c-Jun in Schwann cells promotes the expression of Ret ligands GDNF and Artemin, which leads to enhanced motoneuron survival and axonal regeneration after injury.
Abstract
The AP-1 transcription factor c-Jun is a master regulator of the axonal response in neurons. c-Jun also functions as a negative regulator of myelination in Schwann cells (SCs) and is strongly reactivated in SCs upon axonal injury. We demonstrate here that, after injury, the absence of c-Jun specifically in SCs caused impaired axonal regeneration and severely increased neuronal cell death. c-Jun deficiency resulted in decreased expression of several neurotrophic factors, and GDNF and Artemin, both of which encode ligands for the Ret receptor tyrosine kinase, were identified as novel direct c-Jun target genes. Genetic inactivation of Ret specifically in neurons resulted in regeneration defects without affecting motoneuron survival and, conversely, administration of recombinant GDNF and Artemin protein substantially ameliorated impaired regeneration caused by c-Jun deficiency. These results reveal an unexpected function for c-Jun in SCs in response to axonal injury, and identify paracrine Ret signaling as an important mediator of c-Jun function in SCs during regeneration.
Introduction
The peripheral nervous system (PNS) has the capacity to rebuild itself after damage, contrary to what is observed in the central nervous system (CNS). Although many organs harbor a population of stem cells capable of tissue repair after injury, in the PNS degenerating axons and injury-related signals cause mature Schwann cells (SCs) to de-differentiate to a progenitor-like state. This progenitor-like cell is capable of proliferating and actively supports neuronal regeneration after injury by creating a pro-regenerative microenvironment in terms of tissue organization (Parrinello et al., 2010), myelin debris clearance, and also by providing survival factors (Webber and Zochodne, 2010). Among the injury-induced transcriptionally up-regulated genes in SCs considered of key relevance for axonal regeneration and neuronal survival are multiple members of the neurotrophin family, such as glial-derived neurotrophic factor (GDNF; Naveilhan et al., 1997), Artemin (Artn; Baloh et al., 1998), brain-derived neurotrophic factor (BDNF; Meyer et al., 1992), leukemia inhibitory factor (LIF; Curtis et al., 1994), and nerve growth factor (NGF; Heumann et al., 1987). Better understanding of the mechanisms that contribute to SC plasticity is of importance not only to understand the basic principles governing SC biology and neuronal regeneration, but also because some molecules that have been shown to be involved in repressing myelin gene expression are hyperactivated in demyelinating neuropathies (Jessen and Mirsky, 2008), an example being Notch activation in multiple sclerosis (John et al., 2002) or c-Jun in human neuropathies (Hutton et al., 2011).
Although the network of transcription factors that cooperate to direct SC maturation and myelination is well reported (Svaren and Meijer, 2008), there is scarce knowledge on the intrinsic molecular mechanisms governing SC de-differentiation and response to axonal injury. c-Jun has been shown to be a negative regulator of SC myelination (Parkinson et al., 2008) and is up-regulated in SCs upon injury (Shy et al., 1996). This prompted us to investigate in vivo the role of c-Jun in SCs upon peripheral nerve injury in the context of neuronal regeneration. In this study we selectively deleted Jun in SCs by crossing a strain carrying a floxed Jun allele with a mouse Cre line under the control of the SC-specific promoter myelin protein zero (JunΔSC mice). Neither SC maturation nor the integrity of the nerves was affected. Our findings show that c-Jun controls the injury-induced SC de-differentiation response and it is essential to support motoneuron survival after nerve injury. Upon injury, JunΔSC mice showed strongly impaired functional recovery and an inability to reinnervate their peripheral targets, concomitant with a dramatic decrease in the expression of various neurotrophins. Among those, we identified GDNF and Artn as two novel c-Jun target genes in SCs. GDNF and Artemin belong to the GDNF family of neurotrophins, together with Neurturin and Persephin (Baloh et al., 1998). Both GDNF and Artemin, when bound to their cognate GDNF family α coreceptors (GFRα), signal via a receptor tyrosine kinase encoded by the Ret proto-oncogene (also c-Ret; Durbec et al., 1996; Jing et al., 1996; Treanor et al., 1996).
To further dissect the importance of Jun-mediated transcriptional control of Ret ligands GDNF and Artemin, we also investigated the regenerative potential post-axotomy of motoneurons lacking Ret receptor.
The phenotype described herein is in striking contrast to the one observed by CNS Jun deletion (Raivich et al., 2004). Notably, in this previous study, Jun function in SCs was preserved because the Nestin-Cre line does not show activity in the SC lineage (Kao et al., 2009). Our study describes a novel Jun function in SCs in promoting motoneuron survival after injury and functionally links AP-1 activity and paracrine Ret signaling through the identification of GDNF and Artn as two novel c-Jun target genes in SCs.
Results
Conditional inactivation of Jun in Schwann cells impairs motoneuron survival and axonal regeneration
To investigate the significance of c-Jun expression and function in SCs, floxed c-jun (Junf/f) mice were crossed to P0-Cre transgenic mice previously shown to provide efficient SC-specific Cre activity around E14.5 and peaking at postnatal stages (Feltri et al., 2002). JunΔSC mice were born with Mendelian frequency and were viable and fertile. The overall architecture and histology of the sciatic and facial nerves appeared normal, suggesting that c-Jun function appears to be dispensable in SCs during development (unpublished data; Parkinson et al., 2008).
We investigated c-Jun function in response to axonal injury, and transection of the facial nerve at the stylomastoid foramen level was used as a model system. The facial nerve arises from the facial nucleus located in the brainstem, from where motoneurons project their axons and control facial muscle movement, including whisker hair movement. The effects of SC-specific Jun inactivation on axonal regeneration were assessed by the extent of functional recovery, peripheral target reinnervation, and motoneuron survival, using the same cohorts of control and mutant mice. In a second cohort of mice, we also analyzed the speed of axonal elongation in the early phase of nerve regeneration 4 d after nerve crush.
To assess functional recovery, the overall movement of whisker hair (whisker hair movement, WHM) was scored on a scale of 0 (no movement) to 3 (normal movement; see Materials and methods for details). 28 d after facial nerve cut, both control and mutant mice showed normal movement on the uninjured side. Control Junf/f animals revealed observable recovery at 14 d and improved steadily over the next 2 wk until endpoint at d 28. Recovery in littermate JunΔSC mutants was significantly poorer and did not improve beyond 18 d (Fig. 1 A). These differences are also displayed as WHM recovery index (WHM RI) calculated for each individual animal as the area under the curve for d 0–28 for the time course of functional recovery shown in Fig. 1 A. For the whole group, the WHM RI reached the value of 0.82 ± 0.09 for Junf/f and 0.31 ± 0.03 for JunΔSC (Fig. 1 B).
Figure 1.
Jun SC deletion interferes with neuronal survival and regeneration. (A and B) Recovery of whisker hair movement (WHM). (A) WHM was scored on a scale of 0 (no movement) to 3 (strong, normal movement); see Materials and methods for details. The data points show mean ± SEM for JunΔSC (n = 6) and their littermate controls (n = 9). *, P < 0.05 in unpaired two-tailed t test in this and all the following graphs. (B) WHR recovery index (WHM RI) based on area under the curve (AUC) for each individual animal that was assessed in A. JunΔSC show a very significant poorer overall recovery with 0.82 ± 0.09 for Junf/f animals and 0.31 ± 0.03 for JunΔSC (mean ± SEM); P < 0.05, Student’s t test. (C–G) Peripheral target reinnervation of the whisker pad is reduced in the absence of Jun in SC; same animals as in A and B. (C) Ratio of retrogradely labeled facial motoneurons on the axotomized (Ax) versus contralateral (Co) side after application of FG to both whisker pads, 28 d after facial cut followed by 72 h survival. (D–G) Representative images of retrograde labeling with FG. On the uninjured (contralateral) side, retrogradely labeled facial motoneurons are mainly found in the lateral subnucleus (D and F), but appear randomly distributed throughout the nucleus after cut (E and G). Note the sharp decrease in the number of labeled neurons on the axotomized side in mutant JunΔSC (n = 6) and their littermate controls (n = 9) (G) compared with control (E) animals. (H and I) Nissl-stained brainstem coronal sections 31 d after unilateral axotomy of control (H) and mutant (I) animals. The facial motor nucleus (7n) is circumscribed with a dashed line. Note the sharp decrease in motoneurons in JunΔSC animals in the axotomized side (ax). Bar in H (applies also to I), 0.2 mm. Quantification is shown in J. Motoneuron cell count showing a strong decrease in survival 31 d after nerve cut in the absence of SC c-Jun in JunΔSC (n = 6) and their littermate controls (n = 9), same animals as in A–I. (K) Number of αX+ microglial nodules in the axotomized facial motor nucleus per 20-µm-thick section (n = 5 JunΔSC mutants and n = 5 Junf/f controls). (L) Regeneration distance (in mm, distal from crush) for the fastest growing CGRP+ and galanin+ motor axons, 4 d after facial nerve crush (n = 4 JunΔSC mutants and n = 4 Junf/f controls). (M) Density of αM+ macrophages 4 d after crush, at the facial nerve injury site (0 mm) and 2 and 4 mm distally (n = 4 JunΔSC mutants and n = 4 Junf/f controls). (J–M) *, P < 0.05 in Student’s t test.
To determine the cause of this defect, 28 d after nerve cut the same experimental cohorts were assessed for neuronal muscle reinnervation and motoneuron survival. Whisker pads were labeled on both sides with the fluorescent tracer Fluoro-Gold (FG), followed by 72 h retrograde transport. Motoneurons that successfully reconnected with their targets were identified by the presence of the retrograde tracer in their somas, and counted on every fifth section throughout the facial nucleus (see Materials and methods for details). Although control animals showed retrograde labeling of 66.3 ± 3.6% on the axotomized side compared with the uninjured side, JunΔSC animals showed an almost fivefold decrease with just 13.7 ± 2.0% (Fig. 1, C–G; P < 0.01, Student’s t test).
