Two actin-assembling formins, CYK-1 and FHOD-1, are important for muscle cell growth and maintenance of the contractile lattice in striated muscle cells.
Abstract
Muscle contraction depends on interactions between actin and myosin filaments organized into sarcomeres, but the mechanism by which actin filaments incorporate into sarcomeres remains unclear. We have found that, during larval development in Caenorhabditis elegans, two members of the actin-assembling formin family, CYK-1 and FHOD-1, are present in striated body wall muscles near or on sarcomere Z lines, where barbed ends of actin filaments are anchored. Depletion of either formin during this period stunted growth of the striated contractile lattice, whereas their simultaneous reduction profoundly diminished lattice size and number of striations per muscle cell. CYK-1 persisted at Z lines in adulthood, and its near complete depletion from adults triggered phenotypes ranging from partial loss of Z line–associated filamentous actin to collapse of the contractile lattice. These results are, to our knowledge, the first genetic evidence implicating sarcomere-associated formins in the in vivo organization of the muscle cytoskeleton.
Introduction
The classical experiments of Huxley and Hanson (1954) revealed the sarcomere as the basic organizational unit of striated muscles. The sarcomere ends are defined by two Z lines, each of which anchors one set of actin filaments through association with filament barbed ends. These actin filaments stretch into the sarcomere to interdigitate with myosin filaments, which in turn attach to the M line at the sarcomere center.
It is unclear how actin filaments become incorporated into sarcomeres. De novo assembly of actin filaments from monomers occurs in two steps: nucleation followed by elongation. In vitro, nucleation is rate limiting, as oligomeric nuclei arise from unstable precursors, whereas in vivo, monomer-sequestering proteins, such as profilin, further suppress nucleation (Pollard et al., 2000). Cells overcome these barriers through actin nucleation factors (ANFs), proteins that promote nucleation by mechanisms such as stabilizing or structurally mimicking assembly intermediates (Goode and Eck, 2007). In vitro, once nuclei form, filament elongation occurs rapidly provided that sufficient actin monomers are available but is regulated in vivo by proteins that inhibit or enhance monomer addition. The distinct properties of the two filament ends are such that, under standard cellular conditions, net polymerization occurs at barbed ends, and disassembly is favored at pointed ends (Pollard et al., 2000), although striated muscles with higher actin monomer concentration appear to support polymerization at both ends (Ono, 2010).
Among ANFs, one of the largest and most widespread groups is the formins (Higgs and Peterson, 2005; Chalkia et al., 2008). These dimeric proteins nucleate filaments by stabilizing actin dimers through their FH2 (formin homology-2) domain (Pruyne et al., 2002; Sagot et al., 2002; Pring et al., 2003; Moseley et al., 2004) and by binding additional monomers through their C-terminal diaphanous (DIA) autoregulatory domain (DAD; Gould et al., 2011). Unique among ANFs, formins associate with the barbed ends of actin filaments (Pruyne et al., 2002; Kovar and Pollard, 2004). Unlike most other barbed end–binding proteins, formins generally do not prevent barbed end polymerization but act as processive caps that accommodate the addition of monomers while maintaining contact with the terminal actin subunits, and formins promote elongation by blocking inhibitors such as the barbed end–capping protein CapZ (Kovar et al., 2003; Zigmond et al., 2003; Moseley et al., 2004). Formins also have multiple docking sites for profilin in their conserved FH1 (formin homology-1) domain (Chang et al., 1997; Evangelista et al., 1997; Imamura et al., 1997; Watanabe et al., 1997), and FH1-mediated recruitment of profilin–actin complexes can increase the rate of actin addition to filament ends (Romero et al., 2004; Paul and Pollard, 2008). Thus, formins nucleate actin filaments and enhance the duration and rate of filament elongation.
Formin activity is often regulated through autoinhibition by regions flanking the formin homology domains, as first shown for the mouse formin mDia1 (Nakano et al., 1999; Watanabe et al., 1999). Intramolecular association of an N-terminal DIA inhibitory domain (DID) with the DAD sterically blocks FH2–actin filament interactions (Alberts, 2001; Li and Higgs, 2005; Otomo et al., 2005, 2010; Rose et al., 2005; Nezami et al., 2006, 2010). This inhibitory interaction is broken by several means, the best understood being Rho GTPases, which displace the DAD by binding the DID and a GTPase-binding N-terminal extension (G domain; Watanabe et al., 1999; Li and Higgs, 2005; Otomo et al., 2005; Rose et al., 2005; Nezami et al., 2006). A basic formin structure of G-DID-FH1-FH2-DAD has been replicated with variations early in animal evolution to result in seven subfamilies (Higgs and Peterson, 2005; Chalkia et al., 2008). With regulatory domains of different formins exhibiting different specificities for Rho GTPase isoforms and other regulators, this diversification has allowed coupling of the conserved actin-assembling FH1-FH2 module to a variety of signaling pathways.
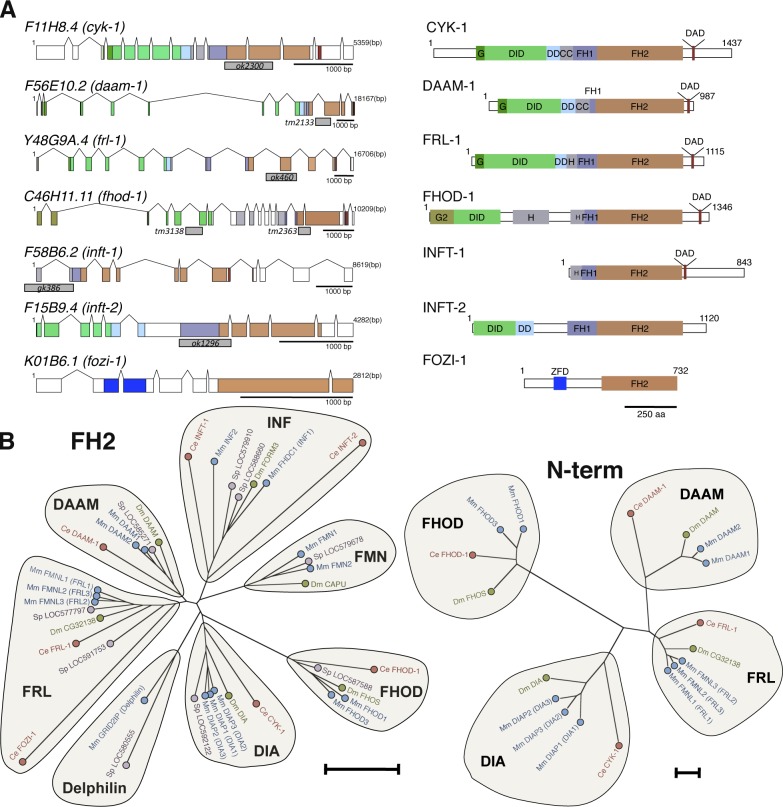
Representative formins from all subfamilies stimulate actin filament assembly when expressed in cultured cells (Goode and Eck, 2007), but in vivo functions for the majority remain largely unknown. Among mammals, formins are particularly diverse with 15 genes, and identifying functions for individual isoforms remains a challenge in the face of potential redundancy within subfamilies, as seen in some studies (Peng et al., 2003; Watanabe et al., 2008). Among invertebrates the situation is simpler, with generally one member of a subfamily per species. The nematode Caenorhabditis elegans provides a particularly attractive model system because of its modest set of seven formins and the availability of deletion alleles for each (Fig. 1 A).
Figure 1.
C. elegans formins. (A) Seven formin genes of C. elegans (left) with protein-coding exons color-coded to match predicted product structural domains (right), including DIA-related formin G domains (dark green), FHOD-specific G domain (G2; olive), DIDs, dimerization domains (DD), helical (H) or coiled-coil (CC) regions, FH1 domains, FH2 domains, core DAD sequences, and zinc finger domain (ZFD). Gray blocks indicate gene sequences deleted by alleles used in this study. (B) Estimated phylogenetic trees based on protein sequence comparisons between mouse (M. Musculus [Mm]; blue), purple sea urchin (S. purpuratus [Sp]; purple), fruit fly (D. melanogaster [Dm]; green), and C. elegans (Ce; red) formin FH2 domains or N termini reveal seven formin subfamilies, five of which (DAAM, DIA, FHOD, FRL, and INF) have C. elegans homologues. N-term, N terminus. Bars, 0.5 substitutions per position.
Previous studies have shown the C. elegans DIA subfamily formin encoded by cyk-1 (cytokinesis defect-1) nucleates and processively caps actin filaments and is required for contractile actin ring assembly (Swan et al., 1998; Severson et al., 2002; Amin et al., 2007; Neidt et al., 2008), whereas the highly divergent fozi-1 (formin homology domain/zinc finger-1) product is a forminlike nuclear protein that influences cell fate choices but does not interact with actin in vitro (Johnston et al., 2006; Amin et al., 2007). For the remaining five worm formins, sequence comparisons with formins of other organisms allow their subfamily assignments (Fig. 1 B). Accordingly, the C. elegans research community has adopted the gene designations: daam-1 (disheveled-associated activator of morphogenesis-1) for the gene F56E10.2, frl-1 (formin-related protein identified in leukocytes-1) for Y48G9A.4, fhod-1 (formin homology domain-containing protein-1) for C46H11.11, and inft-1 and inft-2 (inverted formin/formin three-1 and -2) for F58B6.2 and F15B9.4, respectively (WormBase release WS218, 29 September 2010). We show here that two of these formins, FHOD-1 and CYK-1, are sarcomere components that promote striated muscle development.
Results
FHOD-1 localizes near sarcomere Z lines in striated body wall muscles (BWMs)
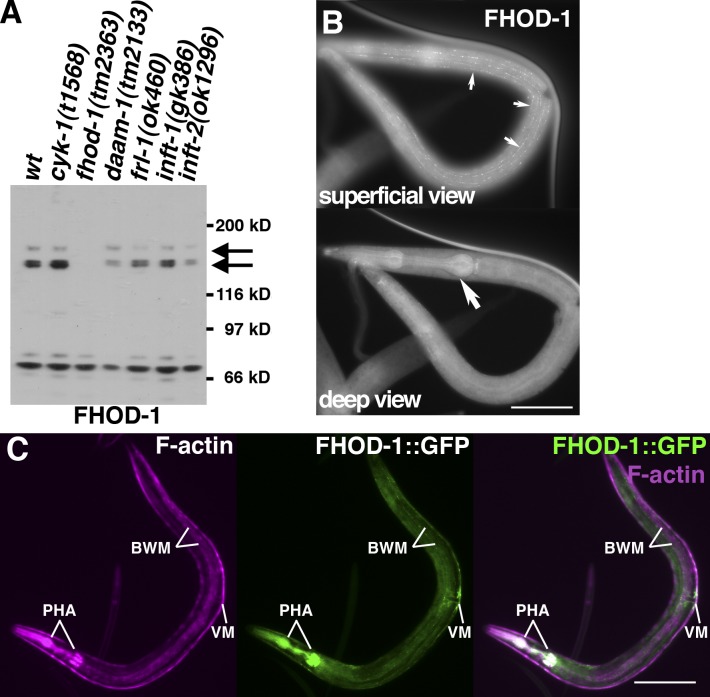
In rough agreement with a predicted 152-kD FHOD-1 formin, polyclonal antibody raised against the FHOD-1 FH2 domain (anti–FHOD-1) recognizes a doublet of proteins with ∼150-kD apparent molecular masses and a third ∼165-kD protein (Fig. 2 A, arrows) in Western blots of extracts from wild-type animals and those bearing mutations in other formin genes but not worms with the fhod-1 deletion allele tm2363. The significance of several apparent molecular masses is unclear, as are the identities of nonspecifically recognized smaller molecular mass proteins. Immunofluorescence staining with anti–FHOD-1 decorates several regions of the worm, including rows of puncta visible in superficial views near the animal surface (Fig. 2 B, small arrows) and the pharynx visible deeper in the head (Fig. 2 B, large arrow). Confirming these represent FHOD-1, C-terminally GFP-tagged FHOD-1 (FHOD-1::GFP) expressed in a genomic context (with the fhod-1 promoter, introns, and downstream sequences) yields a similar fluorescence pattern (Fig. 2 C, green). Fluorescent phalloidin staining of FHOD-1::GFP–expressing animals shows that FHOD-1–expressing tissues are F-actin–rich muscles (Fig. 2 C, magenta), including BWMs, pharyngeal muscles, and vulval muscles.
