Abstract
We devised and tested an in vivo system to monitor the migration of mesenchymal stem cells (MSCs) within the marrow cavity. In vitro studies confirmed that platelet-derived growth factor (PDGF)-AA had the most potent chemotactic effect of the tested factors, and possessed the greatest number of receptors in MSCs. MSCs were labeled with fluorescent nanoparticles and injected into the marrow cavity of nude rats through osteochondral defects created in the distal femur. The defects were sealed with HCF (heparin-conjugated fibrin) or PDGF-AA-loaded HCF. In the HCF-only group, the nanoparticle-labeled MSCs dispersed outside the marrow cavity within 3 days after injection. In the PDGF-AA-loaded HCF group, the labeled cells moved time-dependently for 14 days toward the osteochondral defect. HCF-PDGF in low dose (LD; 8.5 ng/µl) was more effective than HCF-PDGF in high dose (HD; 17 ng/µl) in recruiting the MSCs to the osteochondral defect. After 21 days, the defects treated with PDGF and transforming growth factor (TGF)-β1-loaded HCF showed excellent cartilage repair compared with other groups. Further studies confirmed that this in vivo osteochondral MSCs tracking system (IOMTS) worked for other chemoattractants (chemokine (C-C motif) ligand 2 (CCL2) and PDGF-BB). IOMTS can provide a useful tool to examine the effect of growth factors or chemokines on endogenous cartilage repair.
Introduction
Articular cartilage (AC) has very limited capacity for repair after injury, being poorly supplied by blood vessels, nerves, and the lymphatic system. Recently, several cell-based therapeutic approaches have been developed to repair AC, either with cells alone or with the composite of cells and scaffold, such as poly (ethylene glycol)-based hydrogel, alginate, agarose, and hybrid materials.1,2,3,4 These methods, however, are associated with problems, such as the dedifferentiation of cells with passaging and the donor site morbidity.5,6 Moreover, in these procedures, a culture period of 4–6 weeks is necessary to expand and differentiate the isolated cells before implantation into the defect. For this reason, a method that mobilizes the endogenous pool of mesenchymal stem cells (MSCs) would provide a less costly and less invasive alternative.
One traditional surgical strategy to treat cartilage defects is multiple drilling of the subchondral bone to allow the migration of endogenous pluripotent progenitor cells into the defects.7 Although there is evidence that the resurfaced cartilage may change from fibrous to hyaline cartilage, the tissue generated thereby has very poor mechanical properties compared to normal cartilage,8 eventually leading to failure.9 The reasons for the failure are the inadequate number of available stem cells as well as the inability to contain the migrated cells in situ. A method that directs the migration of a copious number of autologous MSCs in bone marrow toward injury sites would be necessary to enhance the success of this endogenous repair. Although several chemotactic factors that induce the migration of MSCs have been found by in vitro chemotaxis analysis,10,11,12,13,14 it has not yet been examined which factor would most effectively promote the migration of MSCs within bone marrow toward injury sites in an in vivo osteochondral defect model.
The migration and differentiation of transplanted cells in vivo has been principally analyzed by the histological findings.15 However, these techniques are unsuitable for a dynamic evaluation of the proliferation, migration, and the final fate of transplanted cells because only one time point of data can be obtained from each experimental animal. In contrast, live cell imaging allows the real-time tracking of grafted cells and the monitoring of their migration. Recently, cell-labeling methods utilizing fluorescent nanoparticles were developed to overcome the disadvantages of traditional imaging methods, such as viral and nonviral reporter gene systems, and free organic dyes, etc.16,17,18 Quantum dots and a range of other nanomaterials that contain Resovist, a magnetic resonance imaging contrast agent comprising super paramagnetic iron oxide nanoparticles, have been used in stem cell research for in vitro and in vivo bioimaging.19,20,21,22
In this study, we devised and tested an in vivo osteochondral MSCs tracking system (IOMTS) to monitor the migration of bone marrow-derived MSCs. First, the in vitro migration capacity of human MSCs in response to four chemokines and three growth factors were investigated and the best one was determined. The final candidate was loaded in heparin-conjugated fibrin (HCF) and placed in osteochondral defect of athymic nude rat to induce the migration of nanoparticle-labeled MSCs transplanted within the marrow cavity. Then, we examined if the in vivo migration of transplanted human MSCs into the marrow cavity could be detected by this system, and if the migrated MSCs could promote the healing of injured AC. In addition, we also investigated whether in vivo migration effects of other factors could be compared quantitatively using this IOMTS.
Results
Chemotactic activity of chemokines and growth factors in human MSCs
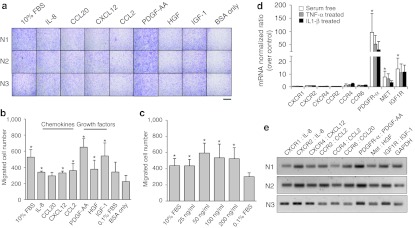
Although most of the tested chemokines and growth factors effectively increased the level of MSC migration above the basal levels (0.1% bovine serum albumin (BSA) only), platelet-derived growth factor (PDGF)-AA had the most potent chemotactic effect (Figure 1a,b). The number of the migrated cells was 3.1 times higher than the negative control (0.1% BSA) (P = 0.0355). When the dose-response effect of PDGF-AA was tested, no additional increase in MSCs migration was observed above a concentration of 50 ng/ml (Figure 1c).
Figure 1.
Comparison of the chemotactic activity of chemokines and growth factors in human MSCs. (a) Migrated cells stained with hematoxylin and eosin. Bar = 400 µm. (b) The number of migrated cells. (c) The dose effect of PDGF-AA on MSCs migration. The bars represent the mean ± SD of migrated cells. n = 3, *P < 0.05 to the negative control. (d) Real-time PCR analysis of receptors. The bars represent the mean ± SD of normalized ratio over the control. n = 3, *P < 0.05 to CXCR2. (e) Gel electrophoresis of the RT-PCR products of each receptor. BSA, bovine serum albumin; CCL2, chemokine (C-C motif) ligand 2; CXCL12, chemokine (C-X-C motif) ligand 12; FBS, fetal bovine serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HGF, hepatocyte growth factor; IGF, insulin-like growth factor; IL, interleukin; mRNA, messenger RNA; MSC, mesenchymal stem cell; PDGF, platelet-derived growth factor; RT-PCR, reverse transcription-PCR; TNF-α, tumor necrosis factor-α.