We explored whether alterations in motoneuron survival contributed to the observed reduced reinnervation and functional recovery. Comparison of motoneuron number on the uninjured and injured sides 31 d after injury revealed a loss of 29.6 ± 3.4% in the control group, injured side (n = 9). However, motoneuron survival was strikingly decreased in JunΔSC animals (n = 6), with loss of 77.5 ± 2.1% (P < 0.01, Student’s t test). Motoneuron death and the ensuing formation of neuronal debris are normally accompanied by the appearance of local phagocytic microglia. These cells aggregate in large 15–30-µm clusters with a peak at 14 d after facial nerve cut, and can be readily identified by αX integrin staining (Kloss et al., 1999). The extent of microglia recruitment correlates with the severity of neuronal cell death (Raivich and Banati, 2004). In agreement with exacerbated neuronal death in JunΔSC facial nucleus, SC Jun deletion was associated with a more than threefold increase in the number of αX integrin-positive microglial nodules within the facial nucleus 14 d after nerve cut (Fig. 1 K). Other neuronal and astrocytic markers were not affected when comparing control and mutant animals; the increase in the total facial nucleus immunoreactivity for the astrocytic activation marker glial fibrillary acidic protein (GFAP) and the microglial αM integrin subunit were unchanged (Fig. S1).
We also examined motoneuron axonal regeneration early on after axotomy, before the occurrence of neuronal death, by measuring the extent of nerve fiber outgrowth 4 d after facial nerve crush. The growth front of the regenerating motor neurites was detected in longitudinally cut facial nerve sections using immunoreactivity for calcitonin gene-related peptide (CGRP) and galanin neuropeptides. The expression of these neuropeptides increases in axotomized facial motoneurons and the extent of neurite outgrowth in the distal facial nerve at d 4 correlates with the speed of reinnervation of the peripheral target (Werner et al., 2000). The axonal growth front advanced beyond the crush site into the distal site 6.8 ± 0.4 mm for the CGRP-positive and 6.5 ± 0.2 mm for the galanin-positive axon in the control Junf/f and was reduced by 30–35% in JunΔSC animals (Fig. 1 L; P < 0.05, Student’s t test).
Compared with the Junf/f animals, JunΔSC injured facial nerves also showed a significant reduction in the number of phagocytic macrophages 2 and 4 mm distal to the site of injury (Fig. 1 M). Thus, absence of SC c-Jun also interferes with local macrophage recruitment within the nerve, in contrast to the increased recruitment of phagocytes in the facial motor nucleus experiencing enhanced cell death (Fig. 1 K).
c-Jun controls axotomy-induced gene expression in Schwann cells
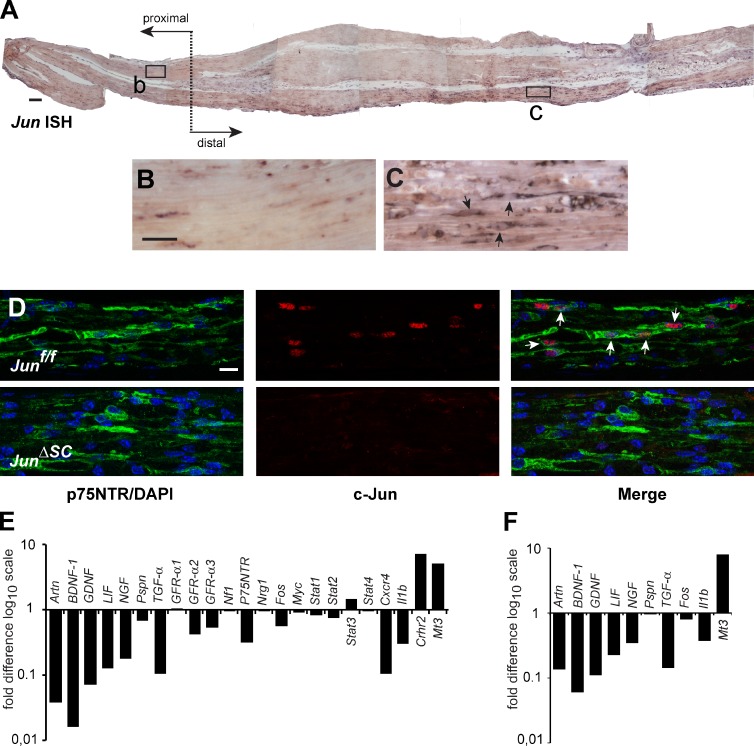
We next characterized the regulation of the Jun gene in the axotomized nerve. In situ hybridization (ISH) revealed low expression in the proximal part of the injured nerve, but Jun mRNA was strongly up-regulated in the distal part (Fig. 2, A–C). The proximal and distal compartments of the injured nerve were readily identifiable by the recruitment of F4/80-positive macrophages in the distal part of the injured nerve (Fig. S2, A–C). Axotomy also dramatically increased the expression of p75 neurotrophin receptor (p75NTR) in the distal nerve, a marker of de-differentiated SCs (Fig. S2, D–F).
Figure 2.
Activation of expression upon nerve injury of neurotrophin genes by denervated SCs depends on Jun up-regulation. (A) Jun ISH on a longitudinal section of a wild-type mouse injured sciatic nerve, 7 d after injury. The approximate crush lesion is indicated by a transverse dotted line. Proximal and distal stump directions are also indicated. Rectangles mark two representative areas, depicted in B and C. (B and C) High magnification from A of the proximal (left) and distal (right) stumps to the lesion correspondingly. (D) Confocal imaging of 7 d post-injury wild-type (Junf/f) and mutant (JunΔSC) longitudinal sciatic nerve sections. Double immunofluorescence for the SC progenitor marker p75 neurotrophin receptor (p75NTR; green, left) and total c-Jun (red, middle). Merge images are shown on the right. DNA (blue) was counterstained with DAPI. Arrows indicate p75NTR+ SC progenitors that coexpress c-Jun. (E) Superarray comparing the neurotrophin gene expression level of sciatic nerves 7 d after injury. Fold change values in the mutant nerves are displayed by normalizing to wild-type levels (baseline value 1; n = 7 for Junf/f mice and n = 6 for JunΔSC mice from a single experiment). (F) Q-PCR confirmation of the results obtained with the superarray shown in E. Fold change values in the mutant nerves are displayed by normalizing to wild-type levels (baseline value 1; mean of three replicates, n = 7 for Junf/f mice and n = 6 for JunΔSC from a single experiment). Bars: (A) 100 µm; (B; also applies to C) 25 µm; (D; bar in top left image applies to the whole panel) 10 µm.
Immunofluorescence showed nuclear c-Jun protein present in the distal injured nerve, which colocalized with p75NTR. Importantly, c-Jun was absent in injured JunΔSC nerves, confirming the efficiency of P0-cre–mediated Jun inactivation and SC-specific expression of c-Jun (Fig. 2 D). Thus, Jun expression is strongly induced in de-differentiated SCs after nerve injury.
To understand the mechanism of c-Jun function in SCs, we sought to identify the target genes involved in motoneuron survival/axonal regeneration. To this end, we performed a superarray analysis that focused on neurotrophic factors and signaling pathway molecules using mRNA isolated from injured control Junf/f and mutant JunΔSC sciatic nerves. The expression of several genes encoding factors with known functions in neuronal survival and axonal growth, such as GDNF, Artn, BDNF1, and LIF, was dramatically reduced in injured JunΔSC nerves, whereas other regeneration-associated molecules remained relatively unchanged (Fig. 2 E). This finding was validated independently by quantitative real-time PCR (qRT PCR; Fig. 2 F).
c-Jun–dependent transcriptional induction of GDNF and Artn genes in Schwann cells after injury
GDNF and Artn both encode ligands for the Ret receptor tyrosine kinase, and both proteins have well-described functions in neuronal survival and axonal elongation. We therefore further investigated a potential role of these genes as mediators of c-Jun function in SCs. ISH revealed low expression of GDNF and Artn in the uninjured and proximal part of the injured nerve 7 d after injury, but GDNF and Artn mRNAs were strongly up-regulated in the distal part in a pattern reminiscent of Jun up-regulation (Figs. 2, A–C, and 3 A). Crucially, injury-induced GDNF and Artn mRNA were barely detectable in JunΔSC nerves (Fig. 3 A), in agreement with the superarray data (Fig. 2 E).
Figure 3.
Transcriptional profile of SCs upon nerve injury or under myelinating conditions after cAMP treatment. (A) ISH for Artn and GDNF genes on Junf/f and JunΔSC uninjured sciatic nerves or 7 d post-crush in proximal and distal stumps from the injury site. Rectangles indicate area shown with higher magnification on the last panel to the right. (B) Q-PCR showing changes in gene expression of injured sciatic nerves 7 d post-crush compared with uninjured wild-type nerves (baseline value uninjured nerve, 1; numbers shown are mean of three replicates, n = 5 injured or control wild-type animals, representative experiment out of two experiments). (C) Q-PCR in primary SC cultures upon treatment with cAMP for 5 d (baseline value untreated, 1). Values plotted are the mean of the means from two independent experiments with three replicates each. Within each experiment, samples from individual pups were pooled. Across the two independent experiments, 10 P2-P5 pups were analyzed for each condition. Error bars represent the SD of the two means. *, P < 0.05 (Student’s t test). (D) Q-PCR of SC primary cultures isolated from wild-type mice. The graph shows fold gene expression when comparing cultures that overexpress c-Jun (adenovirus–c-Jun) versus mock-infected cultures (adenovirus-GFP). Numbers are mean of three replicates, n = 10 P2-P5 pups pooled for each condition, representative experiment out of two experiments. (E) Assessment of Ret activation and downstream signaling in facial motoneurons in axotomized facial nerve of control and c-Jun deleted mutant mice. Cohorts from both genotypes were subjected to facial nerve axotomy and the axotomized facial nucleus was microdissected 7 d after injury. Extracts were then analyzed by Western blot using antibodies for the indicated proteins. Uninjured mice with Ret neuronal deletion were used as a negative control for total and p-Tyr905 Ret Western blots. Bars: (A; bottom left applies to the low magnification three first columns) 150 µm; (high magnification series, top right column) 25 µm.
Having found that genes encoding several neurotrophic factors were expressed at lower levels in injured JunΔSC compared with injured control nerves, we next investigated the regulation of these genes by injury in wild-type animals. qRT PCR showed that the expression of Jun, GDNF, Artn, BDNF-1, and LIF were increased by injury in SCs. Concomitantly, markers for differentiated SCs such as Egr2 and Myelin protein zero (MPZ) were down-regulated, whereas p75NTR expression increased (Fig. 3 B). Thus, GDNF and Artn appeared co-regulated with markers of de-differentiated SCs. To determine the effect of SC differentiation on the expression of these trophic factors, we analyzed changes in gene expression in primary SC cultures after 5 d treatment with cAMP, which triggers differentiation and expression of myelin genes. For most genes, expression showed the opposite pattern compared with injury, as expression of Jun, GDNF, and Artn was strongly repressed (Fig. 3 C). Moreover, ectopic adenoviral expression of c-Jun in primary SC cultures was sufficient to increase mRNA levels of GDNF and Artn (Fig. 3 D), and to decrease the expression of the myelination-related genes Egr2, MPZ, and MBP. Because we found that the up-regulation of GDNF and Artn genes after injury largely depends on SC c-Jun up-regulation, we next wanted to assess whether Ret activation was impaired in JunΔSC mice. We assessed total Ret protein levels and Ret tyrosine phosphorylation as a read-out for Ret receptor activation in axotomized microdissected facial motor nucleus 7 d after injury, as it is known that neurotrophins and their receptors are retrogradely transported. In agreement with failure in the up-regulation of expression of Ret ligands GDNF and Artemin in JunΔSC nerves after injury, Ret receptor activation levels judged by a specific antibody against phospho-Tyr 905 was decreased in mutant animals. However, total Ret protein level was unchanged (Fig. 3 E). We also assessed the status of Akt and ERK signaling in JunΔSC animals during axonal regeneration using phospho-specific antibodies. Although the levels of active Akt appeared unchanged, we detected a decrease in the levels of phosphorylated ERK in the axotomized side of mutant JunΔSC animals.