Figure 2.
FHOD-1 is expressed in muscle cells. (A) Anti–FHOD-1 Western blot of normalized adult worm extracts from wild type (wt) and indicated formin mutants. 150- and 165-kD signals (arrows) are absent from the fhod-1(tm2363) extract. (B) Superficial and deep views of a wild-type larva (worm anterior is shown on the top, and posterior is shown at the bottom) stained with anti–FHOD-1 reveal puncta near the body surface (small arrows) and within the pharynx in the head (large arrow). Bar, 50 µm. (C) FHOD-1::GFP in a larva coincides with fluorescent phalloidin-stained F-actin–rich pharyngeal muscles (PHA) in the head, vulval muscles (VM) near the middle, and body wall muscles (BWM) extending from nose to tail. Bar, 100 µm.
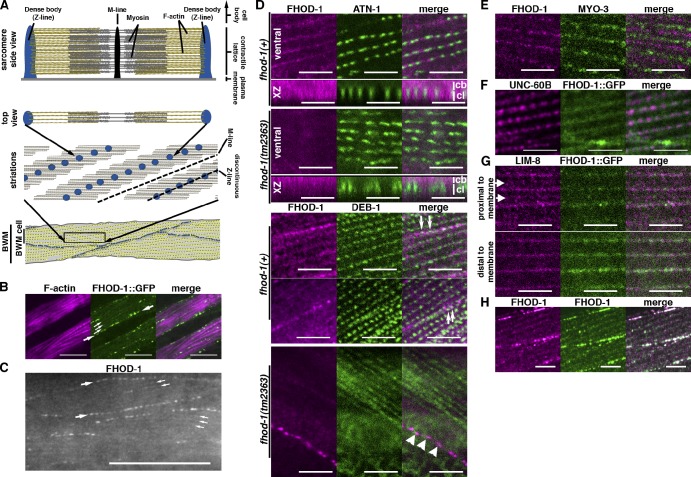
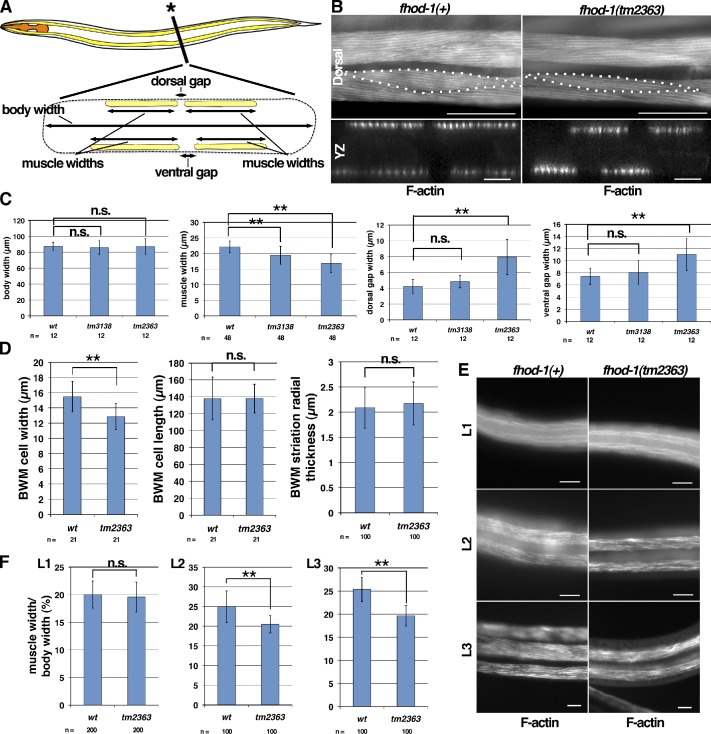
BWMs are the largest muscle group by mass. Flat BWM cells adhere to the body wall with spindle shapes when viewed ventrally/dorsally. Head-to-tail chains of these cells make four long muscles that reach from the nose to the tail, and their flexures bend the worm during crawling and swimming. Their contractile lattices are composed of well-defined sarcomeres arranged in oblique striations when viewed ventrally/dorsally (shown schematically in Fig. 3 A). This lattice is restricted to a layer of cytoplasm interposed between the muscle cell body containing the nucleus and the body wall to which the muscle cell attaches (Waterston, 1988). Focal adhesion–like structures called dense bodies serve as sarcomere Z lines within the lattice, whereas somewhat similar attachment plaques serve that role at cell edges that border adjacent muscle cells (Francis and Waterston, 1985). Integrins associated with dense bodies, attachment plaques, and M lines anchor the contractile lattice to the body wall (Francis and Waterston, 1985; Gettner et al., 1995).
Figure 3.
FHOD-1 localizes close to Z lines in BWM cells. (A) Model of BWM sarcomere organization. Side view of one sarcomere shows that Z-line dense bodies (blue) anchor actin filaments (yellow), and M lines (black) anchor myosin filaments (gray). M lines and dense bodies attach to the plasma membrane, placing the contractile lattice between the membrane and the cell body. In top views, sarcomeres can be seen to combine to form oblique striations, in which rows of dense bodies (blue spots) within the F-actin–rich striations form discontinuous Z lines that alternate with M lines. Striations in turn form the spindle-shaped contractile lattice of BWM cells. At boundaries between BWM cells, attachment plaques (larger dark spots) replace dense bodies (smaller dark spots) as Z-line structures. Myosin filaments have been omitted from striation and BWM models for clarity. (B) Ventral view (anterior to the left) of an FHOD-1::GFP–expressing larva stained with fluorescent phalloidin shows FHOD-1–containing puncta along the edges of F-actin–rich BWM cell contractile lattices (large arrows) and in faint striations across the lattices (small arrows). (C) Ventral view of a wild-type larva stained with anti–FHOD-1 reveals endogenous FHOD-1 in similar puncta (large arrows) and striations (small arrows). (D) Animals double-stained for FHOD-1 and either Z-line marker ATN-1 or DEB-1 show that the formin is closely associated with Z lines. In ventral views or in side views of the discontinuous Z line (xz projection), anti–FHOD-1–stained striations are seen in the contractile lattice (cl) of wild-type but not fhod-1(tm2363) animals, and anti–FHOD-1–stained puncta intermingle with DEB-1–stained attachment plaques (arrows) at the cell edge. Staining of the cell body (cb in xz view) or of an elongated structure that parallels BWMs (bottom, arrowheads) is nonspecific. (E) FHOD-1 striations alternate with MYO-3 striations. (F) Anti-GFP–stained striations in FHOD-1::GFP–expressing animals partially overlap with UNC-60B. (G) Proximal to the plasma membrane, LIM-8 occupies two sets of striations, one near Z lines and a fainter one near M lines (arrows), whereas distal to the membrane, LIM-8 detectable near Z lines strongly comingles with FHOD-1::GFP. (H) Worms stained with anti–FHOD-1 and two secondary antibodies with different fluorophores show extensive, but imperfect, comingling of signal along FHOD-1 striations. Bars: (B) 10 µm; (C) 25 µm; (D–H) 5 µm.
FHOD-1 has two localizations in BWM cells: bright puncta at the muscle cell edges (Fig. 3, B and C, large arrows) and faint striations in the contractile lattice (Fig. 3, B and C, small arrows), both of which are absent from fhod-1(tm2363) BWMs (Fig. 3 D). In animals double stained with anti–FHOD-1 and antibodies for known dense body components α-actinin (ATN-1 in C. elegans) or vinculin/DEB-1 (Francis and Waterston, 1985; Barstead and Waterston, 1989; Moulder et al., 2010), FHOD-1 striations form bands that appear to link dense bodies (Fig. 3 D), placing them near these Z-line structures. Similarly, at the cell edges, where DEB-1 decorates attachment plaques, FHOD-1–containing puncta are situated near, but do not precisely overlap, DEB-1 (Fig. 3 D, arrows). Conversely, striations stained for FHOD-1 and myosin heavy chain A/MYO-3 (Miller et al., 1983) alternate (Fig. 3 E). Muscle cofilin/UNC-60B is another protein that localizes near dense bodies, appearing alternatively as similar indistinct striations or as discrete bodies interposed between dense bodies (Ono and Ono, 2002), and UNC-60B striations show some overlap with FHOD-1, but discrete UNC-60B–containing bodies lack FHOD-1 (Fig. 3 F). Several LIM domain–containing proteins also localize to faint striations near dense bodies and M lines (Qadota et al., 2007). For one, LIM-8, presumptive M line–associated striations near the cell membrane do not overlap with FHOD-1 (Fig. 3 G, proximal to membrane, arrows), but striations deeper in the lattice overlap extensively with FHOD-1 (Fig. 3 G, distal to membrane). Although the two stains do not perfectly match (based on the presence of magenta and green pixels rather than pure white in Fig. 3 G, merge), we suspect this reflects a limitation to our staining, as animals stained with two different secondary antibodies to probe simultaneously for FHOD-1 show a similar degree of intermingling along striations (Fig. 3 H). Thus, FHOD-1 is in bands that appear to envelope and connect BWM Z line structures, although whether these bands represent a defined structure in the lattice is unclear.
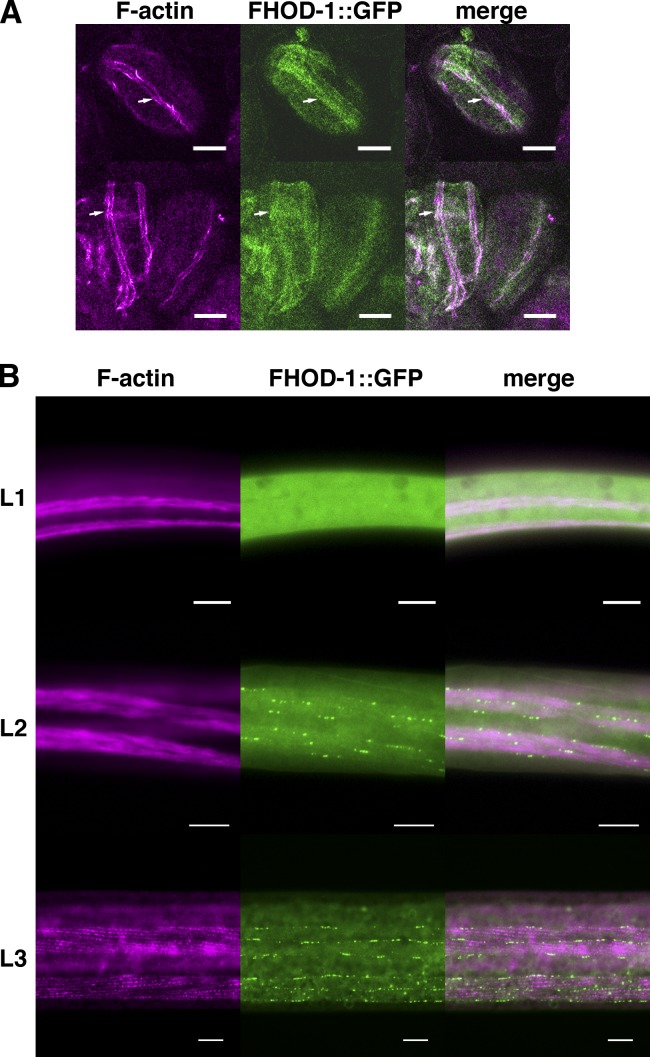
FHOD-1 is seen near BWM Z lines only during particular stages of development. In embryos, the FHOD-1::GFP fusion protein appears diffusely in F-actin–rich embryonic body wall myoblasts (Fig. 4 A, arrows) and is not detectable in BWMs during the first L1 larval stage (Fig. 4 B). Z line–associated FHOD-1 first appears during midlarval development (stage L2; Fig. 4 B) and persists through young adulthood, before vanishing again in older adults. FHOD-1 is also very strongly present in pharyngeal muscles throughout larval development and adulthood and is consistently present in vulval and uterine muscles, but its relationship to sarcomeres in these is unclear (Fig. S1, A–D).
