Abstract
We developed an optical imaging method based on a feedback principle in which the specific scan pattern is adapted according to the shape of the sample. The feedback approach produces nanometer resolved 3D images of very small and moving features in live cells in seconds. We show images of microvilli in live cultured opossum kidney cells expressing NaPi co transporter proteins with different GFP constructs and images of cell protrusions in a collagen matrix with a resolution of about 20 nm. We found that in the microvilli the NaPi proteins can be found clustered. Along cell protrusions in 3D we identified cellular adhesions to the extracellular matrix. Our approach to super resolution and to 3D nanoimaging is different than other proposed methods that break the diffraction limit using non linear effects or are based on single molecule localization.
Keywords: optical microscopy, super resolution, particle tracking, microvilli
Introduction
We describe an optical method capable of producing nanometer resolved 3D images of fluorescent structures in cells. The 3D images are obtained in seconds. The method works on live cells. Several optical microscopy techniques have been developed to “break” the diffraction limit and to produce nanometer resolved images. These techniques can be broadly classified as “physical” techniques in which ingenious approaches are used to break the diffraction limit. For example the STED (stimulated emission depletion) technique uses stimulated emission to reduce the effective size of the PSF (point spread function)1. Other methods use the determination of the center of mass of the fluorescence emission due to single molecules to obtain images with nanometer resolution of cellular features. The PALM and the STORM techniques and their variants use this approach2, 3.
Current methods in laser scanning confocal microscopy are based on moving a laser spot in a predetermined pattern, for example in a raster scan path, to obtain the intensity in each point of a plane of focus. A regular pattern is also used in the STED technique; although in this case the PSF could be in the 20 50 nanometer range. These scanning techniques are inefficient when the features to be imaged are at the nanoscale and sparse since the scanning path crosses the object to be imaged in few points. These objects can be cell protrusions, typical of neuronal spines, long filopodia forming during cell migration or other common cellular structures like microvilli. The efficiency of a predetermined scanning pattern, defined as the ratio of the time the laser beam is on a feature with respect to the total time of scanning, further decreases in 3 dimensions. Crucial biochemical reactions are occurring on or along these nanostructures which are continually remodeled changing shape and position. Current imaging methods including STED or PALM are inadequate to detect the dynamic of chemical reactions in these tiny 3 dimensional structures which occur in the millisecond to second time scale.
We developed an optical imaging method which does not scan the sample with a predetermined pattern. Instead, our method is based on a feedback principle in which the path followed by the laser spot is decided during the scan according to the shape of objects present in the sample. The feedback algorithm is designed to surf a laser spot at a constant distance from a fluorescent surface. Since we know the position of the laser spot and the distance from the surface, we can reconstruct the shape of the object. The uncertainty in the determination of the shape of the object depends on the error in the determination of the position of the laser spot, which is generally in the nanometer range. This feedback imaging approach produces high quality 3D images in seconds and does not require sample fixation.
When an illumination spot approaches a fluorescent surface, the total fluorescence emitted depends on the distance of the spot from the surface but also depends on the density of fluorophores on the surface. Therefore, the fluorescence intensity alone cannot be used to determine the distance of the spot from the fluorescent surface. We then rapidly oscillate the position of the spot close and perpendicularly to the surface. Due to the non linear quasi Gaussian shape of the illumination spot (the point spread function, PSF), the modulation of the fluorescence due to the oscillating spot depends on the distance from the surface. The modulation of the fluorescence is the ratio between the alternating part due to the rapid oscillation and the average part due to the local fluorescence of the surface. This method, modulation tracking (MT), does not use the diffraction of the light and therefore is not breaking the diffraction limit. The sensitivity of MT to the distance from the fluorescent surface depends on the precision of the measurement of the modulation function which is a function of the total number of photons collected at each point of the scan and of the slope of the modulation tracking function. The slope depends on the spatial derivative of the PSF.