PDGF receptors are highly expressed in human MSCs of the serum-free environments
To find a correlation between the chemotactic effect of the tested factors and gene expression levels of receptors on MSCs, real-time PCR was performed for the receptors of the tested factors. Under serum-free conditions containing only 0.1% BSA, human MSCs from the three donors had relatively larger gene expression of receptors for growth factors than those for chemokines (Figure 1d). These findings were in line with the results of in vitro chemotaxis of MSCs, suggesting that the migration effects of growth factors were related to the high levels of their receptors in human MSCs. The PDGF receptor (PDGFR)-α messenger RNA (mRNA) levels were higher than those of the other receptors. In the case of PDGFR-α, the mRNA level was 97 times higher than C-X-C chemokine receptor 2 (CXCR2), receptor of chemokine interleukin (IL)-8 (P = 0.037). To determine if a change in extracellular environment causes a difference in intracellular expression levels of chemokine receptors, an in vitro inflammatory condition was produced by a treatment with tumor necrosis factor (TNF)-α and IL-1β, based on a previous report23 and mRNA levels of the chemokine receptors were reconfirmed. When treated with TNF-α or IL-1β, the mRNA expression levels of all receptors were generally lower than the serum-free state. Under this condition, PDGFR-α still had the highest level of expression. The reverse transcription-PCR products of each receptor were analyzed by agarose gel electrophoresis to confirm the results of real-time PCR. As shown in Figure 1e, the PDGFR-α showed the strongest band intensities in all three donors.
Nanoparticle-labeled human MSCs can be traced to an osteochondral defect of nude rat models
The in vitro PDGF-AA release kinetics of two types of delivery systems (fibrin and HCF) was compared. During the first 3 days, 71.5 ± 6.5% of the loaded PDGF-AA was released from fibrin. From HCF, in contrast, 61 ± 6.4% was released for 14 days (Supplementary Figure S1).
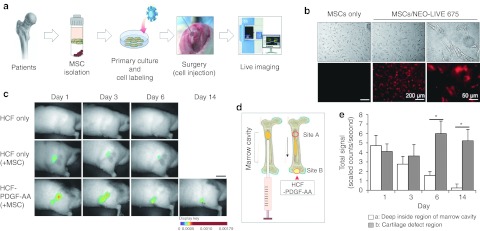
The potential of PDGF-AA to induce MSC migration was examined in vivo in an osteochondral repair model of immunodeficient rats (Figure 2a). The human MSCs were effectively labeled by incubating them with fluorescent nanoparticles. The Cy5.5 fluorescence was strongly detectable in the cultured MSCs after a treatment with nanoparticles (Figure 2b).
Figure 2.
In vivo monitoring of fluorescent nanoparticle-labeled human MSCs in athymic nude rats. (a) The general schema of in vivo experiments. (b) Human MSCs labeled with a fluorescent silica nanoparticle. The left panels are the bright-field images, and the right panels are Cy5.5 fluorescence images. (c) In vivo monitoring of nanoparticle-labeled MSCs. Bar = 20 mm. (d) Two specific sites of signal measurements. Site A represents the most proximal portion of the marrow cavity. Site B represents the osteochondral defect area releasing PDGF-AA. (e) Quantification of the MSCs migration effect by PDGF-AA release from the osteochondral defect in PDGF-AA-loaded HCF group. The bars represent the mean ± SD of fluorescent signal intensity. n = 3, *P < 0.05. HCF, heparin-conjugated fibrin; MSC, mesenchymal stem cell; PDGF, platelet-derived growth factor.
In the HCF-only group without PDGF-AA (the control group), the majority of the nanoparticle-labeled MSCs dispersed outside the marrow cavity within 3 days after the injection despite using matrigel. On the other hand, cells did not disperse in PDGF-AA-loaded HCF group, remaining in the marrow cavity for 14 days. Interestingly, this group showed the time-dependant movement of injected cells toward the osteochondral defect from which PDGF-AA was released. On day 14, all the signals congregated at the osteochondral defect (Figure 2c). When only nanoparticles were injected into marrow cavity and plugged with HCF-PDGF-AA, the signal rapidly disappeared within 24 hours after injection (Supplementary Figure S2). To quantify the migration of MSCs by PDGF-AA released from the osteochondral defect site, the pixels of two specific sites within marrow cavity were counted by densitometric scanning using Maestro EX image software (Cri, Hopkinton, MA). Site A represents the most proximal portion of the marrow cavity. Site B represents the osteochondral defect area releasing PDGF-AA (Figure 2d). The cell densities at site A decreased gradually with time. In contrast, those of site B increased, as expected. This increase reached the maximum levels at day 6 and decreased gradually until day 14 (Figure 2e). The densities of the two areas were significantly different at days 6 and 14 (P = 0.014 and 0.010).
In vivo MSCs tracking patterns differed depending on the PDGF-AA concentration
To determine the optimal concentration of PDGF-AA, another in vivo tracking test of MSCs was performed using the HCFs conjugated with two different PDGF-AA concentrations, 17 ng/µl (HCF-PDGF-high dose (HD)) and 8.5 ng/µl (HCF-PDGF-low dose (LD)). Transforming growth factor (TGF)-β1 was also co-conjugated in both groups, based on previous studies demonstrating that PDGF-BB, one of the PDGF isoforms, had the potential for cartilage regeneration by suppressing endochondral maturation and enhancing chondrocyte proliferation when treated along with TGF-β.24,25
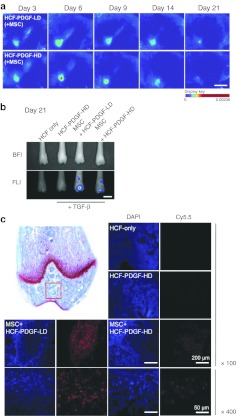
The nanoparticle-labeled MSCs were detected for 21 days using the Maestro image system to investigate the distribution of the migrated human MSCs within osteochondral defect (Figure 3a and Supplementary Figure S3). Three days after injecting the cell–matrigel complex, the fluorescence intensities in the femur of groups HCF-PDGF-LD or HCF-PDGF-HD were similar. On the other hand, both groups showed significant differences since day 6. Quantitative region of interest analysis in the osteochondral defect area revealed ~1.5- and 1.3-fold higher fluorescence signals in the HCF-PDGF-HD group compared to the HCF-PDGF-LD group at days 6 (scaled counts/second = 3.17 ± 0.81, 2.12 ± 0.62) and 9 (scaled counts/second = 2.34 ± 0.26, 1.84 ± 0.43), respectively. On days 14 and 21, however, the fluorescence signals from the osteochondral defect in this group were relatively lower than those in the HCF-PDGF-LD group. So HCF-PDGF-LD was more effective than HCF-PDGF-HD in recruiting the MSCs in the long term. After the animals were sacrificed at day 21, femora were removed from the bodies and the fluorescence signals were detected again to confirm the fluorescence signals of live images from the rats (Figure 3b). Strong fluorescent signals were detected in the osteochondral defect area of both HCF-PDGF-HD and -LD groups, whereas they were not detected in HCF-only or HCF-PDGF-HD groups not injected with labeled MSCs. The total fluorescence signal of HCF-PDGF-LD in the femur was 3.4-fold stronger than that of HCF-PDGF-HD at day 21 (scaled counts/second = 34.83 ± 8.73, 10.21 ± 2.73). This finding demonstrated that the patterns of the fluorescence signal between the postmortem femur and live image were similar, even though the detection ranges of the signal intensities were different. The cause of the decrease in the fluorescent signal in HCF-PDGF-HD group was also investigated by confocal microscopy after paraffin sectioning of each femur. The fluorescent signals were concentrated in high densities within the osteochondral defect area in the HCF-PDGF-LD group (Figure 3c and Supplementary Figure S4b,d). On the other hand, the migrated MSCs in the HCF-PDGF-HD group, unlike the HCF-PDGF-LD group, were spread evenly in the broad area around the defect (Figure 3c and Supplementary Figure S4a,c).