Figure 4.
FHOD-1–containing structures appear in BWMs during mid- and late-larval development. (A) FHOD-1::GFP–expressing embryos stained with fluorescent phalloidin show that FHOD-1 is present diffusely in the elongated bands of F-actin–rich embryonic myoblasts (arrows). (B) In ventral views of FHOD-1::GFP–expressing larvae stained with fluorescent phalloidin, FHOD-1 is not detectable in F-actin–rich BWMs of L1 larvae but appears as puncta along muscle edges and as striations across the BWM contractile lattice in L2 and L3 larvae. Bars, 10 µm.
Mutation of fhod-1 causes modest morphological and functional BWM defects
Worms homozygous for either of two deletion alleles (obtained from the National BioResource Project, Tokyo Women’s Medical University School of Medicine), tm3138 eliminating part of an intron and a DID-encoding exon and tm2363 eliminating part of the FH2 domain and frameshifting the remainder (Fig. 1 A), show no significant lethality or gross morphological defects under our growth conditions (Fig. S2, A and B). However, BWMs of fhod-1(tm3138) and fhod-1(tm2363) mutants are narrower than those of wild type, and the dorsal and ventral pairs of BWMs have wider spaces between paired muscles in the mutants than wild type, with stronger effects from FH2-disrupting tm2363 (Fig. 5, A–C). Confirming these result from loss of FHOD-1, partial fhod-1 knockdown by RNAi partially recapitulates both phenotypes, whereas expression of FHOD-1::GFP partially rescues fhod-1(tm2363) (Fig. S2, C–E). The narrow mutant BWMs reflect narrow muscle cells with fewer striations per cell (Fig. 5, B and D). Additionally, dense bodies of mutant BWM cells appear irregular in shape and spacing (Fig. 3 D), but striations appear normal when stained for F-actin (Fig. 5 B), and fhod-1 mutants show no gross crawling or swimming defects (Fig. S2 F).
Figure 5.
BWM cell growth and positioning are aberrant in fhod-1 mutants. (A) A schematic lateral view of C. elegans (anterior to the left) with a cross section view at the asterisk shows positions of pharyngeal muscles (orange) and BWMs (yellow). The squashed body outline in cross section (dashed line) is consistent with a fixed animal mounted for microscopy, as in B. Dimensions quantified in C are indicated in the cross section schematic. (B) Dorsal and cross section views (yz projections) of fluorescent phalloidin-stained young adults show that fhod-1(tm2363) BWMs are narrower and spaced further apart than wild type. Dotted lines indicate individual BWM cell boundaries. Bars: (dorsal) 50 µm; (yz) 10 µm. (C) Young adult fhod-1 mutants (tm3138 and tm2363) have similar body widths as wild type (wt) but narrower muscles, and fhod-1(tm2363) animals have wider gaps between paired muscles at dorsal and ventral surfaces. Results shown are representative of two independent experiments. (D) Fluorescent phalloidin-stained fhod-1 (tm2363) BWM cells have normal anterior/posterior lengths and radial thicknesses but reduced lateral widths. (E) Ventral views of BWMs in fluorescent phalloidin-stained L1-, L2-, and L3-stage larvae. Bars, 10 µm. (F) Discrepancy between wild-type and fhod-1 mutant (tm2363) BWM widths, expressed as a percentage of total body width, is apparent in L2 and L3 but not L1 stages. Results shown are representative of two independent experiments. For all quantitative results, mean values of n measurements are presented. Error bars indicate one standard deviation. n.s. indicates not significant, P > 0.05. **, P < 0.001.
BWM cells assemble new striations during larval development (Moerman and Williams, 2006). The reduced number of striations in mutant cells suggests Z line–associated FHOD-1 may assist in striation assembly. Consistent with this, the narrow BWM phenotype first appears in L2-stage fhod-1(tm2363) larvae (Fig. 5, E and F), the same time that FHOD-1 normally first appears near BWM Z lines (Fig. 4 B), whereas cessation of BWM growth in adulthood correlates with disappearance of Z line–associated FHOD-1. We had anticipated that the widened gap between paired BWMs of fhod-1 mutants would be a secondary consequence of narrow BWMs, but this phenotype is already present in newly hatched L1-stage fhod-1(tm2363) animals (Figs. 5 E and S2 G), suggesting a muscle-positioning role for FHOD-1 during embryogenesis. Possibly related to this, a significant proportion of fhod-1 mutants grown under alternative conditions experience lethal embryonic body elongation defects (Vanneste, C., and P. Mains, personal communication).
Despite an abundance of FHOD-1 in pharyngeal muscles, we noted no difference in pharynx morphology or F-actin organization for fhod-1 mutants. Vulval muscles are slightly smaller in fhod-1 mutants compared with wild type (Fig. S3, A and B), but this does not cause them to retain embryos in the uterus (20 ± 5 embryos per mutant young adult vs. 20 ± 3 per wild-type young adult; n = 14 and 16 animals, respectively). Mutants for fhod-1 lay eggs at a slower rate and have a smaller total brood size than wild type (Fig. S3, C–G), but it is not clear whether this relates to FHOD-1 functions in the egg-laying muscles or alternative roles in the gonad.
Mutation of cyk-1 but no other formin gene exacerbates fhod-1(tm2363) BWM defects
Considering the mildness of fhod-1 mutant phenotypes, we explored whether additional formins play overlapping roles in BWM actin organization. As FOZI-1 is a divergent nuclear protein that shows no ability to interact biochemically with F-actin (Johnston et al., 2006; Amin et al., 2007), we limited our analysis to the remaining five conventional formins. Based on Western blots using antibodies specific to the appropriate FH2 domains, CYK-1, FRL-1, INFT-1, and INFT-2 do not undergo compensatory up-regulation in response to fhod-1(tm2363) (Fig. S4, A and B). To probe genetically for overlapping BWM functions, we looked for movement defects among strains bearing deletion or deletion/insertion alleles of each formin gene (provided by the National BioResource Project and the C. elegans Gene Knockout Consortium).
Strains homozygous for the formin alleles frl-1(ok460), inft-1(gk386), and inft-2(ok1296) are viable, whereas homozygous daam-1(tm2133) and cyk-1(ok2300) progeny of heterozygous parents are sterile but survive to adulthood and can be identified by PCR. The alleles daam-1(tm2133) and cyk-1(ok2300) delete much of their respective FH2-coding sequences (Fig. 1 A), whereas Western blots show that frl-1(ok460), inft-1(gk386), and inft-2(ok1296) eliminate FH2-containing products (Fig. S4 B), suggesting the products of all five alleles are null for actin assembly activity. Neither frl-1(ok460) nor inft-2(ok1296) causes movement defects alone or in combination with deletion of fhod-1 or any other nonessential formin, and neither causes BWM F-actin defects (Fig. S4 C). Among adult inft-1(gk386) animals, 25% exhibit immobility of the posterior body, a phenotype recapitulated by RNAi targeting inft-1, and a combination of inft-1(gk386) or inft-1(RNAi) with fhod-1(tm2363) results in aberrant rolling movements of mobile adults, but none of these phenotypes correlate with BWM F-actin defects (Fig. S4 C). Homozygous daam-1(tm2133) animals move slowly, but a combination of daam-1(tm2133) with mutation of fhod-1 or any other nonessential formin causes no further movement defects. These results suggest that DAAM-1, FRL-1, INFT-1, and INFT-2 have no direct effect on the BWM F-actin cytoskeleton.
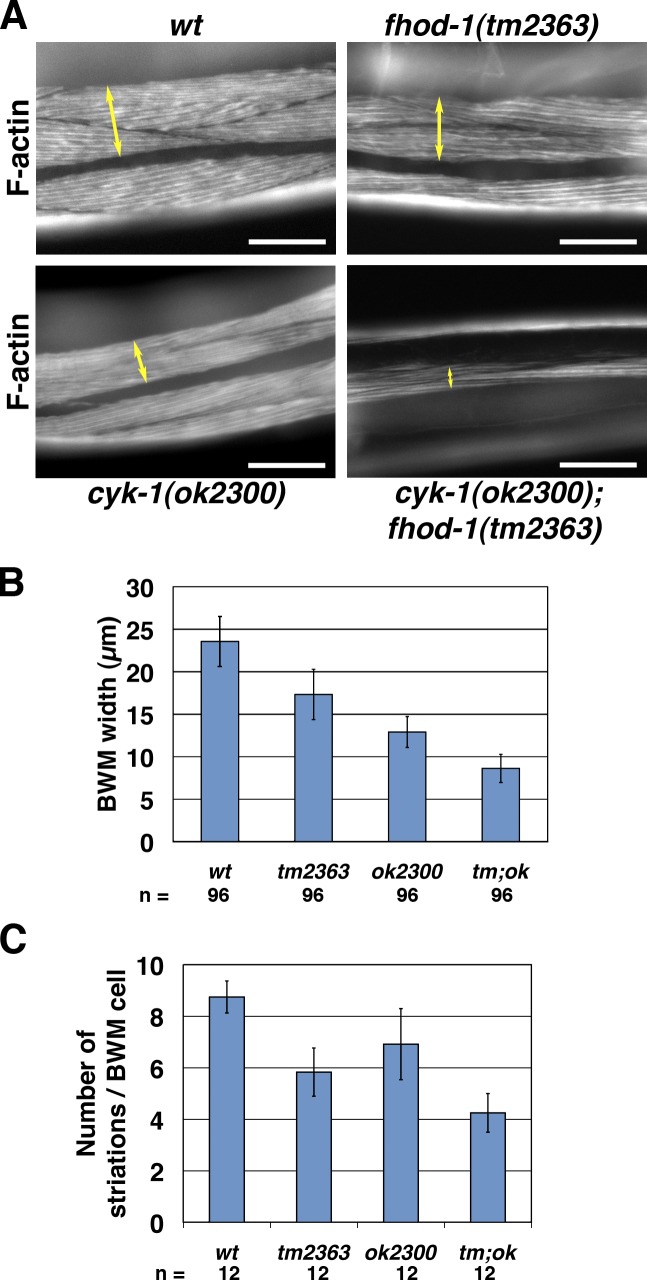
In contrast, cyk-1(ok2300) causes movement and BWM F-actin defects that are exacerbated by fhod-1 mutation. BWMs of homozygous cyk-1(ok2300) animals resemble those of fhod-1 mutants, being spaced further apart and composed of narrow cells with few striations, whereas BWMs of double cyk-1(ok2300); fhod-1(tm2363) homozygotes are profoundly stunted, with even fewer striations per cell (Fig. 6, A–C). Consistent with this, cyk-1(ok2300) animals move slowly, and double cyk-1(ok2300); fhod-1(tm2363) mutants move even more so. Both strains also exhibit limpness in their middle and posterior bodies, although this does not correlate with any specific defect of the BWM cells in these regions, indicating that CYK-1 may also indirectly affect BWM motility. Non-BWM functions for these formins are also suggested by a collection of phenotypes that includes thin cyk-1(ok2300) bodies and even thinner and shorter double cyk-1; fhod-1 mutants, protruding cyk-1(ok2300) vulvas, absence of vulval and uterine muscles from both strains, small cyk-1(ok2300) gonad size exacerbated by fhod-1 mutation, and an arrest of a subset of double cyk-1; fhod-1 mutants as L1 larvae with detached pharynxes (Fig. S5, A–C). Demonstrating that these phenotypes are cyk-1 dependent, expression of C-terminally GFP-tagged CYK-1 from a genomically integrated transgene rescues wild-type motility and body morphology (Fig. S5 A) and even weakly rescues fertility, with ∼2–15 progeny per rescued individual (vs. hundreds for wild type). Thus, CYK-1 is important for several functions, including an overlapping role with FHOD-1 in normal BWM development.
