger population of Myo2 binds Vac17, and thus, transport of vacuoles to the bud is increased. myo2-N1304D, a mutation at the center of both the Vac17 and Mmr1 binding sites, results in decreased inheritance of both vacuoles and mitochondria.
Excess vacuoles or mitochondria inherited by small buds of the myo2 point mutants persisted in large budded and unbudded cells (Fig. 5 B). Note that the FM4-64 present in these cells originated before the 3-h chase period, which shows that the altered organelle volume persists for at least one cell doubling. Therefore, changes in both vacuole and mitochondrial volume that occurred early in the cell cycle in the myo2-I1308A and myo2-D1297N mutants were not corrected to wild-type volumes at the point of cytokinesis. Thus, competition between Mmr1 and Vac17 for access to Myo2 is an important contributor to the modulation of organelle volume.
Additional evidence for competition between Mmr1 and Vac17 came from experiments of wild-type cells that overexpressed either MMR1 or VAC17 (Fig. 5 C). When MMR1 was overexpressed, mitochondria accumulated in the bud (Fig. 5, D and E). This fits with prior studies that Mmr1 elevation causes accumulation of mitochondria in the bud (Itoh et al., 2004; Frederick et al., 2008). Notably, there was a corresponding significant decrease (P < 0.05) in the amount of vacuoles present in the bud (Fig. 5 D). Conversely, overexpression of Vac17 led to higher levels of vacuoles, but fewer mitochondria, in the bud (Fig. 5, C and E).
Overexpression of MMR1 or VAC17 had additional consequences. Overexpression of Vac17 led to abnormal positioning of vacuoles in the mother cell to the anterior portion of the cell (Fig. 5 F). Similarly, overexpression of MMR1 disrupted the cortical distribution and caused an accumulation of mitochondria in the anterior portion of the mother cell (Fig. 5 E). These results provide further support for the hypotheses that Vac17 and Mmr1 link Myo2 to vacuoles and mitochondria, respectively, for transport of these organelles across the mother–bud neck. Together, analysis of mutations in myo2 that block binding of Mmr1 or Vac17 as well as overexpression of Mmr1 or Vac17 strongly suggests that Myo2 transports both mitochondria and vacuoles and, furthermore, that the volume of mitochondria and vacuoles inherited is partially determined by access of Mmr1 and Vac17 to Myo2.
Mmr1 and Vac17 compete for access to Myo2 in vitro
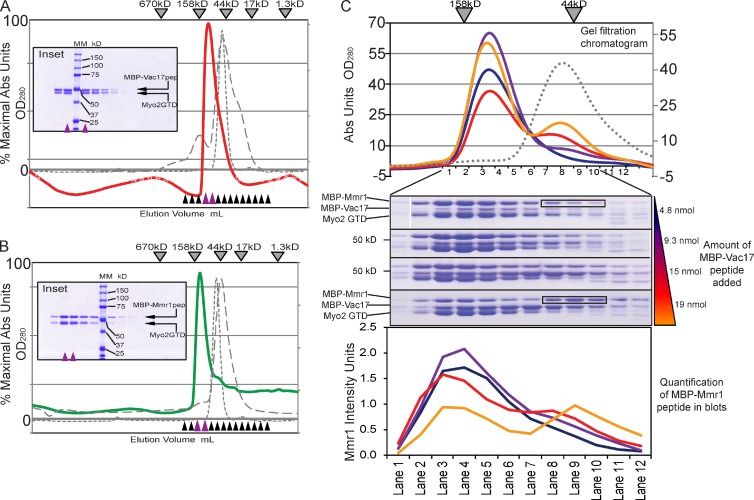
To test whether Myo2 interactions with Vac17 or Mmr1 can be assayed in vitro, we expressed full-length Vac17 and Mmr1 in Escherichia coli; however, neither protein was soluble. We used yeast two-hybrid analysis to identify peptides of Vac17 and Mmr1 that interacted with Myo2 (Fig. S2). Mmr1(378–430) was chosen because it interacted well with wild-type Myo2 but not Myo2-N1304D or Myo2-I1308A, mutants that disrupt full-length Mmr1 binding. Similarly, Vac17(112–157) interacted with Myo2 and Myo2-I1308A but not Myo2-N1304D, which does not bind full-length Vac17. Mmr1 and Vac17 peptides were expressed as fusion proteins with an N-terminal maltose-binding protein (MBP) tag.
Recombinant Myo2 CBD interacted with MBP-Vac17(112–157). Proteins in a 1:1 molar ratio were analyzed by size-exclusion chromatography. Myo2 CBD (50.5 kD) and MBP-Vac17(112–157) (53 kD) eluted as a complex of ∼115 kD (Fig. 6 A). Similarly, Myo2 CBD interacts with MBP-Mmr1(378–430) (55 kD) and migrates as a bimolecular complex of ∼120 kD (Fig. 6 B). MBP and Myo2 CBD in a 1:1 ratio eluted as unbound monomers (Fig. 6, A and B).
Figure 6.
Vac17 competes with Mmr1 for access to Myo2 in vitro. (A and B) Size-exclusion chromatography (Superdex 200, analytical column). Myo2 CBD and MBP-Vac17(112–157) (1:1 molar ratio; red line); MBP-Mmr1(387–430) (1:1 molar ratio; green line). (inset) Fractions on 10% SDS-PAGE gels. Purple arrowheads show the peak. Black and purple arrowheads show fractions tested on SDS-PAGE. Molecular mass is indicated. Elutions of Myo2 CBD monomer (gray dashed line) and MBP (42 kD) with Myo2 CBD (50.5 kD; gray dotted line). (C) MBP-Mmr1 competes with MBP-Vac17 for binding Myo2 CBD. Equal molar ratios of MBP-Mmr1 peptide (11.7 nmol) and Myo2 CBD (11.4 nmol) were added with increasing amounts of MBP-Vac17; 4.8, 9.3, 15, and 19 nmol (colored wedge) in separate runs. (top) Elutions. Gray dotted line shows MBP-Vac17 plus MBP-Mmr1 (1:1 molar ratio) as a control. (middle) Fractions 1–12 correspond with molecular masses from ∼180 to 30 kD. (bottom) Intensity of MBP-Mmr1 bands. Representative of two independent experiments. Black boxes show fractions on SDS-PAGE that correspond to the MBP-Mmr1 monomer peak of ∼50 kD shown in the chromatogram. Abs, absorbance.