In one application of the MT technique we show 3D images of moving microvilli on the surface of opossum kidney (OK) cells (OKP clone)4. The study of microvilli structure and dynamics in vivo is of great importance in biology and physiology since the microvillus is a common motif found in kidneys, intestine and lungs. The microvilli are several micrometer long structures with a diameter of about 100nm. Each cell has many microvilli that appear at the apical membrane. The co transporter proteins NaPi 2a Cerulean and NaPi 2c YFP concentrate at the microvilli giving rise to diffuse fluorescence as seen in confocal images. None of the current nanoimaging optical methods seems capable of measuring the clustering dynamics of proteins on the surface of rapidly moving (on the second time scale) microvilli. Instead the MT method gave high resolution images of the moving microvilli and show transient differential clustering of the NaPi 2a and NaPi 2c proteins using ratio imaging at the microvilli surface. We also measured the diffusion of proteins molecules on the surface of microvilli using fluctuation correlation spectroscopy performed during imaging.
In another illustration of the MT technique we show 3D images of cell protrusions in which cells form adhesion with the extracellular matrix. In this case the protrusions could be very long (20 50 um) and the diameter is variable (0.2um to 1 um). For the images of protrusions we also show simultaneous data acquisition in two colors.
In our implementation of the MT imaging methodology, we first obtain an overall image of the region of interest; which is done using the common raster scan operation of the laser scanning microscope. We then “dive in” the sample to scan a specific structure in 3D. For example, in the case of the imaging of a microvillus, we first obtain a confocal image of the apical membrane and then we start the local 3D imaging at one microvillus.
The Modulation Tracking technique
There are two aspects of the modulation tracking technique that allow us 1) to track the average position of the structure we are imaging and 2) to determine the 3D shape of the object. The feedback orbital technique that allows tracking of rapidly moving fluorescent particles in 3D has been previously described5, 6. For a point particle, we showed that the center of mass can be measured with an error that depends on the waist of the PSF and the total intensity that can be collected along an orbit5, 6. This error is on the order of nanometers for reasonably bright structures. In the case of a microvillus, we have many fluorescent molecules on the membrane and the intensity is about 105 to 106 counts/s which results in nanometer precision determination of the position of the center of mass of the microvilli. In the MT technique the orbital motion is used to track the position of the center of mass of a fluorescence distribution, namely the microvillus center at a given height.
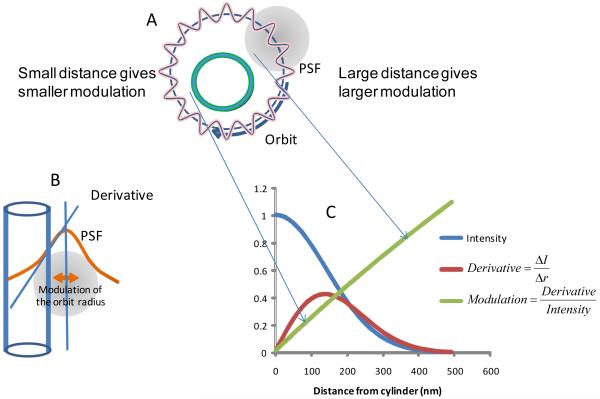
To obtain the shape of the object at one particular orbital plane, we rapidly oscillate the radius of the orbit and then we determine the distance of the object surface from the orbit using the modulation function. Figure 1 shows the principle of the measurement and the physical origin of the modulation function. The modulation changes approximately linearly up to about 500 nm from the surface. At larger distances the intensity becomes too small for the modulation to be useful. This range depends on the size of the PSF. The modulation can be measured with 1% error at the light levels of our experiments, which results in about 10 nm error for the determination of the distance of the fluorescent surface.
Figure 1.
Schematic of the modulation tracking technique. (A) The scanner is programmed to move the laser beam around the microvillus in a circular orbit with radius larger than the radius of the microvillus which is modeled as a cylinder in the figure. The radius of the orbit is modulated at high frequency as shown by the laser path A. (B) When the illumination spot approaches the microvillus surface, due to the modulation of the orbit radius, the fluorescence intensity increases as schematically shown in B. The small oscillation of the radius effectively computes the spatial derivative. (C) The blue curve represents the intensity profile of the spot (the PSF). The red curve represents the spatial derivative of the intensity profile. The green curve is the ratio of the red curve to the blue curve. This ratio is the modulation function which depends on the distance from the surface. The modulation function increases quasi linearly as a function of the distance from the surface. By providing a feedback to the average orbit radius we can maintain the modulation constant, for example at a value of 0.5. The modulation function gives the distance of the surface according to the green curve for every value of the modulation.
