Abstract
Origins of replication are activated throughout S-phase such that some origins fire early and others fire late to ensure that each chromosome is completely replicated in a timely fashion. However, in response to DNA damage or replication fork stalling, eukaryotic cells block activation of unfired origins. Human cells derived from patients with ataxia telangiectasia are deficient in this process due to the lack of a functional ataxia-telegiectasia mutated (ATM) kinase and elicit Radio-resistant DNA synthesis (RDS)1–3 following γ-irradiation2. This effect is conserved in budding yeast, as yeast cells lacking the related kinase Mec1 (ATR) also fail to inhibit DNA synthesis in the presence of DNA damage4. This intra-S-phase checkpoint actively regulates DNA synthesis by inhibiting the firing of late replicating origins, and this inhibition requires both Mec1 and the downstream checkpoint kinase Rad53 (Chk2)5,6. However, the Rad53 substrate(s) whose phosphorylation is required to mediate this function remained unknown. Here, we show that the replication initiation protein Sld3 is phosphorylated by Rad53, and that this phosphorylation, along with phosphorylation of the Cdc7 kinase regulatory subunit Dbf4, blocks late origin firing. Upon exposure to DNA damaging agents, cells expressing nonphosphorylatable alleles of SLD3 and DBF4 (SLD3-m25 and dbf4-m25, respectively) proceed through S-phase faster than wild-type cells by inappropriately firing late origins of replication. SLD3-m25 dbf4-m25 cells grow poorly in the presence of the replication inhibitor hydroxyurea (HU) and accumulate multiple Rad52 foci. Moreover, SLD3-m25 dbf4-m25 cells are delayed in recovering from transient blocks to replication and subsequently arrest at the DNA damage checkpoint. These data suggest that the intra-S-phase checkpoint functions to block late origin firing in adverse conditions to prevent genomic instability and maximize cell survival.
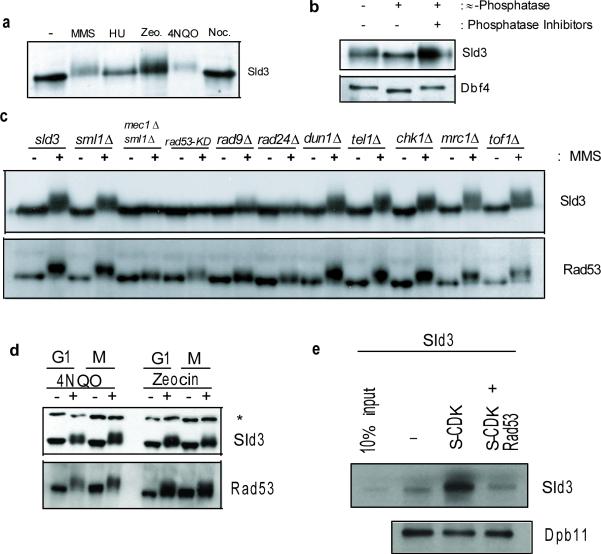
In a whole-genome screen for proteins whose electrophoretic mobility was altered by DNA damage (unpublished results), we found that Sld3, an essential protein required for the initiation of DNA replication7, becomes substantially modified in response to checkpoint activation. Sld3 was hyper-phosphorylated in cells treated with genotoxic drugs, but not in cells arrested at the spindle checkpoint with Nocodazole (Fig. 1a and b). The Sld3 damage-induced phosphorylation depends on Mec1 and Rad53 and is compromised in mutants that reduced Rad53 activation (Fig. 1c). DNA damage-induced Sld3 phosphorylation is not cell cycle regulated, as cells blocked in G1 or mitosis and subsequently treated with 4-nitroquinoline 1-oxide (4NQO) or Zeocin contained phosphorylated Sld3 (Fig. 1d).