Figure 6.
cyk-1 mutation exacerbates BWM defects of fhod-1 mutants. (A) Dorsal views of fluorescent phalloidin-stained animals show the lateral widths of BWMs (double arrows) in fhod-1(tm2363) and cyk-1(ok2300) mutant animals are smaller than wild type (wt), whereas BWM widths in double cyk-1(ok2300); fhod-1(tm2363) animals are even smaller. Bars, 50 µm. (B) BWM lateral widths were measured for wild type, fhod-1 mutants (tm2363), cyk-1 mutants (ok2300), and double mutants (tm;ok). (C) Striations per BWM cell were counted for the same strains. In B and C, mean values of n measurements are presented. Error bars indicate one standard deviation. Results are significantly different in all pairwise comparisons (P < 0.01).
CYK-1 contributes to maintenance of adult BWM actin organization
Despite the requirement for CYK-1 in cytokinesis (Swan et al., 1998; Severson et al., 2002), homozygous cyk-1(ok2300) progeny of heterozygous parents reach adulthood, suggesting CYK-1 protein or cyk-1 mRNA is maternally inherited. To deplete CYK-1 more completely than what may occur in cyk-1(ok2300) animals, we treated worms for RNAi against cyk-1, which should eliminate zygotic and maternal transcripts. We accommodated the essential cytokinetic role of CYK-1 by initiating RNAi on L1 larvae, which have already generated the majority of their BWM cells (Sulston and Horvitz, 1977; Sulston et al., 1983). Affinity-purified antibody raised against the CYK-1 FH2 domain (anti–CYK-1) recognizes in wild-type extracts ∼160- and 170-kD proteins (Fig. 7 A, top arrows), consistent with a predicted size of 160 kD for full-length CYK-1, as well as smaller ∼87-, 75-, and 72-kD products (Fig. 7 A, bottom arrows). Confirming these correspond to FH2 domain–containing CYK-1 products, 3 d of RNAi reduces the putative full-length signal to undetectable levels, whereas the smaller signals are reduced by ∼90%, as compared with control-treated animals.
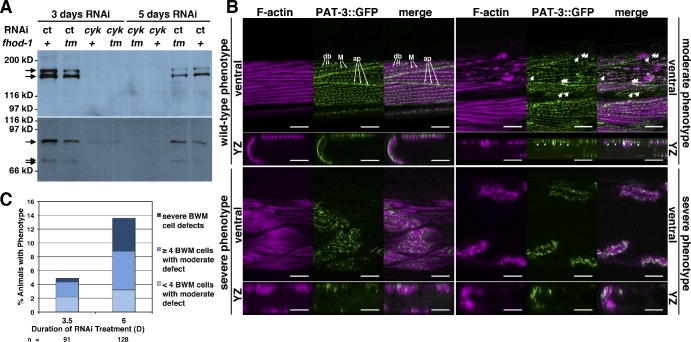
Figure 7.
Prolonged cyk-1 RNAi disrupts adult BWM cytoskeletal organization. (A) Anti–CYK-1 Western blot of normalized worm extracts shows RNAi targeting cyk-1 (cyk) but not control RNAi (ct) reduces detectable CYK-1 (arrows) in wild type (+) and fhod-1 mutants (tm). (B) Ventral and cross section (yz projection) views of cyk-1(RNAi) animals expressing PAT-3::GFP and stained with fluorescent phalloidin show various BWM phenotypes. In wild-type phenotype BWMs, F-actin occupies regular striations in a layer close to the body wall, and PAT-3::GFP decorates dense bodies (db), attachment plaques (ap), and M lines (M). In moderate phenotype BWMs, F-actin is patchy, leaving some GFP-positive bodies free of F-actin (arrows). In severe phenotype BWMs, striated organization is lost, and F-actin is no longer concentrated near the body wall. Bars, 10 µm. (C) The number of animals exhibiting moderate or severe phenotypes increases with longer RNAi treatment. Results shown are typical of two independent experiments.
After reaching adulthood while subject to RNAi (∼2.5 d), adults given control RNAi treatment appear uniformly wild type, whereas some cyk-1(RNAi) animals appear wild type, but others resemble cyk-1(ok2300) animals with thin bodies, protruding vulvas, limp posteriors, and reduced gonads. This variation suggests the rate of CYK-1 depletion varies between animals, with early depletion correlating with cyk-1(ok2300)–like phenotypes. Fluorescent phalloidin staining of cyk-1(RNAi) animals shows that their BWM contractile lattices are normal, with the exception of cyk-1(ok2300)–like animals that have narrow lattices. Furthermore, GFP fluorescence of a functional β integrin/PAT-3::GFP fusion (Plenefisch et al., 2000) present in the RNAi-treated strain shows normal-appearing dense bodies, attachment plaques, and M lines (Fig. 7 B, wild-type phenotype).
With 3 d or longer RNAi treatment, novel phenotypes appear. Beyond the posterior limpness typical of cyk-1(ok2300) mutants, a growing percentage of cyk-1(RNAi), but not control animals, appears fully paralyzed or dead, rising from a few percentages of the population on day 3 of RNAi to roughly one fifth on day 5. Furthermore, BWMs of some cyk-1(RNAi), but not control animals, exhibit F-actin striations that are discontinuous and irregular in width, with the appearance of some presumptive PAT-3::GFP–positive dense bodies lacking F-actin (Fig. 7 B, moderate phenotype, arrows). The number of animals with this phenotype and the number of affected BWM cells per animal increase with further treatment (Fig. 7 C).
With longer RNAi treatment, the BWM contractile lattice in some cyk-1(RNAi), but not control animals, becomes completely disorganized. PAT-3::GFP–associated dense bodies form whorls or clusters at the BWM cell surface or redistribute to the cell interior, and F-actin is no longer confined to a layer near the body wall but reaches deeper into the cytoplasm (Fig. 7 B, severe phenotypes). Typically, 50–100% of BWM cells exhibit this phenotype in affected animals, and the number of affected animals increases with longer treatment (Fig. 7 C).
Consistent with absence of Z line–associated FHOD-1 in adult BWMs, fhod-1(tm2363) does not affect these novel cyk-1(RNAi)–induced BWM defects. As expected, RNAi-treated fhod-1 mutants exhibit irregular dense bodies and thin BWMs typical of fhod-1(tm2363) animals, with the addition of some animals with extremely thin BWMs and the appearance of some larvae with detached pharynxes, as seen among double cyk-1(ok2300); fhod-1(tm2363) mutants (Figs. 6 A and S5 C). However, the frequencies of modest and severe cyk-1(RNAi)–induced BWM phenotypes are unchanged. Considering the rolling phenotype of inft-1(RNAi); fhod-1(tm2363) animals, we also combined inft-1(RNAi) with cyk-1(RNAi) and/or fhod-1(tm2363) but found no effect on cyk-1(RNAi)–induced phenotypes. Neither did we see an effect after knockdown of the related INFT-2 in any combination with these perturbations. Therefore, although CYK-1 and FHOD-1 both contribute to larval BWM development, CYK-1 alone contributes to maintenance of the adult BWM contractile lattice.
CYK-1 associates with sarcomere Z lines in BWM cells
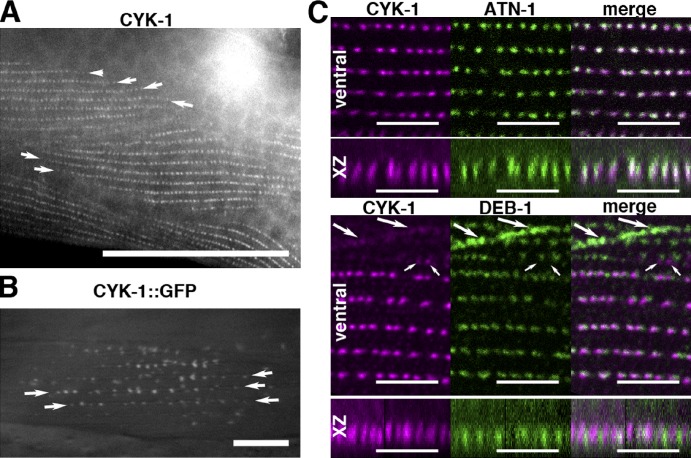
Immunostaining with anti–CYK-1 decorates BWMs in a striated pattern, a distribution confirmed by fluorescence in BWMs of CYK-1::GFP–expressing animals (Figs. 8, A and B; and S5 D). Surprisingly, anti–CYK-1 also decorates striations in cyk-1(ok2300) BWMs, possibly caused by maternal or truncated mutant products (Fig. S5 E). Unlike for FHOD-1 striations that appear as fuzzy continuous bands, CYK-1 striations are rows of puncta (Fig. 8, A and B). Double immunostaining for CYK-1 and the Z-line markers DEB-1 or ATN-1 shows that these puncta are dense bodies, with the strongest signal overlap occurring between CYK-1 and ATN-1 (Fig. 8 C). CYK-1 also weakly decorates DEB-1–stained attachment plaques at the boundaries between muscle cells (Fig. 8 C, large arrows) and small, unidentified patches near the cell edges (Fig. 8 C, small arrows). Consistent with its role in adult BWMs, CYK-1 is present at these structures throughout larval growth and adulthood. Thus, the two formins important for striated muscle development localize near or at sarcomere Z lines.
Figure 8.
CYK-1 is present on BWM Z-line structures. (A) Dorsal view of an adult reveals anti–CYK-1–stained striations formed of puncta. Arrows indicate ends of striations. Bar, 50 µm. (B) Dorsal view of an animal expressing CYK-1::GFP from an extrachromosomal array reveals striations of GFP-positive puncta. Arrows indicate ends of striations. Bar, 10 µm. (C) BWMs of animals double stained for CYK-1 and either ATN-1 or DEB-1, seen in ventral views and side views of the discontinuous Z line (xz projection), show that CYK-1 overlaps most strongly with ATN-1. Large arrows indicate DEB-1–containing attachment plaques with some associated CYK-1. Small arrows indicate CYK-1 patches lacking DEB-1. Bars, 5 µm.
Discussion
The structure of the striated muscle actin cytoskeleton has been studied extensively, but the mechanism by which actin filaments are incorporated into sarcomeres in the contractile apparatus remains unclear. Genetic and microscopic experiments suggest that during striated muscle development, sarcomeric actin organization originates at integrin-based adhesion complexes, with further actin recruitment as sarcomeres mature (Sparrow and Schöck, 2009). For vertebrates, this corresponds to early appearance of stress fiber–like premyofibrils in which actin filament barbed ends are bound to adhesion-associated Z bodies. Later, maturation into striated myofibrils includes fusion of Z bodies into broad Z bands, incorporation of additional actin filaments, and dissociation of most myofibrils from adhesion sites (Sanger et al., 2005, 2010). In contrast, maturing nematode Z lines remain associated with adhesion sites but develop α-actinin–rich extensions to which additional filaments attach, creating hybrid Z line/adhesion dense bodies (Moerman and Williams, 2006). Thus, integrin adhesion structures and maturing Z lines seem likely sites of sarcomeric actin nucleation.
Even with maturation, sarcomeric actin remains dynamic. Actin subunits exchange at both filament ends, despite respective barbed and pointed end–capping by CapZ and tropomodulin (Ono, 2010). These dynamics, particularly at the pointed end, maintain proper filament length (Littlefield et al., 2001; Mardahl-Dumesnil and Fowler, 2001; Bai et al., 2007), but it is unclear to what degree they reflect nucleation of new filaments versus remodeling of old ones.