GDNF and Artn are direct c-Jun target genes
To investigate the regulation of GDNF and Artn by c-Jun in more detail, we used the IMS32 SC line (Watabe et al., 1995). Stimulation of IMS32 with the known JNK activator anisomycin (Anm; 6 h treatment) stimulated JNK activity and c-Jun N-terminal phosphorylation. Conversely, pharmacological JNK inhibition using the SP600125 compound (JNKi, 6 h treatment) resulted in a decrease in c-Jun N-terminal phosphorylation (Fig. 4 A). Anm significantly increased mRNA of Jun, GDNF, Artn, and LIF measured by Q-PCR, whereas JNKi led only to a modest decrease in gene expression, which might be due to low level of basal JNK activity in IMS32 cells (Fig. 4 B). Anm did not lead to profound alterations in the mRNA levels of TGF-α and BDNF1, suggesting that these genes are not directly regulated by c-Jun in IMS32 cells (Fig. 4 B). Thus, the JNK signaling pathway, and the regulation of GDNF and Artn by JNK/c-Jun, appears to be intact in IMS32 cells.
Figure 4.
Molecular analysis of GDNF and Artn promoters. (A) Western blots of IMS32 SC line extracts to detect total and phosphorylated versions of both c-Jun and JNK upon JNK activation (anisomycin, Anm) or JNK inhibition (SP600125) treatment. Actin was used as loading control. (B) Q-PCR from IMS32 SC line samples untreated (white bars) or treated either with anisomycin (black bars) or JNK inhibitor (SP600125; gray bars). Gene values were normalized to the untreated condition (baseline value 1). *, P < 0.05, ANOVA test, n = 3 independent experiments. (C and D) Scheme showing the structure of Mus musculus GDNF (C) and Artn (D) loci. The arrow indicates the transcriptional start site. Exons are shown in black and the UTR regions in gray. The red dot indicates the location of the identified AP-1 site in the promoter in each of these genes. A 5′–3′ species alignment of the AP-1 sequence is shown. The red arrows flanking the AP-1 site indicate the location of the primers used in K. (E, G, and I) Schemes of the pGL3 luciferase reporter constructs into which GDNF (E) or Artn (G) or Jun (I) mouse promoter fragments were cloned. The red dot indicates AP-1–binding sequence. AP-1 sites of GDNF and Artn reporter constructs were mutated (AP-1mut; indicated with a cross). Approximate sizes of the cloned piece of genome, flanking the AP-1, are indicated. (F, H, and J) IMS32 SC cultures were cotransfected with each of these constructs together with ubiquitin-renilla as a control. The graphs show renilla/luciferase ratio of fold induction over pGL3-control (ctrl) transfected cells. Cultures were either untreated or treated with JNK agonist anisomycin. *, P < 0.05, Student’s t test; n = 3 independent experiments. (K) Graph demonstrating phospho-c-Jun in vivo binding to c-Jun, GDNF, and Artn AP-1 containing promoters by chromatin immunoprecipitation using anti–phospho-c-Jun antibody. Location of AP-1 flanking primers used for GDNF and Artn genes is marked in C and D with red arrows. Gapdh gene was used as a control. IMS32 SC line cultures were serum starved overnight and were either untreated or treated with anisomycin or JNK inhibitor for 30 min. The values represent fold enrichment over an isotype matching control immunoglobulin, showing a representative experiment out of three repeats.
The LIF gene has previously been shown to be transcriptionally regulated by c-Jun (Bozec et al., 2008), but the molecular details of GDNF and Artn regulation by c-Jun were not known. In silico analysis revealed the presence of an evolutionarily conserved consensus AP-1 site in the promoters of both GDNF and Artn genes (Fig. 4, C and D). When inserted into a luciferase reporter construct, GDNF and Artn promoter fragments containing the predicted AP-1 sites (pGL3-GDNF-AP1 and pGL3-Artn-AP1, respectively) showed higher reporter activity versus an empty pGL3 control vector, measured as luciferase activity normalized to renilla. Anm stimulation of cultures transfected with GDNF and Artn promoter fragments containing predicted AP-1 sites triggered increase in luciferase activity, indicating increase in reporter gene levels (Fig. 4, F and H). This effect was not seen when cultures were transfected with constructs carrying point mutations in the respective AP-1 binding sites. (Fig. 4, F and H). c-Jun autoregulates its own promoter via two proximal AP-1 binding sites (Angel et al., 1988) and this region of the Jun promoter (pGL3-c-Jun2xAP1) mediated high basal reporter gene activity and Anm inducibility, serving as a positive control for these experiments (Fig. 4, I and J). Chromatin immunoprecipitation (ChIP) using anti–phospho-c-Jun antibody showed binding of c-Jun to its cognate sites in the Jun promoter as well as to the putative AP-1 sites in the GDNF and Artn promoters, but not to a region in the gapdh promoter, used as a negative control (Fig. 4 K). When cultures were treated with Anm (30 min), we detected increased phospho-c-Jun promoter occupancy over untreated cultures, whereas treatment with JNK inhibitor SP600125 (30 min) had the opposite effect. We conclude that GDNF and Artn are direct c-Jun target genes.
The Ret receptor is required in neurons for efficient axonal regeneration
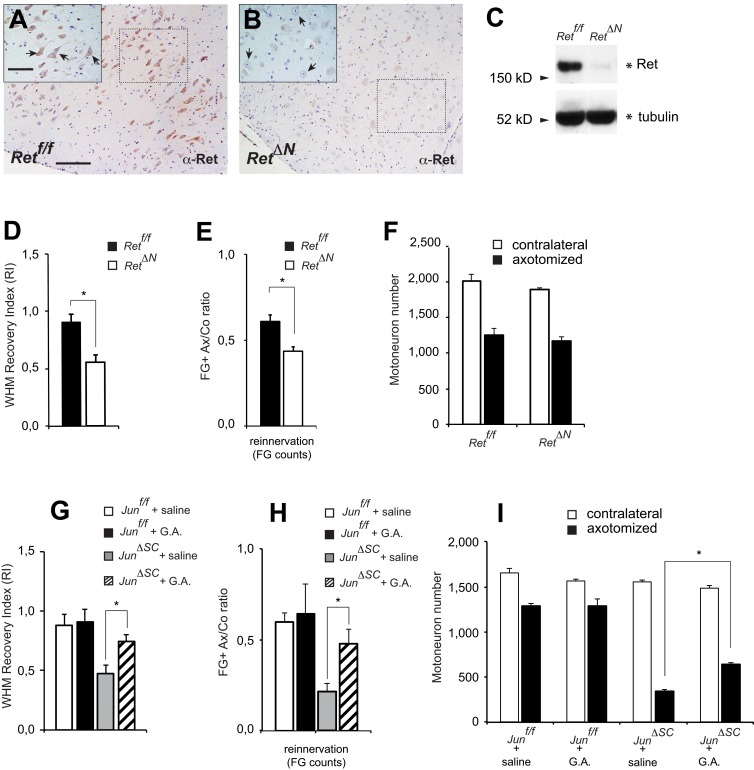
Because both GDNF and Artemin neurotrophins signal through the receptor tyrosine kinase Ret, we tested the significance of paracrine signaling during regeneration by exploring the effects of Ret neuronal-specific deletion. A Ret floxed allele (Kramer et al., 2007) was inactivated using the Synapsin-Cre line, mediating gene deletion in neurons (RetΔN mice; Zhu et al., 2001). IHC for Ret protein revealed high expression in facial motoneurons in control mice, which was absent in RetΔN mutant mice (Fig. 5, A and B). The efficiency of deletion in facial nuclei by Western blot against total protein Ret levels is shown in Fig. 5 C. Other neuronal populations retained Ret expression (unpublished data), as this line causes recombination in neurons in a restricted manner (Hoesche et al., 1993; Heumann et al., 2000). Recovery of WHM was impaired in mice lacking neuronal Ret. The WHM RI, calculated as in Fig. 1 B, was 38% compared with controls (Fig. 5 D). To ascertain whether this functional defect was due to lack of target reinnervation, we performed on the same experimental groups a functional recovery assessment by FG whisker pad retrograde tracing at 28 d after injury. RetΔN animals showed a 30–40% decrease in FG+ Ax/Co ratio motoneurons compared with control littermates (Fig. 5 E). Counts of facial motoneurons revealed similar numbers in the uninjured side of both genotypes, and similar neuronal numbers 31 d after injury, at 61.8 ± 3.2% of uninjured control for Retf/f and 62.7 ± 3.6% for RetΔN (Fig. 5 F). Thus, there was no difference in axotomy-induced cell death in mice lacking neuronal Ret, but the motoneurons showed decreased peripheral target reinnervation, resulting in poor functional recovery.
Figure 5.