In figure 2A the modulation value is converted in distance from the orbit according to the modulation function that was calculated using simulated uniform cylindrical distribution of fluorescence and further verified using empty capsule fluorescent beads (see Methods)
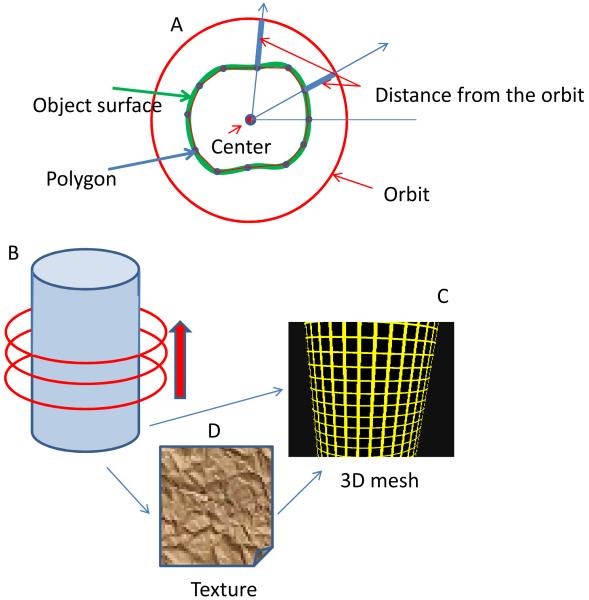
Figure 2.
Schematic of the MT method to construct the 3D shape of the object. (A) As the laser beam goes around the structure, the local modulation function gives the distance of the surface from the average orbit. This distance is calculated at several angles giving a polygon which approximate the shape of the surface at the orbital plane. This polygon is then interpolated to 128 points to produce a smooth line. (B) The orbital plane is then moved along the cylinder axis. For each plane we produce a polygon. (C) The stack of polygons provides a mesh which represents the 3D structure of the object. (D) The 3D mesh is covered by a texture given by specific quantities such as the fluorescence intensity or the intensity ratio of two channels.
This procedure produces a polygonal shape, mapping the fluorescent surface at a given orbital plane. The number of points of the polygon depends on the number of oscillations of the radius during one orbit. We use a number of oscillations between 8 and 32, depending on the size of the object. The polygon is interpolated to 128 points to produce a smooth line. We then move the orbit plane in the direction of the axis of the structure we are examining, which is the vertical axis in the case of microvilli (figure 2B). A new polygon is obtained and the process continues until we reach the end of the microvillus, indicated by a sudden drop of the fluorescence intensity below a given threshold. The stack of polygons obtained form a 3 dimensional mesh that directly corresponds to the shape of the object (Figure 2C).
Once the 3D mesh has been determined with nanometer resolution we add a texture to the mesh according to selected functions such as the fluorescence intensity or fluorescence ratio (figure 2D). We note that although the shape of the microvillus can be determined with nanometer precision, the resolution of molecules (or cluster of molecules) on the microvillus surface is limited by diffraction. In principle we could use the super resolution techniques mentioned in the introduction to increase the resolution of the distribution of the fluorescence at the microvillus surface or use deconvolution techniques, in order to determine the distribution of different molecular species on the surface.
The resolution of the microvillus shape along the z axis (the long axis of the microvillus) depends in part on the size of the illumination spot. We performed simulations of cylinders with a step change in diameter from 100 nm to 180 nm radius. In every case we were able to recover the cylinder diameter within 10 nm, but the transition between the two radii was occurring during a length approximately equal to the axial waist of the PSF. If the microvillus is horizontal, the resolution of the step depends on the radial waist of the PSF, which is generally a factor of 3 smaller than in the axial direction.
The MT method does not require fixed samples. One segment of the microvillus can be measured in about 16 ms in our system. Even fast moving microvilli (in the second range) can be imaged. The 3D image is obtained directly from the mesh given by moving the position of the orbit along the microvillus axis (up and down the microvillus). The scanning of a cylindrical surface is equivalent to the raster scan of a flat surface without retracing. Therefore we apply the principles of raster scan image correlation spectroscopy (RICS) to obtain the diffusion and concentration of fluorescent molecules on the microvillus surface making MT a truly dynamic nanoimaging technique. The MT technique has the capability to identify specific molecules on the microvilli surface, their clustering and dynamics in a millisecond time scale.