To determine whether Vac17 and Mmr1 peptides compete for Myo2 in vitro, we formed a Myo2–Mmr1 complex and added MBP-Vac17. Increasing amounts of MBP-Vac17 resulted in the formation of Myo2–Vac17 complexes with the concomitant displacement of Mmr1 (Fig. 6 C). Thus, Mmr1 and Vac17 compete for access to Myo2 in vitro. MBP-Vac17 did not interact with MBP-Mmr1 and eluted as unbound monomers (Fig. 6 C).
Binding surfaces for Ypt11, Kar9, Inp2, and additional Rab GTPases partially overlap with each other
Ypt31/32 and Sec4 bind a site on Myo2 that is offset 180° from the Vac17/Mmr1 binding site. Inp2, which functions in peroxisome inheritance, binds a subset of these residues plus a few additional residues in this region (Fagarasanu et al., 2009).
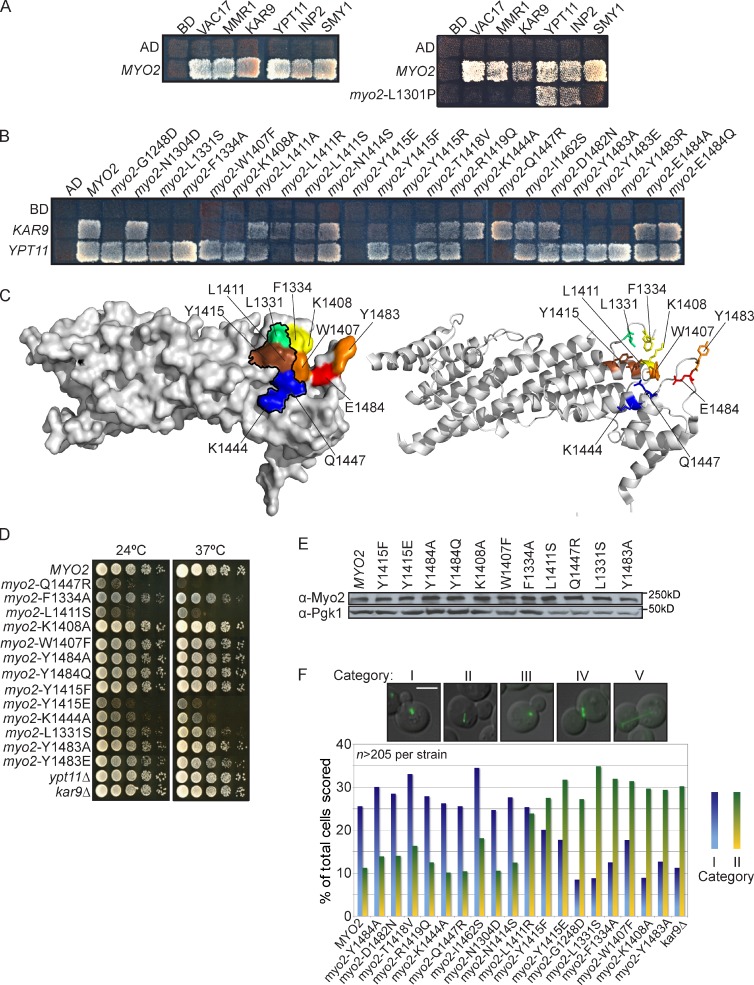
We tested additional proteins whose binding sites were unknown: Kar9, Ypt11, and Smy1 (Fig. 7 A). These proteins bound all Myo2 mutants in the Vac17/Mmr1-binding region. The one exception was myo2-L1301P, which was defective in binding to Smy1 and Kar9 (Fig. 7 A and Table S3). Because other mutations in this region bound to these proteins, it is likely that Smy1 and Kar9 binding are affected because of a conformational change. Some conformational changes affect only a subset of surface residues. For example, mutation of myo2-G1248D, a buried residue, affected Vac17 but not Rab GTPase binding (Fig. S1 D; Catlett and Weisman, 1998; Pashkova et al., 2006).
Figure 7.
Binding site for the post-Golgi Rab GTPases on Myo2 partially overlaps with the Ypt11, Kar9, and Inp2 binding sites. (A, left) Myo2 CBD interacts with Vac17, Mmr1, Kar9, Ypt11, Inp2, and Smy1 in a yeast two-hybrid test. Effects of L1301P are likely caused by conformational changes in the Myo2 CBD. (right) myo2-L1301P, helix 6, affects binding of Vac17, Mmr1, Kar9, and Smy1. Other mutations in this region do not affect Kar9 and Smy1. (B) Surface residues on Myo2 CBD critical for Kar9 and/or Ypt11 binding. (A and B) Incubated at 24°C for 4 d. Top left squares show empty vector controls. (C) Surface (left) and ribbon (right) view of Myo2 CBD indicating surface residues that interact with Kar9, Inp2, Ypt11, and Ypt31/32. Black outline shows Ypt31/32 and Sec4 binding sites. Red: Inp2 only; orange: Kar9 and Inp2; yellow: Kar9 only; green: Kar9, Sec4, and Ypt31/32; blue: Sec4, Ypt11, and Ypt31/32; brown: Kar9, Inp2, Sec4, Ypt11, and Ypt31/32. (D) Myo2 residues L1331, F1334, W1407, K1408, Y1415, and Y1483, the Kar9 binding site, are critical for orientation of the mitotic spindle. G1248D, an internal residue, also affected spindle orientation. myo2Δ cells with wild-type MYO2 or myo2 mutant plasmids transformed with pGFP-Tub1. Orientation of spindle microtubules scored in five categories: (I) Microtubules aligned toward the bud; (II) microtubules improperly aligned away from the mother–bud neck; categories III, IV, and V (Fig. S3) were not informative. Categories I and II were plotted as a percentage of total cells in all five categories. n = 2; ≥215 cells scored per strain. (E) Steady-state levels of Myo2 unchanged in plasmid-containing strains with myo2 point mutations. (F) Myo2 mutants that affect Kar9 and Inp2 interactions had no effect on growth. Mutations in the Rab GTPase binding site affect growth. Strains were grown to log phase, serially diluted, plated on rich media, incubated at 24°C or 37°C for 2–4 d. Bar, 5 µm. AD, activation domain; BD, binding domain.