Figure 3.
In vivo MSCs tracking patterns depending on PDGF-AA concentration in osteochondral defect regions. (a) In vivo monitoring of fluorescent nanoparticle-labeled human MSCs depending on the PDGF-AA concentration in rats. Bar = 20 mm. (b) Quantitative ROI analysis using the removed femurs. BFI and FLI represent bright-field image and fluorescent image, respectively. Bar = 5 mm. (c) CLSM images of femur paraffin sections after DAPI staining. The red square of the upper-left panel indicates common detection regions within each femur section. CLSM, confocal laser scanning microscopy; DAPI, 4′,6-diamidino-2-phenylindole; HCF, heparin-conjugated fibrin; HD, high dose; LD, low dose; MSC, mesenchymal stem cell; PDGF, platelet-derived growth factor; ROI, region of interest; TGF-β, transforming growth factor-β.
Effect of migrated human MSCs on the healing of osteochondral defects
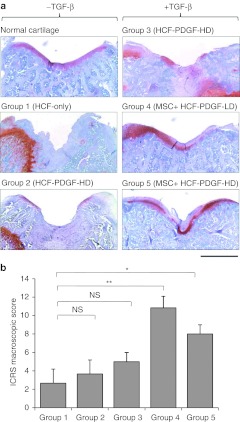
The osteochondral defect was also evaluated histologically by hematoxylin/eosin and Safranin-O/fast green staining 21 days after treatment (Figure 4a). In group 1 (HCF-only without MSCs), the defects were filled with fibrous tissue, the margins clearly demarcated. Their surfaces were totally irregular. In group 2 (HCF-PDGF-HD without MSCs), the defects were largely filled with fibrous tissue as in group 1 and the margin of the defects was also clearly recognizable. In group 3 (HCF-PDGF-HD-TGF-β without MSCs), there were focal islands of cartilage regeneration although the margin of the defects was obviously noticeable and the defects were incompletely reconstituted. On the other hand, in group 5 (HCF-PDGF-HD-TGF-β with MSCs), the defects had become firm and smooth, and the matrices of the repair cartilage were stained with Safranin-O in a similar way to normal cartilage. The defect in the group 4 (HCF-PDGF-LD-TGF-β with MSCs) showed the best healing of all the groups, covered with hyaline cartilage-like tissue. The well-repaired cartilage had a relatively smooth surface with no depression. The thickness and appearance were also similar to that of normal cartilage. Groups 4 and 5 also had significantly higher International Cartilage Repair Society (ICRS) macroscopic score than group 1 (P = 0.0004 and 0.047, Figure 4b).
Figure 4.
Effect of the migrated human MSCs on the healing of an osteochondral defect. (a) Histological findings of the defect from Safranin-O staining. Bar = 1 mm. (b) ICRS macroscopic score of each group. Bars represent the mean ± SD of the score. n = 3, *P < 0.05, **P < 0.01, NS = not significant. HCF, heparin-conjugated fibrin; HD, high dose; ICRS, International Cartilage Repair Society; LD, low dose; MSC, mesenchymal stem cell; PDGF, platelet-derived growth factor; TGF-β, transforming growth factor-β
Other chemoattractants can be investigated using IOMTS
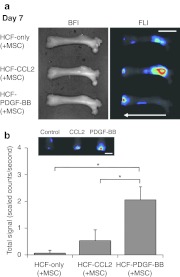
To examine the effect of chemoattractants other than PDGF-AA in this system, further in vivo experiment was performed under the same surgery condition as the preceding study except that only the fluorescence images of the removed femora were obtained 7 days after cell injection. Two chemoattractants were used in this experiment. The first was chemokine (C-C motif) ligand 2 (CCL2), which was the most effective of all chemokines in our in vitro chemotaxis screening. The second was PDGF-BB, which was one of three isotypes of PDGF. In the case of the HCF-only femur, most of the nanoparticle-labeled MSCs were spread out of the marrow cavity, showing reduced fluorescence intensity. On the other hand, strong fluorescence intensity was detected in the femora treated with CCL2 or PDGF-BB. This finding demonstrates that the diffusion of MSCs outside was prevented by CCL2 or PDGF-BB released from HCF. Interestingly, most of the fluorescence signals of the CCL2 femur were detectable in the head region of femur, whereas those of the PDGF-BB femur were only detectable in the osteochondral defect region after 7 days of cell injection. This shows that PDGF-BB-HCF has a stronger chemotactic activity than CCL2-HCF (Figure 5a). In addition, the chemotactic activities of CCL2 and PDGF-BB were compared quantitatively by reading the total fluorescence signals of the osteochondral region. As shown in Figure 5b, the total fluorescence signal in the osteochondral defect region from the PDGF-BB-HCF femur was 3.9 times stronger than that from the CCL2-HCF femur (scaled counts/second = 2.06 ± 0.48, 0.53 ± 0.41; P = 0.019). This shows that the chemotactic ability of various cytokines can be examined by IOMTS.
Figure 5.
The effects of chemokine CCL2 and growth factor PDGF-BB treated in an osteochondral defect on the migration of MSC. (a) Fluorescence image of migrated MSCs in the removed femurs at 7 days post-cell injection. BFI and FLI represent bright-field image and fluorescent image, respectively. White arrow represents the migratory direction of labeled MSCs. Bar = 10 mm. (b) The total fluorescent intensity of a defect region in the removed femur. Bar = 5 mm. Bar graphs represents the mean ± SD of fluorescent signal intensity. n = 3, *P < 0.05. CCL2, chemokine (C-C motif) ligand 2; HCF, heparin-conjugated fibrin; MSC, mesenchymal stem cell; PDGF, platelet-derived growth factor.