Based on demonstrated or predicted nucleation activity, three factors have been proposed as ANFs in striated muscles. Tropomodulin-related leiomodin-2 is a mammalian cardiac muscle protein that nucleates actin filaments (Chereau et al., 2008). However, leiomodin-2 appears in cardiomyocytes after myofibril formation, arguing against an early nucleation role but consistent with a suggested role in regulating filament length through its ability to compete with tropomodulin and, thus, uncap pointed ends (Tsukada et al., 2010). A second striated muscle ANF is formed by a complex of the sarcomeric protein nebulin with N-WASP, a protein better known for partnering with the Arp2/3 complex ANF. In murine skeletal muscle, N-WASP–nebulin complexes appear at Z lines in response to IGF-1 signaling, triggering new filament assembly and muscle hypertrophy, indicating this path is at least important for actin assembly in preformed sarcomeres (Takano et al., 2010). Finally, FHOD3 is a mammalian formin found in cardiomyocyte sarcomeres and whose absence from cultured cells blocks formation of striated myofibrils, suggesting an early role in myofibrillogenesis (Taniguchi et al., 2009; Iskratsch et al., 2010). We provide here genetic evidence demonstrating that sarcomere-associated formins promote growth and maintenance of the striated muscle contractile lattice in vivo in the context of an intact animal.
The nature of formin–actin interactions suggests that filaments assembled by Z line–associated formins would be predisposed for incorporation into sarcomeres. That is, processive capping of filaments by formins would retain barbed ends of assembling filaments near Z lines, where they must ultimately attach. Filament elongation, either at formin-bound barbed ends or at free pointed ends, would drive pointed ends toward the sarcomere center to intermingle with myosin filaments. Two formins of C. elegans, CYK-1 and FHOD-1, are present in sarcomeres of developing striated BWMs on or near Z-line dense bodies (Figs. 3 D and 8 C). Demonstrating their importance here, the BWM contractile lattices of worms lacking the nonessential FHOD-1 or expressing low levels of the essential CYK-1 grow slowly and assemble fewer striations (and therefore fewer sarcomeres) than wild type, whereas worms bearing mutations affecting both formins have profound reductions in lattice growth (Figs. 5 and 6). Importantly, BWM cells do not arise through fusion of myoblasts, whose proliferation would depend on CYK-1 for its role in cytokinesis (Swan et al., 1998; Severson et al., 2002), arguing against cell division defects as an indirect cause for diminished mutant muscle cells. Rather, we suggest these formins contribute to contractile lattice growth by nucleating new sarcomeric actin filaments.
The BWM contractile lattice grows through gradual addition of new striations plus the growth of preexisting striations by increases in actin filament length and number (Moerman and Williams, 2006). Interestingly, the two formins differ in their effects on these. Compared with the effects of diminished CYK-1, the loss of FHOD-1 causes greater reduction of striation number but lesser reduction of overall contractile lattice width (Fig. 6, B and C). Thus, FHOD-1 may act primarily during formation of new striations, whereas CYK-1 may function preferentially within preformed striations. It remains to be determined whether these differences relate to the relative stronger presences of FHOD-1 along developing BWM cell edges and CYK-1 among internal striations (compare Figs. 3 C and 8 A).
Surprisingly, although the total F-actin content of formin mutant BWM cells is reduced, the F-actin density in remaining striations in both formin mutants appears normal (Fig. 6 A), and even thin-section electron microscopy reveals no significant differences in myosin to actin filament ratios (not depicted). Thus, reduction of total F-actin during BWM development may be matched by reduction in organization of other sarcomere components. Also notable, double cyk-1; fhod-1 mutants retain some F-actin striations, but the nature of the cyk-1 mutant used (homozygous progeny rescued maternally for viability) prevents us from determining whether these result from an additional ANF or from maternally derived CYK-1.
Although both formins are present through much of BWM growth, only CYK-1 is retained at adult Z lines. Consistent with a later role for CYK-1, its depletion from adults results in formation of gaps in BWM F-actin striations and subsets of putative dense bodies lacking associated F-actin (Fig. 7 B, moderate phenotype, arrows). With longer periods of CYK-1 depletion, BWMs of some animals lose their striated organization and redistribute their F-actin from a position near membrane-associated Z lines to a cytoplasmic mass (Fig. 7 B, severe phenotypes). The moderate phenotype is consistent with loss of actin assembly from a subset of Z-line structures, with remaining Z lines continuing to organize actin, whereas the more severe phenotype might be expected from complete loss of Z line–associated assembly, leaving the large pool of muscle actin to polymerize at inappropriate locations in the cytoplasm. However, these phenotypes occur with low frequency and only after prolonged CYK-1 depletion (Fig. 7 C), suggesting that dynamics of adult muscle actin are likely to be predominated by formin-independent remodeling of filaments, with only infrequent formin-dependent filament replacement.
An interesting question is why two formins have adopted similar locations near the Z lines of the same muscle cell. Although cyk-1 and fhod-1 mutations cause similar BWM phenotypes, it remains possible that CYK-1 and FHOD-1 make distinct contributions to actin organization because of differing biochemical properties. Like other DIA homologues, CYK-1 nucleates and processively caps actin filaments in vitro (Amin et al., 2007; Neidt et al., 2008). For FHOD-1–related formins, only mammalian FHOD3 has been characterized in vitro, and its ability to nucleate has yet to be demonstrated. Rather, FHOD3 inhibits barbed end elongation in pyrene actin assembly assays, even in the presence of profilin, suggesting it caps barbed ends (Taniguchi et al., 2009). However, in striated muscles, where pointed-end dynamics often determine filament length, such effects on barbed-end dynamics by a nucleating formin may be irrelevant.
We offer an alternative suggestion that the presence of two formins near the same Z line results from dual mechanical demands placed on BWMs: to contract in a rapid, coordinated manner and to adhere tightly to the body wall. The ability of muscles to contract rapidly correlates with the presence of muscle-specific isoforms of sarcomeric proteins and, frequently, a striated sarcomere organization. Localization of FHOD-1 near BWM Z lines resembles a reported localization of FHOD3 near cardiomyocyte Z bands (Iskratsch et al., 2010), suggesting that association with muscle sarcomeres may be conserved among formin homology domain (FHOD) subfamily proteins. This might be through binding to a muscle-specific sarcomere component positioned near Z lines or perhaps even to the molecular scaffold that organizes Z lines. Regarding this last possibility, fairly large distances separate nematode Z-line dense bodies, and how dense bodies align into oblique striations is unknown, but the appearance of FHOD-1 in bands that intersect and intermingle with dense bodies (Fig. 3 D) resembles what might be expected for a striation-forming scaffold.
Adhesion of BWMs is mediated in part through integrin-based interactions of CYK-1–containing dense bodies with the ECM. Formins have not previously been similarly associated with adhesion sites in vertebrate muscles, but nonmuscle cells have sarcomere-containing stress fibers associated with dense body–like focal adhesions, and their formation depends on CYK-1–related DIA subfamily formins (Peng et al., 2003; Tojkander et al., 2011). Mammalian DIA-dependent activity copurifies with integrin adhesion complexes (Butler et al., 2006), suggesting that organization of sarcomeric actin from adhesion structures may be a conserved function for DIA subfamily proteins. Thus, the worm BWM contractile lattice may result from the marriage of two conserved sarcomere-organizing systems, a DIA-based system associated with adhesion structures and an FHOD-based system associated with muscle-specific components.
It will be interesting to see whether DIA and FHOD isoforms play dual roles in muscles of other organisms. It is unknown how actin filaments are assembled at membrane-associated Z bodies in vertebrate premyofibrils, but our results suggest that DIA formins could function here. When Z bodies then fuse into myofibril Z bands and muscle-specific isoforms are incorporated into the sarcomeres, FHOD formins would be recruited, and as Z bands dissociate from adhesion structures, DIA formins would be lost. Interestingly, although cardiomyocytes depleted for FHOD3 lose mature myofibrils, they retain FHOD3-independent stress fiber–like structures that might be premyofibrils (Iskratsch et al., 2010). Such a maturation-based transformation may be partially recapitulated in C. elegans, as CYK-1 appears at immature dense bodies in L1 larvae, followed later by the appearance of FHOD-1 near maturing dense bodies. However, as dense bodies retain their nature as adhesion sites, CYK-1 remains present, explaining the relatively mild effect of loss of FHOD-1 in C. elegans.
In summary, the importance of sarcomere-associated formins to growth and maintenance of the striated muscle contractile lattice in C. elegans provides the first in vivo demonstration that formins help organize this structure. Considering the strong conservation of formins, as well as the conserved nature of the muscle sarcomere, such a role is likely to be widespread across diverse species.
Materials and methods
Sequence analysis and structure predictions
Seven worm genes predicted to encode FH2-containing proteins were identified through tBLASTn searches (Altschul et al., 1990) of the C. elegans genome, using the budding yeast Saccharomyces cerevisiae formin Bni1p FH2 domain as a reference. Searches referencing the conserved Bni1p G domain or DID, the nonconserved N-terminal portions of Mus musculus FMN2 or Drosophila melanogaster CAPU, or the C-terminal portions of M. musculus INF1 and INF2 or D. melanogaster FORM3 revealed no additional worm formins. Formins from M. musculus, D. melanogaster, the purple sea urchin Strongylocentratus purpuratus, and the parasitic nematodes Ascaris suum and Brugia malayi were similarly identified by tBLASTn and BLASTp.
Coding sequences for full-length fhod-1 and frl-1, bases C1389 to STOP of cyk-1 isoform b, and bases A344 to STOP of inft-2 were confirmed by sequencing of cDNA clones yk850e7(fhod-1), yk1009g02(frl-1), yk576g9(cyk-1), and yk1693g10(inft-2), all provided as gifts by Y. Kohara (National Institute of Genetics, Mishima, Japan). The 5′ portions of cyk-1 and inft-2 and full-length daam-1 and inft-1 coding sequences were confirmed by sequencing cDNAs isolated by PCR amplification from the ProQuest C. elegans cDNA library for yeast two hybrid screening (Invitrogen) using primers homologous to predicted formin cDNA sequences with primers homologous to the pPC86 library plasmid. START codons for inft-1, inft-2, and daam-1 were verified by identification of upstream in-frame STOP codons in PCR-derived cDNAs. All cDNA-derived sequences match those currently predicted by WormBase, with the exceptions of the following: cyk-1 differed by G472A (Glu158Lys) and A571T (Thr191Ser); inft-2 differed by T723C (silent) and absence of the predicted third exon; and inft-1 differed by absence of the predicted last 11 bp of exon 3 and the first 103 bp of exon 4. All of these changes were recovered repeatedly in our isolated cDNAs.
To compare formin sequences, homologous amino acid sequences were aligned using the MegAlign module of Lasergene 8.0 (DNASTAR, Inc.). With the exception of INFT-2 N-terminal domains, conserved homology domains of C. elegans formins were identified based on alignments to M. musculus DIAP1/mDia1 and DIAP2/mDia3, human FHOD1, and S. cerevisiae Bni1p, for which G domain, DID, dimerization domain, and/or FH2 domain structures have been determined (Shimada et al., 2004; Xu et al., 2004; Otomo et al., 2005; Rose et al., 2005; Schulte et al., 2008). Approximate boundaries for conserved domains were based on structural predictions generated by PHYRE 2.0 (Kelley and Sternberg, 2009). The INFT-2 N terminus shows no obvious homology to mouse, human, or yeast formins by BLAST (basic local alignment search tool) but is predicted by PHYRE 2.0 to fold into a partial DID. Furthermore, BLAST searches reveal homology to B. malayi (18% identity) and A. suum (27%) INFT-2 homologues in regions where those homologues are predicted to adopt DID and dimerization domain folds based on BLAST and PHYRE 2.0 predictions, allowing a tentative identification of the C. elegans INFT-2 DID and dimerization domains.
For generation of phylogenetic trees, the following programs of the PHYLIP 3.65 suite (copyright University of Washington and J. Felsenstein, University of Washington, Seattle, WA) were used: PROTDIST to compute distance matrices for aligned sequences using the Jones, Taylor, and Thornton model of amino acid change (Jones et al., 1992), FITCH to construct unrooted phylogenetic tree estimations using the Fitch-Margoliash criterion, and DRAWTREE to render trees graphically. FH2 domain and N-terminal trees (Fig. 1 B) were based on alignments of core 130–amino acid sequences of the FH2 domain or amino acid sequences N terminal to the FH1 domain, respectively.
