In contrast to what has been reported previously, endo-lysosomal dysfunction in presenilin-deficient cells does not arise from improper glycosylation of the lysosomal V-ATPase but rather from defective lysosomal calcium homeostasis.
Abstract
Presenilin (PSEN) deficiency is accompanied by accumulation of endosomes and autophagosomes, likely caused by impaired endo-lysosomal fusion. Recently, Lee et al. (2010. Cell. doi: http://dx.doi.org/10.1016/j.cell.2010.05.008) attributed this phenomenon to PSEN1 enabling the transport of mature V0a1 subunits of the vacuolar ATPase (V-ATPase) to lysosomes. In their view, PSEN1 mediates the N-glycosylation of V0a1 in the endoplasmic reticulum (ER); consequently, PSEN deficiency prevents V0a1 glycosylation, compromising the delivery of unglycosylated V0a1 to lysosomes, ultimately impairing V-ATPase function and lysosomal acidification. We show here that N-glycosylation is not a prerequisite for proper targeting and function of this V-ATPase subunit both in vitro and in vivo in Drosophila melanogaster. We conclude that endo-lysosomal dysfunction in PSEN−/− cells is not a consequence of failed N-glycosylation of V0a1, or compromised lysosomal acidification. Instead, lysosomal calcium storage/release is significantly altered in PSEN−/− cells and neurons, thus providing an alternative hypothesis that accounts for the impaired lysosomal fusion capacity and accumulation of endomembranes that accompanies PSEN deficiency.
Introduction
Presenilin (PSEN) deficiency delays the turnover of long-lived proteins, a defect postulated to arise from impaired lysosomal fusion and disruption of macroautophagy-mediated degradation (Annaert et al., 2001; Esselens et al., 2004; Wilson et al., 2004; Neely et al., 2011). Recently, Lee et al. (2010) explained these phenomena through a novel role of PSEN1 in chaperoning the transfer of oligosaccharide precursors to V0a1 by interacting with the oligosaccharyl–transferase (OST) complex during cotranslational translocation in the rough ER. V0a1 is one isoform of the largest subunit of the vacuolar ATPase (V-ATPase) complex, required for acidifying intracellular compartments including endosomes and lysosomes (Toei et al., 2010). According to Lee et al. (2010), PSEN1 deficiency leads to failed transfer of N-glycans to V0a1, resulting in its ER retention, improper targeting to lysosomes causing decreased lysosomal acidification, and subsequent defects in protein turnover. However, the present reinvestigation failed to support this hypothesis. Instead, we demonstrate that Ca2+ homeostasis, rather than lysosomal acidification, is affected. This may explain the observed endolysosomal defects.
Results and discussion
PSEN deficiency does not lead to acidification defects
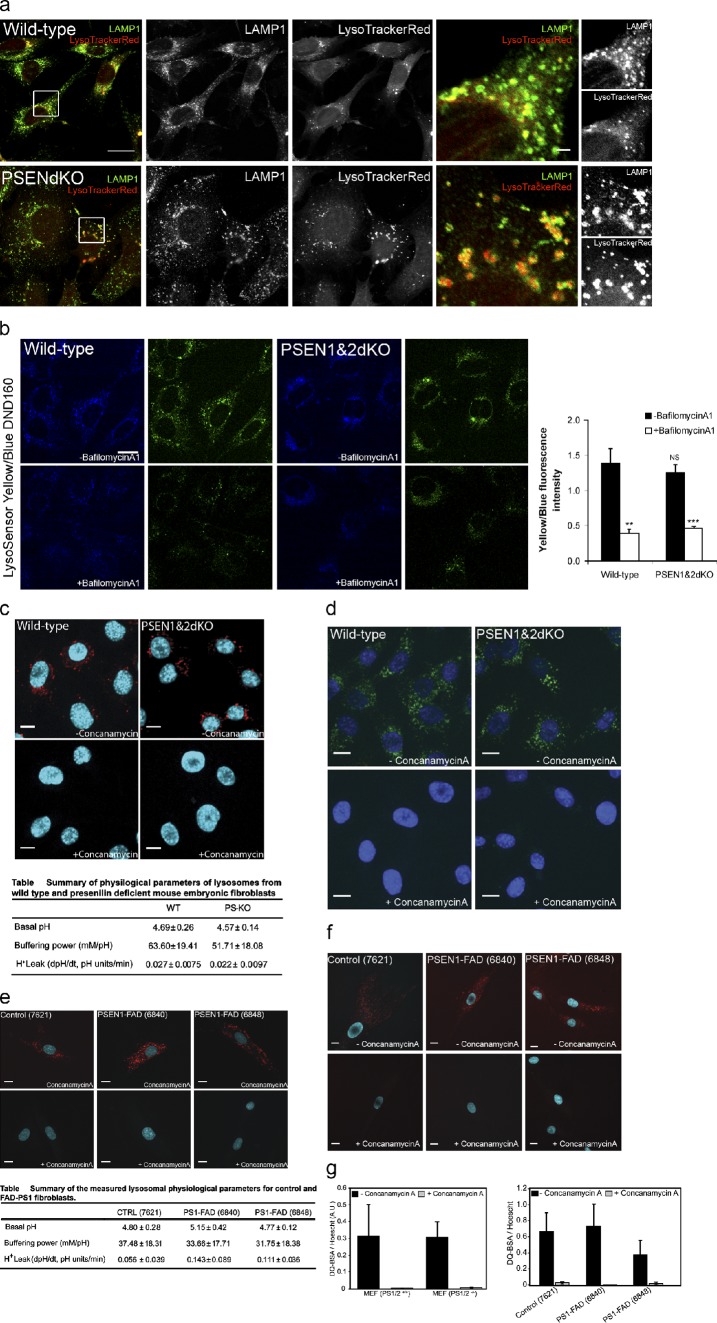
Unlike Lee et al. (2010), we failed to detect any differences in vesicular acidification between wild-type (WT) and PSEN1&2 knockout (PSENdKO) mouse embryonic fibroblasts (MEFs), in either fixed (Fig. 1 a) or live cells (Fig.1 c). LAMP1 immunostaining indicated that LysoTracker-positive vesicles are late endosomal/lysosomal in nature. The loss of staining after treatment with concanamycin A confirmed that LysoTracker accumulation was in all cases caused by functional V-ATPases (Fig. 1 c). We were similarly unable to detect significant differences between WT, PSEN1−/−, and PSENdKO murine blastocysts (Fig. S1, a and b), the cells used by Lee et al. (2010), ruling out cell type-specific differences as an explanation for the apparent discrepancy.
Figure 1.
Lysosomal acidification is not affected in PSENdKO MEFs. (a) LysoTracker and LAMP1 immunostaining showed an accumulation of enlarged degradative organelles in PSENdKO MEFs (indicated by the boxed regions). (b) Lysosomal pH, determined using LysoSensor, is not affected in PSENdKO MEFs (n = 3). Bafilomycin A1 was used to suppress V-ATPase–driven acidification (n = 2; error bars indicate mean ± SEM; >75 cells/experiment). (c) WT and PSENdKO cells were treated with vehicle (top) or concanamycin A (bottom) and stained with LysoTracker and Hoechst. The table summarizes the physiological properties and pH of lysosomes (n = 6; ≥20 lysosomes from multiple cells/experiment, unpaired t test). (d) MEFs were loaded with self-quenching probe DQ-BSA green or red, and cells treated without (top) and with concanamycin A (bottom) are shown. (e) Control and FAD-PSEN1 mutant fibroblasts treated with vehicle (top) or concanamycin A (bottom), stained with LysoTracker and Hoechst. The table summarizes the physiological properties and pH of lysosomes (n = 4; ≥20 lysosomes from multiple cells/experiment, ANOVA followed by Bonferroni’s contrast test). (f) Control and FAD fibroblasts loaded with DQ-BSA red and treated with vehicle (top) or concanamycin A (bottom). (g) Quantitative analysis of DQ-BSA fluorescence normalized to nuclear staining (error bars indicate mean ± SEM, n = 3, >12 cells). Bars: (a) 10 µm; (a, insets) 1 µm; (c, d, e, and f) 7.2 µm.
Because LysoTracker provides only a qualitative assessment of pH, we used two ratiometric approaches to independently quantify lysosomal acidification. First, the acidotropic dye LysoSensor yellow/blue DND160 did not reveal significant differences in acidification between PSENdKO and WT MEFs (Fig. 1 b). The V-ATPase inhibitor, bafilomycin A, caused a significant and comparable drop in the fluorescence ratio for both cell lines, implying that the observed alkalinization was caused by inhibition of functional V-ATPases. A second live-cell imaging approach using membrane-impermeant pH-sensitive fluorescein derivatives (Steinberg et al., 2010; Carraro-Lacroix et al., 2011) enabled us to monitor organellar pH in situ and study the effects of reversible alkalinizing agents or pump inhibitors. The initial rate of alkalinization, recorded immediately after adding concanamycin A—fully inhibiting V-ATPase function (Fig. 1 c)—was not different, which indicates that the rate of H+ pumping at steady state was indistinguishable between WT and PSEN-deficient MEFs. As lysosomal pH is influenced by the luminal buffering power, we measured this parameter by pulsing the cells with a defined amount of the weak base NH4Cl and found similar values in both cell types (Fig. 1 c and Fig. S1, a–c).