Discussion
In this study, we first assessed the in vitro chemotactic potential of human bone-marrow MSCs. Seven factors were selected according to previous reports on the chemotactic activity to MSCs,10,11,12,13,14 and the Boyden chamber assay was used to choose a factor inducing the most effective migration of MSCs, which was PDGF-AA. PDGF-AA is a potent chemoattractant for fibroblasts, neutrophils and monocytes activated by lymphocytes or cytokines as well as for MSCs.26,27,28 Although the chemotactic responses of MSC to several migration factors had been compared previously,10,29 we thought that for a possible clinical application, further study was necessary to determine the best chemotactic factor.
Several studies have reported the key role of chemokines and their corresponding receptors in the migration of MSC.30,31 MSCs express the receptors for growth factors at a moderate to high level, including PDGFR and insulin-like growth factor (IGF)-1R.26,32 A previous study reported that the chemotactic response of MSCs to hepatocyte growth factor (HGF) could be regulated significantly by HGF-c-met signaling.11 That is, the endogenous level of receptor can be crucial for the chemotactic response of MSC by each ligand. Our results demonstrated that mRNA of PDGFR-α, an endogenous receptor for PDGF-AA, was expressed at high levels from the MSCs either under serum-free conditions or in presence of TNF-α while the gene expression of CXCR4, the receptor for chemokine (C-X-C motif) ligand 12 (CXCL12)/stromal cell-derived factor (SDF)-1, was not. This finding also additively suggested that PDGF-AA was the best chemotactic factor for MSCs among the seven tested chemoattractants.
The osteochondral defect model is a well-known in vivo system that has been used mostly to examine the effect of specific scaffolds, stem cells or both on cartilage regeneration.33,34,35 In this study, IOMTS was applied to identify the in vivo migration of human MSCs as well as the repair of injured AC by the migration of cells into the defect. There are three essential components in this system. The first is the use of athymic nude animals to investigate migration of human MSCs. Unusual athymic nude rats were needed because the femur of nude mouse was too small to create an osteochondral defect, and nude rabbits were unavailable to date. This animal has recently been used for in vivo osteochondral defect studies in spite of the high cost.36,37 The second is HCF, which had been reported previously as a successful controlled release system for cytokines.38 The constant release of PDGF-AA for at least 14 days was possible with the application of PDGF-AA-loaded HCF. The period was sufficient to induce a chemotactic response from the MSCs injected inside the marrow cavity of rats. In addition, these results demonstrated that the patterns of MSCs migration to an osteochondral defect area differ according to the dose of PDGF-AA. This suggests that the present system can also be applied to determine the optimum in vivo dose of specific migration factor. The third is the labeling of MSCs with silica-coated magnetic nanoparticles incorporating Cy5.5. Previous labeling methods, including reporter gene systems composed of viral and nonviral vectors, have several disadvantages and may draw confusing results that are unsuitable for the long-term monitoring of stem cells.39,40,41 In addition, fluorescent materials, such as GFP (green fluorescent protein), have photo-bleaching problems.42 Therefore, the fluorescence intensity and labeling specificity of these dyes are inappropriate for the long-term monitoring of stem cells after cell division. The performance of imaging systems has been steadily improved with the advancements of cell-labeling techniques.43 Recently, Park et al. reported that silica-coated magnetic nanoparticles can be taken up by human MSCs without cytotoxicity. Fluorescence was maintained up to passage 7, and the nanoparticles did not affect the expression of cell surface markers.22 However, to date, no group has attempted to monitor the movement of labeled cells inside the bone marrow cavity. We detected the time-dependent migration of MSC within the bone marrow to a defect as well as their destination after cell tracking for 21 days.
As we used human PDGF-AA for the chemotaxis of human MSCs in the nude rats, most of osteochondral healing is attributed to injected human MSCs although host cells certainly contributed, too. This point can be elucidated when we compared the much better quality of osteochondral repair in group 5 (HCF-PDGF-HD-TGF-β with MSCs) compared with group 3 (HCF-PDGF-HD-TGF-β without MSCs), which successively showed better healing than group 1 (HCF-only without growth factors or MSCs). As the nanoparticle can be maintained in cells after division or differentiation and is nontoxic to the cells, it provides a useful tool for stem cell imaging. We had first anticipated to find many labeled cells inside the repaired osteochondral defect area on the patellar groove. However, contrary to our expectation, we found most of the migrated cells around defect area after 21 days. Nevertheless, good healing of the osteochondral defects was achieved with the quality of AC approaching that of normal cartilage. Therefore, we currently speculate that the migrated human MSCs may have induced the regeneration of rat cartilage in other ways, i.e., releasing growth factors or matrix required in tissue regeneration, instead of direct differentiation into AC tissue.
This IOMTS also has some technical problems that require an elaborate in vivo works to solve. First, the high costs of each source including athymic nude rats, commercial nanoparticles, and in vivo image device etc., can impose a heavy burden on the users of this system. The required number of animals for statistical analysis would be very expensive. Second, the fluorescence detecting device used in this study, the Maestro system, has fluorescence detection limits. An equal fluorescence can be detected with different signal intensities depending on the individual difference in bone and skin thickness, and spreading status of labeled cells in the detection area. Sometimes, no fluorescence signals were observed in the animals after the labeled MSCs were injected into the marrow cavity. On that occasion, the animals needed to be sacrificed, and the fluorescence images were taken directly from the denuded bone. Third, as in any experiment using MSCs isolated from patients, there are issues of inhomogeneity and batch-to-batch variations of MSCs as well as the need to define the responsible subpopulation that mediates the therapeutic effect in the given model. In addition, the off-target effects of a tested factor need to be considered, and separately evaluated depending on the model.
Although many in vivo monitoring studies have been performed to detect the tracking of labeled cells from the subcutaneous, intravenous, and intraperitoneal route of the animal to another target site,22,44,45 this report is the first to show the in vivo monitoring of labeled MSCs from the marrow cavity to an osteochondral defect region using Cy5.5 tagged nanoparticles. Notwithstanding the limitations, the significance of this study lies in establishing a unique model to examine the migration of MSCs in response to a chemotactic signal. This study also showed that migrated MSCs could successfully promote the healing of injured AC and that the chemotactic effect of several migration factors can be compared directly with each other using the IOMTS.
In conclusion, the IOMTS can provide a very useful tool to investigate the effect of growth factors or chemokines on endogenous cartilage repair. This system is expected to have clinical applications and make a valuable contribution to basic research.
Materials and Methods
Procurement of samples, cell isolation, and cultivation. The bone marrow samples used to isolate the MSCs were obtained from three patients (mean age: 55 years, range: 38–73 years) undergoing a total hip replacement due to osteoarthritis. Informed consent was obtained from all donors. The MSCs were isolated from fresh bone marrow samples, and expanded as described previously.46 Passage 3 cells were used for experiments.