To test whether Smy1 or Kar9 bind near the Ypt31/32, Sec4, or Inp2 binding sites, we tested 16 mutations near Myo2-Y1415, a surface residue critical for binding these proteins (Fig. 7 B, Fig. S3 A, and Table S3). Smy1 bound to all of the mutants tested; thus, the Smy1-binding region remains to be determined. Ypt11 binding was disrupted by most mutations that disrupt Myo2 interaction with the Rab GTPases Ypt31/32 and Sec4 (Fig. 7 C). However, Myo2-L1331, important for binding Ypt31/32 and Sec4, is not important for Ypt11 interaction with Myo2 (Fig. 7 C). myo2-L1331S expressed from a plasmid as the sole source of MYO2 causes a modest growth defect (Fig. 7d), consistent with a role for Myo2-L1331 in binding Ypt31/32 and/or Sec4. The growth defect was not caused by altered steady-state levels of Myo2-L1331S (Fig. 7 E). Thus, Myo2-L1331 may uniquely bind Ypt31/32 and Sec4, whereas residues L1411, Y1415, and K1444 bind the Rab GTPases that interact with Myo2. Deletion of Ypt11 does not affect yeast growth (Fig. 7 D).
In a yeast two-hybrid test, Kar9 failed to interact with two mutants defective in binding the Rab GTPases myo2-L1331S and myo2-Y1415E. Kar9 also failed to interact with myo2-F1334A, myo2-K1408A, and myo2-Y1483A. To test the functional significance of this interaction, we measured spindle orientation. In the absence of Kar9, there is only a partial defect in spindle orientation because of a parallel dynein-dependent pathway (Grava et al., 2006).
Myo2 mutants defective in binding to Kar9 had defects in orientation of the mitotic spindle. Asynchronous myo2Δ cells expressing a plasmid with wild-type or mutant myo2 were scored according to five categories. Orientations assigned to categories III, IV, and V were not informative and, while recorded (Fig. S3 B), were excluded from the analysis. Categories I and II were informative, and ∼35–50% of cells in both the mutants and wild type were assigned to these two groups (Fig. 7 F). Wild-type MYO2 plasmid–containing cells had a 2.5-fold higher percentage of correctly oriented spindle microtubules compared with kar9Δ cells. There was a direct correlation between mutations that disrupted Myo2–Kar9 interaction in the yeast two-hybrid test and those that cause misorientation of spindle microtubules in vivo. Mutations of residues adjacent to these mutations did not affect spindle orientation or Kar9 binding (Fig. S3 and Table S3). A mutation that disrupts vacuole and mitochondria inheritance, myo2-N1304D, also did not affect spindle orientation. These results demonstrate that Kar9, Inp2, Sec4, Ypt11, and Ypt31/32 bind to overlapping regions on the Myo2 CBD. Thus, of nine proteins known to interact with the Myo2 CBD, six overlap in this region.
Discussion
Characterization of organelle-specific cargo adaptors, which directly link cargoes to myosin V motors, revealed that organelle transport occurs via direct regulation of cargo adaptor proteins (Moore and Miller, 2007; Peng and Weisman, 2008; Fagarasanu et al., 2009). For example, initiation of vacuole movement requires Cdk1-dependent phosphorylation of the adaptor Vac17 (Peng and Weisman, 2008), which promotes Vac17 binding to Myo2 and vacuole movement. Vac17 regulation is also required for termination of vacuole movement (Tang et al., 2003). When the vacuole reaches the bud, a pathway that utilizes the Vac17 PEST sequence promotes the degradation of Vac17. In addition, Vac17 expression is coordinated with the cell cycle and peaks when the vacuole moves into the bud (Tang et al., 2003; Peng and Weisman, 2008).
Multiple modes of regulation of Vac17 suggested that cargo adaptors regulate cargo movement, whereas the motor provides an inert platform for cargo binding. In support of this postulate, two cargo adaptors, Vac17 and the secretory vesicle-specific Rabs Ypt31/32, bound distinct sites on Myo2. Thus, it was assumed that binding of each Myo2 adaptor would be independently regulated.
Evidence that the CBD of myosin V plays a role cargo selection came from discoveries that Xenopus laevis myosin V and yeast Myo2 undergo reversible phosphorylation (Rogers et al., 1999; Karcher et al., 2001; Legesse-Miller et al., 2006). Moreover, phosphorylation of the myosin V CBD in Xenopus oocytes regulates cargo interactions. Thus, phosphorylation of the myosin V CDB may facilitate the attachment or detachment of cargoes.
Further evidence that the CBD of Myo2 may regulate cargo selection was the finding that the Inp2 and Rab GTPase binding sites overlap (Fagarasanu et al., 2009). Kar9 and Ypt11 also interact with residues at this region (Fig. 7). Similarly, Mmr1 binds to a second site, which overlaps with the Vac17 binding site (Fig. 1). Overlap of eight cargo adaptors at two sites suggested that the overlap has a function.
While this paper was in revision, a new study showed that Pex19 binds to Myo2 at residue I1232, adjacent to the Mmr1/Vac17 binding site, and at I1440, adjacent to the Rab GTPase/Inp2/Kar9 site (Otzen et al., 2012). Thus, the complexity of interactions between cargo adaptors is even greater than described here.
We postulated that physical constraints allow only Mmr1 or Vac17 to occupy Myo2 at a given time. Indeed, this overlap contributes to the regulation of the inheritance of mitochondria and vacuoles (Fig. 5). One possibility was that competition between Mmr1 and Vac17 provided a temporal order for the inheritance of these organelles. However, mitochondria and vacuoles move into the bud at similar times (Fig. 3). Moreover, either the vacuole or mitochondria moved first, or both crossed the mother–bud neck simultaneously. Thus, Mmr1 and Vac17 have equal access to Myo2.