There are other optical techniques capable of measuring the height of structures based on interferometric measurements and optical profilometry7, 8. These techniques have very high resolution in the direction parallel to the light propagation while the resolution in the x y plane is limited by diffraction. Interferometric techniques don’t allow the reconstruction of 3D objects like microvilli or cell protrusion at the nanometer scale. Also, interferometry is based on reflectivity and suffers from lack of selectivity attained by fluorescence.
Results
Figure 3 shows images obtained using the Zeiss LSM510 microscope taken in the x y plane and x z plane of OK cells transfected with NaPi 2a Cerulean and the 3D reconstruction using the MT technique.
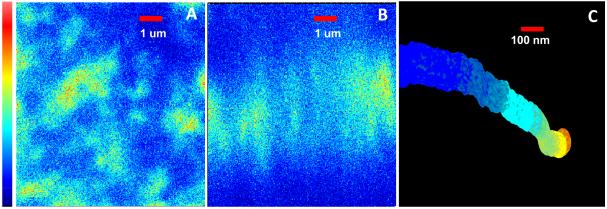
Figure 3.
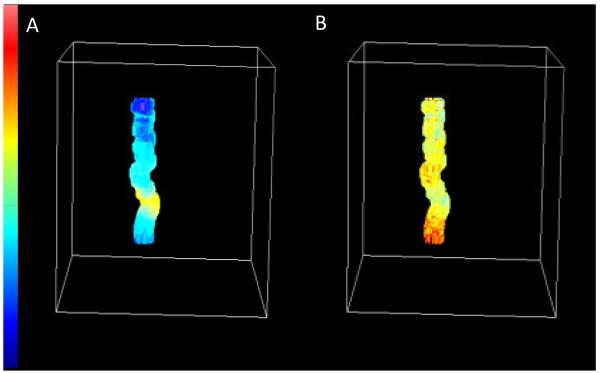
Imaging microvilli. (A) x y raster scan image of OK cell expressing NaPi 2a Cerulean. The microvilli appear as the roundish bright areas in this projection. (B) Detail of the same cell imaged in the x z plane showing adjacent microvilli. The color scale at left for panel A and B corresponds to 0 to 255 levels of the analog output of the LSM 510 microscope. (C) 3D reconstruction of a microvillus using the MT technique. The region reconstructed is about 2 um long. The image is painted according to the fluorescence intensity. The color scale at right corresponds to 0 256KHz of the photon counting acquisition electronics of the M5 microscope. The brightest part is toward the base of the microvillus. Note the non uniform distribution of the fluorescence along the microvillus.
The microvilli can be barely identified in the confocal images since they are too small to be resolved and also they are defocused because they are moving (Figure 3A). The x z image shows the microvilli extending for about 5 um (Figure 3B). For the 3D MT imaging the scanner was centered at one microvillus, in a region with few sparse microvilli. Then the tracking routine was started. The z position of the orbit was moved for a height of about 2 um in 10 s starting at the base of the villus. Figure 3C shows the reconstructed 3D image of one microvillus. The surface of the recovered image is painted according to the fluorescence intensity. The surface was reconstructed at a resolution of 2 nm for the center of the microvillus and about 20 nm for the radius. This nanometer 3D reconstruction of the microvillus shows a convoluted tortuous structure. The average diameter of the microvillus is about 100nm. The protein concentration, as judged from the fluorescence intensity, changes along the microvillus.