Chemotaxis assay. Chemotaxis assays were performed in a Boyden chamber using 48-well chemotaxis chamber plates (Neuro Probe, Gaithersburg, MD) with polycarbonate PVP free membranes with 8 µm pores (Neuro Probe). The inserts were prepared before assay by incubating for 2 hours at room temperature with 3 µg/ml fibronectin (Sigma, St Louis, MO). Growth factors and chemokines (IL-8, CXCL12, CCL2, CCL20, PDGF-AA, IGF-1, and HGF: R&D System, Minneapolis, MN) were respectively diluted to 50 ng/ml in Dulbecco's modified Eagle's medium/F12 supplemented with 0.1% BSA (Sigma) which was also used as nonspecific chemoattractant, and 30 µl of the final dilution was placed in the lower chamber; 1 × 104 cells were allowed to attach to the top wells of the insert. The chambers were incubated in a humidified incubator at 37 °C in an atmosphere of 5% CO2 for 17 hours. After incubation, inserts were removed carefully from the chamber and non-migrated cells on the upper side of inserts were removed with a rubber wiper. The migrated cells on the lower side of the inserts were stained with Diff-Quick staining kit (Sysmex, Kobe, Japan). The migrated cells were counted from three random fields per well under a light microscope at ×100 magnification.
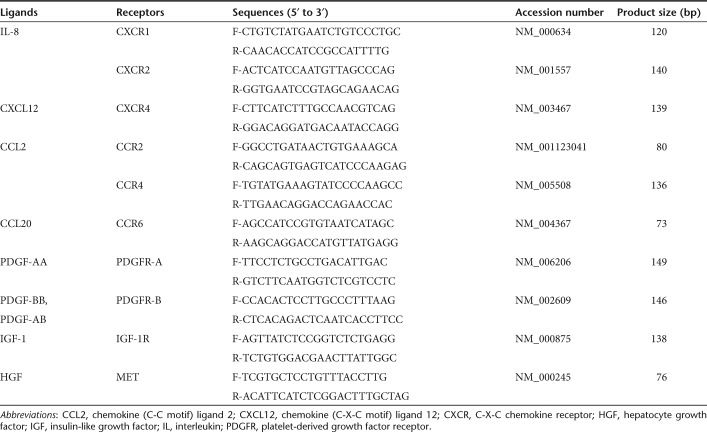
Reverse transcription and real-time PCR. The MSCs were incubated for 4 hours in serum-free Dulbecco's modified Eagle's medium/F12 supplemented with 0.1% BSA to examine the expression levels of the receptors under serum-free culture conditions. The MSCs were also incubated for 4 hours in Dulbecco's modified Eagle's medium/F12 containing 1 ng/ml TNF-α or 0.5 ng/ml IL-1β in order to simulate an in vitro inflammatory condition.23 The total RNA was isolated using an RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instruction and quantified using the Quant-iT RNA assay kit and Qubit Fluorometer system (Invitrogen, Carlsbad, CA). Using Sprint RT Complete-Oligo(dT)18 (Clontech, Mountain View, CA) for reverse transcription-PCR, 500 ng of the total RNA was reverse transcribed. Real-time PCR analysis were performed on a LightCycler 480 system (Roche Diagnostics, Mannheim, Germany) in standard 15 µl reactions. The threshold cycle (Ct) value of each gene was normalized to that of GAPDH. The values thus obtained were again normalized against the control (CXCR2, serum free). The control PCR was performed with RNA extracts that had not undergone reverse transcription (Supplementary Figure S5). The PCR primers are listed in Table 1.
Table 1. Primers used for real-time PCR.
Fluorescent microscopy for labeled MSCs. Fluorescent silica nanoparticles for NEO-LIVET Magnoxide 675 cell labeling kit (Biterials, Seoul, Korea) were used to label the human MSCs. Biocompatible magnetic nanoparticles contained Cy5.5 within a silica shell (50 nm size, biocompatible silica-coated magnetic nanoparticles (MNP@SiO2)). The MSCs were treated with a solution containing 0.4 mg/ml of nanoparticles for 36 hours, as recommended by the manufacturer. Labeling was stopped by washing the cells three times with phosphate-buffered saline (PBS). The cells were examined under a fluorescent microscope (Leica, Wetzlar, Germany) to detect the intracellular localization of MNPs@SiO2(Cy5.5).
Synthesis of HCF. Since normal fibrin releases cytokines only for a short term, heparin was conjugated to fibrin to prolong the release period of the specific cytokine. Heparin (molecular weight 4,000–6,000; Sigma) was covalently bonded to plasminogen-free fibrinogen (Sigma) as previously described.38 HCF was formed by mixing HCF (40 mg/ml) and normal fibrinogen (60 mg/ml) with factor XIII, aprotinin (100 KIU/ml), calcium chloride (6 mg/ml), and thrombin (500 IU/mg).
In vitro PDGF-AA release kinetics. The kinetics of PDGF-AA release from PDGF-AA-loaded HCF was determined with an enzyme-linked immunosorbent assay. Polyester cylindrical tubes (4.5 mm in inner diameter and 3 mm in height) were used for fibrin gel formation. PDGF-AA (10 µg)-loaded HCF or fibrin were injected into the void space (50 µl) in the cylindrical tubes38 through a device designed for simultaneous injection of HCF and thrombin solutions. Each HCF or fibrin gel containing PDGF-AA (n = 3 per group) was immerged in a 2 ml microcentrifuge tube containing 1.5 ml PBS, and the tubes were incubated at 37 °C with continuous agitation. At various time points, the supernatant was collected, and the microcentrifuge tubes were replenished with fresh buffer. The amounts of PDGF-AA in the supernatants were determined with an Quantikine human/mouse PDGF-AA immunoassay kit (R&D System).