Ret signaling is involved in the regeneration of facial nucleus motoneurons after axotomy. (A and B) Total Ret IHC in wild-type (A, Retf/f) and mutant (B, RetΔN) healthy animals. Sections were counterstained with hematoxylin & eosin. Facial nucleus (7n) motoneurons express Ret at high levels (A, arrows in inset). The deletion of Ret in these motoneurons by crossing with a Synapsin-Cre deleter mouse was confirmed by the lack of Ret staining in B. Dotted squares indicate the high magnification inset at the top left corner. (C) Facial nuclei from Retf/f and RetΔN mice were microdissected and Ret levels were assessed by Western blot. (D) WHM RI, as in Fig. 1 B, based on Area Under the Curve (AUC) data for each individual animal. RetΔN mice show a significantly poorer response compared with the Retf/f control group (P < 0.01, Student’s t test; n = 6 for each genotype). (E) Target reinnervation of the whisker pad is reduced in RetΔN mice. The graph shows the ratio of retrogradely labeled facial motoneurons on the axotomized (Ax) versus contralateral (Co) side after application of FG to both whisker pads, 28 d after facial nerve cut, followed by 72 h survival; *, P < 0.01, Student’s t test; n = 6 for each genotype; same animals as in D. (F) Motoneuron cell count shows no decrease in survival 31 d after nerve cut in the absence of neuronal Ret when comparing control and mutant animals (n = 6 for each genotype; same animals as in D and E. (G) WHM RI of the four experimental cohorts. Treatment with GDNF and Artemin partially improves functional recovery in the injured JunΔSC mice. *, P < 0.05 in ANOVA and post-hoc Tukey test. In Junf/f mice, treatment with GDNF and Artemin does not have a significant effect over saline (P = 0.49). Group size: n = 10 Junf/f + saline, n = 2 Junf/f + G.A., n = 5 JunΔSC + saline, and n = 8 JunΔSC + G.A. mice. (H) Treatment with GDNF and Artemin also significantly improves target reinnervation (whisker pad) in JunΔSC mice. The graph shows the axotomized versus contralateral ratio for FG-labeled facial motoneurons using the same animals as in G. As in G, the neurotrophin treatment did not significantly improve target reinnervation in Junf/f mice (P = 0.78 in post-hoc Tukey test). (I) Total motoneuron cell count showing a significant but partial rescue of motoneuron survival just in the JunΔSC mice 31 d after nerve cut. Group size in G–I: n = 6 Junf/f + saline, n = 3 Junf/f + G.A., n = 5 JunΔSC + saline, and n = 8 JunΔSC + G.A. mice. Bars: (A; also applies to B) low magnification, 200 µm; high magnification insets, 100 µm.
Exogenous delivery of recombinant GDNF and Artemin proteins partially rescues impaired axonal regeneration in JunΔSC animals
GDNF and Artn were only two of several neurotrophins that were not induced by axotomy in JunΔSC animals. Because our data demonstrates that they are direct c-Jun target genes, we wanted to assess directly their contribution to PNS regeneration. Ret receptor has been shown to not be expressed in SCs (Widenfalk et al., 1997, 1998), and we confirmed this by the distinct staining patterns of Ret, which labeled axonal fibers (Fig. S3, A and B). Using Junf/f control and JunΔSC mutant mice, we tested whether the exogenous administration of GDNF and Artemin would ameliorate the observed defects in axonal regeneration or survival in mutant mice. Recombinant GDNF and Artemin proteins (+G.A.), or saline-loaded mock solution, were locally delivered at the site of injury, where the growth cones are located, starting immediately after facial nerve axotomy. Osmotic pumps coupled to a catheter were subcutaneously implanted for sustained delivery over a period of 28 d (see Materials and methods and Fig. S3 C).
Administration of GDNF and Artemin proteins substantially increased WHM RI in JunΔSC animals, whereas saline mock-treated JunΔSC animals showed, as expected, very poor recovery (P < 0.05; ANOVA, Tukey). The saline-treated JunΔSC group had a WHM RI of 0.47 ± 0.09, significantly lower than that observed for saline-treated Junf/f 0.84 ± 0.13 and +G.A.-treated JunΔSC 0.71 ± 0.05 (P < 0.05; ANOVA, post-hoc Tukey followed by Bonferroni multiple comparison correction). In control animals, GDNF and Artemin administration did not cause a significant increase in the WHM RI over saline-treated control animals (Fig. 5 G; 0.88 ± 0.11 compared with 0.84 ± 0.13, respectively).
FG retrograde tracing at 28 d after axotomy revealed that target reconnection was significantly increased in +G.A.-treated (47.9 ± 8.2%) compared with saline-treated JunΔSC mice (21.4 ± 4.5%). However, the reinnervation efficiency in +G.A.-treated JunΔSC mice was still lower compared with saline-treated wild-type animals (59.8 ± 4.9%). +G.A. treatment in control animals led to a nonsignificant increase (64.3 ± 16.4%) relative to saline control administration (Fig. 5 H). Motoneuron number in response to axotomy was increased by GDNF and Artemin administration in JunΔSC mice, from 21.4 ± 2.8% (saline) to 44.9 ± 5.8 (+G.A.), but it was substantially lower than in control Junf/f 77.9 ± 2.6% (saline) and 83.1 ± 6.8% (+G.A.) when comparing motoneuron number on the uninjured and injured sides 31 d after injury (Fig. 5 I).
Taken together, these data identify GDNF and Artemin production by SCs and thus paracrine Ret signaling as an important mediator of c-Jun function in regeneration.
Discussion
Although previous scientific evidence seconds the concept that the injury-induced generation of the denervated SCs is critical for successful nerve regeneration and functional repair, very few genetic mouse models that directly test this idea in an in vivo approach exist to date. Furthermore, the molecular pathways involved are incompletely understood.
We describe a molecular mechanism by which c-Jun controls the ability of SCs to support motoneuron survival and axonal regeneration after peripheral nerve injury. Specifically, the expression of neurotrophin genes Artn, BDNF-1, GDNF, LIF, and NGF was severely down-regulated in the absence of c-Jun, causing combined severe defects in motoneuron survival and slight decrease in axonal growth, resulting in very poor functional recovery. We further identified Ret ligands GDNF and Artemin as two novel target genes in SCs, characterizing their transcriptional regulation by c-Jun, expression upon injury, and their role in supporting nerve regeneration in the adult.
The lack of Jun in SCs from the stage when the P0 promoter drives Cre-mediated recombination (around E14.5, peaking at postnatal stages; Feltri et al., 2002) has no discernible effect on SC development or the ability of SCs to myelinate axons (unpublished data). However, we found that Jun up-regulation in SCs upon facial motoneuron (FMN) axotomy appears indispensable to allow substantial neuronal survival and axonal regeneration. The absence of Jun in SCs leads to dramatically increased neuronal death, paralleled with an increase in the number of αX phagocytic microglial clusters in the facial motor nucleus at 14 d after nerve cut.
Many of the neuronal molecular changes induced by axotomy depend on the presence of neuronal c-Jun (Raivich et al., 2004). In JunΔSC nerves, the injury-induced increase in neuronal immunoreactivity for the adhesion molecules CD44, neuropeptides CGRP and galanin was unaffected (Fig. S1). This observation is consistent with the fact that neuronal c-Jun up-regulation and phosphorylation caused by injury was not affected in JunΔSC FMN somas, 4 d (Fig. S4, A–D) or 7 d (Fig. S4 E) after nerve axotomy. However, although neurons up-regulate correctly pro-regenerative molecules, they show slower axonal growth rate early on after axotomy. Therefore, cell-autonomous neuronal up-regulation of these pro-regenerative molecules occurs independently of SC c-Jun but is not sufficient to support normal axonal growth rates after crush.
Our study describes for the first time that the transcription factor c-Jun is required for the expression of several neurotrophin genes in SCs after nerve injury; in its absence in mutant nerves, the up-regulation was largely abolished. Although the requirements of neurotrophins for neuronal regeneration are widely known (Baloh et al., 2000; Andres et al., 2001; Airaksinen and Saarma, 2002; Barati et al., 2006), our current knowledge on the signaling pathways and the transcription factors involved in their gene expression is very limited. This is of special relevance because neurotrophins are considered as emerging drugs suitable to treat a variety of neuropathies, as in the case of GDNF and Artemin (Bespalov and Saarma, 2007).
We identified GDNF and Artn Ret ligands as two novel direct c-Jun transcriptional targets genes. Other pathways have been identified to be of importance in driving SC de-differentiation such as Notch (Woodhoo et al., 2009) and MAPK (Napoli et al., 2012). However, we have not seen alterations in mRNA levels of Notch target genes Hes1 and Hes5 (unpublished data), suggesting that Notch activity is normal in the absence of Jun. Further studies will be required to assess whether MAPK functions in promoting neuronal regeneration are c-Jun dependent.
GDNF and Artemin are known to induce phosphorylation of four key Ret tyrosine residues (Coulpier et al., 2002). Adaptor proteins then bind to Ret tyrosine residues and trigger a variety of different signaling pathways such as Ras/MAPK and PI3K/Akt (Kaplan and Miller, 2000). Our study supports the idea that axonal growth potential and facial motoneuron survival largely depend on SC-derived trophic support. The link between c-Jun transcription in SCs and paracrine signaling in neurons through the neurotrophin receptor Ret constitutes a novel cross talk that is essential for efficient target reconnection and functional recovery after nerve injury of facial nucleus motoneurons. Ret-specific neuronal deletion caused delayed functional recovery as well as reduced muscle target reinnervation, reinforcing the idea that Ret signaling is an important effector of AP-1 function. However, it is important to note that Ret inactivation only partially mimics the defects seen in JunΔSC mice; and the defects observed in the JunΔSC facial nerves after injury are by far more pronounced than those observed in the RetΔN, especially regarding motoneuron survival.
Thus, c-Jun expression in SCs controls several regeneration-relevant signaling pathways other than Ret. In agreement with this, the number of surviving projecting facial motoneurons was similar in RetΔN than in wild-type counterparts, most likely due to the trophic support provided by many other neurotrophins, such as BDNF1, LIF, and NGF, whose expression also depends on c-Jun, but use receptors different than Ret. Indeed, mice with deletion of Ret in the CNS display abnormal innervation of the hindlimb during development, without affecting neuronal survival (Kramer et al., 2006), implicating Ret receptor in axonal outgrowth and pathfinding. Recently, Ret has been shown to be a multifunctional receptor able to provide topographical cues during development to growing axons through the integration of EphA and GDNF signaling (Bonanomi et al., 2012).
Restoration of GDNF and Artemin to injured JunΔSC nerves was sufficient to significantly enhance nerve regeneration as judged by target reinnervation and functional recovery. To minimize the possibility of supra-physiological effects, we chose relatively low doses of GDNF and Artemin for our study (Boyd and Gordon, 2003; Wang et al., 2008). The lack of neuronal Ret does not cause an increase in FMN death, but impairs target reconnection; hence, GNDF and Artemin must exert their effects primarily by stimulating axonal plasticity and outgrowth, thereby facilitating target reinnervation. GDNF and Artemin have been previously shown to induce axonal outgrowth both in vivo and in vitro (Markus et al., 2002; Wang et al., 2008).
Therefore, the increased neuronal death and dramatic decrease in nerve regeneration in the axotomized facial nucleus in JunΔSC mice is most likely due to the combined effects of diminished expression of several neurotrophins, such as BDNF1, NGF, and LIF, and a variety of other growth factors, cytokines, and chemokines, in addition to the Ret ligands. Such a poor neurotrophic environment has an impact on the short-term ability of axons to regrow after crush, ultimately leading to massive motoneuron death and permanent deficit in nerve function.