Even though we failed to detect differences in the ability of WT or PSEN−/− cells to acidify endosomes and lysosomes, we considered that lysosomal proteolysis by acid hydrolases may have nevertheless been compromised as a consequence of impaired PSEN function. To assess lysosomal proteolytic activity, we allowed cells to internalize DQ-BSA, a self-quenched substrate of lysosomal proteases that, upon pH-dependent cleavage, releases fluorescent peptides; this approach failed to reveal any differences in proteolytic activity between WT and PSENdKO cells (Fig. 1, d and g). Treatment with concanamycin A suppressed the release of fluorescent peptides, indicating that DQ-BSA hydrolysis was V-ATPase (acidification) dependent. Moreover, we found no differences in the maturation of cathepsin D between WT, PSEN1−/−, and PSENdKO blastocysts (Fig. S1 e, and MEFs, not depicted).
Finally, we expanded these quantitative analyses to the same human fibroblasts carrying familial Alzheimer’s disease (FAD)-related mutations of PSEN1 used by Lee et al. (2010), but were unable to observe any significant differences in V-ATPase function or pH-dependent proteolysis (Fig. 1, e–g). Thus, impaired V-ATPase–mediated acidification is not responsible for the observed endo-lysosomal abnormalities in PSEN-deficient cells.
V0a1 is not essential for lysosomal acidification
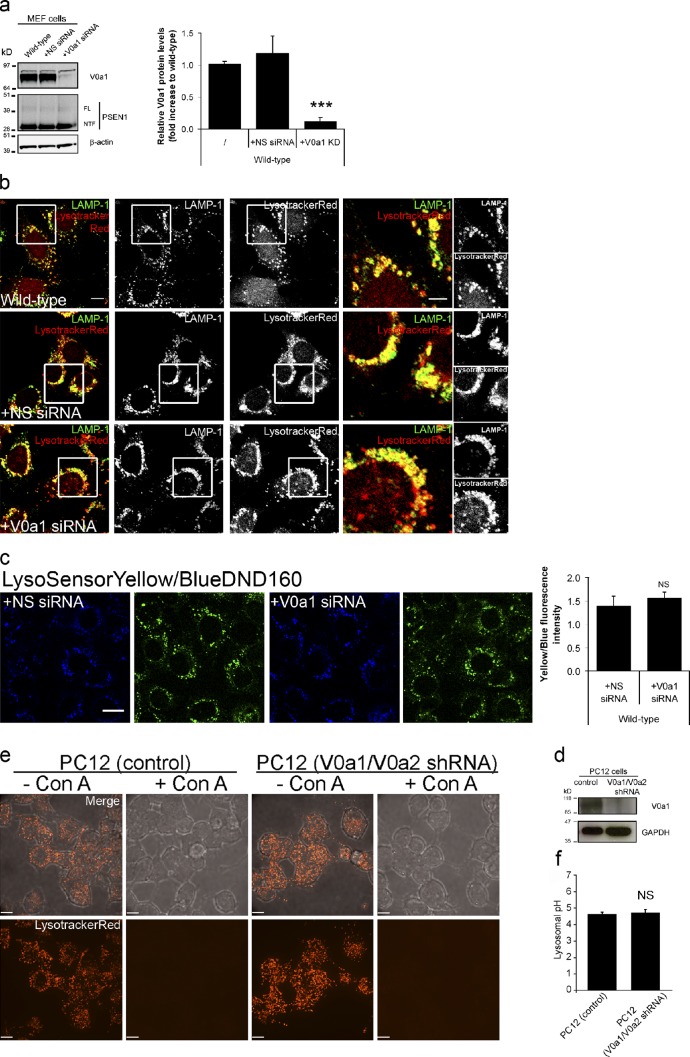
We next reinvestigated the maturation/targeting of V0a1, and its requirement for acidification in the late degradative pathway. V0a1 has three orthologous genes—V0a2, V0a3, and V0a4—whose products have highly specific tissue and subcellular distribution patterns. V0a1 is mostly neuronal (Nishi and Forgac, 2000; Hiesinger et al., 2005), which suggests a localization to specialized organelles, rather than a general role in lysosomal acidification. Indeed, V0a1 is confined predominantly to synaptic vesicles and endosomal compartments (Hiesinger et al., 2005; Williamson et al., 2010). V0a2 does not colocalize with late endosomal/lysosomal markers, but is localized to early endosomes and Golgi (Toyomura et al., 2003; Saw et al., 2011), corresponding to its proposed ARF6/ARNO-dependent endosomal function (Hurtado-Lorenzo et al., 2006). In contrast, V0a3 does colocalize with LAMP2 in macrophages and NIH3T3 fibroblasts (Toyomura et al., 2003), making it perhaps a better candidate to account for late endosomal/lysosomal acidification than V0a1. Using RT-qPCR, we detected only RNA transcripts for the V0a1, -a2, and -a3 subunits in MEFs, and none of them were affected by PSEN deficiency (Fig. S2 b).
Knockdown of V0a1 in WT MEFs (∼85–90%; Fig. 2 a) had no effect on acidification assessed by LysoTracker (Fig. 2 b) and LysoSensor (Fig. 2 c). Furthermore, LysoTracker accumulation (Fig. 2 e) and lysosomal pH measurements (Fig. 2 f) in neuroendocrine PC12 cells where both V0a1 and V0a2 were stably knocked down were indistinguishable from control cells (Fig. 2 d; Saw et al., 2011). The effect of concanamycin A indicates that these cells still express functional V-ATPases, most likely using V0a3. Overall, these data strongly support the notion that V0a1 is not essential for V-ATPase–dependent lysosomal acidification in MEFs or neuronal PC12 cells. This corroborates findings in Drosophila melanogaster, where V0a1 is only required in neurons, and where its loss results in only partial reduction of acidification, presumably because of the presence of other ubiquitous V-ATPase subunits (Williamson et al., 2010). It is therefore difficult to envisage how inactivation of V0a1 could account for V-ATPase–mediated endo-lysosomal dysfunction, as proposed by Lee et al. (2010).
Figure 2.
Knockdown of V0a1 is not essential for lysosomal acidification. (a) Western blot and quantification of V0a1 knockdown in WT MEFs (error bars indicate mean ± SEM; n = 3). (b) WT MEFs with/without V0a1 down-regulation, stained with LysoTracker and immunolabeled for LAMP1. The boxed regions are shown enlarged on the right. (c) Lysosomal pH (using LysoSensor) is not affected in control and V0a1 knockdown cells (n = 2; error bars indicate mean ± SEM; >60 cells/experiment). (d) Western blot for V0a1 and GAPDH in control and V0a1/V0a2-depleted PC12 cell lysates. (e) Z-stack projections of cells stained with LysoTracker (bottom) and merged with differential interference contrast (top). Concanamycin A (Con A) was used to suppress acidification. (f) pH (measured using fluorescein-dextran) of lysosomes from WT and V0a1/V0a2-depleted PC12 cells was not significantly different (error bars indicate mean ± SEM, n = 4, >40cells/experiment). Bars: (b) 10 µm; (b, insets) 5 µm; (c and e) 10 µm.
Post-Golgi trafficking and N-glycosylation of V0a1 are not affected in PSEN−/− cells
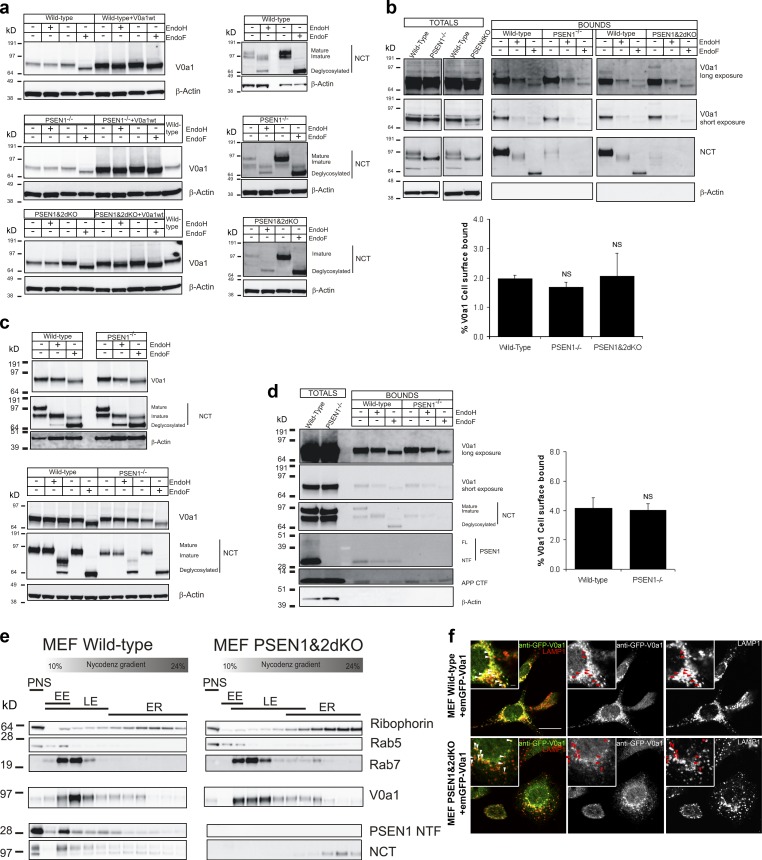
Because we did not detect any V-ATPase–dependent acidification defects in the absence of PSEN, we felt that the role of PSEN in V0a1 glycosylation, a key aspect of the hypothesis of Lee et al. (2010), required reanalysis. As a control for glycosylation, we used endogenous Nicastrin (NCT) that is synthesized as an endoglycosidase-H (endoH)-sensitive immature species and further complex glycosylated in the Golgi to mature NCT (Herreman et al., 2003). N-glycosylation of V0a1 in PSEN−/− MEFs and neurons or brain cortex turned out to be reminiscent of their WT counterparts (Fig. 3, a and c; and Fig. S3, a and b). Overexpressed V0a1 remained fully EndoF-sensitive in PSENdKO MEFs, indicating that the initial ER-associated N-glycosylation persisted in the absence of PSEN. Of note, we used electrophoresis conditions that were distinct from those of Lee et al. (2010) to better resolve the subtle mobility shifts between glycosylated and unglycosylated V0a1 forms. Knockdown and overexpression of V0a1 confirmed the specificity of the antibodies used to detect endogenous V0a1 (Fig. S3, a and b).