In vivo injection of labeled hMSCs into marrow cavity of nude rats and the application of PDGF-AA-loaded HCF. The animal experiments conducted as a part of this study were approved by the Animal Research and Care Committee at our institution. Twelve-week-old male nude rats (Athymic Nude Hsd: RH-Foxn1_rnu; Harlen Laboratory, Indianapolis, IN) were anesthetized with zoletil and xylazine. A 1.5 mm outer diameter trephine drill was employed to create osteochondral defects (2.0 × 2.0 mm) in the trochlear groove of the femur. A hole was created inside the defect using a 26-gauge needle, and deepened until blood gushed out; 1.0 × 106 NEO-LIVET-675 nanoparticle-labeled human MSCs suspended in 10 µl PBS was mixed with BD Matrigel (BD Bioscience, Mississauga, Ontario, Canada) of the same volume to prevent the spreading of the injected cells by blood flow into entire body of the animal, thus allowing prolonged exposure of the injected MSCs to the released migration factors. The mixture was then injected slowly into the marrow cavity through the hole using a Hamilton syringe with a 30 gauge needle. Two in vivo experiments were designed to examine the function of PDGF-AA-loaded HCF. In the first experiment, the osteochondral defects were managed using one of the following methods: filling with 3 µl of HCF only (group 1, n = 3) and filling with 3 µl of PDGF-AA (R&D system)-loaded HCF (group 2, n = 3). An equal volume of thrombin solution was injected simultaneously with HCF in all groups. A PDGF-AA dose of 17 ng/µl HCF was chosen based on the results of in vitro chemotaxis analysis. Two control groups without the injection of MSCs were also employed: filling with 3 µl of HCF only (control 1), and filling with 3 µl of PDGF-AA-loaded HCF after the injection of nanoparticles into the marrow cavity (control 2). In the second experiment, the defects were also managed using one of the following methods: filling with 3 µl of HCF-only without labeled MSCs (group 1, n = 3); filling with 3 µl of PDGF-HD-loaded HCF without labeled MSCs (group 2, n = 3); filling with 3 µl of PDGF-HD and TGF-β1 (R&D system)-loaded HCF without labeled MSCs (group 3, n = 3); filling with 3 µl of PDGF-LD and TGF-β1-loaded HCF after injection of labeled MSCs (group 4, n = 3); and filling with 3 µl of PDGF-HD and TGF-β1-loaded HCF after injection of labeled MSCs (group 5, n = 3). In this study, PDGF-AA dose of PDGF-HD was 17 ng/µl HCF. PDGF-LD contained PDGF-AA of 8.5 ng/µl HCF. TGF-β1 dose of 8 ng/µl HCF was also chosen for groups 3–5.
Detection of in vivo fluorescence. After implantation of the PDGF-HCF or PDGF/TGF-β1-HCF complex, the animals were anesthetized with zoletil (40 mg/kg) and xylazine (10 mg/kg) for fluorescence imaging. The rats were scanned using the Maestro in vivo imaging system (Cri). Optical imaging was performed using a green filter set (excitation: 640–680 nm, emission: 700 nm approximately, long-pass filter). A camera was used to acquire captured images at constant exposure times (1 second). Each Cy5.5 fluorescence spectrum in the rat carrying the NEO-LIVE-675 nanoparticles was acquired by subtracting the autofluorescence spectrum from the mixed spectrum for the rat with the NEO-LIVE-675 nanoparticles.
Gross and histological analysis. The distal femurs were dissected and scored according to ICRS macroscopic score.47 After fixation in 10% neutral formalin, the tissue was decalcified, embedded in paraffin, and sectioned. Sections (4 µm thick) were stained with hematoxylin/eosin and Safranin-O/fast green for microscopic evaluation.
Confocal laser scanning microscopy. Three-dimensional image reconstructions of the human MSCs labeled with NEO-LIVE-675 nanoparticles were obtained using a Zeiss LSM 510 CLS microscope (Zeiss, Maple Grove, MN) equipped with a computer-controlled scan stage. For cell imaging, an argon laser was used for Cy5.5 excitation at 633 nm. For each cell, more than 10 optical planes were scanned by changing the focus to identify the nanoparticles at a variety of sites within the cell. For nuclear counterstaining, the sections were incubated for 10 minutes with 10 µg/ml 4′,6-diamidino-2-phenylindole (DAPI) reagent (Sigma) in PBS.
In vivo chemotaxis assay of CCL2 and PDGF-BB. CCL2 and PDGF-BB (R&D system)–fused HCF were also synthesized for another in vivo experiment. Nine nude rats (n = 3, for each group) were used in this study. After injecting the nanoparticle-labeled MSCs, the osteochondral defects of each rat were sealed with HCF only, CCL2-fused HCF, and PDGF-BB fused HCF, respectively. The CCL2 and PDGF-BB doses were 17 ng/µl, as reported in the preceding animal experiments. At day 7, their distal femurs were dissected and scanned using the Maestro in vivo imaging system. Their total fluorescence intensities at the osteochondral defect region were compared with one another.
Statistics. Descriptive statistics were used to determine the group means and standard deviations for numerical data, and the analysis was performed using analysis of variance (ANOVA) followed by Bonferroni's correction for multiple comparisons. A P value <0.05 was considered significant.
SUPPLEMENTARY MATERIAL Figure S1. Profiles of PDGF-AA released from HCF and fibrin. Figure S2. In vivo tracking of fluorescent nanoparticle-only controls. Figure S3. Live imaging of fluorescent nanoparticle-labeled human BMSCs depending on PDGF-AA concentration in an osteochondral defect of athymic rats. Figure S4. Additional CLSM images of femur paraffin sections from HCF-PDGF-LD and HCF-PDGF-HD rats. Figure S5. The control PCR of RNA extracts that had not undergone reverse transcription.
Acknowledgments
This study was supported by a grant from the National Research Foundation of Korea (2011-0029952). The authors declared no conflict of interest.
Supplementary Material
Profiles of PDGF-AA released from HCF and fibrin.
In vivo tracking of fluorescent nanoparticle-only controls.
Live imaging of fluorescent nanoparticle-labeled human BMSCs depending on PDGF-AA concentration in an osteochondral defect of athymic rats.
Additional CLSM images of femur paraffin sections from HCF-PDGF-LD and HCF-PDGF-HD rats.
The control PCR of RNA extracts that had not undergone reverse transcription.