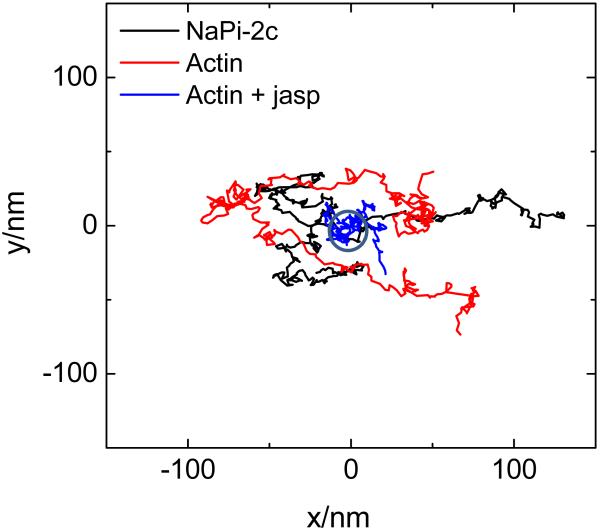
Not all microvilli have the same tortuous structure. Some of the microvilli appear straighter. When we scanned the same microvillus back and forth, we found that the microvillus moved during the scan. The time scale of movement also changed among the microvilli. Some of the microvilli show regions of higher fluorescence intensity along the surface, indicating that clustering of the protein was occurring. The typical dynamics of the center of mass of microvilli is shown in figure 4. In this experiment the z coordinate was kept constant and the feedback mechanism kept the orbit centered on the microvillus at a given height. Figure 4 also shows that we can track different proteins present in the microvilli (Napi_2C and actin). The microvilli move about 100 nm in the x direction and 50 nm in the y direction over the course of 30 s, demonstrating the relatively fast motion of the microvillus and the recovery of the center of mass of the microvillus position with nm resolution. Treatment with Jasplakinolide, which stabilizes actin polymerization, strongly reduced the motion of the microvilli as seen by the actin signal. The circle in figure 4 corresponds to a standard deviation of 5 nm for the determination of the average position of the actin filled microvillus. Measurement were repeated for 12 microvilli, both treated and untreated with Jasplakinolide, giving similar results, The treated microvilli stop moving in all cases.
Figure 4.
Dynamics of the center of mass of microvilli. The z position of the scanner was kept constant while the feedback mechanism followed the motion of the center of mass of the microvilli. Each measurement is done different microvilli according to the legend color. Two different proteins were imaged, Cerulean NaPi 2C which resides on the microvilli surface and Actin EGFP which presumably is in the cytoplasm. Stabilization of actin polymerization was obtained by treatment with Jasplakinolide.
The MT method allows imaging of the protein distribution on the microvillar membrane. We imaged microvilli in OK cells co transfected with NaPi 2a Cerulean and NaPi 2c YFP. For the example reported in Fig.5, the length of the orbit is 1.256 um. We can clearly distinguish that the fluorescence distribution of NaPi 2a Cerulean is not uniform along this microvillus (figure 5A). Along the microvillus, the fluorescence is distributed along bands that seem to concentrate in only one side of the microvillus at the microvillus base.
Figure 5.
Display of the intensity and normalized ratio along the surface of the microvillus. The size of the box is 1 um. The smaller structures in the surface are about 200 nm in size. (A) Painting according to intensity in channel 1 (colors scale 0 256KHz). (B) Painting according to normalized ratio (color scale 1 to +1). The bottom of the image corresponds to the microvillus base.
The size of the protein patches shown in figure 5 is determined by the PSF. Figure 5B shows the image of the same microvillus painted according to the normalized ratio function of the intensities in the two channels or GP. The GP image is relatively uniform along the circumference of the microvillus but has bands along the microvillus length showing that the two proteins are not equally distributed along the microvillus. The width of the structures along the microvillus is on the order of 250 nm, consistent with the size of the illumination spot. These structures are dynamic, as they can disappear or appear at different location when we image a microvillus at subsequent times (Δt=20s, data not shown in this paper). This example, as well as the tracking of the center of mass in figure 4 shows the dynamic capability of the MT method for displaying nanometer structural changes occurring in sub second time scale.
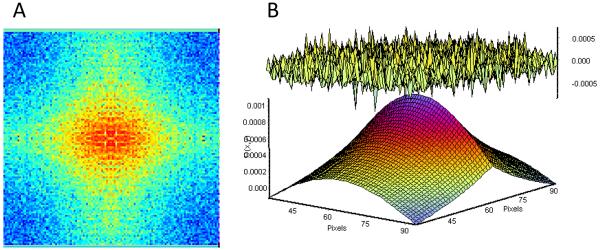
We are performing a circular scanning of the cylinder at relatively high speed. Every pixel is measured for about 62.5 μs and pixels along the orbit are separated by 0.01 μm. We performed a dynamic RICS (Raster scan Image Correlation Spectroscopy) analysis to observe the motion of molecules on the microvilli membrane surface. In Figure 6 we show such analysis and the calculation of the diffusion coefficient according to the equations and methods described by Digman et al9. We found that Cerulean NaPi2c has a local diffusion coefficient of about 0.02 μm2/s using a model for 2D diffusion.
Figure 6.