Figure 3.
Post-golgi trafficking and N-glycosylation of V0a1 is not affected by PSEN deficiency. (a) Western blot of endogenous and overexpressed V0a1 in WT, PSEN1−/−, and PSENdKO MEFs after EndoH or -F treatment. Overexpressed V0a1 was EndoH-sensitive, underscoring N-glycosylation even in the absence of PSENs. In all cell lines, endogenous V0a1 was insensitive to EndoH and thus mature N-glycosylated. NCT was used as control (right). Same observations in PSEN1−/− brain cortex (c, top) and hippocampal neurons (3-div; c, bottom). Cell surface biotinylated V0a1 was EndoH resistant in WT, PSEN1−/−, and PSENdKO MEFs (b) and in PSEN1−/− neurons (d). Quantification showed no differences in V0a1 trafficking to the cell surface for PSENdKO, PSEN1−/− versus WT MEFs (b, bottom; error bars indicate mean ± SEM), and PSEN1−/− versus WT neurons (d, right). NCT was used as control (error bars indicate mean ± SEM; n = 3–6). (e) Nycodenz gradient centrifugation of a PNS of WT and PSENdKO MEFs. For both cell lines, V0a1 codistributed with late endosomes (Rab7) and was devoid in ER-enriched fractions (Ribophorin) at the bottom fractions. (f) EmGFP-V0a1 stably expressed in WT and PSENdKO colocalizes with LAMP1 in late endosomal compartments. Bars: 10 µm; (insets) 1 µm.
We proceeded to analyze the traffic of V0a1 in PSENdKO cells. The abundance of V0a1 at the cell surface of PSEN−/− MEFs and neurons was comparable to that of the corresponding WT (Fig. 3, b and d). Importantly, only the mature glycosylated V0a1 was detected at the surface using biotinylation, underscoring the lack of any measurable biochemical deficit in the trafficking to post-Golgi sites. Additionally, immunostaining of endogenous V0a1 in WT, PSEN1−/−, and PSENdKO blastocysts failed to detect colocalization with the ER-marker protein protein disulfide isomerase (PDI), indicating that V0a1 does not accumulate in the ER of PSEN-deficient blastocysts (Fig. S1 d). Finally, subcellular fractionation of WT or PSENdKO postnuclear supernatant (PNS) revealed no changes in the distribution of endogenous V0a1 with respect to ribophorin-positive ER membranes. V0a1 codistributed with endosomal markers in PSENdKO as seen in WT MEFs (Fig. 3 e).
N-glycosylation is not required for V0a1 trafficking
The hypothesis put forth by Lee et al. (2010) assigned a fundamental role to PSEN1 in initial protein N-glycosylation in the rough ER, as part of the early quality control steps (Helenius and Aebi, 2001; Annaert and De Strooper, 2010). If valid, this function should affect more than a single protein. Yet, genuine defects in protein glycosylation have not been described for PSEN-deficient cells or in FAD mutants. Additionally, the chaperoning role for PSEN1 in the cotranslocational transfer of the oligosaccharide to the native V0a1 is confined to the (low abundance, short-lived) PSEN1 holoprotein. In an in vitro ER-budding assay, we confirmed previous findings (Kim et al., 2005b; Kim et al., 2007) that FL-PSEN1 is mobile and exits the ER in a manner reminiscent of p58/ERGIC-53, a lectin receptor that cycles between the ER and cis-Golgi (Fig. S3 c). Using the same assay, we found that V0a1 exited the ER in both WT and PSENdKO MEFs (Fig. S3, c and d). In contrast, components of the OST complex like ribophorin I remain ER resident. Thus, PSEN deficiency does not prevent ER exit of V0a1, which supports our findings of its normal post-Golgi localization.
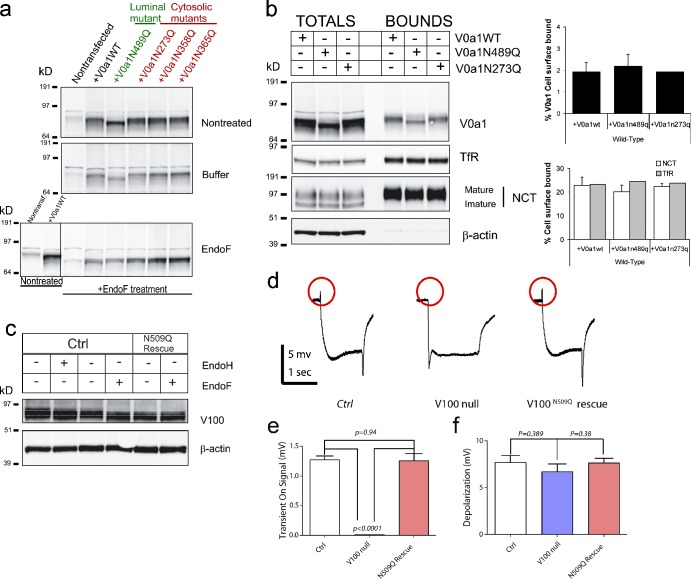
Third, N-glycosylation occurs on NxT/S motifs (Helenius and Aebi, 2001). In silico predictions revealed four potential N-glycosylation sites in the mouse V0a1 sequence (N273QTE, N358QTP, N365KTN, and N489WTE; Fig. S2 a). The first three sites fall within the cytosolically oriented N terminus of V0a1 (Forgac, 2007), and are not accessible to the luminally located glycosylation enzymes, leaving only N489xT available for glycosylation (Fig. S2 a). We therefore constructed a site-directed V0a1 mutant (N489Q) defective for N-glycosylation, and three additional V0a1 mutations (N273Q, N358Q, and N365Q) in the predicted inaccessible motifs. Removal of N489 caused a mobility shift, not observed for the cytosolic mutants, and migrated with identical mobility as the overexpressed, EndoF-treated WT V0a1 (Fig. 4 a). EndoF did not cause a further mobility change of V0a1-N489Q, in contrast to the cytosolic mutants that shifted exactly like WT V0a1. Together, these data prove that V0a1 has a single N-glycosylation site at N489xT. Overexpression of WT, N-glycosylation–deficient, and cytosolic N273Q mutant in WT MEFs resulted in identical levels of V0a1 at the cell surface (Fig. 4 b, right), indicating that the N-glycan chain is not essential for the ER quality control and traffic of V0a1; this was finally confirmed using the in vitro ER-budding assay (Fig. S3 e).
Figure 4.
V0a1 has one N-glycosylation site, not required for trafficking and in vivo function. (a) Western blot analysis of V0a1 WT versus N489Q and three cytosolic mutants (N273Q, N358Q, and N365Q) overexpressed in WT MEFs. Extracts were directly analyzed (nontreated) or incubated in buffer with or without EndoF. Only N489Q shifted in mobility in the absence of EndoF. EndoF resulted in identical mobility shifts for WT V0a1 and cytosolic mutants, indicating that the shift for N489Q is caused by the absence of an N-glycan. NCT was used as control. (b) Cell surface biotinylation does not reveal a significant change in transport to the cell surface between WT and mutant V0a1. (b, right) quantification with NCT and TfR as positive and β-actin as negative controls; mean ± SEM (error bars; n = 4, except for V0a1N273Q, where n = 1). (c) Western blot analysis of Drosophila v100 WT and v100N509Q rescued adult fly heads demonstrated a slight mobility shift in V100N509Q, caused by the lack of a single N-glycan, as treatment of WT extracts results in the same mobility shift. (d) ERG recordings in flies (WT, v100 null mutant, and v100N509Q rescue) in which only photoreceptor neurons are rendered mutant. Red circles, postsynaptic responses of photoreceptor neurons to light stimulation (transient On signal). (e and f) Quantification of transient On signal (e) and depolarization (f) of indicated genotypes (error bars indicate mean ± SEM; n = 10).
Mutating the N-glycosylation site of Drosophila V0a1 does not affect its function in vivo
Complete retention of the V0a1 subunit in the ER of PSEN−/−, as postulated by Lee et al. (2010), would be expected to cause physiological effects resembling those of a gene knockout. Knockout of the Drosophila V0a1 orthologue, v100, is embryonic-lethal, as it is required for organellar acidification as well as endosomal/synaptic vesicle exocytosis (Hiesinger et al., 2005; Williamson et al., 2010). Importantly, neuron-specific expression of WT v100 is sufficient to rescue the mutant Drosophila phenotypes. The single N-glycosylation site is conserved in Drosophila v100 (N509; Fig. S2 a), allowing us to test whether N-glycosylation is essential for v100 function in vivo. We mutated Asn509 to Gln509 (v100N509Q) and generated transgenic flies with cell-specific expression of v100N509Q. Western blot analysis of fly head extracts showed the expected shift for the v100N509Q mutant, which was reminiscent of the mobility change induced by EndoF treatment of WT v100 (Fig. 4 c). EndoF of v100N509Q flies did not alter mobility, proving that there is a single N-glycosylation site in the fly V0a1 orthologue as well. To assess functionality of v100N509Q, we generated flies in which only the photoreceptors are homozygous mutant in otherwise heterozygous flies. Loss of v100 in photoreceptors causes loss of neurotransmission, shown by loss of “on” transients in electroretinogram (ERG) recordings (red circles in Fig. 4 d; Hiesinger et al., 2005; Williamson et al., 2010). Photoreceptor-specific expression of v100N509Q fully rescues this defect (Fig. 4, d–f). Correspondingly, pan-neuronal expression of v100N509Q rescues lethality of v100 null mutants indistinguishable from WT v100. These data indicate that N-glycosylation is not required for v100 function in Drosophila in vivo.