We conclude that c-Jun–dependent transcriptional regulation of GNDF and Artn in SCs, together with other neurotrophins that act in a paracrine manner in neurons, is essential for motoneuron survival and successful peripheral target reinnervation.
Materials and methods
Mouse lines and animal surgery
All experiments involving mice received approval from the London Research Institute (LRI) Animal Ethics Committee, following Home Office guidelines.
To obtain SCs lacking c-Jun (JunΔSC), Junf/f mice were crossed with P0-CRE mice. Junf/f mice are described in Behrens et al. (2002). In brief, the genomic c-jun locus derived from a λ FIX 129 library (Agilent Technologies) was cloned and the 5′ loxP site was inserted into an EcoRI site present in the 5′ untranslated region (UTR) of c-jun. A floxed neomycin resistance and thymidine kinase gene selection cassette was inserted into 3′ flanking sequence 2.5 kb downstream of the translation stop codon. A diphtheria toxin (DTα) gene was inserted for selection against random integrants. The linearized targeting construct was electroporated into E14.1 ES cells and the homologous recombinants were identified by PCR. The neomycin and thymidine kinase genes were deleted by transient transfection of a vector expressing cre recombinase. ES cell clones carrying a floxed allele of c-jun were injected into C57BL/6 blastocysts and several chimeras from one ES cell clone transmitted the mutant allele to their offspring. P0-CRE mice have been described previously (Feltri et al., 2002), and contain a complete mouse P0 gene with 6 kilobases of promoter, in which the ATG start of translation was mutated and substituted with Cre recombinase.
Resulting Junf/f ; P0-CRE+ mice were crossed back to Junf/f animals to obtain experimental cohorts. To obtain mice with neurons lacking Ret (Ret proto-oncogene; RetΔN), Retf/f mice were crossed with Synapsin-Cre mice (Hoesche et al., 1993). Resulting Retf/f:Synapsin-Cre+ mice were crossed back to Retf/f animals to obtain experimental cohorts. Retf/f mice have been described previously (Kramer et al., 2006). The conditional floxed Ret allele Retf was generated by flanking exon 12 with loxP sites; exon 12 encodes the ATP-binding site of the Ret kinase domain. The DNA fragments for the construct were amplified by PCR and the ES cells screened by Southern blotting with a 750-bp probe encoding exon 15. The neo cassette in the targeted Ret locus was removed by crossing Retf mice with FLPe mice.
We performed facial nerve axotomies in 2–5-mo-old animals of these four genotypes: Junf/f , JunΔSC, Retf/f and RetΔN mice. 1 mo later, mice were culled by anesthetic overdose, and transcardially perfused for 5 min with PBS followed by 10 min with a solution of fixative containing 4% (wt/vol) paraformaldehyde (PFA).
For the experiments shown in Fig. 5, G–I, injured Junf/f and JunΔSC mice were treated with growth factors or saline solution using osmotic minipumps (model 2004; Alzet). Osmotic pumps were loaded according to the manufacturer’s instructions with saline solution or a cocktail of recombinant GDNF (eBioscience) and Artemin (R&D systems) at a concentration of 25 µg/ml. This provided growth factor delivery during ∼30 d at a rate of 150 ng a day. Similar doses have been previously used in in vivo regeneration assays, (Boyd and Gordon, 2003; Wang et al., 2008). A polyurethane mouse jugular catheter (Alzet) was coupled to the osmotic pump. For the surgical procedure, Junf/f and JunΔSC adult animals were given analgesia 1 h before i.p. anesthesia with Avertin (Sigma-Aldrich). The area around the right ear was shaved and disinfected. A small 1-cm infra-auricular incision was performed to access to the facial nerve. The right facial nerve fibers (including the retroauricular branch) were completely transected at the stylomastoid foramen. A small subcutaneous pocket was opened with forceps on the back to insert the pump, through the same incision used to cut the facial nerve. A loop was created with the catheter, and the proximity of the tip was fixed to the neck muscle with 4-0 Mersilk nonabsorbable suture (Ethicon) at a distance of ∼5 mm from the cut region of the facial nerve. The wound was closed and the animals were given a second dose of analgesia and allowed to regain consciousness in a temperature-controlled environment. Upon recovery, the lack of whisker and eyelid movement was verified.
Functional recovery of whisker hair movement after facial nerve axotomy
1 wk after facial nerve transection, two blinded observers three times a wk for the following 3 wk assessed whisker hair movement (WHM). Complete lack of movement in the ipsilateral vibrissae was assigned a score of 0, complete recovery 3, with 0.5-step intervals. The presence of asymmetric, fibrillation-related movement of the ipsilateral vibrissae received a score of 0.5. 1 was given for symmetrical but weak vibrissae sweeping, and 2 for moderately good sweep. Each animal showed an individual recovery time course (e.g., from “0” at 7 d to “2.5” at 28 d). For each individual animal, WHM recovery index was calculated using the linearly interpolating area under the curve (AUC) function on the 0–28-d time course, and then divided by 28 d to produce the index number used for Fig. 1 B and Fig. 5, D and G. Each data point (Tx, WHMx) is connected to the next data point (Tx+1, WHMx+1) with a straight line, starting with the (0,0) data point. For each segment, the trapezoid area under the connecting line is calculated using the formula (Tx+1 – Tx) * (WHMx + WHMx+1)/2 and then all segment areas are added together for each individual animal
Target reinnervation and neuronal cell counts
Whisker pad reinnervation was assessed by applying 15 µl of a 4% solution of the fluorescent retrograde tracer Fluoro-Gold (Fluorochrome) into both whisker pads, 28 d after unilateral nerve cut. Animals were killed 72 h later. The brain tissue was postfixed for 2 d in 4% (wt/vol) PFA, cryoprotected in PBS/30% sucrose overnight, and frozen on dry ice. Tissue was cut in a cryostat at −15°C, and the 25-µm frontal tissue sections—normally 35–40 sections in total—were collected throughout both facial nuclei (contralateral and axotomized). Every fifth section (#1, #6, #11, etc.) was used to count the number of fluorescently labeled motoneurons in the injured and contralateral facial motor nuclei. The remaining sections (approximately 30) were stained with Toluidine blue (Nissl), counted, and the numbers for “missing” sections taken for Fluoro-Gold fluorescence replaced by linear interpolation from adjacent counted sections (i.e., average of #5 and #7 inserted for #6).
As in previous studies (Raivich et al., 2004), total neuronal cell counts were corrected for neuronal cell body size using the Abercrombie correction (Abercrombie, 1946). In brief, this method corrects for counting overestimation derived from cellular structures being present in adjacent sections. We followed the formula N = n x {D/(D + d)}, where N is the corrected neuronal number; n+ is the counted number of neurons; D is the section thickness and d is the mean neuronal diameter, which was determined from the size of nucleated neurons in tissue sections of axotomized and contralateral facial nuclei from the same animal.
Cell culture and transfection
Schwann cell primary cultures were prepared from sciatic nerve and brachial plexus from P3 to P5 mice as described previously (Morgan et al., 1991). In brief, fresh tissue was digested in trypsin/collagenase and seeded onto poly-l-lysine/laminin–coated dishes. Fibroblasts were selectively removed with AraC (Sigma-Aldrich) treatment during 2–3 d. Purified SCs were cultured in expansion medium containing 0.5% normal horse serum, 20 ng/ml β-neuregulin (R&D Systems), and 100 mM cAMP (Sigma-Aldrich) and switched to 10 ng/ml and 2 mM cAMP for the myelination induction experiment shown in Fig. 3 C. The IMS32 SC line was cultured in DME (Invitrogen) supplemented with 10% (wt/vol) FCS in the presence of 1% (wt/vol) penicillin/streptomycin (10,000 U/ml; Invitrogen). For starvation, cells were kept overnight in Opti-MEM (Invitrogen) medium. IMS32 cell cultures were transfected with Lipofectamine 2000 (Invitrogen).
Western blot analysis
IMS32 cells were subjected to lysis buffer supplemented with protease inhibitor cocktail (Sigma-Aldrich) and phosphatase inhibitors (Cell Signaling Technology). Western blot analysis was performed as described previously (Nateri et al., 2005). For immunoblotting, we used antibodies against total c-Jun (BD), phospho-c-Jun (KM1; Santa Cruz Biotechnology, Inc.), and phospho c-Jun Ser73, phospho-SAPK/JNK, SAPK/JNK, phospho-Ret Tyr 405, phosphor-Akt473, total Akt, phospho-ERK 1,2, and total ERK1,2 from Cell Signaling Technology. We also used β-actin (Sigma-Aldrich) and total Ret (Epitomics).
Luciferase assays and constructs
Jun +300/-460, Artn −1082/−1533, and GDNF −2004/−1506 AP-1 containing mouse promoter regions were cloned into a luciferase reporter construct pGL3 containing a TATA box (Promega) using primers: Jun: forward 5′-TGCTTTCTGGATCCCCGCGTCTC-3′, reverse 5′-CCCGGACTTGTGAGCTTCTTCTC-3′; Artn: forward 5′-GGTGGCGCACAACTTTAATCCAGC-3′, reverse 5′-CATCTGTAATGGGATCTGATGCCCT-3′; GDNF: forward 5′-ATCCTTGCTTCCCTGCAAGGTGTTG-3′, reverse 5′-ACAAGAGGAGGACAGGCTCTTG-3′. For the AP1 mutated constructs these sets of primers were used: Artn-AP1-Mut: forward 5′-TACCTGCCCACCCCAAGAGTGTCGTTGTGTTTGGGATGTGGCCCAGAT-3′, reverse 5′-ATCTGGGCCACATCCCAAACACAACGACACTCTTGGGGTGGGCAGGTA-3′; GDNF-AP1-Mut: forward 5′-ACATACTCCCCAACAACTGGCCTAGCTGTAAGAGGAGAGGGGTGTA-3′, reverse 5′-TACACCCCTCTCCTCTTACAGCTAGGCCAGTTGTTGTTGGGGAGTATGT-3′.
IMS32 cell cultures were cotransfected with each of these constructs together with a ubiquitin-renilla luciferase construct. 16 h after transfection, cultures were either left untreated or treated with anisomycin (Anm) during 6 h. Cultures were lysed and luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega). Data are expressed as fold induction over parental pGL3 control–transfected cultures after being normalized using the ubiquitin-renilla luciferase construct.