RICS analysis for Cerulean NaPi2c. (A) RICS autocorrelation function. (B) Fit (lower surface) and residues (upper surface) according to a model for 2D diffusion. The recovered diffusion coefficient for Cerulean NaPi2c is 0.02Qm2/s.
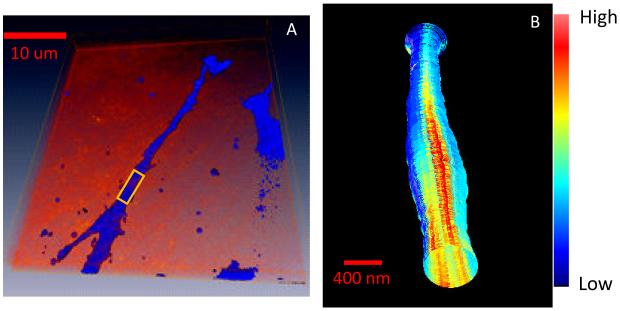
Next we show that the MT technique can measure cellular features extending several tenths of microns and with variable shape. Figure 7A shows the conventional z stack 3D image of a protrusion of a MB231 cell expressing Paxillin EGFP growing in a 3D collagen matrix. A region of interest is selected as shown by the rectangle in figure 7A. Then we “dive in” to image the 3D protrusion at the nanoscale (Figure 7B). The protrusion has a larger diameter than a microvillus and also the diameter changes along the protrusion. The fluorescence intensity is larger at some specific regions along the cell body, presumably where the cell contacts the collagen.
Figure 7.
(A) 3D raster scan image of a protrusion of MB231 cell growing in a 3D collagen matrix. (B) MT image of a small portion of the protrusion indicate in (A). The diameter of the protrusion changes along the filopodium. The fluorescence is not uniform on the cell surface but clusters at specific direction where contacts are made with the collagen matrix.
Discussion/Conclusions
In the MT technique the position of a fluorescent surface is determined by the modulation of the fluorescence signal as a function of the distance from the surface. Since the intensity as a function of the distance from the surface changes non linearly, using the modulation of the light caused by the rapid oscillation of the laser spot we effectively measure the derivative of the fluorescence intensity as a function of the distance from the surface with relatively high precision. We then produce a topographical map of the fluorescent surface using the modulation function. In the case of microvilli imaging, we measure the deviation of the center of mass and the deviations of the radius with nanometer accuracy. This is possible because we transformed the measurement of the shape of the microvilli in the measurement of the topography of its surface. Instead, in the direction along the microvillus surface the topographical resolution cannot be any better than the size of the spot used. However, established methods as these based on deconvolution or super resolution can be used to locate fluorophores on the surface of the membrane with higher resolution. Since the MT technique is based on the measurement of the modulation function (ratio of the alternate to the constant part), bleaching of the fluorescence at the surface has a minimal effect on the determination of the 3D shape.
We have applied the MT technique to quasi cylindrical structures. For this symmetrical shape the MT feedback method is particularly efficient. However, the method could be applied to arbitrarily shaped surfaces since the feedback based on the modulation function moves the spot at a constant distance from the surface. There is a limit in the speed of scanning of the surface, which depends on the galvo scanner used in our instrument for the modulation in the xy plane and of the piezo that we use to move the laser spot along the z axis. In our instrument the maximum scanning frequency we can achieve is about 2 kHz in the x y direction and 200Hz in the z direction, respectively. Another consideration is that the shape of the PSF is different in the radial and in the axial direction of the microscope. Since the shape of the PSF only determines the slope of the PSF and this slope is measured independently, the shape of the PSF is taken into account in the feedback algorithm and therefore the shape of the object is determined accurately in the 3 directions. However, as noted before, the error in the determination of the surface position is larger in the radial direction. Interestingly, the resolution of steps in shape changes is better when the cylindrical object is laying horizontally since the PSF is narrow in the radial direction.
The MT imaging approach can be applied to live cells. The motion of the microvilli or any other structure is tracked and the assignment of the fluorescence intensity to a voxel in space and time is accurate with respect to the object surface. Scanning along the surface provides direct visualization of how proteins are distributed on a 3D membrane structure like the microvillus or cell protrusions, a feature that can be important for the study of the NaPi transporters regulation and more in general for studying membrane micro domain organization. More importantly, the orbital scanning along the membrane surface allows fluorescence image correlation techniques to be applied. Since the microvillus (or any other cellular feature) is always at the center of the orbit, if we keep the z axis constant, we can measure the fluorescence along the orbit and the fluorescence fluctuations due to the diffusion of molecules. This is equivalent of the RICS scanning experiment. It provides information about the diffusion and the brightness of the fluorescent molecules in the membrane. This is a unique possibility offered by the MT approach.