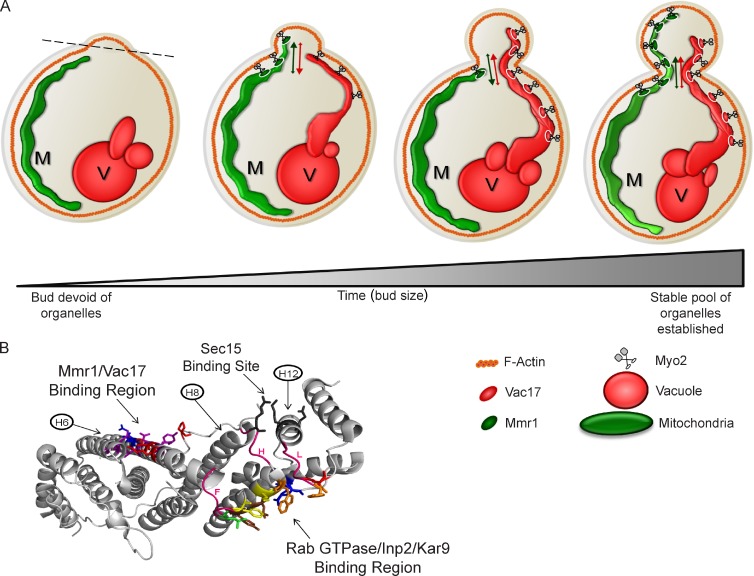
Despite similar access to Myo2, Mmr1 and Vac17 compete with each other in vivo and in vitro. The major significance of this competition is regulation of the volume of inherited vacuoles and mitochondria. Little is known about the regulation of organelle volume (Chan and Marshall, 2010). It had been assumed that organelle volume is regulated via a combination of organelle biogenesis and autophagy/turnover. Our experiments reveal an additional determinant based on the competition between cargo adaptor proteins for Myo2. A mutant that blocks mitochondria inheritance has an increased vacuole volume in the bud. Conversely, a mutant that blocks vacuole inheritance has an increased mitochondrial volume in the bud. This greater volume of mitochondria or vacuoles persists and is observed in large budded and unbudded cells. Thus, competition between Mmr1 and Vac17 for access to Myo2 plays a major role in regulating the volume of mitochondria and vacuoles (Fig. 8 A).
Figure 8.
Model for competition between Vac17 and Mmr1 for access to Myo2. (A) Myo2 transports vacuoles (V) and mitochondria (M). Mmr1/Myo2 localizes to the leading portion of the mitochondrial tubule in the mother near the mother–bud neck and on mitochondria in the bud. Mitochondria move across the mother–bud neck and retract into the mother several times then are stably anchored in the bud. Vac17/Myo2 localizes to the leading portion of vacuoles before entering the bud and on vacuoles in the bud. Vacuoles move across the mother–bud neck and retract into the mother several times and then are anchored in the bud. (B) All cargo-binding regions may be interconnected via conformational changes that modulate the ability of Myo2 to interact with each cargo adaptor. Myo2 CBD showing binding sites for Vac17/Mmr1, Rab GTPase/Inp2/Kar9, and Sec15. Three loops that connect these regions (pink) are labeled F, H, or L according to the 14 loops that appear in the CBD structure, from A to N, beginning at the N terminus. Vac17/Mmr1 binding site was predominantly on helix 6. Myo2-L1331 (lime green) lies within the Rab binding site at the C terminus of helix 6, in loop F. Two loops connect the Sec15 binding site with the Rab GTPase/Inp2/Kar9 binding region and extend from helix 8 (loop H) and helix 12 (loop L) to the Rab/Inp2/Kar9 region.
Both mitochondria and vacuoles undergo retrograde movement across the mother–bud neck. One possibility is that Myo2 stochastically releases cargoes through a loss of Myo2 attachment to the cargo adaptor. If true, there would be less rocking in mutants that affect only one cargo adaptor. Indeed, in both the myo2-D1297N and myo2-I1308 mutants, there are fewer retrograde movements of the inherited organelle. One interpretation is that the probability of reinitiation of Myo2 binding to Vac17 (for myo2-I1308A) or Mmr1 (for myo2-D1297N) is higher when there is not competition from the other cargo adaptor.
Mutants with defects in moving mitochondria or vacuoles often had portions of each organelle poised in the mother at the mother–bud neck. This suggests that attachment of the organelle to Myo2 is not completely blocked. Furthermore, a higher level of force might be needed to move organelles through the mother–bud neck. Perhaps fewer motors are required to bring a portion of the organelle to the mother–bud neck, and additional motors are required for movement through the mother–bud neck. The environment in the neck may contribute to the rocking motion.
Eventually, stable pools of mitochondria and vacuoles are established in the bud. Tethering proteins likely anchor these organelles. Mmr1 and Ypt11, as well as ERMES tethering of mitochondria to the endoplasmic reticulum, may perform this function for mitochondria. Similarly, there are likely tethers for the vacuole. Interestingly, we observed that overexpression of either VAC17 or MMR1 overcomes a tethering mechanism in the mother cell for vacuoles or mitochondria, respectively (Fig. 5).
That mitochondria and vacuoles move across the mother–bud neck at similar times and that Mmr1 and Vac17 appear to have equal access to a common binding region on Myo2 suggest that the cell cycle–dependent regulation of Vac17 and Mmr1 may be similar. Mmr1 may be the target of the same Cdk1–cyclin complexes that target Vac17 (Peng and Weisman, 2008); Mmr1 has seven Cdk1 consensus sites. Along similar lines, we predict that detachment of Mmr1 from Myo2 may use a similar mechanism to that observed with Vac17. Vac17 detachment from Myo2 requires the Vac17 PEST sequence. Similarly, Mmr1 contains two predicted PEST sequences (Fig. S2). Moreover, like Vac17, Myo2 mutations that block mitochondria inheritance result in an elevation of Mmr1.
Determination of the functional significance of the overlap of six cargo adaptors at the Rab GTPase binding site will be challenging. In addition to the Rab GTPase Ypt11 and the secretory vesicle Rab GTPases Ypt31/32 and Sec4, Kar9 and Inp2 bind at this region. Moreover, all cargo adaptor binding sites may modulate each other through structural changes in the Myo2 CBD.
Binding of the Rab GTPases Ypt31/32 and Sec4 may impact the Sec15 binding site and vice versa (Fig. 8 B). Loop H in the CBD structure contains a Sec15-binding residue and connects to helix 9, which has several residues critical for interaction with Kar9, Inp2, and the Rab GTPases. Helix 12 contributes residues to Sec15 binding, and loop L, which extends from this helix, contains residues important for Kar9 and Inp2 binding. Thus, binding of Inp2, Kar9, and/or Rab GTPases may change the Sec15 binding site or vice versa. This may be critical to the regulation of Myo2 attachment to secretory vesicles.