PSEN1−/− compromises lysosomal Ca2+ homeostasis
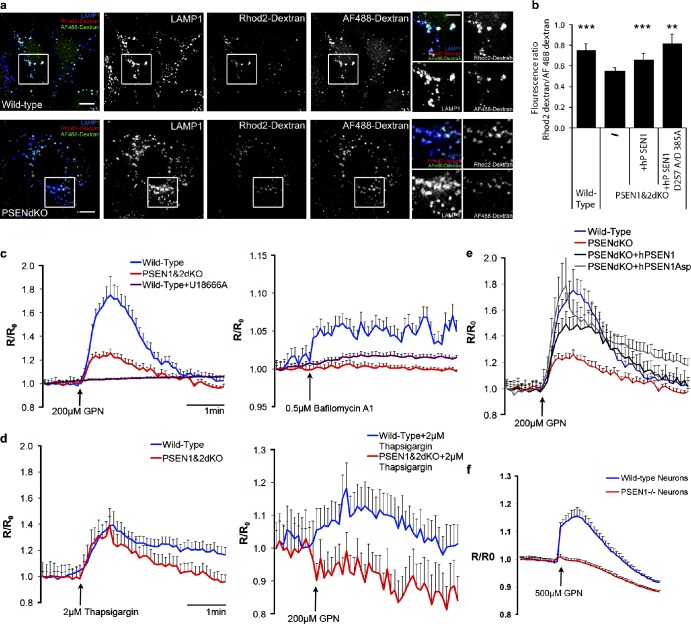
In PSENdKO cells, LAMP1-positive compartments appear enlarged and more dispersed, which indicates that late endosome/lysosome function is defective (Fig. 1 a). This could occur as a consequence of impaired lysosomal fusion (Esselens et al., 2004; Wilson et al., 2004). However, neither lysosomal proteolysis nor acidification is impaired in PSENdKO MEFs. As fusion depends on both luminal acidification and local lysosomal Ca2+ release (Luzio et al., 2007; Saftig and Klumperman, 2009), we explored whether lysosomal Ca2+ homeostasis was perturbed by PSEN deficiency, using two approaches. First, we loaded lysosomes with a low-affinity rhodamine2-dextran Ca2+ probe and a Ca2+-insensitive Alexa Fluor 488–dextran. Their ratio reflects the Ca2+ storage in degradative organelles, and this was significantly decreased in PSENdKO MEFs (Fig. 5, a and b). Second, lysosomal Ca2+ release was monitored indirectly by treating Fura-2–loaded MEFs with Gly–Phe–β-naphtylamide (GPN), a tripeptide causing osmotic lysis of cathepsin C–positive lysosomes. In WT MEFs, GPN induces a steep increase in the cytosolic Ca2+ levels, which was fully blocked by pretreating cells with the cholesterol transport blocker U18666A (Fig. 5 c, left; Lloyd-Evans et al., 2008). GPN-induced lysosomal Ca2+ release was greatly reduced in Fura-2–loaded PSENdKO MEFs. Comparable results were obtained when MEFs were treated with bafilomycin A1 (Fig. 5 c, right) or pretreated with thapsigargin before GPN (Fig. 5 d, right), ruling out a contribution from ER-associated Ca2+ stores. This defect was rescued by hPSEN1 and hPSEN1D257A/D385A, underscoring a γ-secretase–independent function (Fig. 5 e). PSEN1−/− hippocampal neurons also displayed greatly decreased lysosomal Ca2+ release, demonstrating a cell type–independent function for PSEN1 in lysosomal Ca2+ homeostasis (Fig. 5 f). Such a significantly diminished Ca2+ release may define the primary defect in lysosomal fusion and protein clearance in PSEN-deficient cells and neurons.
Figure 5.
PSEN gene deletion affects lysosomal Ca2+ homeostasis. (a) Lysosomal Ca2+ is decreased in PSENdKO MEFs and can be rescued by hPSEN1 and hPSEN1D257A/D385A. The boxed regions are shown enlarged on the right. Bars: 10 µm; (insets) 5 µm. (b) Quantified graphs represent red/green fluorescence intensities (error bars indicate mean ± SEM; one-way ANOVA; n = 3, >45 cells/experiment). (c) Intracellular Ca2+ recording (340/380nm) in Fura-2AM loaded MEFs after unloading with GPN (left) or Bafilomycin A1 (right) indicated a considerable lower level in PSENdKO MEFs, only exceeded by U18666A. (d) ER Ca2+ stores were emptied with 2 µM thapsigargin, before GPN induction of Ca2+ release from acidic stores. WT MEFs showed a small elevation while no effect was seen for PSENdKOs. (e) Human PSEN1 and D257A/D385A mutant rescue the Ca2+ defect in PSENdKO MEFs (n = 3–10; ≥60 cells/Ca2+ trace). (f) Lysosomal Ca2+ release using GPN was significantly reduced in PSEN1−/− hippocampal neurons (4 div). n = 4; 5–15 cells/Ca2+ trace.
In summary, our studies show that in PSENdKO MEFs and PSEN1−/− neurons/brain, the V0a1 subunit is mature, complex-glycosylated, and properly targeted to post-Golgi compartments, indistinguishable from what is observed in WT. Furthermore, N-glycosylation of V0a1 is not required for its localization, or its in vivo role in neurons. Accordingly, we could not detect any changes in V-ATPase function, as assessed by lysosomal pH measurements and hydrolytic activity. Importantly, these lysosomal functions were also unperturbed in human fibroblasts carrying FAD-PSEN1 mutations.
These data make it difficult to attribute the endo/lysosomal abnormalities observed in PSEN-deficient cells to loss of V-ATPase–dependent acidification, as claimed by Lee et al. (2010). Instead, the accumulation of proteins and lipids (Thimiri Govinda Raj et al., 2011) in seemingly poorly degradative but acidified compartments is linked to a significant deficiency in lysosomal Ca2+ storage/release, which is required for efficient lysosomal fusion (Saftig and Klumperman, 2009). This conclusion is also consistent with the proposed γ-secretase–independent function for PSEN in a late step in autophagy, at the level of lysosomal fusion (Esselens et al., 2004; Neely et al., 2011). For instance, it is conceivable that the reported impaired capability of autophagosomes to fuse with degradative organelles in PSEN1−/− neurons or PSENdKO cells prevents autophagosomes from acquiring degradative enzymes and maturing into autophagolysosomes. This is corroborated by the fact that PSEN-deficient cells have constitutively elevated levels for key autophagic proteins like LC3-II and phosphorylated mTOR, concomitant with defective clearance of long-lived proteins and accumulation of autophagosomes (Esselens et al., 2004; Lee et al., 2010; Neely et al., 2011). Here we identified lysosomal Ca2+ homeostasis as an alternative molecular mechanism that is corrupted in PSEN-deficient cells and neurons, explaining consequent lysosomal fusion impairments. Our data extend the documented role for PSEN in ER Ca2+ homeostasis (Bezprozvanny, 2009; De Strooper and Annaert, 2010) to a contribution of a γ-secretase–independent function of PSEN in lysosomal Ca2+ homeostasis and fusion.
Materials and methods
Antibodies and chemicals
The following antibodies were used: rabbit polyclonal antibodies (pAb) to PSEN1-NTF (B19.2; peptide antigen residues, CSQERQQQHGRQRLDN), APP (B63.1; peptide antigen, CNGYENPTYKFFEQMQN), and monoclonal antibody (mAb) to NCT (9C3; peptide antigen, CAKADVLFIAPREPGAVSY); all have been described previously (Annaert et al., 1999; Esselens et al., 2004; Spasic et al., 2006, 2007). Commercially available antibodies include: mAbs 1D4B (anti-LAMP1), F-6 (anti-V-ATPase subunit V1B1; Santa Cruz Biotechnology, Inc.), AC15 (anti–β-actin; Sigma-Aldrich), FlagM2 (Sigma-Aldrich), RAB5 (Synaptic Systems), TfR (Invitrogen), 1D3 (anti-PDI I; Enzo Life Sciences), and pAbs to V0a1 V-ATPase subunit (Santa Cruz Biotechnology, Inc.; H-140; Synaptic Systems); and Ribophorin I (BD). pAb to RAB7 was obtained from P. Chavrier (Centre National de la Recherche Scientifique, Paris, France).
Thapsigargin was from EMD Millipore. Bafilomycin A1 and U18666A were from Tebu-Bio. GPN was from Sigma-Aldrich.