References
- Alhadlaq A., and, Mao JJ. Tissue-engineered neogenesis of human-shaped mandibular condyle from rat mesenchymal stem cells. J Dent Res. 2003;82:951–956. doi: 10.1177/154405910308201203. [DOI] [PubMed] [Google Scholar]
- Rahfoth B, Weisser J, Sternkopf F, Aigner T, von der Mark K., and, Bräuer R. Transplantation of allograft chondrocytes embedded in agarose gel into cartilage defects of rabbits. Osteoarthr Cartil. 1998;6:50–65. doi: 10.1053/joca.1997.0092. [DOI] [PubMed] [Google Scholar]
- Steinert A, Weber M, Dimmler A, Julius C, Schütze N, Nöth U.et al. (2003Chondrogenic differentiation of mesenchymal progenitor cells encapsulated in ultrahigh-viscosity alginate J Orthop Res 211090–1097. [DOI] [PubMed] [Google Scholar]
- Li WJ, Tuli R, Okafor C, Derfoul A, Danielson KG, Hall DJ.et al. (2005A three-dimensional nanofibrous scaffold for cartilage tissue engineering using human mesenchymal stem cells Biomaterials 26599–609. [DOI] [PubMed] [Google Scholar]
- Benya PD, Padilla SR., and, Nimni ME. Independent regulation of collagen types by chondrocytes during the loss of differentiated function in culture. Cell. 1978;15:1313–1321. doi: 10.1016/0092-8674(78)90056-9. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Peeters-Joris C., and, Vaes G. Production of collagens, collagenase and collagenase inhibitor during the dedifferentiation of articular chondrocytes by serial subcultures. Biochim Biophys Acta. 1990;1051:266–275. doi: 10.1016/0167-4889(90)90132-w. [DOI] [PubMed] [Google Scholar]
- Beiser IH., and, Kanat IO. Subchondral bone drilling: a treatment for cartilage defects. J Foot Surg. 1990;29:595–601. [PubMed] [Google Scholar]
- Shapiro F, Koide S., and, Glimcher MJ. Cell origin and differentiation in the repair of full-thickness defects of articular cartilage. J Bone Joint Surg Am. 1993;75:532–553. doi: 10.2106/00004623-199304000-00009. [DOI] [PubMed] [Google Scholar]
- Wei X, Gao J., and, Messner K. Maturation-dependent repair of untreated osteochondral defects in the rabbit knee joint. J Biomed Mater Res. 1997;34:63–72. doi: 10.1002/(sici)1097-4636(199701)34:1<63::aid-jbm9>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Rüster B, Grace B, Seitz O, Seifried E., and, Henschler R. Induction and detection of human mesenchymal stem cell migration in the 48-well reusable transwell assay. Stem Cells Dev. 2005;14:231–235. doi: 10.1089/scd.2005.14.231. [DOI] [PubMed] [Google Scholar]
- Son BR, Marquez-Curtis LA, Kucia M, Wysoczynski M, Turner AR, Ratajczak J.et al. (2006Migration of bone marrow and cord blood mesenchymal stem cells in vitro is regulated by stromal-derived factor-1-CXCR4 and hepatocyte growth factor-c-met axes and involves matrix metalloproteinases Stem Cells 241254–1264. [DOI] [PubMed] [Google Scholar]
- Ponte AL, Marais E, Gallay N, Langonné A, Delorme B, Hérault O.et al. (2007The in vitro migration capacity of human bone marrow mesenchymal stem cells: comparison of chemokine and growth factor chemotactic activities Stem Cells 251737–1745. [DOI] [PubMed] [Google Scholar]
- Forte G, Minieri M, Cossa P, Antenucci D, Sala M, Gnocchi V.et al. (2006Hepatocyte growth factor effects on mesenchymal stem cells: proliferation, migration, and differentiation Stem Cells 2423–33. [DOI] [PubMed] [Google Scholar]
- Zhang F, Tsai S, Kato K, Yamanouchi D, Wang C, Rafii S.et al. (2009Transforming growth factor-beta promotes recruitment of bone marrow cells and bone marrow-derived mesenchymal stem cells through stimulation of MCP-1 production in vascular smooth muscle cells J Biol Chem 28417564–17574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Yang HN, Woo DG, Jeon SY., and, Park KH. Chondrogenesis of human mesenchymal stem cells in fibrin constructs evaluated in vitro and in nude mouse and rabbit defects models. Biomaterials. 2011;32:1495–1507. doi: 10.1016/j.biomaterials.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Kim HS, Cho HR, Choi SH, Woo JS., and, Moon WK. In vivo imaging of tumor transduced with bimodal lentiviral vector encoding human ferritin and green fluorescent protein on a 1.5T clinical magnetic resonance scanner. Cancer Res. 2010;70:7315–7324. doi: 10.1158/0008-5472.CAN-10-0241. [DOI] [PubMed] [Google Scholar]
- Huang M, Chen Z, Hu S, Jia F, Li Z, Hoyt G.et al. (2009Novel minicircle vector for gene therapy in murine myocardial infarction Circulation 120suppl. 11S230–S237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Peng J, Guo Q, Huang J, Zhang L, Yao J.et al. (2008A cartilage ECM-derived 3-D porous acellular matrix scaffold for in vivo cartilage tissue engineering with PKH26-labeled chondrogenic bone marrow-derived mesenchymal stem cells Biomaterials 292378–2387. [DOI] [PubMed] [Google Scholar]
- Shah BS, Clark PA, Moioli EK, Stroscio MA., and, Mao JJ. Labeling of mesenchymal stem cells by bioconjugated quantum dots. Nano Lett. 2007;7:3071–3079. doi: 10.1021/nl071547f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CW, Hung Y, Hsiao JK, Yao M, Chung TH, Lin YS.et al. (2007Bifunctional magnetic silica nanoparticles for highly efficient human stem cell labeling Nano Lett 7149–154. [DOI] [PubMed] [Google Scholar]
- He G, Zhang H, Wei H, Wang Y, Zhang X, Tang Y.et al. (2007In vivo imaging of bone marrow mesenchymal stem cells transplanted into myocardium using magnetic resonance imaging: a novel method to trace the transplanted cells Int J Cardiol 1144–10. [DOI] [PubMed] [Google Scholar]
- Park KS, Tae J, Choi B, Kim YS, Moon C, Kim SH.et al. (2010Characterization, in vitro cytotoxicity assessment, and in vivo visualization of multimodal, RITC-labeled, silica-coated magnetic nanoparticles for labeling human cord blood-derived mesenchymal stem cells Nanomedicine 6263–276. [DOI] [PubMed] [Google Scholar]
- Ding J, Ghali O, Lencel P, Broux O, Chauveau C, Devedjian JC.et al. (2009TNF-alpha and IL-1beta inhibit RUNX2 and collagen expression but increase alkaline phosphatase activity and mineralization in human mesenchymal stem cells Life Sci 84499–504. [DOI] [PubMed] [Google Scholar]
- Cassiede P, Dennis JE, Ma F., and, Caplan AI. Osteochondrogenic potential of marrow mesenchymal progenitor cells exposed to TGF-beta 1 or PDGF-BB as assayed in vivo and in vitro. J Bone Miner Res. 1996;11:1264–1273. doi: 10.1002/jbmr.5650110911. [DOI] [PubMed] [Google Scholar]