Plasmids
FH2-encoding sequences were amplified from cDNAs of cyk-1 (bases 2,413–3,603), fhod-1 (2,443–3,618), frl-1 (1,831–3,036), inft-1 (382–1,515), and inft-2 (1,780–2,979), and subcloned into the expression vector pGEX-6p-3 (GE Healthcare), yielding pGEX-6p-(CYK-1, FHOD-1, FRL-1, INFT-1, or INFT-2)FH2 for production of the recombinant protein. For production of double-stranded RNA (dsRNA) for RNAi experiments, two nonoverlapping fhod-1 cDNA sequences (bases 561–1,184 and 2,853–3,257) and one each of cyk-1 (2,413–3,603), inft-1 (382–1,515), and inft-2 (720–1,543) were amplified by PCR and subcloned into L4440 (Timmons and Fire, 1998) to generate L4440–fhod-1A, L4440–fhod-1B, L4440–cyk-1, L4440–inft-1, and L4440–inft-2.
Genomic sequences containing the fhod-1 and cyk-1 loci, including 5 kb upstream and 2 kb downstream of their predicted coding sequences, were obtained from the yeast artificial chromosomes Y55F5 for fhod-1 and Y51H12 for cyk-1 (provided by the Wellcome Trust Sanger Institute) and subcloned into plasmid pRS315 (Sikorski and Hieter, 1989) by the method of Gene CATCHR (gene cloning and tagging of C. elegans genes using yeast homologous recombination; Sassi et al., 2005). DNA encoding GFP (with three synthetic introns) was amplified from pPD95.75 (provided by A. Fire, Carnegie Institution of Washington, Washington, DC), appended with a Gly-Ala-Gly-Ala-Gly–coding linker at its 5′ end by PCR, and introduced immediately upstream of the fhod-1 and cyk-1 STOP codons by continuation of the Gene CATCHR method, yielding fhod-1::gfp and cyk-1::gfp. Yeast strain ASHSY2 used for Gene CATCHR was a gift provided by A. Spence (University of Toronto, Toronto, Ontario, Canada). The 18.4-kb fhod-1::gfp and 13.4-kb cyk-1::gfp sequences were each introduced into the worm transformation vector pJKL702 (a gift from J.K. Liu, Cornell University, Ithaca, NY) by standard subcloning to generate pJKL702-fhod-1::gfp and pJKL702-cyk-1::gfp.
C. elegans strains and growth conditions
Worms were maintained and manipulated using standard techniques (Brenner, 1974) and grown at 20°C unless indicated otherwise. For synchronization of worms, gravid animals were washed from dishes and treated with alkaline bleach for 5–6 min to release embryos, which were then hatched overnight in M9. Resultant starved L1-stage larvae were reintroduced to food to resume growth as synchronous populations. An alternative method used when fewer animals were required allowed gravid worms to lay eggs on fresh bacterial lawns for 3–4 h before removal, yielding semisynchronized progeny.
Strains FX02133 (daam-1(tm2133)/(+) V), FX02363 (fhod-1(tm2363) I), and FX03138 (fhod-1(tm3138) I) were gifts from S. Mitani (National BioResource Project for the Experimental Animal Nematode C. elegans, Tokyo Women’s Medical University School of Medicine, Tokyo, Japan). Strains N2 (wild-type Bristol worms), GE2722 (cyk-1(t1568) unc-32(e189)/qC1[dpy-19(e1259) glp-1(q339)] III; him-3(e1147) IV), RB696 (frl-1(ok460) III), RB1280 (inft-2(ok1296) V/nT1[qIs51] (IV;V)), VC909 (inft-1(gk386) III), and VC1895 (cyk-1(ok2300) III/mT1[dpy-10(e128)] (II;III)) were supplied by the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN). Deletion alleles were outcrossed six times to N2 before use in this study. Nonlethal fhod-1(tm2363), fhod-1(tm3138), frl-1(ok460), inft-1(gk386), and inft-2(ok1296) alleles were bred to homozygosity. Allele daam-1(tm2133) was balanced with the recessively lethal chromosomal rearrangement nT1[qIs51] (IV;V) to yield XA8002 (daam-1(tm2133) V/nT1[qIs51] (IV;V)). Homozygosity for daam-1(tm2133), identified by single-worm PCR or by the absence of qIs51-encoded pharyngeal GFP, correlates with sterility and an absence of dark yolk granules in the intestine. Homozygosity for cyk-1(ok2300), identified by single-worm PCR, correlates with thinness, sterility, protruding vulvas, and limp posteriors. Heterozygous cyk-1(ok2300)/(+) populations were maintained through propagation of single worms whose progeny included such thin, sterile animals. All pairwise combinations of nonlethal formin deletions and of daam-1(tm2133)/nT1 with nonlethal formin deletions plus double fhod-1(tm2363); cyk-1(ok2300) mutants were generated through crosses of appropriate outcrossed single mutants.
The FHOD-1::GFP–expressing strain DWP3 (qaIs8001[unc-119(+) fhod-1::gfp]) was generated by microparticle bombardment (Praitis et al., 2001) of pJKL702-fhod-1::gfp into unc-119(ed4) III animals, with recovery of transformants at 25°C followed by six outcrosses to N2. Further outcross to fhod-1(tm2363) animals yielded DWP10 (fhod-1(tm2363) I; qaIs8001[unc-119(+) fhod-1::gfp]). DWP13 (unc-119(ed4) III; upsIs3[unc-119(+) cyk-1::gfp]) was similarly generated by microparticle bombardment of pJKL702-cyk-1::gfp into unc-119(ed4) III animals but without outcross to N2. DWP22 (cyk-1(ok2300) III; upsIs3[unc-119(+) cyk-1::gfp]) was isolated through cross of DWP13 to cyk-1(ok2300)/(+) animals and identifying cyk-1::gfp-bearing cyk-1(ok2300) homozygotes in the F2 progeny through PCR of single animals. Bombardment of pJKL702-cyk-1::gfp also generated worms in which cyk-1::gfp did not stably integrate but was present as an extrachromosomal array. The presumed high copy number of cyk-1::gfp in these allowed easier observation of CYK-1::GFP in BWMs (Fig. 8 B).
Worms bearing rhIs2[pat-3::HA::gfp] I to express β integrin/PAT-3 tagged with HA and GFP were a gift from J. Plenefisch (University of Toledo, Toledo, OH; Plenefisch et al., 2000). DWP20 (fhod-1(tm2363) rhIs2[pat-3::HA::gfp] I) was generated through crosses of these to fhod-1(tm2363) animals. The RNAi-sensitive strain GR1373 (eri-1(mg366) IV) was a gift from S.S. Lee (Cornell University, Ithaca, NY). GR1373 crossed to DWP20 generated in its F2 progeny DWP28 (rhIs2[pat-3::HA::gfp] I; eri-1(mg366) IV) and DWP29 (fhod-1(tm2363) rhIs2[pat-3::HA::gfp] I; eri-1(mg366) IV). RNAi-sensitive strains were maintained at 16°C.
To induce fhod-1(RNAi), inft-1(RNAi), and double fhod-1(RNAi); inft-1(RNAi), GR1373 animals were grown for several generations on dsRNA-producing Escherichia coli strain HT115 as per standard protocols (Wang and Barr, 2005). In brief, cultures of HT115 bearing L4440 plasmids were grown overnight in 2×YT medium (with 12.5 µg/ml tetracycline and 100 µg/ml ampicillin) before 1:100 dilution with fresh medium and further growth for 3 h at 37°C. IPTG was then added to a 0.4-mM final concentration to induce production of dsRNA for 2–4 h at 37°C. Cultures were concentrated fivefold, and 150 µl of concentrated, induced culture of HT115 bearing L4440 control plasmid (lacking worm DNA), L4440–fhod-1A, L4440–fhod-1B, or L4440–inft-1 or a 1:1 mixture of appropriate cultures was seeded onto plates before addition of L4-stage GR1373 animals. For prolonged RNAi treatments of adults, starved L1 DWP28 and DWP29 were placed on plates with 150 µl IPTG-induced HT115 bearing L4440, L4440–cyk-1, L4440–inft-1, L4440–inft-2, or equal pairwise or three-way mixtures of the L4440 formin constructs at 25°C. Fresh induced bacteria was added to adult animals every 3 d. Sterility caused by eri-1(mg366) at 25°C allowed RNAi-treated animals to grow without being overcrowded by their progeny.
Antigen and antibody production and Western blot analysis
Recombinant FHOD-1FH2, CYK-1FH2, FRL-1FH2, INFT-1FH2, and INFT-2FH2 were produced by expression of GST-FH2 fusions in the E. coli strain BL21 bearing the appropriate pGEX-6p-forminFH2 with affinity purification and cleavage from GST as described previously (Amin et al., 2007). Antigens were injected into two rabbits each, and final crude sera were verified for specificity through Western blot analysis of bacterial and whole-worm extracts. Sera with the least nonspecific signals were selected for affinity purification. Affinity-purified anti–CYK-1 (DPMSP1), anti–FHOD-1 (DPMSP2), anti–FRL-1 (DPMSP4), anti–INFT-1 (DPMSP5), and anti–INFT-2 (DPSMP6) were generated by incubation of crude sera with a recombinant FH2 antigen covalently coupled to CNBr-Sepharose followed by washing and elution as performed previously for Bni1pFH1FH2C (Pruyne et al., 2004).
For Western blot analysis of mixed-stage populations of animals, worms were washed from nonstarved plates and resuspended in M9 to 50% (vol/vol) worms. For analysis of adult animals, ∼1,000 adult animals of the appropriate strain were individually picked from plates to M9 before washing and volume reduction to 50% (vol/vol) worm slurry. To pick homozygotes of the sterility-causing allele daam-1(tm2133), progeny of XA8002 that lacked pharyngeal GFP (encoded by the nT1 balancer) and dark intestine granules were selected. To pick homozygotes for the sterility-causing allele cyk-1(t1568), progeny of GE2722 that were coiled and crawled poorly because of homozygosity of the linked unc-32(e189) were selected. To obtain crude worm extracts for Western blot analysis, worm slurries were diluted with 2× reducing sample buffer, boiled for 3 min, subjected to disruption with a 1.7-ml tube tissue homogenizer (VWR International), and boiled again for 3 min. Genomic DNA was sheared in worm extract samples immediately before gel loading by eight passages through an insulin syringe. Samples were first resolved by SDS-PAGE and stained with Coomassie brilliant blue, and intensities of two major protein bands (200 and 180 kD) were quantified to allow normalization of loads for Western blot analysis. For Western blot analysis, affinity-purified formin antibodies were in 1% milk/Tris-buffered saline, pH 8.3/0.1% Tween 20 at 1:100 for DPMSP5, 1:175 for DPMSP6, 1:200 for DPMSP1 and DPMSP2, or 1:500 for DPMSP4. We found that optimal antiformin signals required blots be incubated for 5 h at room temperature with the primary antibody.
Fluorescence and differential interference contrast (DIC) microscopy
For high resolution GFP fluorescence or DIC microscopy of live animals, worms were anaesthetized with 0.1% levamisole/0.01% tricaine or just 0.1% levamisole in M9 for 20 min and then mounted on 2% agarose pads. Immunostaining was performed by the method of Finney and Ruvkun (1990). F-actin was visualized in whole worms by a modification of the same method, with animals suspended in M9 and fixed by the addition of 1 vol of 2× phalloidin mix (160 mM KCl, 40 mM NaCl, 1 mM EGTA, 30 mM Pipes, pH 7.3, 100 mM spermidine-HCl, and 1:125 Alexa Flour 568–phalloidin) and 0.1 vol of 20% formaldehyde (made fresh from paraformaldehyde) followed immediately by three rapid freezes in liquid nitrogen and thaws to near completion in a 65°C bath, with the final thaw followed by fixation for 1 h on ice. For visualization of F-actin in GFP-expressing worms, fixation time was reduced to 30 min to preserve GFP fluorescence. After fixation, worms were washed twice for 1 min with 1 ml Tris–Triton X-100 (100 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 1% Triton X-100) and once for 15 min with 1 ml PBST-B (PBS, 1 mM EDTA, 5 mM NaN3, 0.5% Triton X-100, and 0.1% BSA). Small aliquots of fixed/washed animals were mixed overnight at room temperature in PBST-B/1:250 Alexa Fluor 568–phalloidin and washed the following day three times for 25 min with 0.5 ml PBST-B, once for 25 min with 0.5 ml 1 µg/ml DAPI in PBST-B, and once briefly with 0.5 ml PBST-B before mounting.