Similarly, the Mmr1/Vac17 binding region on helix 6 is connected to loop F, which contains residues in the Rab GTPase/Kar9/Inp2 binding site. Thus, binding of a Rab or other proteins to this region may affect the binding of Vac17 and Mmr1 and vice versa. In support of this hypothesis, mutation of myo2-L1301 on helix 6, which disrupts Myo2 interaction with Mmr1 and Vac17, also disrupts the binding of Kar9 and Smy1, which suggests that this mutation causes a distant conformational change of the surface residues in the Kar9-binding region.
Secretory vesicles are the only known essential cargo of Myo2. Thus, it is tempting to speculate that Ypt31/32 and Sec4, required for secretory vesicle interaction with Myo2, may have priority to occupy the CBD. Thus, cargo transport may be coordinated in part through structural changes in the myosin V CBD that occur through long distance communication between each of the adaptor binding sites.
Materials and methods
Yeast strains and plasmids
Yeast strains and plasmids are listed in Tables S1 and S2. Strains were grown at 24°C unless indicated. The mmr1Δ strain was constructed using PCR amplification of the endogenous kanamycin cassette from the YLR190W haploid knockout strain (Thermo Fisher Scientific), transformed into a diploid wild-type strain LWY9087, and sporulated to obtain the haploid knockout. ypt11Δ was constructed by PCR amplification of the endogenous kanamycin cassette from the YNL304W haploid knockout strain (Thermo Fisher Scientific) and transformed into the haploid wild-type yeast strain LWY7235. Vac17-3×GFP was constructed using PCR with the plasmid PB1960 (obtained from D. Pellman, Harvard University, Cambridge, MA) and transformed into diploid yeast, sporulated, and dissected (Jin et al., 2009). Vac17-3×GFP was used because endogenous Vac17 is of low abundance: ∼20 copies per cell (Tang et al., 2006). Mmr1-GFP was constructed using PCR amplification of the endogenous GFP-HIS3 cassette containing flanking ends from the YLR190W ORF strain (Invitrogen) and transformed into the haploid wild-type yeast strain (Jin et al., 2009). Mmr1-GFP is functional; cells expressing Mmr1-GFP expressed from the endogenous locus have normal inheritance of their mitochondria. In the Mmr1-GFP strain (LWY8867) transformed with mitoRFP (2µ and URA3), 80% of small and medium buds inherit mitochondria versus wild-type cells transformed with mitoRFP (2µ and URA3), which inherit mitochondria in 84% of small and medium buds (three independent experiments; a minimum of 33 cells analyzed per strain per experiment). mitoRFP and mitoGFP plasmids were obtained from J. Shaw (University of Utah, Salt Lake City, UT). Microtubules were visualized using the Tub1-GFP plasmid pBJ1333 obtained from J. Cooper (Washington University, St. Louis, MO).
For generation of pRS416 MMR1-V5, a 3.1-kb KpnI–SacII fragment of MMR1 was subcloned into pBlueScript SK+ (pBS) at KpnI and SacII sites to generate pBS MMR1. A BglII site was generated at the C terminus of MMR1 by PCR using primers 5′-GGAGAAGAAGGAAAAAaGAtctGTCAACTTCAAATTAAATTAAC-3′ and 5′-GTTAATTTAATTTGAAGTTGACagaTCtTTTTTCCTTCTTCTCC-3′ to generate pBS MMR1-BglII (lowercase letters indicate that there are deletions or mutations within the gene). To insert a V5 tag into the BglII site of pBS MMR1-BglII, annealed V5 primers encoded the V5 tag were used, 5′-GATCTGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGTGAG-3′ and 5′-GATCCTCACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCA-3′, to generate pBS MMR1-V5. The resulting KpnI–SacII fragment was subcloned from pBS MMR1-V5 into pRS416 at the KpnI and SacII sites.
Strains were grown in rich media (1% yeast extract, 2% peptone, and 2% dextrose), synthetic media (2% dextrose) lacking the indicated amino acids, or 5-fluoroorotic acid. myo2 mutations were created with the site-directed mutagenesis kit (QuikChange II; Agilent Technologies) following the manufacturer’s protocol. For myo2 plasmid–carrying strains, myo2Δ cells containing the MYO2 (URA2 and centromere [CEN]) plasmid were transformed with mutant myo2 plasmids (HIS3 and CEN) and grown on 5-fluoroorotic acid. myo2 plasmids in myo2Δ cells were maintained on rich media. Crystal structure representations of the Myo2 CBD (Protein Data Bank accession no. 2F6H; Pashkova et al., 2006) were made using PyMOL Molecular Graphics System, Version 1.3 (Schrödinger, LLC) using the cartoon view.
Microscopy
Cultures were maintained at or below 5 × 106 cells/ml for a minimum of six doubling times. Mitochondria were visualized by transformation of a mitochondrial-specific targeting peptide fused to GFP or RFP: mitoGFP/mitoRFP (Frederick et al., 2008). For vacuoles, cells were incubated with 60 µg FM4-64 for 45 min to 1 h in 2.5 ml media, washed twice, and grown in 5 ml media for one doubling time (1.5–3 h). Images were acquired on a DeltaVision Restoration system (Applied Precision) using an inverted epifluorescence microscope (IX-71; Olympus) with a charge-coupled device camera (CoolSNAP HQ; Photometrics). Wide-field epifluorescence images were acquired without deconvolution using a 100× Plan Apochromat objective (Olympus). After acquisition, fluorescence intensity was adjusted and applied to all images using softWoRx suite 3.5.1 (DeltaVision). Images converted to TIF format were overlaid and cropped in Photoshop (Adobe). All budded cells in a given field were analyzed/scored if the bud was in focus using differential interference contrast, and the mother cell contained visible mitochondria and vacuoles that were in focus using fluorescence. Mitochondria inheritance was scored in small and medium budded cells according to four categories: (I) mitochondria in the mother cell progressed to the mother–bud neck only; (II) a small amount of mitochondria concentrated to the bud tip but not elsewhere in the bud; (III) mitochondria progressed into the bud but no more than halfway to the bud tip; and (IV) mitochondria distributed throughout the cortex up to the bud tip. Categories III and IV were scored as wild-type phenotypes. Small bud diameter was greater than one third the diameter of the mother cell. Medium buds were one third to one half the diameter of the mother cell. Large buds were greater than one half the diameter of the mother. The large budded cell population included both budded and unbudded cells. The larger diameter cell in large budded cells was assumed to be the mother. For quantitative fluorescence analysis, the softWoRx Data Inspector tool was used concurrently with the Measure Distance tool. For each channel, individual measurements of bud cell fluorescence were divided by mother cell fluorescence after subtracting the background. Background was defined for each cell per channel by measuring detectable fluorescence next to the cell (Waters, 2009).