Cloning
To generate the constructs of V0a1 N273Q, N358Q, N365Q, and N489Q mutants, WT V0a1 splice variant 3 in pFLAG-CMV-5a (obtained from U. Kornak, Universitätsmedizin Berlin, Germany) was used as a template for site-directed mutagenesis using QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies) with the following primers: 5′-GCCAATGTTCACTCAAGGGCAGTGGACGGAGGAG-3′ and its reverse complement. V0a1 WT and mutants were sequenced to confirm the presence of designed mutation. cDNAs of human WT PSEN1 and PSEN1D257A/D385A were cloned in the retroviral vector pMSCV*-puromycin (Takara Bio Inc.; Nyabi et al., 2003).
Cell culture and primary hippocampal neuron culture
MEF cell lines and human fibroblasts (Coriel Institute) were maintained in DME-F12 (Sigma-Aldrich) containing 10% (vol/vol; heat inactivated) FCS and maintained in a humidified chamber with 5% CO2 at 37°C. Human fibroblasts included AGO7621 (control), AGO6840, and AGO6848 (carrying the FAD-A246E mutation in PSEN1). Blastocysts from WT, PSEN1−/−, and PSEN1&2dKO mice were obtained from B. Yankner (Harvard University, Cambridge, MA) and cultured in RPMI-1640 (Sigma-Aldrich) containing 20% (vol/vol) FCS, penicillin/streptomycin, 1 mM sodium-pyruvate, 2 mM glutamax, and 100 µM β-mercaptoethanol, and maintained in a humidified chamber with 5% CO2 at 37°C. WT and PSEN1−/− primary hippocampal neurons were derived from embryonic day 17 embryos from heterozygous crosses and were cocultured with a glial feeder layer (Esselens et al., 2004). Single cell suspensions obtained from hippocampi of individual embryos were plated on a poly-l-lysine–coated surface in minimal essential medium (MEM) supplemented with 10% (vol/vol) horse serum. 4 h after plating, culture medium was replaced by serum-free neurobasal medium supplemented with B27 (Gibco). Hippocampal neurons were maintained at 37°C and 5% CO2; for long term cultures, neurons (>4 d in vitro [div]) were maintained in the presence of glial feeder layer after 1 div, to allow for full differentiation and polarization. PC12 cells (small hairpin RNA [shRNA] V0a1/V0a2 and nontargeting shRNA control; Saw et al., 2011) were cultured in DME with 4 mM l-glutamine and 4,500 mg/L glucose, 5% (vol/vol) bovine calf serum, 5% (vol/vol) equine serum, and 2.5 µg/ml puromycin for continuous selection of the shRNA-expressing plasmid.
Generation of v100 mutant rescue constructs and transgenic lines
Site-directed mutagenesis (Agilent Technologies) was used to introduce N509Q mutation in v100 by using full-length v100 cDNA in a pOT2 vector (Berkeley Drosophila Genome Project clone LD21248) with the following primers: 5′-GCACTTATCCTACCAAAAGTCCACCGTGAT-3′ and its reverse complement. The resultant construct was introduced into the pUAST vector by conventional PCR and ligation procedures and sequenced to confirm the presence of the mutation. Transgenic lines bearing this construct were generated by Rainbow Transgenics, Inc., and maintained at room temperature. The genotypes of flies used in ERG assay were as follows: ey3.5FLP;GMR>Gal4/UAS-v100WT;FRT82B,CL/FRT82B,V100null (Ctrl), ey3.5FLP;;FRT82B,CL/FRT82B,v100null (v100 null), and ey3.5FLP;GMR>Gal4/UAS-V100N509Q;FRT82B,CL/FRT82B, v100null (v100N509Q rescue).
The embryonic lethality of v100 null was also able to be rescued through pan-neuronal expression of a UAS-v100N509Q construct driven by Elav-Gal4. The genotypes of flies used for lethality rescue experiments were Elav>Gal4;;v100 Def/TM3,sb (Ctrl) and Elav>Gal4; UAS-v100N509Q/+; v100 Def/v100null (v100N509Q rescue).
The total protein extracted from the above control and rescue fly heads was used to conduct glycosylation analysis.
Transient transfection
Overexpression of WT and N489Q V0a1 was performed using Fugene6 (Promega) via reverse transfection. Knockdown of V0a1 was performed using the mouse-specific ON-TARGETplus SMARTpool L-040215-01-0010 (Thermo Fisher Scientific). Nontargeting siRNA oligos (NS, RAETA-000031; Thermo Fisher Scientific) were used as a control.
Retroviral and lentiviral transduction
cDNAs of human PSEN1 and PSEN1D257A/D385A mutant were cloned in the retroviral vector pMSCV*-puromycin (Takara Bio Inc.; Nyabi et al., 2003). Different pMSCV* constructs were used for generation of retroviral particles via cotransfection with the helper plasmid pIκ (Ecopac) in HEK293 cells for packaging of retroviruses. Viruses were harvested, snap-frozen, and stored at −80°C. PSENKO MEFs were transduced with retrovirus for 24–48 h, selected using puromycin (5 µg/ml in DME-F12/10% FCS), and subcloned to obtain single stable rescue clones.
Stable WT and PSENdKO MEFs expressing V0a1-emerald GFP were generated as described previously (Saw et al., 2011) using the lentiviral pLVX-IRES-V0a1-emerald GFP-blasticidin vector (IMAGE clone ID 5195776). Lentiviral vectors were produced at the Leuven Viral Vector Core (Division of Molecular Medicine, Katholieke Universiteit Leuven) as described previously (Ibrahimi et al., 2009): HIV-1–derived vector particles encoding the gene of interest, pseudotyped with the envelope of vesicular stomatitis virus, were produced via triple transfection of HEK293T cells in serum-free OPTIMEM I medium (Gibco; Invitrogen). After concentration with a Vivaspin 15 column (Vivascience), the vector particles were snap-frozen and stored at −80°C. Virus particles were 1/25 diluted and supplemented in MEFs for 72 h, then selected using blasticidin (5 µg/ml in DME-F12/10% [vol/vol] FCS).
Stable knockdown of V0a1 in WT MEFs was obtained with SMARTvector Lentiviral shRNA particles (SH-040215-01-10: 5′-GAGTCAATACCAGGATCGA-3′; Table S1). SMARTvector Non-Targeting Control Particles were used as a control. Knockdown cells were selected on puromycin and subcloned.
Stable knockdown of Voa1 and V0a2 in PC12 cells was obtained (Saw et al., 2011) by specifically targeting the 21-nucleotide sequence 5′-GCTGCTTATTGTTGTGTCAGT-3′ (residues 61–81, Voa1KD) of rat Voa1, and 5′-GGTGGAGCTCAGAGAAGTCAC-3′ (residues 315–335, Voa2KD) of rat Voa2. We used CTCGAG as a linker sequence. 58-bp oligos containing sense and antisense of the target sequences were annealed and subcloned into the AgeI–EcoRI sites of pLKO-puro or pLKO-neo, generating the VOa1 (pLKO-puro-Voa1KD) and Voa2 knockdown plasmids (pLKO-neo-Voa2KD). Inserted sequences were verified by sequencing. Recombinant lentiviruses were obtained by cotransfecting each of pLKO-puro and pLKO-neo (control, 9 µg), 9 µg pLKO-puro-Voa1KD, or 9 µg pLKO-neo-Voa2KD with two other plasmids (3 µg pMD2G; 4.8 µg pCMV-dR8.74) into HEK-293FT (Invitrogen) cells using 40 µl of polyethylenimine (1.2 mg/ml, pH 7.2). To generate the stable Voa1 and Voa2 dKD cells, PC12 cells were sequentially infected with lentiviruses generated from pLKO-puro-Voa1KD and pLKO-neo-Voa2KD. Cells were selected with puromycin (2.5 µg/ml) and G418 (0.7 mg/ml), and maintained their phenotypes for 2–3 mo.
ERG recording
ERG recordings were performed as described previously (Fabian-Fine et al., 2003; Williamson et al., 2010). Light stimulus was provided in 1-s pulses by a computer-controlled white light–emitting diode (MC1500; Schott). Data were recorded using Clampex (version 10.1; Axon Instruments) and measured using Clampfit (version 10.2; Axon Instruments).
Confocal microscopy
Cells were fixed (4% paraformaldehyde/4% sucrose in PBS−/−, 20 min at RT), permeabilized (0.1% Triton X-100 in PBS−/−, 5 min at RT), blocked (2% BSA, 2% FBS, 1% gelatin, and 2% goat serum in PBS−/−, 1 h at RT), and processed for indirect immunofluorescence. Overnight incubation (4°C) with primary antibodies diluted in blocking buffer was followed by a 1-h (at RT) incubation with Alexa Fluor 488–, Alexa Fluor 568–, or Alexa Fluor 647–conjugated secondary antibodies (Invitrogen) and mounting in Mowiol. Images were captured at room temperature on a confocal laser microscope system (A1R; Nikon) connected to an inverse microscope (Ti-2000; Nikon) or on a Radiance 2100 (Carl Zeiss) connected to an upright microscope (Eclipse E800; Nikon) using oil-immersion Plan-Apochromat 60× A/1.40 NA objective lenses. Data were collected using Nikon Imaging Software or Lasersharp 3.0 and processed in PhotoshopCS 8.0 (Adobe)/ImageJ. The Pearson coefficient was determined with the colocalization threshold plug-in (JACoP).
Voa1and PDI immunostaining.