The MT method can be implemented in microscopes that produce a small illumination spot such as confocal and 2 photon excitation microscopes and where the spot can be rapidly moved in 3D. The implementation of the method only requires the appropriate software to move the scanner along specific trajectories in response to a measured modulation at a given position. The 3D structure of the surface of a microvillus is obtained with much higher resolution than techniques based on the localization of fluorescent molecules. The method works in the presence of many molecules and it does not require a particular hardware or high illumination power. All common fluorescent labels can be used.
Strength of the MT technique is its capability to follow molecular events such as diffusion and interactions directly in 3D at a time and spatial scale that cannot be obtained with current optical techniques. There is growing interest in live cell imaging to display cellular processes at the millisecond to second time scale. The MT imaging technique is exquisitely dynamic and therefore ideal for the study of such processes. Since MT is essentially a tracking technique, it is very efficient for imaging objects that are spatially isolated, while it is difficult to use in a crowded field.
Methods
Cell culture and transfections
Opossum kidney (OK) cell culture was performed as previously reported10, 11. Briefly, cells were grown in Dulbecco’s modified Eagle’s medium (DMEM/F12) supplemented with 10% fetal bovine serum, penicillin, streptomycin and L glutamine in a humidified 5% CO2 95% air atmosphere incubator at 37°C. Transfections and cotransfections were achieved with Lipofectamine 2000 (Invitrogen) and cells at 90% confluency, following the manufacturer’s instructions. OK cells expressing the fluorescent fusion proteins were grown on poly L lysine coated eight well Lab Tek chambered coverglass (Nunc). Measurements were performed 24–48 h after transfection. Type I rat tail collagen was prepared with 10× PBS and H2O at 2.5mg/ml final concentration and adjusted to pH=7.4 using 10N NaOH. MDA MB231 stable cells lines expressing paxillin EGFP at a concentration of 0.25×106 were mixed with the collagen and allowed to polymerize for 1 hour at 25°C without FBS supplemented media. Then they were incubated at 37°C overnight before imaging.
Microscope systems
A Zeiss 510 LSM microscope (Jena, Germany), equipped with a Confocor 3 unit and the META detector was used for the xy and xz images of figure 3A and 3B. The argon ion laser with excitation at 456 nm (2% power) and 514 nm (0.5% power) was used to excite the cerulean and the YFP constructs, respectively. Images where obtained in the 256×256 format with a pixel dwell time of 12.5 us and a zoom of 12. Data were saved in the lsm format and further processed by the SimFCS software (www.lfd.uci.edu, UCI, Irvine). For the xz images the “fast z” acquisition mode of the LSM 510 was used. The modulation tracking system was implemented in the M5 microscope at the LFD previously described5, 6. Briefly, this microscope is build around an Olympus X71 body. The light source is a Chameleon Ultra II laser (Coherent, CA) that produces 2 photon excitation of the cerulean and the YFP constructs. The laser power was set between 20 and 30 mW, measured before entering the microscope. The laser wavelength was set at 820 and 940 nm for cerulean and YFP excitation, respectively. Two GaAS detector H7241P (Hamamatsu, Japan) were used to acquire the fluorescence in the 380 480 nm range and 500 550nm for the two channels, respectively. The galvano scanner (Cambridge Technology, MA) was driven by the ISS 3axis card (ISS, Champaign IL) as well as the piezo z axis (Phisik Instrument, Germany). The circular orbital pattern and the modulation pattern was stored in the memory of the ISS 3axis card and the center offset in the x y position was updated according to the tracking mechanism every 16 ms. The fluorescence intensity at 128 points along the orbit was collected at a sampling rate of 8 kHZ. The modulation of the orbit was set at a period of 2 ms. We determined that the 3dB point of the scanner response in the x y plane was 3 ms. We used an amplitude modulation for the modulation tracking that gives 10% modulation of the orbit radius. The orbit radius was initially set at different values between 200 nm and 400 nm, depending on the diameter of the microvillus. The feedback mechanism changed the radius until a constant modulation of 10% was obtained along the orbit. Under this