Live-cell time-lapse imaging
FM4-64 labeling was modified. Cells were labeled with 125 µg FM4-64 in 3 ml of fresh media for 1 h. Cells were centrifuged once, supernatant was aspirated completely, and cells were resuspended in 5 ml of fresh media (without washing). Cells were grown for one doubling period (2–3 h). Glass-bottom chambers (Lab-Tek II; Thermo Fisher Scientific) were treated overnight at 4°C with concanavalin A dissolved at 1 mg/ml in 50-mM Hepes, pH 7.5, 20 mM calcium acetate, and 1 mM manganese sulfate and then air dried for ≥30 min. Cells adsorbed to concanavalin A–treated chambers for 2 min. Unbound cells were removed by aspiration, and 250 µl of fresh media was added. Cells were imaged at 23–24.5°C. Three criteria were applied to the cells imaged: (1) at the start, both organelles were absent from the bud; (2) both organelles were visible in the mother; and (3) the cell remained in focus during imaging.
Yeast two-hybrid assays
S. cerevisiae strain PJ69-4A (James et al., 1996) was cotransformed with LEU2 and TRP1 plasmids containing the GAL4 transcription factor activation domain and binding domain, respectively. Colonies were grown for 3–4 d at 24°C on synthetic complete (SC)-Leu-Trp media and patched onto SC-Leu-Trp agar plates before replica plating to selection media: SC-Leu-Trp, SC-Leu-Trp-Ade-His, and SC-Leu-Trp-Ade-His + 3AT (3 mM 3-aminotriazole). Plates were imaged after 3–10 d. Contrast and intensity settings were adjusted in Photoshop using the same settings for all images from a single experiment.
Western blot
TCA precipitation/NaOH method of protein extraction from whole cells was used. In brief, 4 × 108 cells were collected, washed with distilled, deionized H2O, and resuspended in ice-cold 0.2 M NaOH. 50 µl of 100% TCA was added, and the sample was centrifuged. 50–200 µl of 2×SDS loading buffer was added to the pellet, which was incubated at 70°C, 5 min before loading on SDS-PAGE. Goat anti-Myo2 antibodies were affinity purified and used at 1:1,500. Mouse monoclonal antibodies, anti-GFP (Roche), and anti-HA (Covance) were used at 1:5,000 and 1:500, respectively. Mouse monoclonal anti-Pgk1 (Invitrogen) was used at 1:10,000. Mouse anti-V5 antibody (Invitrogen) was used at 1:1,000. Sheep anti-Vac17 antibody was used at 1:500 (Tang et al., 2003). HRP-conjugated donkey anti–goat antibody IgG, HRP-conjugated goat anti–sheep antibody IgG, and HRP-conjugated goat anti–mouse IgG (Jackson ImmunoResearch Laboratories, Inc.) were used at 1:5,000.
In vitro purification and binding and competition analysis with size-exclusion chromatography
Plasmids pMBP-Vac17(112–157)p, pMBP-Mmr1(378–430)p, and pGST-Myo2(1,131–1,574)p were transformed into E. coli strain BL21/DE3/STAR and grown to an Abs600nm of 1.00 ± 0.1 at 37°C in Terrific broth (Invitrogen) containing ampicillin and spectinomycin, each 100 µg/ml. Cells were cooled for 15 min and then incubated at 20°C for 14 h in 1 mM isopropyl-β-d-thio-galactoside to induce expression. Cells were harvested at 4°C. All subsequent steps took place at 0–4°C. Cells were resuspended in 40 ml lysis buffer containing 20 mM Hepes, pH 7.9, 200 mM NaCl, 1 mM EDTA, and 0.05% β-mercaptoethanol. Lysis was performed using a probe sonicator with six to eight repetitions of 10–15 s, 50% intensity, in the presence of protease inhibitor cocktail (without EDTA; one tablet per 40 ml lysis buffer; Roche) and1 µM Pefabloc SC (Roche). Lysates were centrifuged at 40,000 rpm for 90 min in a rotor (Ti45; Beckman Coulter). Lysate supernatants were incubated with the affinity matrix.
For GST-Myo2 CBD, the following was performed: Supernatant was incubated with 2.5 ml glutathione–Sepharose 4B resin (GE Healthcare) for 1 h. GST was cleaved from the Myo2 CBD via incubation with 5 µl biotinylated thrombin (Thrombin Cleavage Capture kit; EMD) for 3.5 h in 1.5 ml thrombin buffer. Biotinylated thrombin was removed with streptavidin-linked agarose (EMD). For MBP-conjugated peptides the following was performed: Supernatants were incubated with 3.0 ml amylose resin (New England BioLabs, Inc.) for 1 h. Resin was centrifuged, collected in 10 ml Polyprep columns (Bio-Rad Laboratories), and washed in lysis buffer. MBP peptides were eluted using lysis buffer containing 10 mM maltose.
Myo2 CBD and MBP peptides were purified on a prep-grade exclusion column (HiLoad 16/60 Superdex 200; GE Healthcare). Purified proteins were analyzed on SDS–10% PAGE and visualized with Coomassie. Both bicinchoninic acid (Thermo Fisher Scientific) and Bradford (Bio-Rad Laboratories) assays were used to verify protein concentrations; a BSA standard curve was generated for each analysis. Myo2 CBD and MBP peptide–binding assays and competition assays were performed on an analytical column (HiLoad 16/60 Superdex 200).
Visualization of filamentous actin in fixed cells
Actin structures were visualized in fixed cells using rhodamine-phalloidin. In brief, 5 × 106 cells/ml in log phase were resuspended in 3.7% formaldehyde (final concentration) and incubated at RT for 10 min, washed, and resuspended in PBS, pH 7.4, containing 3.7% formaldehyde. Rhodamine-phalloidin (Invitrogen) was dissolved in methanol, and100 µl of fixed cells was mixed with 6.1 µM rhodamine-phalloidin (final concentration).