Blastocysts were cultured on 18-mm glass coverslips and fixed with 4% (vol/vol) PFA. Cell permeabilization was achieved by a 15-min incubation with 0.1% (wt/vol) SDS, 0.4% (wt/vol) saponin, 1% (vol/vol) normal goat serum (NGS), and 1% (wt/vol) BSA in PBS. Next, the rabbit anti-Voa1 (1:1,000) and mouse anti-PDI (1:200) antibodies were incubated with the cells overnight in PBS containing 0.4% saponin, 1% NGS, and 1% BSA. After washing with PBS containing 0.4% saponin, 1% NGS, and 1% BSA, the secondary antibodies, goat anti–rabbit Cy3, and goat anti–mouse dyLight-488 (Jackson ImmunoResearch Laboratories) were diluted 1:1,000 in PBS containing 0.4% saponin, 1% NGS, and 1% BSA, and incubated with the cells for 1 h before washing. After staining, the cells were immediately imaged on an Axiovert 200M microscopes (Carl Zeiss, Inc.) outfitted with 63× (NA 1.4) and 100× (NA 1.45) oil immersion objective lenses. Images were acquired using back-thinned, electron-multiplied cameras (model C9100-13 ImagEM; Hamamatsu Photonics), and image acquisition was controlled by the software Volocity (PerkinElmer). The Pearson’s coefficient of colocalization was determined using the software Volocity (PerkinElmer).
Protein determination and Western blotting
A DC protein assay (Bio-Rad Laboratories) was used for protein measurements. Cell extracts were run on 4–12% Bis-Tris NuPAGE gels in MOPS running buffer (Invitrogen) and transferred onto nitrocellulose; after blocking in 5% nonfat milk, membranes were processed for immunoblotting. Immunodetection was done using Western Lightning Plus ECL reagent (PerkinElmer), and digitally captured and quantified on a MiniLAS 3000 imager (Fujifilm) using Aida software (Raytest).
Cell surface biotinylation
MEFs were placed on ice, washed in PBS, pH 8, and next incubated in the presence of 0.25 mg/ml sulfo-NHS-SS-Biotin (Thermo Fisher Scientific) for 15 min at 4°C. After washing excess biotin, MEFs were incubated with 100 mM glycine and 0.5% BSA in PBS (15 min at 4°C), followed by protein extraction in lysis buffer (50 mM Hepes, pH 7.2, 100 mM NaCl, 1% Triton X-100, and protease inhibitor cocktail; Sigma-Aldrich). Biotinylated proteins were pulled down from equal amounts of extracts using streptavidin Sepharose (Thermo Fisher Scientific) at 4°C overnight. Bound material was eluted in 2× loading buffer supplemented with 2% β-mercaptoethanol (70°C, 10 min), separated on SDS-NuPAGE gels, and processed for quantitative Western blotting.
Semipermeabilization and in vitro COPII budding reaction
MEFs grown to 90% confluency were trypsinized. Next, 0.05 mg/ml soybean trypsin inhibitor was added and cells were further diluted with ice-cold KHM buffer (110 mM KOAC, 20 mM Hepes, pH 7.2, and 2 mM Mg(OAc)2). Cells were harvested (1,000 g, 3 min, 4°C) and resuspended in KHM buffer. Next, cells were permeabilized for by adding 40 µg/ml of digitonin and mixed by inversion. Permeabilization was stopped by diluting cells in KHM and centrifuging at 1,000 g for 3 min (4°C). Semipermeabilized cell pellets were resuspended in KHM buffer. The in vitro COPII budding reactions were performed as described by Kim et al. (2005a), with slight modifications. Each reaction contained 0.026–0.031 OD units of semi-permeabilized cells in a final reaction of 300 µl. The full budding reaction contained an ATP generating system (40 mM creatine phosphate, 0.2 mg/ml creatine phosphokinase, and 1 mM ATP), 0.2 mM GTP, and concentrated cytosol. As negative controls, either the ATP/GTP regenerating system or cytosol were omitted from the reaction mixture. As a control for COPII-dependent vesicle formation, 0.3 µg of purified recombinant Sar1a H79G was added to block ER exit. The budding reaction was initiated at 30°C and terminated by transferring the tubes to ice after 45 min. The vesicle fraction was separated from the donor semipermeabilized cells by centrifugation at 14,000 g for 25 min at 4°C. The vesicles were collected by centrifugation at 55,000 rpm at 4°C in a TLA100/TLA100.4 rotor for 30 min. Vesicle and semi-permeabilized cell pellets were solubilized with 250 mM sucrose, 5 mM Tris-HCl, pH 7.4, and 1 mM EGTA, and SDS loading buffer was added. The samples were heated at 37°C for 10 min 4.2% of the starting membranes, and 83% of the vesicle fraction was separated by SDS-PAGE and analyzed by immunoblotting.
RNA isolation and RT-qPCR
Total RNA was extracted using the RNeasy Plus Mini kit (QIAGEN) and quantified on a NanoDrop 1000 (Thermo Fisher Scientific). 2 µg of template RNA was reverse transcribed using the RevertAid H Minus First Strand cDNA Synthesis kit (Thermo Fisher Scientific). Primers were designed with PrimerExpress software (Table S2; Applied Biosystems), and the housekeeping genes gusb and hprt1 were used for normalization. For relative quantization, the reaction mixtures consisted of quantitative PCR (qPCR) MasterMix Plus for SYBR Green I dNTP (Eurogentec), with 500 nM of each primer and 10 ng cDNA in a total volume of 50 µl. After an initial denaturation step for 10 min at 95°C, the thermal profile of the reaction was 15 s at 95°C and 1 min at 60°C for 40 cycles. Dissociation curves were determined for each reaction; samples were run in duplicate on an ABI PRWASM 7000 and final analysis was done in Excel via the comparative ΔΔCt method (Sequence Detection System, bulletin no. 2; Applied Biosystems).
Subcellular fractionation
Confluent monolayers of MEFs were harvested in PBS−/− and pelleted. Cell pellets were resuspended in a small volume of homogenization buffer (10 mM triethanolamine, 10 mM acetic acid, 250 mM sucrose, 1 mM EDTA, and 1 mM DTT, pH 7.4) and homogenized using a ball-bearing cell cracker (clearance, 10 µm). Protease inhibitor cocktail (Sigma-Aldrich) was added to the homogenates, followed by 10 min centrifugation at 425 g. The PNS was collected and loaded on top of a preformed 10–24% nycodenz gradient (Hammond and Helenius, 1994). After ultracentrifugation (SW41; 1 h and 30 min at 169,044 g, 37,000 rpm), 11 fractions were collected from the top and analyzed by SDS-PAGE and Western blotting. The bottom fraction 12 containing the gradient cushion was discarded.
Analysis of lysosomes
LysoTracker Red DND-99 labeling.
Cells being analyzed were grown on coverslips, placed in Hepes-buffered solution RPMI, and preequilibrated without CO2 for 10 min at 37°C. Cells were then stained for 8 min with LysoTracker Red DND-99 at a final concentration of 250 nM. When Hoechst dye was used, it was incubated with LysoTracker at a final concentration of 20 µM. Where indicated, the cells were pretreated with 500 nM concanamycin A for 1 h at 37°C. Vehicle alone was used as a control. Each coverslip was labeled individually, washed three times, and the live cells were imaged immediately on an Axiovert 200M microscopes (Carl Zeiss) outfitted with 63× (NA 1.4) and 100× (NA 1.45) oil immersion objectives. These spinning disk confocal systems (Quorum Technologies) are equipped with diode-pump solid state lasers (440, 491, 561, 638, and 655 nm; Spectral Applied Research) and a motorized XY stage (Applied Scientific Instrumentation) with a heated stage insert. Images were acquired using back-thinned, electron-multiplied cameras (model C9100-13 ImagEM; Hamamatsu Photonics), and image acquisition was controlled with Volocity software (PerkinElmer).
Quantitative assessment of lysosomal acidification and pH using LysoSensor and fluorescein-dextran.
Both are ratiometric procedures and eliminate uncertainties introduced by variations in focal plane, excitation light intensity, and photobleaching.
LysoSensor labeling.
MEFs grown on coverslips were stained for 10 min with LysoSensor yellow/blue DND160 at a final concentration of 1 µM in normal growth medium. Where indicated, the cells were pretreated with 1 µM bafilomycin A1 for 15 min at 37°C. Each coverslip was labeled individually and washed three times; the live cells were then imaged immediately at 37°C in normal growth medium on a confocal laser microscope system (A1R) connected to an inverse microscope (Ti-2000; Nikon) using oil-immersion Plan-Apochromat 60× A/1.40 NA objective lenses. Data were collected using Nikon Imaging Software and processed in Photoshop CS 8.0 (Adobe/ImageJ.
Lysosomal pH.