condition, the laser spot is travelling at a constant distance from the fluorescent surface. Then the plane of the orbit is moved along the surface cylindrical axis. The local direction of the surface axis is obtained by comparing the center of the orbit at two different positions along the axis. If the object starts to bend, then the direction of the subsequent orbit is changed so that the orbit is always perpendicular to the axis of the object and centered on the axis. The PSF of our instrument at any given experimental condition (wavelength of excitation, laser power, etc) was measured prior to each tracking measurement using 20 nm beads fixed on a slide. The size of the PSF only influences the amount of modulation of the MT technique but it does not affect the size of the objects that were imaged. The calibration of the modulation tracking function was performed using fluorescent beads shells of known diameter, shown in Supplemental Figure S1 (Invitrogen Carlsbad CA). Most of the measurements were done using an air objective (Olympus, 0.9NA x60 W). The position of the orbit plane along the object axis was moved with a ramp function that is variable in amplitude and period. The amplitude was set between 1Qm to 3 Qm and the period between 1s and 20 s, depending on the experiments. The initial position of the circular orbit was set by first acquiring an x y image at a given z position and then pointing with the mouse to one feature that we recognized to be a microvillus. Then the tracking of the center of the microvillus and the ramping along the microvillus height are done. In live samples, the center of mass of the microvillus moves during the time needed to acquire the 3D image. Repeating the scan up down of the same microvillus many times provided a movie of the 3D position of the villus (not shown in this paper). The intensity along the cylindrical surface is modulated according to the number of oscillations of the radius used during one orbit (generally 8). This modulation of the intensity is easily removed using a notch filter of the appropriate frequency. At the laser power used, bleaching of the fluorescence was minimal (Supplemental Figure S2).
Supplementary Material
Acknowledgements
Work supported in part by NIH RO1 DK066029 (ML, EG), NIH P41 RRO3155 (EG, PF and LL), P50 GM076516 (EG, PF and LL) and NIH U54 GM064346 Cell Migration Consortium (MD and EG).
References
- 1.Willig KI, Harke B, Medda R, Hell SW. STED microscopy with continuous wave beams. Nat Methods. 2007;4:915–918. doi: 10.1038/nmeth1108. [DOI] [PubMed] [Google Scholar]
- 2.Manley S, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 3.Shroff H, Galbraith CG, Galbraith JA, Betzig E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat Methods. 2008;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole JA, Forte LR, Krause WJ, Thorne PK. Clonal sublines that are morphologically and functionally distinct from parental OK cells. Am J Physiol. 1989;256:F672–679. doi: 10.1152/ajprenal.1989.256.4.F672. [DOI] [PubMed] [Google Scholar]
- 5.Levi V, Ruan Q, Gratton E. 3-D particle tracking in a two-photon microscope: application to the study of molecular dynamics in cells. Biophys J. 2005;88:2919–2928. doi: 10.1529/biophysj.104.044230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levi V, Ruan Q, Kis-Petikova K, Gratton E. Scanning FCS, a novel method for three-dimensional particle tracking. Biochem Soc Trans. 2003;31:997–1000. doi: 10.1042/bst0310997. [DOI] [PubMed] [Google Scholar]
- 7.Lee CH, Mong HY, Lin WC. Noninterferometric wide-field optical profilometry with nanometer depth resolution. Optics Letters. 2002;27:1773–1775. doi: 10.1364/ol.27.001773. [DOI] [PubMed] [Google Scholar]
- 8.Lee CH, Wang JP. Noninterferometric differential confocal microscopy with 2-nm depth resolution. Optics Communications. 1997;135:233–237. [Google Scholar]
- 9.Digman MA, et al. Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys J. 2005;89:1317–1327. doi: 10.1529/biophysj.105.062836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blaine J, et al. PTH-induced internalization of apical membrane NaPi2a: role of actin and myosin VI. Am J Physiol Cell Physiol. 2009;297:C1339–1346. doi: 10.1152/ajpcell.00260.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breusegem SY, et al. Acute and chronic changes in cholesterol modulate Na-Pi cotransport activity in OK cells. Am J Physiol Renal Physiol. 2005;289:F154–165. doi: 10.1152/ajprenal.00331.2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.