RNA extraction
RNA extraction from adult sciatic nerves for qRT-PCR and Superarray analysis of gene expression was as follows. Sciatic nerves were dissected, and the epineurium was removed. Tissue was snap-frozen in liquid nitrogen isolated according to the manufacturer’s instructions, using Trizol reagent (Invitrogen), followed by chloroform/isopropanol purification.
Superarray
1 µg of total isolated RNA from sciatic nerves of Junf/f (n = 6) and JunΔSC (n = 7) adult mice, 7 d post-injury, was reverse transcribed to cDNA according to the company guidelines. PCR Array PAMM-031 for mouse neurotrophin and receptors (SABiosciences) was used to detect changes in gene expression.
In situ hybridization
In situ hybridization (ISH) was performed by the Cancer Research UK in situ hybridization service at the LRI as described previously (Haigh et al., 2000), with minor modifications. The Artn in situ probe corresponded to the 5′ UTR and was amplified using the primers: forward 5′-ATCTATCTCCAAGCCTTTGCAC-3′, reverse 5′-ATAAATAGGGCTTCGGCTTCC-3′. The GDNF in situ probe corresponded to the last exon, and the primers to generate it were: forward 5′-AGGTCACCAGATAAACAAGCGGCAGC-3′, reverse 5′-CTAAAAACGACAGGTCGTCGTCG-3′. Jun in situ probe hybridizes with the exonic sequence and primers used were: forward 5′-AGCCTACGGCTACAGTAACCCTAAG-3′, reverse 5′-CCGGGTTGAAGTTGCTGAGGTTG-3′. 7-d post-injury or uninjured sciatic nerves from both Junf/f and JunΔSC adult mice were transcardially perfused for 10 min with PBS followed by a solution of fixative containing 4% (wt/vol) PFA. Sciatic nerves were postfixed overnight and the tissue was embedded in an Agarose block before paraffin embedding and sectioning. ISH images (Figs. 2, A–C, and 3 A) were acquired with a camera (AxioCam HRC; Carl Zeiss) and microscope (model AX10; Carl Zeiss) with Axiovision software CD29, using a 10×/0.3 NA EC Plan-NeoFluor objective, a 40×/0.95 NA Plan-Apochromat, and 2.5x/0.075 NA EC Plan-NeoFluor, all from Carl Zeiss.
Chromatin immunoprecipitation (ChIP)
ChIP analysis was performed as described previously (Alberts et al., 1998) using the IMS32 SC line. In some cases cells were treated for 2 h or 30 min with 50 µM JNK inhibitor II (Merck Biosciences) or 25 ng/ml anisomycin (Sigma-Aldrich), respectively, before collection. Immunoprecipitation with monoclonal anti-phospho c-Jun Ser63 (Santa Cruz Biotechnology) or isotype matching IgG1 anti-HA (Cancer Research UK) was performed overnight at 4°C with rotation with the indicated antibodies. Immunocomplexes were collected by rotating for 2 h at 4°C with ChIP-grade Protein G–Sepharose beads (Cell Signaling Technology). The oligonucleotide sequences used to amplify the DNA fragments were Jun: forward 5′-GGGTGACATCATGGGCTATTTT-3′, reverse 5′-TCCAGCCTGAGCTCAACACTT-3′; Artn: forward 5′-GCCAAAATCACAGGTGAAAACC-3′, reverse 5′-GAATGGAAGAAAGGAAGAAAAGAACTAC-3′; GDNF: forward 5′-CCACACACATTACCCCTATCTATTGT-3′, reverse 5′-CTCTCCTCTTATGACTCATCCAGTTG-3′; Gapdh: forward 5′-GGTCCTCAGTGTAGCCCAAG-3′, reverse 5′-AATGTGTCCGTCGTGGATCT-3′.
qRT-PCR was done with SYBR Green incorporation (Platinum Quantitative PCR SuperMix-UDG w/ROX; Invitrogen) using an ABI7900HT system (Applied Bioscience), and the data were analyzed using SDS 2.3 software. Enrichment from the IP was quantified with reference to the input and results are presented as fold enhancement over untreated cells.
qRT-PCR analysis
For qRT-PCR analysis, total mRNA was isolated from adult sciatic nerves of both Junf/f and JunΔSC. cDNA was synthesized using Superscript III reagents according to the manufacturer’s instructions (Invitrogen). qRT-PCR was performed measuring SYBR Green incorporation (Platinum Quantitative PCR SuperMix-UDG w/ROX; Invitrogen) on an ABI7900HT system (Applied Bioscience). Data were analyzed using the SDS 2.3 software. The following primers were used for mouse cell qRT-PCR analysis: Artn: forward 5′-TTGCCCATCTGGAGAAGTCA-3′, reverse 5′-GTGCTTGGCAGGACCTGTTC-3′; BDNF 1: forward 5′-CCTGGCGACAGGGAAATCT-3′, reverse 5′-GCACGCCGATCCTTTGC-3′; BDNF: forward 5′-GGTATCCAAAGGCCAACTGA-3′, reverse 5′-GCAGCCTTCCTTGGTGTAAC-3′; GDNF: forward 5′-TCTCGAGCAGGTTCGAATGG-3′, reverse 5′-AAGAACCGTCGCAAACTTTACC-3′; LIF: forward 5′-ACGCGTGTCAGCGACAAA-3′, reverse 5′-GGCAGCCAGTCCTGAGATGA-3′; NGFb: forward 5′-TGGAGCTGGCCTTATATTTGGAT-3′, reverse 5′-TTGAGGCACAGCATGTTCACTA-3′; Psph: forward 5′-CCTGGACGCCCCATCAG-3′, reverse 5′-GCTCCACAGTCGGCATGAA-3′: TGF-α: forward 5′-GAGCCTGTGTGGGCACCTA-3′, reverse 5′-TCACGAGGACGCTAATCCACTT-3′; Fos: forward 5′-GGGAGGACCTTACCTGTTCGT-3′, reverse 5′-CAGATGTGGATGCTTGCAAGTC-3′; Il1b: forward 5′-CTACAGGCTCCGAGATGAACAAC-3′, reverse 5′-TCCATTGAGGTGGAGAGCTTTC-3′; Mt3: forward 5′-GACCTGCCCCTGTCCTACTG-3′, reverse 5′-CATTTGCAGCCCTTGCACTT-3′; Jun: forward 5′-AATGGGCACATCACCACTACAC-3′, reverse 5′-TGCTCGTCGGTCACGTTCT-3′; Cxcr4: forward 5′-ACCTCTACAGCAGCGTTCTCATC-3′, reverse 5′-TGTTGGTGGCGTGGACAATA-3′; MBP: forward 5′-GGCGGTGACAGACTCCAAG-3′, reverse 5′-GAAGCTCGTCGGACTCTGAG-3′; p75NTR: forward 5′-TGATGGAGTCGGGCTAATGTC-3′, reverse 5′-AGATTCATCCCTCCACAAATGC-3′; MPZ: forward 5′-CGGACAGGGAAATCTATGGTGC-3′, reverse 5′-TGGTAGCGCCAGGTAAAAGAG-3′; Egr2: forward 5′-GCCAAGGCCGTAGACAAAATC-3′, reverse 5′-CCACTCCGTTCATCTGGTCA-3′; Gapdh: forward 5′-TGTGTCCGTCGTGGATCTGA-3′, reverse 5′-CCTGCTTCACCACCTTCTTGA-3′.
Immunohistochemistry and imaging for c-Jun, p75NTR, and Ret
Mice were perfused and tissue fixed and paraffin embedded or cryoprotected with 30% buffered sucrose and frozen on dry ice as described previously (Raivich et al., 2004). Paraffin sections of injured Junf/f and JunΔSC sciatic nerves (longitudinally cut, 8-µm thickness) from 2-mo-old mice were co-stained using total c-Jun (BD) and p75NTR (Cancer Research UK antibody facility; Fig. 2 D). Subsequently, TRITC- and FITC-coupled secondary antibodies (Invitrogen) were used, respectively. Sections were mounted on VectorShield Hardset mounting media with DAPI (Vector Laboratories). Images were acquired at room temperature with a confocal microscope (TCS SP5; Leica) and an HCX PL-APO 40× 1.25 NA objective (Leica). The software used was Application Suite 2.6.0.7266 (Leica). For Fig. 5 (A and B), paraffin sections of uninjured control or mutant mice were stained with DAB for total Ret antibody (Epitomics). Images were acquired with a camera (AxioCam HRC; Carl Zeiss) and microscope (model AX10; Carl Zeiss) with Axiovision software CD29, using a 10×/0.3 NA EC Plan-NeoFluor objective, a 40×/0.95 NA Plan-Apochromat, and a 2.5x/0.075 NA EC Plan NeoFluor, all from Carl Zeiss. For Fig. S4, we used total c-Jun (H-79; Santa Cruz Biotechnology, Inc.) and secondary antibody biotinylated anti–rabbit Ig (goat polyclonal antibody; Vector Laboratories). Images were obtained with a microscope (Eclipse E600, 10x/0.45 NA; Nikon) at room temperature, using a camera (S1 Alpha; Leica).
Quantitative immunohistochemistry
All quantitative assessment was done on sections from cryoprotected/frozen tissue. 20-µm coronal brainstem sections at the level of the facial motor nucleus (brainstem) were cut in a cryotome, stored at −80°C, and then processed for immunohistochemistry as described previously (Möller et al., 1996). The sections were incubated overnight with primary antibodies against integrin subunits αX integrin (1:400; Endogen) and αM integrin (1:5,000; BD) or CD44 (Millipore), CGRP, Galanin (1:100 and 1:1,000, respectively; Peninsula Laboratories), or GFAP (Dako) for 2 h with a biotinylated goat anti–rabbit, –rat (1:100, Vector Laboratories), or –hamster (1:100; Dianova) secondary antibody, then with Avidin-biotinylated horseradish peroxidase complex (ABC; Vector Laboratories) and visualized with diaminobenzidine/H2O2 (Sigma-Aldrich). 8-bit digital images were obtained using a color video camera (3CCD; Sony AVT-Horn) with a microscope (Eclipse E600; Nikon). Images of both control and axotomized nuclei and surrounding glass were captured at 10x/0.45 NA at room temperature, using Optimas 6.2 software (Bothell) to obtain the mean and standard deviation (SD) values for optical luminosity. SD was subtracted from the mean for each image (Mean-SD algorithm) and the resulting values were subtracted from those of the surrounding glass to obtain the specific values for the control and axotomized sides, shown in Fig. S1. Values for each animal were calculated based on two sections per animal, spaced 340 µm apart. Microglial αX integrin immunoreactivity was not diffusely distributed over the whole nucleus but concentrated over few previously described phagocytic microglia that adhere to each other in the form of 15–30-µm large clusters. In Fig. 1 I, we therefore counted the number of these αX+ clusters in the axotomized motor nucleus 14 d after facial nerve cut. Values for each animal were calculated based on two sections spaced 340 µm apart.