To measure lysosomal pH, MEFs grown on glass coverslips were incubated for 3 h with fluorescein-dextran (10,000 MW, 500 µg/ml) and chased for 1 h in dye-free solution to load lysosomes. Coverslips were placed in a thermo-regulated chamber at 37°C in normal growth medium, mounted on the stage of an Axiovert 200M microscope (Carl Zeiss) outfitted with 63× (NA 1.4) and 100× (NA 1.45) oil immersion objectives. Images were acquired using back-thinned, electron-multiplied cameras (model C9100-13 ImagEM; Hamamatsu Photonics), and image acquisition was controlled by the software Volocity (PerkinElmer). Light from a XFO 120 lamp was transmitted alternately through 485- and 438-nm excitation filters mounted on a filter wheel (Sutter Instrument), and directed to the sample using a 505-nm dichroic mirror. At the end of each experiment, an in situ calibration was performed. The cells were sequentially bathed in isotonic K+ solutions (145 mM KCl, 10 mM glucose, 1 mM MgCl2, and 20 mM Hepes or MES, as appropriate) buffered to pH ranging from 4.2 to 7.2 and containing 10 µM nigericin and 5 µM monensin. For calibration, images were acquired 4 min after the addition of each solution to ensure equilibration of pH across compartments. The resulting fluorescence intensity ratios (485 nm/438 nm, corrected for background) were converted to pH values by curve-fitting. Buffering capacity (βi) was determined according to Boyarsky et al. (1988) and calculated as described by Weintraub and Machen (1989). In brief, cells were consecutively perfused with a NaHCO3-free Ringer solution containing 10, 5, 2.5, 1, and 0 mM NH4Cl. Fluorescence changes were recorded and the data were converted to pH using the nigericin–high potassium method as described previously (Steinberg et al., 2010; Carraro-Lacroix et al., 2011). Cells were exposed to Hepes-buffered solutions containing NH4Cl (5–10 mM). Calculation of buffer capacity was performed according to the formula βi = [ΔNH4+]i/ΔpHi, where the intracellular NH4+ concentration ([NH4+]i) was calculated from the Henderson-Hasselbach equation on the assumption that [NH3]i = [NH3]o). Total buffer capacity of the cells was calculated as βTotal = βi + βCO2, where βCO2 = 2.3 [HCO3−]. Proton equivalent flux (JH+) was calculated as JH+ = (ΔpHi/dt) × BTotal. All incubations were at 37°C.
DQ-BSA labeling.
MEFs or human fibroblasts were pretreated with vehicle alone or with 500 nM concanamycin A for 30 min before the addition of DQ-BSA at a final concentration of 50 µM. The cells were further incubated at 37°C for 150 min. Individual coverslips were washed and incubated in HPMI for 10 min at 37°C with and without concanamycin A. Coverslips were individually stained with 20 µM Hoechst for 8 min and live cells were imaged immediately on Axiovert 200M microscopes (Carl Zeiss) outfitted with 63× (NA 1.4) and 100× (NA 1.45) oil immersion objective lenses. Images were acquired using back-thinned, electron-multiplied cameras (model C9100-13 ImagEM; Hamamatsu Photonics), and image acquisition was controlled with Volocity software (PerkinElmer).
Calcium measurements
Cytosolic Ca2+.
Fura-2-AM (Invitrogen) was used to detect cytosolic Ca2+ responses resulting from incubation of lysosomal ligands in Ca2+-free solution with or without thapsigargin pretreatment. Cells were incubated with 2.5 µM Fura-2-AM for 30 min at RT in Ringer’s solution (155 mM NaCl, 5 mM KCl, 1 mM MgCl2, 2 mM CaCl2 [or 2 mM EGTA in case of Ca2+-free solution], 10 mM glucose, 10 mM Hepes, and 2 mM NaH2PO4 × 2H2O, pH 7.3). Cells were examined with an inverted microscope (IX81; Olympus) using an UApo/340 40×/1.35 NA oil objective lens and FvII camera (Olympus soft imaging solution). The setup was operated by Cell^R software (Olympus). Recordings were done using 340 and 380 nm excitation and 530 nm emission filters. Experiments were done at 37°C in Ca2+-free Ringer’s solution, and frames were taken every 6 s for 5 min.
Lysosomal Ca2+.
Lysosomal Ca2+ was measured using low-affinity Rhodamine2 dextran 10 kD in conjunction with Ca2+-insensitive Alexa Fluor 488 dextran (0.25 mg/ml; Invitrogen), as described previously (Lloyd-Evans et al., 2008).
Determination of low-affinity Rhodamine2 dextran Kd at lysosomal pH was performed in vitro in a medium that mimics the intracellular environment (140 mM KCl, 10 mM NaCl, 1 mM MgCl2, and 10 mM acetate) at pH 4.5 at 20–22°C. Rhodamine2 dextran was used at 5 µM and its fluorescence (excitation/emission, 544/590 nm) was monitored at increasing free [Ca2+] generated by increasing the total added [Ca2+] (0–4 mM) in the presence of 5 mM of the Ca2+ chelator BAPTA tetrasodium salt (Invitrogen; free [Ca2+] was calculated using Winmaxchelator 3.2, C. Patton, Stanford University). To generate 1–50 mM free [Ca2+], BAPTA was omitted and the medium was supplemented with these concentrations of Ca2+ alone. The data were plotted in Excel, with each point representing the mean of two experiments. The Kd was determined according to the following equation: Kd = [Ca2+]Free × ([Fmax − F]/[F − Fmin]). Fluorescence intensity was indicated by F at 590 nm using a spectrofluorometer (VictorX3; PerkinElmer).
Fluorescent labeling of lysosomes in MEFs was performed by loading cells with low-affinity Rhodamine2 dextran 10 kD (Kd = 550.65 ± 65.44 µM) in conjunction with Ca2+-insensitive Alexa Fluor 488 dextran for 15 h at 37°C at a concentration of 0.25 mg/ml, followed by a 10-h chase at 37°C to label lysosomes, both in normal growth medium. After fixation (4% paraformaldehyde; 20 min at RT), cells were immunostained for LAMP1. Images were captured using oil-immersion plan-Apochromat 60× A/1.40 NA objective lenses on a confocal laser microscope system (A1R) connected to an inverse microscope (Ti-2000; Nikon). Data were collected using Nikon Imaging Software and processed in Photoshop CS 8.0/ImageJ. To obtain an organelle Ca2+ ratio, the background-corrected 488-nm image was divided by the background corrected 568-nm image. Next, to restrict the analysis to the organelles of interest, a mask was applied based on the thresholded signal from the LAMP1 antibody staining recorded in the 647 nm channel. Values were exported to Excel (Microsoft) for further data processing.
Statistics
Comparison between data points was done using the one-way analysis of variance (ANOVA) test on at least three independent experiments. Data were plotted as mean ± SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.005.
Online supplemental material
Fig. S1 shows characterization of V-ATPase function in blastocyst-derived extraembryonic endoderm cells that lack PSENs. Fig. S2 shows that sequence alignment of V0a1 subunit orthologues predicts one conserved N-glycosylation site. Fig. S3 shows Western blot analysis of the V-ATPase subunit V0a1 on different gel types and running buffers as well as in vitro ER exit of V0a1 in WT and PSENdKO MEFs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201201076/DC1.
Supplementary Material
Acknowledgments
W. Annaert is financially supported by VIB and Katholieke Universiteit Leuven (GOA/11/009), the Federal Government of Belgium (IAP-P7 2012-17), the Alzheimer’s Association [IIRG-08–91535], SAO/FRMA (no. 10010, cycle 2010), the FWO-Vlaanderen (G.0754.10), and the Hercules Foundation (AKUL/09/037). K. Coen holds an IWT fellowship. R.S. Flannagan was supported by a Canadian Institutes of Health Research fellowship and is supported by a RESTRACOMP fellowship. L.R. Carraro-Lacroix was a recipient of a CAPES fellowship. S. Grinstein was the current holder of the Pitblado Chair in Cell Biology. P.R. Hiesinger was supported by grants from the National Institutes of Health (RO1EY018884 and RO1GM088803) and the Welch Foundation (I-1657).
K. Coen, S. Grinstein, and W. Annaert designed experiments. W. Vermeire, C. Michiels, V. Baert, and S. Munck contributed with confocal imaging and glycosylation data. R.S. Flannagan, L.R. Carraro-Lacroix, and S. Sugita performed lysosomal function assays, D. Wang and P.R. Hiesinger performed Drosophila work, and S. Baron and F. Wuytack performed lysosomal Ca2+ assays. K. Coen and W. Annaert wrote the paper; all authors provided valuable criticisms and input.