The monoclonal antibodies MH24 (anti–DEB-1/vinculin) generated by R.H. Waterston (Washington University School of Medicine, St. Louis, MO; Francis and Waterston, 1985) and 5-6 (anti–MYO-3/myosin A heavy chain) generated by H.F. Epstein (Baylor College of Medicine, Houston, TX; Miller et al., 1983) were obtained through the Developmental Studies Hybridoma Bank (University of Iowa). Monoclonal antibody MH35 (anti–ATN-1/α-actinin) generated by R.H. Waterston (Francis and Waterston, 1985) was a gift from P. Hoppe (Western Michigan University, Kalamazoo, MI), rabbit anti–UNC-60B/cofilin (Ono and Ono, 2002) was a gift from S. Ono (Emory University, Atlanta, GA), and Benian-7 (anti–LIM-8; Qadota et al., 2007) was a gift from G. Benian (Emory University). Mouse anti-GFP (Rockland Immunochemicals) was used to detect GFP fusion proteins by immunostaining. Antibodies were diluted in PBST-A (PBST-B with 1% BSA) at 1:200 for DPMSP1, DPMSP2, MH24, and anti–UNC-60B, 1:500 for all secondary antibodies (Texas red–conjugated goat anti–rabbit and FITC-conjugated goat anti–mouse or reversed fluorophore/species combinations; Rockland Immunochemicals), 1:1,000 for mouse anti-GFP, 1:5 × 106 for 5-6, and 1:107 for MH35. Before use, DPMSP2 was precleared of nonspecific antibodies by incubation with fixed fhod-1(tm2363) animals, and anti-GFP was precleared by incubation with fixed N2 animals, both for 1 h at room temperature. Secondary antibodies were precleared by incubation with fixed N2 animals overnight at room temperature.
For single-label immunostains (Figs. 2 B, 3 C, 8 A, and S5 E), Texas red–conjugated antibodies were used. For double-label immunostains (Figs. 3, D–H; and 8 C), magenta images show fluorescence from Texas red–conjugated antibodies, and green images show fluorescence form FITC-conjugated antibodies. For all F-actin stains (Figs. 2 C, 3 B, 4, A and B; 5, B and E; 6 A, 7 B, S1, B–D; S3 A, S4 C, and S5, B and C), fluorescence is from Alexa Fluor 568–labeled phalloidin.
Wide-field images were acquired at room temperature (∼21–22°C) on a research upright microscope (Eclipse 90i; Nikon) through CFI Achro Flat Field dark low 10×/NA 0.25 (Figs. 2 C and S2 A), CFI Plan Apochromat 40×/NA 1.0 oil immersion (Figs. 2, B and C; 4 B, L3 images; 5, B [dorsal view] and E [L3 images]; 6 A, S3 B, and S5, A–C and E), or CFI Plan Apochromat violet-corrected 60×/NA 1.4 oil immersion (Figs. 4, A and B, L1 and L2 images; 5 E, L1 and L2 images; S3 A, and S5 B) objectives with a digital monochrome charge-coupled device camera (CoolSNAP HQ2; Photometrics) driven by NIS-Elements AR acquisition and analysis software (version 3.1; Nikon). Confocal images (Figs. 3, B and D–G; 5 B, yz view; 7 B, 8 C, S1, B–D; and S5 D) were acquired at 37°C through an HCX Plan Apochromat 63×/NA 1.4 oil λ objective on a laser-scanning confocal microscope (SP5; Leica) driven by LAS AF software (version 2.2.0, build 4758; Leica). Cross-sectional views (Figs. 3 D, xz views; 5 B, yz views; 7 B, yz views; and 8 C, xz views) were generated from three-dimensional reconstructions derived from z stacks of confocal images using LAS AF. Measurements of image dimensions were performed using NIS-Elements AR or LAS AF. Images were linearly processed to enhance contrast through Photoshop CS4 (Adobe).
Behavioral and viability assays
The viability of worm strains was tested as follows. 20 young adult hermaphrodites, defined as having been adults for ≤24 h, were placed singly on separate, fresh bacterial lawns and permitted to lay eggs for 3 h before removal. Each plate was scored for the number of eggs laid, the number of unhatched eggs left the following day, and the number of viable worms each of the following 2 d.
To determine brood sizes, 20 L4 hermaphrodite animals were placed singly on separate, fresh bacterial lawns at 20°C and then transferred to new plates with fresh bacterial lawns every 12 h, and eggs were counted on vacated plates. This was repeated until no animal was laying any further eggs. For determination of morphology of newly laid embryos, young adult hermaphrodites were set to fresh bacterial lawns at 20°C to permit egg laying for 3 h and then removed, and plates were inspected under an a stereoimaging microscope (SZ61; Olympus) to score embryos within eggs using the morphological categories: before morphogenesis, lima bean, comma, and 1.5-fold, twofold, or threefold (Wood, 1988). Worms were tested for their ability to lay eggs after pharmacological stimulation of the vulval muscles by the method of Trent et al. (1983).
Motility of worms was tested by immersion of animals in M9 and recording videos of swimming animals at room temperature through a stereoimaging microscope (SZ61TR; Olympus) using an digital camera (DP-20; Olympus) driven by DP2-BSW software (Olympus). Animals were scored for whether after 5 min they were thrashing and for their rate of thrashing by counting the number of full oscillations over a 5-s period. Motility of wild-type and fhod-1(tm2363) animals was so observed, but cyk-1(ok2300) or cyk-1(ok2300); fhod-1(tm2363) mutants were not similarly scored, as these animals exhibit flaccid posteriors and coordination defects, which seemed likely to interfere with swimming movements.
Statistical analysis
Numerical results of experiments comparing two groups (Figs. 5, D and F; S2, B, F, and G; and S3 F) were subjected to unpaired, two-tailed Student’s t tests, with P < 0.05 considered statistically significant. Results comparing three or more groups (Figs. 5 C, S2, D and E; and S3 B) were subjected to one-factor analysis of variance, whereas those comparing groups that differ by time and strain (Fig. 3 C) were subjected to two-factor analysis of variance, both procedures being followed by Fisher’s least significant difference posthoc testing to identify differences between groups. Groups were considered different when differences between their means exceeded the 95% confidence interval. Numerical data in the main text and figures are presented as means ± one standard deviation.
Online supplemental material
Fig. S1 shows FHOD-1::GFP localization in pharyngeal, vulval, and uterine muscles. Fig. S2 shows further BWM effects of fhod-1 perturbation, including RNAi against fhod-1 and mutant rescue by FHOD-1::GFP expression as well as effects on motility and muscle positioning. Fig. S3 shows that fhod-1 mutants have partial defects in vulval muscle morphology and in egg laying. Fig. S4 shows that disruption of frl-1, inft-1, and inft-2 did not alter BWM F-actin organization. Fig. S5 shows additional features of cyk-1(ok2300) and cyk-1(ok2300); fhod-1(tm2363) mutants, including morphological, gonad, and pharyngeal defects, localization of CYK-1::GFP in mutants, and staining of mutant BWMs by anti–CYK-1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201202053/DC1.
Supplementary Material
Acknowledgments
We would like to thank Jun Kelly Liu, Diane Morton, Michel Labouesse, Shoichiro Ono, Vladimir Sirotkin, Mira Krendel, and Jeffrey Amack for helpful advice and comments and Yuji Kohara, Jun Kelly Liu, John Plenefisch, Andrew Spence, Siu Sylvia Lee, Shoichiro Ono, Guy Benian, Pamela Hoppe, the Fire Laboratory, and the Wellcome Trust Sanger Institute for strains, antibodies, and constructs. Some nematodes used in this work were provided by the National BioResource Project for the Experimental Animal Nematode C. elegans, Tokyo Women’s Medical University School of Medicine, and by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. Monoclonal antibodies MH24 developed by Robert H. Waterston and 5-6 developed by Henry F. Epstein were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by The University of Iowa, Department of Biology.
This work was supported by National Institutes of Health grant GM36066 (to A. Bretscher) and the American Heart Association (to D. Pruyne).
Footnotes
Abbreviations used in this paper:
- ANF
- actin nucleation factor
- BWM
- body wall muscle
- DAD
- DIA autoregulatory domain
- DIA
- diaphanous
- DIC
- differential interference contrast
- DID
- DIA inhibitory domain
- dsRNA
- double-stranded RNA
- FHOD
- formin homology domain
References
- Alberts A.S. 2001. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem. 276:2824–2830 10.1074/jbc.M006205200 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- Amin N.M., Hu K., Pruyne D., Terzic D., Bretscher A., Liu J. 2007. A Zn-finger/FH2-domain containing protein, FOZI-1, acts redundantly with CeMyoD to specify striated body wall muscle fates in the Caenorhabditis elegans postembryonic mesoderm. Development. 134:19–29 10.1242/dev.02709 [DOI] [PubMed] [Google Scholar]
- Bai J., Hartwig J.H., Perrimon N. 2007. SALS, a WH2-domain-containing protein, promotes sarcomeric actin filament elongation from pointed ends during Drosophila muscle growth. Dev. Cell. 13:828–842 10.1016/j.devcel.2007.10.003 [DOI] [PubMed] [Google Scholar]
- Barstead R.J., Waterston R.H. 1989. The basal component of the nematode dense-body is vinculin. J. Biol. Chem. 264:10177–10185 [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler B., Gao C., Mersich A.T., Blystone S.D. 2006. Purified integrin adhesion complexes exhibit actin-polymerization activity. Curr. Biol. 16:242–251 10.1016/j.cub.2005.12.033 [DOI] [PubMed] [Google Scholar]
- Chalkia D., Nikolaidis N., Makalowski W., Klein J., Nei M. 2008. Origins and evolution of the formin multigene family that is involved in the formation of actin filaments. Mol. Biol. Evol. 25:2717–2733 10.1093/molbev/msn215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Drubin D., Nurse P. 1997. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J. Cell Biol. 137:169–182 10.1083/jcb.137.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D., Boczkowska M., Skwarek-Maruszewska A., Fujiwara I., Hayes D.B., Rebowski G., Lappalainen P., Pollard T.D., Dominguez R. 2008. Leiomodin is an actin filament nucleator in muscle cells. Science. 320:239–243 10.1126/science.1155313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M., Blundell K., Longtine M.S., Chow C.J., Adames N., Pringle J.R., Peter M., Boone C. 1997. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 276:118–122 10.1126/science.276.5309.118 [DOI] [PubMed] [Google Scholar]
- Finney M., Ruvkun G. 1990. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 63:895–905 10.1016/0092-8674(90)90493-X [DOI] [PubMed] [Google Scholar]
- Francis G.R., Waterston R.H. 1985. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J. Cell Biol. 101:1532–1549 10.1083/jcb.101.4.1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gettner S.N., Kenyon C., Reichardt L.F. 1995. Characterization of β pat-3 heterodimers, a family of essential integrin receptors in C. elegans. J. Cell Biol. 129:1127–1141 10.1083/jcb.129.4.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B.L., Eck M.J. 2007. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76:593–627 10.1146/annurev.biochem.75.103004.142647 [DOI] [PubMed] [Google Scholar]
- Gould C.J., Maiti S., Michelot A., Graziano B.R., Blanchoin L., Goode B.L. 2011. The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr. Biol. 21:384–390 10.1016/j.cub.2011.01.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs H.N., Peterson K.J. 2005. Phylogenetic analysis of the formin homology 2 domain. Mol. Biol. Cell. 16:1–13 10.1091/mbc.E04-07-0565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley H., Hanson J. 1954. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature. 173:973–976 10.1038/173973a0 [DOI] [PubMed] [Google Scholar]