Axonal regeneration and macrophage influx after facial nerve crush
The speed of facial axonal regeneration was determined as described in Werner et al. (2000), 4 d after facial nerve crush. The facial nerve was fixed in situ by 60 min perfusion with 1% PFA/PBS, dissected, and frozen on dry ice, and 15–20-mm-long pieces of nerve were longitudinally sectioned at 10-µm thickness. The regenerating axons were visualized by immunostaining for galanin (1:100; Peninsula Laboratories) or calcitonin gene-related peptide (CGRP, 1:1,000; Peninsula Laboratories) in every fifth section, and the average distance for each animal was calculated from 4–5 tissue sections for each animal and neuropeptide.
Macrophage influx into the injured facial nerve 4 d after crush was assessed at the crush site or 2 and 4 mm distally. As in previous studies (Galiano et al., 2001), the number of macrophages was determined by counting the αM+ cells crossing a 0.5-mm-long vertical microscopic grid line, perpendicular to the nerve’s longitudinal axis. Again, the numbers for each animal were also calculated from 4–5 tissue sections, spaced 50 µm apart.
Statistical analysis
When only two groups were present, statistical analysis for axonal regeneration distance, neuronal cell counts, macrophage number in the injured nerve, and immunohistochemical staining intensity between mutant mice and wild-type controls was performed using a two-tailed, unpaired Student’s t test. When more than two groups were present, statistical analysis was done using one-way ANOVA followed by post-hoc Tukey test. For all three tests, the α significance level was set at 0.05.
In the case of αX+ phagocytic macrophage clusters in the axotomized facial motor nucleus, statistical analysis was performed on a derived, semi-logarithmic function, f, defined as: f = log (x + 1), with x being the mean number of macrophage clusters in the facial nucleus for each individual animal. The number of clusters in brackets was increased by 1 to avoid negative infinite numbers for those nonaxotomized facial nuclei that did not contain clusters. The function led to a strong reduction in the normally extensive variation (factor 2–3) for the cluster number in axotomized facial nuclei.
Online supplemental material
Fig. S1 shows quantitative immunohistochemistry of standard markers of neuronal and glial response in the facial motor nucleus after injury in Junf/f and JunΔSC mice. Fig. S2 shows macrophage infiltration and up-regulation of SC immature marker p75NTR in the distal compartment to the injury in longitudinal sections of wild-type sciatic nerve 7 d after injury. Fig. S3 shows differential staining patterns for Ret, found in sciatic nerve axonal fibers, and p75NTR, expressed by SC progenitors. The figure also depicts an in vivo mode of growth factor delivery via i.p. osmotic pump. Fig. S4 shows unaltered c-Jun neuronal activation after injury by immunohistochemistry and Western blot in JunΔSC facial nucleus compared with Junf/f. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201205025/DC1.
Supplementary Material
Acknowledgments
We are grateful to the Animal Unit, Equipment Park, and the Experimental Pathology Laboratory at the London Research Institute for technical help and advice on histology. The IMS32 line was a kind gift of Kazuhiko Watabe (Tokyo Metropolitan Institute for Neuroscience, Tokyo, Japan).
X. Fontana is funded by a Marie Curie Intraeuropean Fellowship (PIEF-GA-2009-236307). The London Research Institute is funded by Cancer Research UK. G. Raivich would like to acknowledge support by BBSRC (S20299, B/D009537/1), the Wellcome Research Trust (WT088646MA), and SPARKS Medical.
The authors declare no conflict of interest.
Footnotes
Abbreviations used in this paper:
- +G.A.
- recombinant GDNF and Artemin proteins
- Anm
- anisomycin
- BDNF
- brain derived neurotrophic factor
- CGRP
- calcitonin gene-related peptide
- CNS
- central nervous system
- FG
- Fluoro-Gold
- FMN
- facial motoneuron
- GDNF
- glial-derived neurotrophic factor
- ISH
- in situ hybridization
- JunΔSC
- Jun deletion in Schwann cells
- Junf/f
- floxed Jun allele
- LIF
- leukemia inhibitory factor
- MPZ
- myelin protein zero
- p75NTR
- p75 neurotrophin receptor
- PNS
- peripheral nervous system
- qRT-PCR
- quantitative real-time PCR
- RetΔN
- Ret deletion in neurons
- SC
- Schwann cell
- WHM
- whisker hair movement
- WHM RI
- whisker hair movement recovery index
References
- Abercrombie M. 1946. Estimation of nuclear population from microtome sections. Anat. Rec. 94:239–247 10.1002/ar.1090940210 [DOI] [PubMed] [Google Scholar]
- Airaksinen M.S., Saarma M. 2002. The GDNF family: signalling, biological functions and therapeutic value. Nat. Rev. Neurosci. 3:383–394 10.1038/nrn812 [DOI] [PubMed] [Google Scholar]
- Alberts A.S., Geneste O., Treisman R. 1998. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell. 92:475–487 10.1016/S0092-8674(00)80941-1 [DOI] [PubMed] [Google Scholar]
- Andres R., Forgie A., Wyatt S., Chen Q., de Sauvage F.J., Davies A.M. 2001. Multiple effects of artemin on sympathetic neurone generation, survival and growth. Development. 128:3685–3695 [DOI] [PubMed] [Google Scholar]
- Angel P., Hattori K., Smeal T., Karin M. 1988. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 55:875–885 10.1016/0092-8674(88)90143-2 [DOI] [PubMed] [Google Scholar]
- Baloh R.H., Tansey M.G., Lampe P.A., Fahrner T.J., Enomoto H., Simburger K.S., Leitner M.L., Araki T., Johnson E.M., Jr, Milbrandt J. 1998. Artemin, a novel member of the GDNF ligand family, supports peripheral and central neurons and signals through the GFRalpha3-RET receptor complex. Neuron. 21:1291–1302 10.1016/S0896-6273(00)80649-2 [DOI] [PubMed] [Google Scholar]
- Baloh R.H., Enomoto H., Johnson E.M., Jr, Milbrandt J. 2000. The GDNF family ligands and receptors - implications for neural development. Curr. Opin. Neurobiol. 10:103–110 10.1016/S0959-4388(99)00048-3 [DOI] [PubMed] [Google Scholar]
- Barati S., Hurtado P.R., Zhang S.H., Tinsley R., Ferguson I.A., Rush R.A. 2006. GDNF gene delivery via the p75(NTR) receptor rescues injured motor neurons. Exp. Neurol. 202:179–188 10.1016/j.expneurol.2006.05.027 [DOI] [PubMed] [Google Scholar]
- Behrens A., Sibilia M., David J.P., Möhle-Steinlein U., Tronche F., Schütz G., Wagner E.F. 2002. Impaired postnatal hepatocyte proliferation and liver regeneration in mice lacking c-jun in the liver. EMBO J. 21:1782–1790 10.1093/emboj/21.7.1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bespalov M.M., Saarma M. 2007. GDNF family receptor complexes are emerging drug targets. Trends Pharmacol. Sci. 28:68–74 10.1016/j.tips.2006.12.005 [DOI] [PubMed] [Google Scholar]
- Bonanomi D., Chivatakarn O., Bai G., Abdesselem H., Lettieri K., Marquardt T., Pierchala B.A., Pfaff S.L. 2012. Ret is a multifunctional coreceptor that integrates diffusible- and contact-axon guidance signals. Cell. 148:568–582 10.1016/j.cell.2012.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd J.G., Gordon T. 2003. Glial cell line-derived neurotrophic factor and brain-derived neurotrophic factor sustain the axonal regeneration of chronically axotomized motoneurons in vivo. Exp. Neurol. 183:610–619 10.1016/S0014-4886(03)00183-3 [DOI] [PubMed] [Google Scholar]
- Bozec A., Bakiri L., Hoebertz A., Eferl R., Schilling A.F., Komnenovic V., Scheuch H., Priemel M., Stewart C.L., Amling M., Wagner E.F. 2008. Osteoclast size is controlled by Fra-2 through LIF/LIF-receptor signalling and hypoxia. Nature. 454:221–225 10.1038/nature07019 [DOI] [PubMed] [Google Scholar]
- Coulpier M., Anders J., Ibáñez C.F. 2002. Coordinated activation of autophosphorylation sites in the RET receptor tyrosine kinase: importance of tyrosine 1062 for GDNF mediated neuronal differentiation and survival. J. Biol. Chem. 277:1991–1999 10.1074/jbc.M107992200 [DOI] [PubMed] [Google Scholar]
- Curtis R., Scherer S.S., Somogyi R., Adryan K.M., Ip N.Y., Zhu Y., Lindsay R.M., DiStefano P.S. 1994. Retrograde axonal transport of LIF is increased by peripheral nerve injury: correlation with increased LIF expression in distal nerve. Neuron. 12:191–204 10.1016/0896-6273(94)90163-5 [DOI] [PubMed] [Google Scholar]
- Durbec P., Marcos-Gutierrez C.V., Kilkenny C., Grigoriou M., Wartiowaara K., Suvanto P., Smith D., Ponder B., Costantini F., Saarma M., et al. 1996. GDNF signalling through the Ret receptor tyrosine kinase. Nature. 381:789–793 10.1038/381789a0 [DOI] [PubMed] [Google Scholar]
- Feltri M.L., Graus Porta D., Previtali S.C., Nodari A., Migliavacca B., Cassetti A., Littlewood-Evans A., Reichardt L.F., Messing A., Quattrini A., et al. 2002. Conditional disruption of beta 1 integrin in Schwann cells impedes interactions with axons. J. Cell Biol. 156:199–209 10.1083/jcb.200109021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiano M., Liu Z.Q., Kalla R., Bohatschek M., Koppius A., Gschwendtner A., Xu S., Werner A., Kloss C.U., Jones L.L., et al. 2001. Interleukin-6 (IL6) and cellular response to facial nerve injury: effects on lymphocyte recruitment, early microglial activation and axonal outgrowth in IL6-deficient mice. Eur. J. Neurosci. 14:327–341 10.1046/j.0953-816x.2001.01647.x [DOI] [PubMed] [Google Scholar]
- Haigh J.J., Gerber H.P., Ferrara N., Wagner E.F. 2000. Conditional inactivation of VEGF-A in areas of collagen2a1 expression results in embryonic lethality in the heterozygous state. Development. 127:1445–1453 [DOI] [PubMed] [Google Scholar]