The authors declare that they have no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- div
- days in vitro
- endoH
- endoglycosidase-H
- ERG
- electroretinogram
- FAD
- familial Alzheimer’s disease
- GPN
- Gly–Phe–β-naphtylamide
- MEF
- mouse embryonic fibroblast
- NCT
- Nicastrin
- NGS
- normal goat serum
- OST
- oligosaccharyl–transferase
- pAb
- polyclonal antibody
- PDI
- protein disulfide isomerase
- PNS
- postnuclear supernatant
- PSEN
- presenilin
- qPCR
- quantitative PCR
- shRNA
- small hairpin RNA
- V-ATPase
- vacuolar ATPase
- WT
- wild type
References
- Annaert W., De Strooper B. 2010. Alzheimer’s disease neurons fail the acid test. Cell. 141:1112–1114 10.1016/j.cell.2010.06.009 [DOI] [PubMed] [Google Scholar]
- Annaert W.G., Levesque L., Craessaerts K., Dierinck I., Snellings G., Westaway D., George-Hyslop P.S., Cordell B., Fraser P., De Strooper B. 1999. Presenilin 1 controls gamma-secretase processing of amyloid precursor protein in pre-golgi compartments of hippocampal neurons. J. Cell Biol. 147:277–294 10.1083/jcb.147.2.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annaert W.G., Esselens C., Baert V., Boeve C., Snellings G., Cupers P., Craessaerts K., De Strooper B. 2001. Interaction with telencephalin and the amyloid precursor protein predicts a ring structure for presenilins. Neuron. 32:579–589 10.1016/S0896-6273(01)00512-8 [DOI] [PubMed] [Google Scholar]
- Bezprozvanny I. 2009. Calcium signaling and neurodegenerative diseases. Trends Mol. Med. 15:89–100 10.1016/j.molmed.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyarsky G., Ganz M.B., Sterzel R.B., Boron W.F. 1988. pH regulation in single glomerular mesangial cells. II. Na+-dependent and -independent Cl(-)-HCO3- exchangers. Am. J. Physiol. 255:C857–C869 [DOI] [PubMed] [Google Scholar]
- Carraro-Lacroix L.R., Jaumouillé V., Fairn G.D., Grinstein S. 2011. A weak base-generating system suitable for selective manipulation of lysosomal pH. Traffic. 12:1490–1500 10.1111/j.1600-0854.2011.01266.x [DOI] [PubMed] [Google Scholar]
- De Strooper B., Annaert W. 2010. Novel research horizons for presenilins and γ-secretases in cell biology and disease. Annu. Rev. Cell Dev. Biol. 26:235–260 10.1146/annurev-cellbio-100109-104117 [DOI] [PubMed] [Google Scholar]
- Esselens C., Oorschot V., Baert V., Raemaekers T., Spittaels K., Serneels L., Zheng H., Saftig P., De Strooper B., Klumperman J., Annaert W. 2004. Presenilin 1 mediates the turnover of telencephalin in hippocampal neurons via an autophagic degradative pathway. J. Cell Biol. 166:1041–1054 10.1083/jcb.200406060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian-Fine R., Verstreken P., Hiesinger P.R., Horne J.A., Kostyleva R., Zhou Y., Bellen H.J., Meinertzhagen I.A. 2003. Endophilin promotes a late step in endocytosis at glial invaginations in Drosophila photoreceptor terminals. J. Neurosci. 23:10732–10744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgac M. 2007. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8:917–929 10.1038/nrm2272 [DOI] [PubMed] [Google Scholar]
- Hammond C., Helenius A. 1994. Quality control in the secretory pathway: retention of a misfolded viral membrane glycoprotein involves cycling between the ER, intermediate compartment, and Golgi apparatus. J. Cell Biol. 126:41–52 10.1083/jcb.126.1.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A., Aebi M. 2001. Intracellular functions of N-linked glycans. Science. 291:2364–2369 10.1126/science.291.5512.2364 [DOI] [PubMed] [Google Scholar]
- Herreman A., Van Gassen G., Bentahir M., Nyabi O., Craessaerts K., Mueller U., Annaert W., De Strooper B. 2003. gamma-Secretase activity requires the presenilin-dependent trafficking of nicastrin through the Golgi apparatus but not its complex glycosylation. J. Cell Sci. 116:1127–1136 10.1242/jcs.00292 [DOI] [PubMed] [Google Scholar]
- Hiesinger P.R., Fayyazuddin A., Mehta S.Q., Rosenmund T., Schulze K.L., Zhai R.G., Verstreken P., Cao Y., Zhou Y., Kunz J., Bellen H.J. 2005. The v-ATPase V0 subunit a1 is required for a late step in synaptic vesicle exocytosis in Drosophila. Cell. 121:607–620 10.1016/j.cell.2005.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado-Lorenzo A., Skinner M., El Annan J., Futai M., Sun-Wada G.H., Bourgoin S., Casanova J., Wildeman A., Bechoua S., Ausiello D.A., et al. 2006. V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat. Cell Biol. 8:124–136 10.1038/ncb1348 [DOI] [PubMed] [Google Scholar]
- Ibrahimi A., Vande Velde G., Reumers V., Toelen J., Thiry I., Vandeputte C., Vets S., Deroose C., Bormans G., Baekelandt V., et al. 2009. Highly efficient multicistronic lentiviral vectors with peptide 2A sequences. Hum. Gene Ther. 20:845–860 10.1089/hum.2008.188 [DOI] [PubMed] [Google Scholar]
- Kim D.Y., Ingano L.A., Carey B.W., Pettingell W.H., Kovacs D.M. 2005a. Presenilin/gamma-secretase-mediated cleavage of the voltage-gated sodium channel beta2-subunit regulates cell adhesion and migration. J. Biol. Chem. 280:23251–23261 10.1074/jbc.M412938200 [DOI] [PubMed] [Google Scholar]
- Kim J., Hamamoto S., Ravazzola M., Orci L., Schekman R. 2005b. Uncoupled packaging of amyloid precursor protein and presenilin 1 into coat protein complex II vesicles. J. Biol. Chem. 280:7758–7768 10.1074/jbc.M411091200 [DOI] [PubMed] [Google Scholar]
- Kim J., Kleizen B., Choy R., Thinakaran G., Sisodia S.S., Schekman R.W. 2007. Biogenesis of gamma-secretase early in the secretory pathway. J. Cell Biol. 179:951–963 10.1083/jcb.200709012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Yu W.H., Kumar A., Lee S., Mohan P.S., Peterhoff C.M., Wolfe D.M., Martinez-Vicente M., Massey A.C., Sovak G., et al. 2010. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 141:1146–1158 10.1016/j.cell.2010.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Evans E., Morgan A.J., He X., Smith D.A., Elliot-Smith E., Sillence D.J., Churchill G.C., Schuchman E.H., Galione A., Platt F.M. 2008. Niemann-Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat. Med. 14:1247–1255 10.1038/nm.1876 [DOI] [PubMed] [Google Scholar]
- Luzio J.P., Bright N.A., Pryor P.R. 2007. The role of calcium and other ions in sorting and delivery in the late endocytic pathway. Biochem. Soc. Trans. 35:1088–1091 10.1042/BST0351088 [DOI] [PubMed] [Google Scholar]
- Neely K.M., Green K.N., LaFerla F.M. 2011. Presenilin is necessary for efficient proteolysis through the autophagy-lysosome system in a γ-secretase-independent manner. J. Neurosci. 31:2781–2791 10.1523/JNEUROSCI.5156-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi T., Forgac M. 2000. Molecular cloning and expression of three isoforms of the 100-kDa a subunit of the mouse vacuolar proton-translocating ATPase. J. Biol. Chem. 275:6824–6830 10.1074/jbc.275.10.6824 [DOI] [PubMed] [Google Scholar]
- Nyabi O., Bentahir M., Horré K., Herreman A., Gottardi-Littell N., Van Broeckhoven C., Merchiers P., Spittaels K., Annaert W., De Strooper B. 2003. Presenilins mutated at Asp-257 or Asp-385 restore Pen-2 expression and Nicastrin glycosylation but remain catalytically inactive in the absence of wild type Presenilin. J. Biol. Chem. 278:43430–43436 10.1074/jbc.M306957200 [DOI] [PubMed] [Google Scholar]
- Saftig P., Klumperman J. 2009. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10:623–635 10.1038/nrm2745 [DOI] [PubMed] [Google Scholar]
- Saw N.M., Kang S.Y., Parsaud L., Han G.A., Jiang T., Grzegorczyk K., Surkont M., Sun-Wada G.H., Wada Y., Li L., Sugita S. 2011. Vacuolar H(+)-ATPase subunits Voa1 and Voa2 cooperatively regulate secretory vesicle acidification, transmitter uptake, and storage. Mol. Biol. Cell. 22:3394–3409 10.1091/mbc.E11-02-0155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spasic D., Tolia A., Dillen K., Baert V., De Strooper B., Vrijens S., Annaert W. 2006. Presenilin-1 maintains a nine-transmembrane topology throughout the secretory pathway. J. Biol. Chem. 281:26569–26577 10.1074/jbc.M600592200 [DOI] [PubMed] [Google Scholar]
- Spasic D., Raemaekers T., Dillen K., Declerck I., Baert V., Serneels L., Füllekrug J., Annaert W. 2007. Rer1p competes with APH-1 for binding to nicastrin and regulates gamma-secretase complex assembly in the early secretory pathway. J. Cell Biol. 176:629–640 10.1083/jcb.200609180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg B.E., Huynh K.K., Brodovitch A., Jabs S., Stauber T., Jentsch T.J., Grinstein S. 2010. A cation counterflux supports lysosomal acidification. J. Cell Biol. 189:1171–1186 10.1083/jcb.200911083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimiri Govinda Raj D.B., Ghesquière B., Tharkeshwar A.K., Coen K., Derua R., Vanderschaeghe D., Rysman E., Bagadi M., Baatsen P., De Strooper B., et al. 2011. A novel strategy for the comprehensive analysis of the biomolecular composition of isolated plasma membranes. Mol. Syst. Biol. 7:541 10.1038/msb.2011.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toei M., Saum R., Forgac M. 2010. Regulation and isoform function of the V-ATPases. Biochemistry. 49:4715–4723 10.1021/bi100397s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomura T., Murata Y., Yamamoto A., Oka T., Sun-Wada G.H., Wada Y., Futai M. 2003. From lysosomes to the plasma membrane: localization of vacuolar-type H+ -ATPase with the a3 isoform during osteoclast differentiation. J. Biol. Chem. 278:22023–22030 10.1074/jbc.M302436200 [DOI] [PubMed] [Google Scholar]
- Weintraub W.H., Machen T.E. 1989. pH regulation in hepatoma cells: roles for Na-H exchange, Cl-HCO3 exchange, and Na-HCO3 cotransport. Am. J. Physiol. 257:G317–G327 [DOI] [PubMed] [Google Scholar]
- Williamson W.R., Wang D., Haberman A.S., Hiesinger P.R. 2010. A dual function of V0-ATPase a1 provides an endolysosomal degradation mechanism in Drosophila melanogaster photoreceptors. J. Cell Biol. 189:885–899 10.1083/jcb.201003062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C.A., Murphy D.D., Giasson B.I., Zhang B., Trojanowski J.Q., Lee V.M. 2004. Degradative organelles containing mislocalized alpha-and beta-synuclein proliferate in presenilin-1 null neurons. J. Cell Biol. 165:335–346 10.1083/jcb.200403061 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.