Online supplemental material
Fig. S1 shows surface residues on Myo2 that comprise the Vac17 and Mmr1 binding sites. Fig. S2 shows Mmr1 and Vac17 peptides that bind Myo2 CBD. Fig. S3 shows additional experiments of the binding surface near the Rab GTPase binding site. Table S1 shows yeast strains used in this study. Table S2 shows plasmids used in this study. Table S3 shows a summary of yeast two-hybrid and in vivo analyses. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201201024/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Natasha Pashkova for initial studies that myo2-L1229R and myo2-E1293K have defects in binding Vac17 and vacuole inheritance. We thank Drs. Janet Shaw for pmitoGFP and pmitoRFP plasmids and John Cooper for pTub1-GFP plasmids. We thank members of the Weisman Laboratory for helpful discussions.
This work was supported by National Institutes of Health grant R37-GM066261 to L.S. Weisman. P.T. Eves was supported in part through National Institutes of Health grant T32-GM007315.
Footnotes
Abbreviations used in this paper:
- CBD
- cargo-binding domain
- CEN
- centromere
- ERMES
- ER–mitochondrial encounter structure
- MBP
- maltose-binding protein
- pBS
- pBlueScript SK+
- SC
- synthetic complete
References
- Altmann K., Frank M., Neumann D., Jakobs S., Westermann B. 2008. The class V myosin motor protein, Myo2, plays a major role in mitochondrial motility in Saccharomyces cerevisiae. J. Cell Biol. 181:119–130 10.1083/jcb.200709099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S., Noda Y., Kainuma S., Wada I., Yoda K. 2008. Ypt11 functions in bud-directed transport of the Golgi by linking Myo2 to the coatomer subunit Ret2. Curr. Biol. 18:987–991 10.1016/j.cub.2008.06.028 [DOI] [PubMed] [Google Scholar]
- Beach D.L., Thibodeaux J., Maddox P., Yeh E., Bloom K. 2000. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr. Biol. 10:1497–1506 10.1016/S0960-9822(00)00837-X [DOI] [PubMed] [Google Scholar]
- Boldogh I.R., Yang H.C., Pon L.A. 2001. Mitochondrial inheritance in budding yeast. Traffic. 2:368–374 10.1034/j.1600-0854.2001.002006368.x [DOI] [PubMed] [Google Scholar]
- Boldogh I.R., Ramcharan S.L., Yang H.C., Pon L.A. 2004. A type V myosin (Myo2p) and a Rab-like G-protein (Ypt11p) are required for retention of newly inherited mitochondria in yeast cells during cell division. Mol. Biol. Cell. 15:3994–4002 10.1091/mbc.E04-01-0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot Frei S., Rahl P.B., Nussbaum M., Briggs B.J., Calero M., Janeczko S., Regan A.D., Chen C.Z., Barral Y., Whittaker G.R., Collins R.N. 2006. Bioinformatic and comparative localization of Rab proteins reveals functional insights into the uncharacterized GTPases Ypt10p and Ypt11p. Mol. Cell. Biol. 26:7299–7317 10.1128/MCB.02405-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett N.L., Weisman L.S. 1998. The terminal tail region of a yeast myosin-V mediates its attachment to vacuole membranes and sites of polarized growth. Proc. Natl. Acad. Sci. USA. 95:14799–14804 10.1073/pnas.95.25.14799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett N.L., Duex J.E., Tang F., Weisman L.S. 2000. Two distinct regions in a yeast myosin-V tail domain are required for the movement of different cargoes. J. Cell Biol. 150:513–526 10.1083/jcb.150.3.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.H., Marshall W.F. 2010. Scaling properties of cell and organelle size. Organogenesis. 6:88–96 10.4161/org.6.2.11464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A., Fagarasanu M., Eitzen G.A., Aitchison J.D., Rachubinski R.A. 2006. The peroxisomal membrane protein Inp2p is the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev. Cell. 10:587–600 10.1016/j.devcel.2006.04.012 [DOI] [PubMed] [Google Scholar]
- Fagarasanu A., Mast F.D., Knoblach B., Jin Y., Brunner M.J., Logan M.R., Glover J.N., Eitzen G.A., Aitchison J.D., Weisman L.S., Rachubinski R.A. 2009. Myosin-driven peroxisome partitioning in S. cerevisiae. J. Cell Biol. 186:541–554 10.1083/jcb.200904050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förtsch J., Hummel E., Krist M., Westermann B. 2011. The myosin-related motor protein Myo2 is an essential mediator of bud-directed mitochondrial movement in yeast. J. Cell Biol. 194:473–488 10.1083/jcb.201012088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick R.L., McCaffery J.M., Cunningham K.W., Okamoto K., Shaw J.M. 2004. Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway. J. Cell Biol. 167:87–98 10.1083/jcb.200405100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick R.L., Okamoto K., Shaw J.M. 2008. Multiple pathways influence mitochondrial inheritance in budding yeast. Genetics. 178:825–837 10.1534/genetics.107.083055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan B., Bowser R., Novick P. 1995. The role of Myo2, a yeast class V myosin, in vesicular transport. J. Cell Biol. 128:1055–1068 10.1083/jcb.128.6.1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grava S., Schaerer F., Faty M., Philippsen P., Barral Y. 2006. Asymmetric recruitment of dynein to spindle poles and microtubules promotes proper spindle orientation in yeast. Dev. Cell. 10:425–439 10.1016/j.devcel.2006.02.018 [DOI] [PubMed] [Google Scholar]
- Hill K.L., Catlett N.L., Weisman L.S. 1996. Actin and myosin function in directed vacuole movement during cell division in Saccharomyces cerevisiae. J. Cell Biol. 135:1535–1549 10.1083/jcb.135.6.1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D., van den Berg M., Philippsen P., Tabak H.F., Hettema E.H. 2001. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155:979–990 10.1083/jcb.200107028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K., Catlett N.L., Novak J.L., Tang F., Nau J.J., Weisman L.S. 2003. Identification of an organelle-specific myosin V receptor. J. Cell Biol. 160:887–897 10.1083/jcb.200210139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Watabe A., Toh-E A., Matsui Y. 2002. Complex formation with Ypt11p, a rab-type small GTPase, is essential to facilitate the function of Myo2p, a class V myosin, in mitochondrial distribution in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:7744–7757 10.1128/MCB.22.22.7744-7757.