- Lohmann CH, Schwartz Z, Niederauer GG, Carnes DL, Jr, Dean DD., and, Boyan BD. Pretreatment with platelet derived growth factor-BB modulates the ability of costochondral resting zone chondrocytes incorporated into PLA/PGA scaffolds to form new cartilage in vivo. Biomaterials. 2000;21:49–61. doi: 10.1016/s0142-9612(99)00132-5. [DOI] [PubMed] [Google Scholar]
- Shure D, Senior RM, Griffin GL., and, Deuel TF. PDGF AA homodimers are potent chemoattractants for fibroblasts and neutrophils, and for monocytes activated by lymphocytes or cytokines. Biochem Biophys Res Commun. 1992;186:1510–1514. doi: 10.1016/s0006-291x(05)81577-3. [DOI] [PubMed] [Google Scholar]
- Ricono JM, Wagner B, Gorin Y, Arar M, Kazlauskas A, Choudhury GG.et al. (2009PDGF receptor-{beta} modulates metanephric mesenchyme chemotaxis induced by PDGF AA Am J Physiol Renal Physiol 296F406–F417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson A, Siegbahn A, Westermark B, Heldin CH., and, Claesson-Welsh L. PDGF alpha- and beta-receptors activate unique and common signal transduction pathways. EMBO J. 1992;11:543–550. doi: 10.1002/j.1460-2075.1992.tb05085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y., and, Lotz M. Chemotaxis of human articular chondrocytes and mesenchymal stem cells. J Orthop Res. 2008;26:1407–1412. doi: 10.1002/jor.20668. [DOI] [PubMed] [Google Scholar]
- Ringe J, Strassburg S, Neumann K, Endres M, Notter M, Burmester GR.et al. (2007Towards in situ tissue repair: human mesenchymal stem cells express chemokine receptors CXCR1, CXCR2 and CCR2, and migrate upon stimulation with CXCL8 but not CCL2 J Cell Biochem 101135–146. [DOI] [PubMed] [Google Scholar]
- Fox JM, Chamberlain G, Ashton BA., and, Middleton J. Recent advances into the understanding of mesenchymal stem cell trafficking. Br J Haematol. 2007;137:491–502. doi: 10.1111/j.1365-2141.2007.06610.x. [DOI] [PubMed] [Google Scholar]
- Fiedler J, Brill C, Blum WF., and, Brenner RE. IGF-I and IGF-II stimulate directed cell migration of bone-marrow-derived human mesenchymal progenitor cells. Biochem Biophys Res Commun. 2006;345:1177–1183. doi: 10.1016/j.bbrc.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Toh WS, Lee EH, Guo XM, Chan JK, Yeow CH, Choo AB.et al. (2010Cartilage repair using hyaluronan hydrogel-encapsulated human embryonic stem cell-derived chondrogenic cells Biomaterials 316968–6980. [DOI] [PubMed] [Google Scholar]
- Wang W, Li B, Yang J, Xin L, Li Y, Yin H.et al. (2010The restoration of full-thickness cartilage defects with BMSCs and TGF-beta 1 loaded PLGA/fibrin gel constructs Biomaterials 318964–8973. [DOI] [PubMed] [Google Scholar]
- Emans PJ, van Rhijn LW, Welting TJ, Cremers A, Wijnands N, Spaapen F.et al. (2010Autologous engineering of cartilage Proc Natl Acad Sci USA 1073418–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Cooper GM, Gharaibeh B, Meszaros LB, Li G, Usas A.et al. (2009Cartilage repair in a rat model of osteoarthritis through intraarticular transplantation of muscle-derived stem cells expressing bone morphogenetic protein 4 and soluble Flt-1 Arthritis Rheum 601390–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Mitsuru A, Igura K, Takahashi K, Ichinose S, Yamaguchi S.et al. (2006Mesenchymal progenitor cells derived from chorionic villi of human placenta for cartilage tissue engineering Biochem Biophys Res Commun 340944–952. [DOI] [PubMed] [Google Scholar]
- Yang HS, La WG, Bhang SH, Jeon JY, Lee JH., and, Kim BS. Heparin-conjugated fibrin as an injectable system for sustained delivery of bone morphogenetic protein-2. Tissue Eng Part A. 2010;16:1225–1233. doi: 10.1089/ten.TEA.2009.0390. [DOI] [PubMed] [Google Scholar]
- Howard DB, Powers K, Wang Y., and, Harvey BK. Tropism and toxicity of adeno-associated viral vector serotypes 1, 2, 5, 6, 7, 8, and 9 in rat neurons and glia in vitro. Virology. 2008;372:24–34. doi: 10.1016/j.virol.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin E, Obolensky A, Piontek E, Falk H, Pikarsky E, Pe'er J.et al. (2003Gene delivery by viral vectors in primary cultures of lacrimal gland tissue Invest Ophthalmol Vis Sci 441529–1533. [DOI] [PubMed] [Google Scholar]
- Stone D, David A, Bolognani F, Lowenstein PR., and, Castro MG. Viral vectors for gene delivery and gene therapy within the endocrine system. J Endocrinol. 2000;164:103–118. doi: 10.1677/joe.0.1640103. [DOI] [PubMed] [Google Scholar]
- Greenbaum L, Rothmann C, Lavie R., and, Malik Z. Green fluorescent protein photobleaching: a model for protein damage by endogenous and exogenous singlet oxygen. Biol Chem. 2000;381:1251–1258. doi: 10.1515/BC.2000.153. [DOI] [PubMed] [Google Scholar]
- Li SC, Tachiki LM, Luo J, Dethlefs BA, Chen Z., and, Loudon WG. A biological global positioning system: considerations for tracking stem cell behaviors in the whole body. Stem Cell Rev. 2010;6:317–333. doi: 10.1007/s12015-010-9130-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingtgen SD, Kasmieh R, van de Water J, Weissleder R., and, Shah K. A novel molecule integrating therapeutic and diagnostic activities reveals multiple aspects of stem cell-based therapy. Stem Cells. 2010;28:832–841. doi: 10.1002/stem.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Yoon TJ, Yu KN, Kim BG, Park SJ, Kim HW.et al. (2006Toxicity and tissue distribution of magnetic nanoparticles in mice Toxicol Sci 89338–347. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Kim HJ., and, Im GI. PTHrP promotes chondrogenesis and suppresses hypertrophy from both bone marrow-derived and adipose tissue-derived MSCs. Biochem Biophys Res Commun. 2008;373:104–108. doi: 10.1016/j.bbrc.2008.05.183. [DOI] [PubMed] [Google Scholar]
- van den Borne MP, Raijmakers NJ, Vanlauwe J, Victor J, de Jong SN, Bellemans J.et al. (2007International Cartilage Repair Society (ICRS) and Oswestry macroscopic cartilage evaluation scores validated for use in Autologous Chondrocyte Implantation (ACI) and microfracture Osteoarthr Cartil 151397–1402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Profiles of PDGF-AA released from HCF and fibrin.
In vivo tracking of fluorescent nanoparticle-only controls.
Live imaging of fluorescent nanoparticle-labeled human BMSCs depending on PDGF-AA concentration in an osteochondral defect of athymic rats.
Additional CLSM images of femur paraffin sections from HCF-PDGF-LD and HCF-PDGF-HD rats.
The control PCR of RNA extracts that had not undergone reverse transcription.