- Imamura H., Tanaka K., Hihara T., Umikawa M., Kamei T., Takahashi K., Sasaki T., Takai Y. 1997. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 16:2745–2755 10.1093/emboj/16.10.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskratsch T., Lange S., Dwyer J., Kho A.L., dos Remedios C., Ehler E. 2010. Formin follows function: a muscle-specific isoform of FHOD3 is regulated by CK2 phosphorylation and promotes myofibril maintenance. J. Cell Biol. 191:1159–1172 10.1083/jcb.201005060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J., Jr, Copeland J.W., Fasnacht M., Etchberger J.F., Liu J., Honig B., Hobert O. 2006. An unusual Zn-finger/FH2 domain protein controls a left/right asymmetric neuronal fate decision in C. elegans. Development. 133:3317–3328 10.1242/dev.02494 [DOI] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275–282 [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Sternberg M.J.E. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- Kovar D.R., Pollard T.D. 2004. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl. Acad. Sci. USA. 101:14725–14730 10.1073/pnas.0405902101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D.R., Kuhn J.R., Tichy A.L., Pollard T.D. 2003. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 161:875–887 10.1083/jcb.200211078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Higgs H.N. 2005. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J. Biol. Chem. 280:6986–6992 10.1074/jbc.M411605200 [DOI] [PubMed] [Google Scholar]
- Littlefield R., Almenar-Queralt A., Fowler V.M. 2001. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat. Cell Biol. 3:544–551 10.1038/35078517 [DOI] [PubMed] [Google Scholar]
- Mardahl-Dumesnil M., Fowler V.M. 2001. Thin filaments elongate from their pointed ends during myofibril assembly in Drosophila indirect flight muscle. J. Cell Biol. 155:1043–1053 10.1083/jcb.200108026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.M., III, Ortiz I., Berliner G.C., Epstein H.F. 1983. Differential localization of two myosins within nematode thick filaments. Cell. 34:477–490 10.1016/0092-8674(83)90381-1 [DOI] [PubMed] [Google Scholar]
- Moerman D.G., Williams B.D. 2006. Sarcomere assembly in C. elegans muscle. WormBook. 16:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.B., Sagot I., Manning A.L., Xu Y., Eck M.J., Pellman D., Goode B.L. 2004. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell. 15:896–907 10.1091/mbc.E03-08-0621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder G.L., Cremona G.H., Duerr J., Stirman J.N., Fields S.D., Martin W., Qadota H., Benian G.M., Lu H., Barstead R.J. 2010. α-actinin is required for the proper assembly of Z-disk/focal-adhesion-like structures and for efficient locomotion in Caenorhabditis elegans. J. Mol. Biol. 403:516–528 10.1016/j.jmb.2010.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K., Takaishi K., Kodama A., Mammoto A., Shiozaki H., Monden M., Takai Y. 1999. Distinct actions and cooperative roles of ROCK and mDia in Rho small G protein-induced reorganization of the actin cytoskeleton in Madin-Darby canine kidney cells. Mol. Biol. Cell. 10:2481–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidt E.M., Skau C.T., Kovar D.R. 2008. The cytokinesis formins from the nematode worm and fission yeast differentially mediate actin filament assembly. J. Biol. Chem. 283:23872–23883 10.1074/jbc.M803734200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezami A.G., Poy F., Eck M.J. 2006. Structure of the autoinhibitory switch in formin mDia1. Structure. 14:257–263 10.1016/j.str.2005.12.003 [DOI] [PubMed] [Google Scholar]
- Nezami A., Poy F., Toms A., Zheng W., Eck M.J. 2010. Crystal structure of a complex between amino and carboxy terminal fragments of mDia1: insights into autoinhibition of diaphanous-related formins. PLoS ONE. 5:e12992 10.1371/journal.pone.0012992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. 2010. Dynamic regulation of sarcomeric actin filaments in striated muscle. Cytoskeleton (Hoboken). 67:677–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S., Ono K. 2002. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J. Cell Biol. 156:1065–1076 10.1083/jcb.200110013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T., Otomo C., Tomchick D.R., Machius M., Rosen M.K. 2005. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol. Cell. 18:273–281 10.1016/j.molcel.2005.04.002 [DOI] [PubMed] [Google Scholar]
- Otomo T., Tomchick D.R., Otomo C., Machius M., Rosen M.K. 2010. Crystal structure of the Formin mDia1 in autoinhibited conformation. PLoS ONE. 5:e12896 10.1371/journal.pone.0012896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A.S., Pollard T.D. 2008. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol. 18:9–19 10.1016/j.cub.2007.11.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Wallar B.J., Flanders A., Swiatek P.J., Alberts A.S. 2003. Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr. Biol. 13:534–545 10.1016/S0960-9822(03)00170-2 [DOI] [PubMed] [Google Scholar]
- Plenefisch J.D., Zhu X., Hedgecock E.M. 2000. Fragile skeletal muscle attachments in dystrophic mutants of Caenorhabditis elegans: isolation and characterization of the mua genes. Development. 127:1197–1207 [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Blanchoin L., Mullins R.D. 2000. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29:545–576 10.1146/annurev.biophys.29.1.545 [DOI] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 157:1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pring M., Evangelista M., Boone C., Yang C., Zigmond S.H. 2003. Mechanism of formin-induced nucleation of actin filaments. Biochemistry. 42:486–496 10.1021/bi026520j [DOI] [PubMed] [Google Scholar]
- Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A., Boone C. 2002. Role of formins in actin assembly: nucleation and barbed-end association. Science. 297:612–615 10.1126/science.1072309 [DOI] [PubMed] [Google Scholar]
- Pruyne D., Gao L., Bi E., Bretscher A. 2004. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell. 15:4971–4989 10.1091/mbc.E04-04-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H., Mercer K.B., Miller R.K., Kaibuchi K., Benian G.M. 2007. Two LIM domain proteins and UNC-96 link UNC-97/pinch to myosin thick filaments in Caenorhabditis elegans muscle. Mol. Biol. Cell. 18:4317–4326 10.1091/mbc.E07-03-0278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero S., Le Clainche C., Didry D., Egile C., Pantaloni D., Carlier M.F. 2004. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 119:419–429 10.1016/j.cell.2004.09.039 [DOI] [PubMed] [Google Scholar]
- Rose R., Weyand M., Lammers M., Ishizaki T., Ahmadian M.R., Wittinghofer A. 2005. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature. 435:513–518 10.1038/nature03604 [DOI] [PubMed] [Google Scholar]
- Sagot I., Rodal A.A., Moseley J., Goode B.L., Pellman D. 2002. An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4:626–631 [DOI] [PubMed] [Google Scholar]
- Sanger J.W., Kang S., Siebrands C.C., Freeman N., Du A., Wang J., Stout A.L., Sanger J.M. 2005. How to build a myofibril. J. Muscle Res. Cell Motil. 26:343–354 10.1007/s10974-005-9016-7 [DOI] [PubMed] [Google Scholar]
- Sanger J.W., Wang J., Fan Y., White J., Sanger J.M. 2010. Assembly and dynamics of myofibrils. J. Biomed. Biotechnol. 2010:1–8 10.1155/2010/858606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi H.E., Renihan S., Spence A.M., Cooperstock R.L. 2005. Gene CATCHR—gene cloning and tagging for Caenorhabditis elegans using yeast homologous recombination: a novel approach for the analysis of gene expression. Nucleic Acids Res. 33:e163 10.1093/nar/gni164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte A., Stolp B., Schönichen A., Pylypenko O., Rak A., Fackler O.T., Geyer M. 2008. The human formin FHOD1 contains a bipartite structure of FH3 and GTPase-binding domains required for activation. Structure. 16:1313–1323 10.1016/j.str.2008.06.008 [DOI] [PubMed] [Google Scholar]
- Severson A.F., Baillie D.L., Bowerman B. 2002. A Formin Homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr. Biol. 12:2066–2075 10.1016/S0960-9822(02)01355-6 [DOI] [PubMed] [Google Scholar]
- Shimada A., Nyitrai M., Vetter I.R., Kühlmann D., Bugyi B., Narumiya S., Geeves M.A., Wittinghofer A. 2004. The core FH2 domain of diaphanous-related formins is an elongated actin binding protein that inhibits polymerization. Mol. Cell. 13:511–522 10.1016/S1097-2765(04)00059-0 [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow J.C., Schöck F. 2009. The initial steps of myofibril assembly: integrins pave the way. Nat. Rev. Mol. Cell Biol. 10:293–298 10.1038/nrm2634 [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Horvitz H.R. 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56:110–156 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Schierenberg E., White J.G., Thomson J.N. 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100:64–119 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Swan K.A., Severson A.F., Carter J.C., Martin P.R., Schnabel H., Schnabel R., Bowerman B. 1998. cyk-1: a C. elegans FH gene required for a late step in embryonic cytokinesis. J. Cell Sci. 111:2017–2027 [DOI] [PubMed] [Google Scholar]
- Takano K., Watanabe-Takano H., Suetsugu S., Kurita S., Tsujita K., Kimura S., Karatsu T., Takenawa T., Endo T. 2010. Nebulin and N-WASP cooperate to cause IGF-1-induced sarcomeric actin filament formation. Science. 330:1536–1540 10.1126/science.1197767 [DOI] [PubMed] [Google Scholar]
- Taniguchi K., Takeya R., Suetsugu S., Kan-O M., Narusawa M., Shiose A., Tominaga R., Sumimoto H. 2009. Mammalian formin fhod3 regulates actin assembly and sarcomere organization in striated muscles. J. Biol. Chem. 284:29873–29881 10.1074/jbc.M109.059303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Fire A. 1998. Specific interference by ingested dsRNA. Nature. 395:854 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Tojkander S., Gateva G., Schevzov G., Hotulainen P., Naumanen P., Martin C., Gunning P.W., Lappalainen P. 2011. A molecular pathway for myosin II recruitment to stress fibers. Curr. Biol. 21:539–550 10.1016/j.cub.2011.03.007 [DOI] [PubMed] [Google Scholar]
- Trent C., Tsuing N., Horvitz H.R. 1983. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics. 104:619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada T., Pappas C.T., Moroz N., Antin P.B., Kostyukova A.S., Gregorio C.C. 2010. Leiomodin-2 is an antagonist of tropomodulin-1 at the pointed end of the thin filaments in cardiac muscle. J. Cell Sci. 123:3136–3145 10.1242/jcs.071837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Barr M.M. 2005. RNA interference in Caenorhabditis elegans. Methods Enzymol. 392:36–55 10.1016/S0076-6879(04)92003-4 [DOI] [PubMed] [Google Scholar]
- Watanabe N., Madaule P., Reid T., Ishizaki T., Watanabe G., Kakizuka A., Saito Y., Nakao K., Jockusch B.M., Narumiya S. 1997. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 16:3044–3056 10.1093/emboj/16.11.3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N., Kato T., Fujita A., Ishizaki T., Narumiya S. 1999. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1:136–143 10.1038/11056 [DOI] [PubMed] [Google Scholar]
- Watanabe S., Ando Y., Yasuda S., Hosoya H., Watanabe N., Ishizaki T., Narumiya S. 2008. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol. Biol. Cell. 19:2328–2338 10.1091/mbc.E07-10-1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R.H. 1988. Muscle. In The Nematode Caenorhabditis elegans. Wood W.B., editor Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York: 281–335 [Google Scholar]
- Wood W.B. 1988. Embryology. In The Nematode Caenorhabditis elegans. Wood W.B., editor Cold Spring Harbor Laboratory Press, Cod Spring Harbor, NY: 215–241 [Google Scholar]
- Xu Y., Moseley J.B., Sagot I., Poy F., Pellman D., Goode B.L., Eck M.J. 2004. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 116:711–723 10.1016/S0092-8674(04)00210-7 [DOI] [PubMed] [Google Scholar]
- Zigmond S.H., Evangelista M., Boone C., Yang C., Dar A.C., Sicheri F., Forkey J., Pring M. 2003. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr. Biol. 13:1820–1823 10.1016/j.cub.2003.09.057 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.