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Toh-E A., Matsui Y. 2004. Mmr1p is a mitochondrial factor for Myo2p-dependent inheritance of mitochondria in the budding yeast. EMBO J. 23:2520–2530 10.1038/sj.emboj.7600271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E.A. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 144:1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Taylor Eves P., Tang F., Weisman L.S. 2009. PTC1 is required for vacuole inheritance and promotes the association of the myosin-V vacuole-specific receptor complex. Mol. Biol. Cell. 20:1312–1323 10.1091/mbc.E08-09-0954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Sultana A., Gandhi P., Franklin E., Hamamoto S., Khan A.R., Munson M., Schekman R., Weisman L.S. 2011. Myosin V transports secretory vesicles via a Rab GTPase cascade and interaction with the exocyst complex. Dev. Cell. 21:1156–1170 10.1016/j.devcel.2011.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher R.L., Roland J.T., Zappacosta F., Huddleston M.J., Annan R.S., Carr S.A., Gelfand V.I. 2001. Cell cycle regulation of myosin-V by calcium/calmodulin-dependent protein kinase II. Science. 293:1317–1320 10.1126/science.1061086 [DOI] [PubMed] [Google Scholar]
- Korinek W.S., Copeland M.J., Chaudhuri A., Chant J. 2000. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science. 287:2257–2259 10.1126/science.287.5461.2257 [DOI] [PubMed] [Google Scholar]
- Kornmann B., Walter P. 2010. ERMES-mediated ER-mitochondria contacts: molecular hubs for the regulation of mitochondrial biology. J. Cell Sci. 123:1389–1393 10.1242/jcs.058636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., Walter P. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325:477–481 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B., Osman C., Walter P. 2011. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc. Natl. Acad. Sci. USA. 108:14151–14156 10.1073/pnas.1111314108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legesse-Miller A., Zhang S., Santiago-Tirado F.H., Van Pelt C.K., Bretscher A. 2006. Regulated phosphorylation of budding yeast’s essential myosin V heavy chain, Myo2p. Mol. Biol. Cell. 17:1812–1821 10.1091/mbc.E05-09-0872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z., Tokarev A.A., Jin Y., Mulholland J., Weisman L.S., Segev N. 2008. Direct interaction between a myosin V motor and the Rab GTPases Ypt31/32 is required for polarized secretion. Mol. Biol. Cell. 19:4177–4187 10.1091/mbc.E08-02-0220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast F.D., Rachubinski R.A., Dacks J.B. 2012. Emergent complexity in Myosin V-based organelle inheritance. Mol. Biol. Evol. 29:975–984 10.1093/molbev/msr264 [DOI] [PubMed] [Google Scholar]
- Miller R.K., Cheng S.C., Rose M.D. 2000. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol. Biol. Cell. 11:2949–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.K., Miller R.K. 2007. The cyclin-dependent kinase Cdc28p regulates multiple aspects of Kar9p function in yeast. Mol. Biol. Cell. 18:1187–1202 10.1091/mbc.E06-04-0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otzen M., Rucktäschel R., Thoms S., Emmrich K., Krikken A.M., Erdmann R., van der Klei I.J. 2012. Pex19p contributes to peroxisome inheritance in the association of peroxisomes to Myo2p. Traffic. [DOI] [PubMed] [Google Scholar]
- Pashkova N., Jin Y., Ramaswamy S., Weisman L.S. 2006. Structural basis for myosin V discrimination between distinct cargoes. EMBO J. 25:693–700 10.1038/sj.emboj.7600965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Weisman L.S. 2008. The cyclin-dependent kinase Cdk1 directly regulates vacuole inheritance. Dev. Cell. 15:478–485 10.1016/j.devcel.2008.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peraza-Reyes L., Crider D.G., Pon L.A. 2010. Mitochondrial manoeuvres: latest insights and hypotheses on mitochondrial partitioning during mitosis in Saccharomyces cerevisiae. Bioessays. 32:1040–1049 10.1002/bies.201000083 [DOI] [PubMed] [Google Scholar]
- Rogers S.L., Karcher R.L., Roland J.T., Minin A.A., Steffen W., Gelfand V.I. 1999. Regulation of melanosome movement in the cell cycle by reversible association with myosin V. J. Cell Biol. 146:1265–1276 10.1083/jcb.146.6.1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossanese O.W., Reinke C.A., Bevis B.J., Hammond A.T., Sears I.B., O’Connor J., Glick B.S. 2001. A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J. Cell Biol. 153:47–62 10.1083/jcb.153.1.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Tirado F.H., Legesse-Miller A., Schott D., Bretscher A. 2011. PI4P and Rab inputs collaborate in myosin-V-dependent transport of secretory compartments in yeast. Dev. Cell. 20:47–59 10.1016/j.devcel.2010.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard K.A., Gerber A.P., Jambhekar A., Takizawa P.A., Brown P.O., Herschlag D., DeRisi J.L., Vale R.D. 2003. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. USA. 100:11429–11434 10.1073/pnas.2033246100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne T.C., Zhou C., Boldogh I.R., Charalel J.K., McFaline-Figueroa J.R., Thoms S., Yang C., Leung G., McInnes J., Erdmann R., Pon L.A. 2011. Role for cER and Mmr1p in anchorage of mitochondria at sites of polarized surface growth in budding yeast. Curr. Biol. 21:1994–1999 10.1016/j.cub.2011.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Kauffman E.J., Novak J.L., Nau J.J., Catlett N.L., Weisman L.S. 2003. Regulated degradation of a class V myosin receptor directs movement of the yeast vacuole. Nature. 422:87–92 10.1038/nature01453 [DOI] [PubMed] [Google Scholar]
- Tang F., Peng Y., Nau J.J., Kauffman E.J., Weisman L.S. 2006. Vac8p, an armadillo repeat protein, coordinates vacuole inheritance with multiple vacuolar processes. Traffic. 7:1368–1377 10.1111/j.1600-0854.2006.00458.x [DOI] [PubMed] [Google Scholar]
- Waters J.C. 2009. Accuracy and precision in quantitative fluorescence microscopy. J. Cell Biol. 185:1135–1148 10.1083/jcb.200903097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman L.S. 2006. Organelles on the move: insights from yeast vacuole inheritance. Nat. Rev. Mol. Cell Biol. 7:243–252 10.1038/nrm1892 [DOI] [PubMed] [Google Scholar]
- Yin H., Pruyne D., Huffaker T.C., Bretscher A. 2000. Myosin V orientates the mitotic spindle in yeast. Nature. 406:1013–1015 10.1038/35023024 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.