- Heumann R., Korsching S., Bandtlow C., Thoenen H. 1987. Changes of nerve growth factor synthesis in nonneuronal cells in response to sciatic nerve transection. J. Cell Biol. 104:1623–1631 10.1083/jcb.104.6.1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heumann R., Goemans C., Bartsch D., Lingenhöhl K., Waldmeier P.C., Hengerer B., Allegrini P.R., Schellander K., Wagner E.F., Arendt T., et al. 2000. Transgenic activation of Ras in neurons promotes hypertrophy and protects from lesion-induced degeneration. J. Cell Biol. 151:1537–1548 10.1083/jcb.151.7.1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoesche C., Sauerwald A., Veh R.W., Krippl B., Kilimann M.W. 1993. The 5′-flanking region of the rat synapsin I gene directs neuron-specific and developmentally regulated reporter gene expression in transgenic mice. J. Biol. Chem. 268:26494–26502 [PubMed] [Google Scholar]
- Hutton E.J., Carty L., Laurá M., Houlden H., Lunn M.P., Brandner S., Mirsky R., Jessen K., Reilly M.M. 2011. c-Jun expression in human neuropathies: a pilot study. J. Peripher. Nerv. Syst. 16:295–303 10.1111/j.1529-8027.2011.00360.x [DOI] [PubMed] [Google Scholar]
- Jessen K.R., Mirsky R. 2008. Negative regulation of myelination: relevance for development, injury, and demyelinating disease. Glia. 56:1552–1565 10.1002/glia.20761 [DOI] [PubMed] [Google Scholar]
- Jing S., Wen D., Yu Y., Holst P.L., Luo Y., Fang M., Tamir R., Antonio L., Hu Z., Cupples R., et al. 1996. GDNF-induced activation of the ret protein tyrosine kinase is mediated by GDNFR-alpha, a novel receptor for GDNF. Cell. 85:1113–1124 10.1016/S0092-8674(00)81311-2 [DOI] [PubMed] [Google Scholar]
- John G.R., Shankar S.L., Shafit-Zagardo B., Massimi A., Lee S.C., Raine C.S., Brosnan C.F. 2002. Multiple sclerosis: re-expression of a developmental pathway that restricts oligodendrocyte maturation. Nat. Med. 8:1115–1121 10.1038/nm781 [DOI] [PubMed] [Google Scholar]
- Kao S.C., Wu H., Xie J., Chang C.P., Ranish J.A., Graef I.A., Crabtree G.R. 2009. Calcineurin/NFAT signaling is required for neuregulin-regulated Schwann cell differentiation. Science. 323:651–654 10.1126/science.1166562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D.R., Miller F.D. 2000. Neurotrophin signal transduction in the nervous system. Curr. Opin. Neurobiol. 10:381–391 10.1016/S0959-4388(00)00092-1 [DOI] [PubMed] [Google Scholar]
- Kloss C.U., Werner A., Klein M.A., Shen J., Menuz K., Probst J.C., Kreutzberg G.W., Raivich G. 1999. Integrin family of cell adhesion molecules in the injured brain: regulation and cellular localization in the normal and regenerating mouse facial motor nucleus. J. Comp. Neurol. 411:162–178 [DOI] [PubMed] [Google Scholar]
- Kramer E.R., Knott L., Su F., Dessaud E., Krull C.E., Helmbacher F., Klein R. 2006. Cooperation between GDNF/Ret and ephrinA/EphA4 signals for motor-axon pathway selection in the limb. Neuron. 50:35–47 10.1016/j.neuron.2006.02.020 [DOI] [PubMed] [Google Scholar]
- Kramer E.R., Aron L., Ramakers G.M., Seitz S., Zhuang X., Beyer K., Smidt M.P., Klein R. 2007. Absence of Ret signaling in mice causes progressive and late degeneration of the nigrostriatal system. PLoS Biol. 5:e39 10.1371/journal.pbio.0050039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus A., Patel T.D., Snider W.D. 2002. Neurotrophic factors and axonal growth. Curr. Opin. Neurobiol. 12:523–531 10.1016/S0959-4388(02)00372-0 [DOI] [PubMed] [Google Scholar]
- Meyer M., Matsuoka I., Wetmore C., Olson L., Thoenen H. 1992. Enhanced synthesis of brain-derived neurotrophic factor in the lesioned peripheral nerve: different mechanisms are responsible for the regulation of BDNF and NGF mRNA. J. Cell Biol. 119:45–54 10.1083/jcb.119.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller J.C., Klein M.A., Haas S., Jones L.L., Kreutzberg G.W., Raivich G. 1996. Regulation of thrombospondin in the regenerating mouse facial motor nucleus. Glia. 17:121–132 [DOI] [PubMed] [Google Scholar]
- Morgan L., Jessen K.R., Mirsky R. 1991. The effects of cAMP on differentiation of cultured Schwann cells: progression from an early phenotype (04+) to a myelin phenotype (P0+, GFAP-, N-CAM-, NGF-receptor-) depends on growth inhibition. J. Cell Biol. 112:457–467 10.1083/jcb.112.3.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I., Noon L.A., Ribeiro S., Kerai A.P., Parrinello S., Rosenberg L.H., Collins M.J., Harrisingh M.C., White I.J., Woodhoo A., Lloyd A.C. 2012. A central role for the ERK-signaling pathway in controlling Schwann cell plasticity and peripheral nerve regeneration in vivo. Neuron. 73:729–742 10.1016/j.neuron.2011.11.031 [DOI] [PubMed] [Google Scholar]
- Nateri A.S., Spencer-Dene B., Behrens A. 2005. Interaction of phosphorylated c-Jun with TCF4 regulates intestinal cancer development. Nature. 437:281–285 10.1038/nature03914 [DOI] [PubMed] [Google Scholar]
- Naveilhan P., ElShamy W.M., Ernfors P. 1997. Differential regulation of mRNAs for GDNF and its receptors Ret and GDNFR alpha after sciatic nerve lesion in the mouse. Eur. J. Neurosci. 9:1450–1460 10.1111/j.1460-9568.1997.tb01499.x [DOI] [PubMed] [Google Scholar]
- Parkinson D.B., Bhaskaran A., Arthur-Farraj P., Noon L.A., Woodhoo A., Lloyd A.C., Feltri M.L., Wrabetz L., Behrens A., Mirsky R., Jessen K.R. 2008. c-Jun is a negative regulator of myelination. J. Cell Biol. 181:625–637 10.1083/jcb.200803013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello S., Napoli I., Ribeiro S., Digby P.W., Fedorova M., Parkinson D.B., Doddrell R.D., Nakayama M., Adams R.H., Lloyd A.C. 2010. EphB signaling directs peripheral nerve regeneration through Sox2-dependent Schwann cell sorting. Cell. 143:145–155 10.1016/j.cell.2010.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivich G., Banati R. 2004. Brain microglia and blood-derived macrophages: molecular profiles and functional roles in multiple sclerosis and animal models of autoimmune demyelinating disease. Brain Res. Brain Res. Rev. 46:261–281 10.1016/j.brainresrev.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Raivich G., Bohatschek M., Da Costa C., Iwata O., Galiano M., Hristova M., Nateri A.S., Makwana M., Riera-Sans L., Wolfer D.P., et al. 2004. The AP-1 transcription factor c-Jun is required for efficient axonal regeneration. Neuron. 43:57–67 10.1016/j.neuron.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Shy M.E., Shi Y., Wrabetz L., Kamholz J., Scherer S.S. 1996. Axon-Schwann cell interactions regulate the expression of c-jun in Schwann cells. J. Neurosci. Res. 43:511–525 [DOI] [PubMed] [Google Scholar]
- Svaren J., Meijer D. 2008. The molecular machinery of myelin gene transcription in Schwann cells. Glia. 56:1541–1551 10.1002/glia.20767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treanor J.J., Goodman L., de Sauvage F., Stone D.M., Poulsen K.T., Beck C.D., Gray C., Armanini M.P., Pollock R.A., Hefti F., et al. 1996. Characterization of a multicomponent receptor for GDNF. Nature. 382:80–83 10.1038/382080a0 [DOI] [PubMed] [Google Scholar]
- Wang R., King T., Ossipov M.H., Rossomando A.J., Vanderah T.W., Harvey P., Cariani P., Frank E., Sah D.W., Porreca F. 2008. Persistent restoration of sensory function by immediate or delayed systemic artemin after dorsal root injury. Nat. Neurosci. 11:488–496 10.1038/nn2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe K., Fukuda T., Tanaka J., Honda H., Toyohara K., Sakai O. 1995. Spontaneously immortalized adult mouse Schwann cells secrete autocrine and paracrine growth-promoting activities. J. Neurosci. Res. 41:279–290 10.1002/jnr.490410215 [DOI] [PubMed] [Google Scholar]
- Webber C., Zochodne D. 2010. The nerve regenerative microenvironment: early behavior and partnership of axons and Schwann cells. Exp. Neurol. 223:51–59 10.1016/j.expneurol.2009.05.037 [DOI] [PubMed] [Google Scholar]
- Werner A., Willem M., Jones L.L., Kreutzberg G.W., Mayer U., Raivich G. 2000. Impaired axonal regeneration in alpha7 integrin-deficient mice. J. Neurosci. 20:1822–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widenfalk J., Nosrat C., Tomac A., Westphal H., Hoffer B., Olson L. 1997. Neurturin and glial cell line-derived neurotrophic factor receptor-beta (GDNFR-beta), novel proteins related to GDNF and GDNFR-alpha with specific cellular patterns of expression suggesting roles in the developing and adult nervous system and in peripheral organs. J. Neurosci. 17:8506–8519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widenfalk J., Tomac A., Lindqvist E., Hoffer B., Olson L. 1998. GFRalpha-3, a protein related to GFRalpha-1, is expressed in developing peripheral neurons and ensheathing cells. Eur. J. Neurosci. 10:1508–1517 10.1046/j.1460-9568.1998.00192.x [DOI] [PubMed] [Google Scholar]
- Woodhoo A., Alonso M.B., Droggiti A., Turmaine M., D’Antonio M., Parkinson D.B., Wilton D.K., Al-Shawi R., Simons P., Shen J., et al. 2009. Notch controls embryonic Schwann cell differentiation, postnatal myelination and adult plasticity. Nat. Neurosci. 12:839–847 10.1038/nn.2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Romero M.I., Ghosh P., Ye Z., Charnay P., Rushing E.J., Marth J.D., Parada L.F. 2001. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 15:859–876 10.1101/gad.862101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.