Figure 1. Rad53-dependent phosphorylation of Sld3.
a, Immunoblotting of asynchronous cells expressing Sld3-3Flag in the absence (−) or presence of MMS (alkylating agent-0.05%), HU (ribonucleoside reductase inhibitor-200 mM), zeocin (radiation mimetic-200 μg/mL), 4NQO (UV-mimetic-2 μg/mL), or nocodazole (10 μg/mL) for 90 minutes. b, Phosphatase assays of Sld3-3Flag and Dbf4-TAP purified from MMS-treated cells. c, Immunoblot of checkpoint deletion mutants expressing Sld3-9myc. Samples were taken from cells arrested in G1 with α-factor and released in the presence or absence of MMS for 180 minutes. d, Immunoblot of cells expressing Sld3-9myc arrested in G1 with α-factor or metaphase with nocodazole for 120 minutes and then treated with either 4NQO or Zeocin, as in a, for 60 minutes. * Denotes a background band. e, In vitro binding assay. Recombinant Sld3-3Flag mock phosphorylated (−), Clb5-TAP CDK phosphorylated or Clb5-TAP CDK phosphorylated followed by Rad53 phosphorylation. 10% of the untreated Sld3-3Flag is shown as input. Immobilized Dpb11-TAP was used to pull down the differentially phosphorylated Sld3 species.
Origin firing requires both the phosphorylation of Sld3 by the S-phase cyclin-dependent kinase (S-CDK)8,9, and the activity of the Dbf4-dependent kinase (DDK)10,11. The S-CDK phosphorylation of Sld3 allows binding to Dpb11 and promotes origin firing8,9. In vitro binding assays using recombinant full-length Sld3 and immobilized Dpb11, revealed that S-CDK phosphorylation of Sld3 is required to bind Dpb11, as previously seen. However, subsequent Rad53 phosphorylation of S-CDK-phosphorylated Sld3 eliminated binding to Dpb11 (Fig. 1e). This indicates that Sld3 is a Rad53 substrate and suggests that, by antagonizing Sld3 binding to Dpb11, Rad53 can inhibit late origin firing.
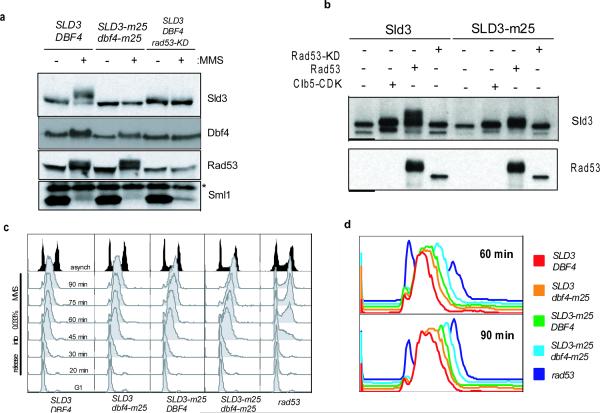
To address this hypothesis directly, we constructed an unphosphorylatable SLD3 allele. Using mass spectrometry, twenty-eight phosphorylation sites were identified (Supplemental Table 1a). Twenty-five potential phosphorylation sites were mutated to generate the SLD3-m25 allele. When SLD3-m25 was introduced into cells, damage-induced Sld3 phosphorylation was not detected (Fig. 2a and Supplemental Fig. 2). In vitro kinase assays revealed that activated Rad53, but not a catalytic mutant (Rad53-K227A), phosphorylated Sld3 strongly, but phosphorylated the Sld3-m25 mutant less efficiently (Fig. 2b). Additional mass spectrometry analysis of Sld3 phosphorylated in vitro by Rad53 identified nine phosphorylation sites, five of which were mutated in the SLD3-m25 allele (Supplemental Table 1a).
Figure 2. SLD3-m25 and dbf4-m25 cells are intra-S-phase checkpoint defective.
a, Immunoblots from asynchronous cells grown in the presence or absence of 0.05% MMS for 90 minutes. rad53-KD (K227A) is a hypomorphic allele that is checkpoint-defective, but able to perform the essential function rescued by Sml1 deletion. b, Immunoblots of an in vitro kinase assay on recombinant full-length Sld3 or Sld3-m25 substrates treated with yeast clb5-TAPCDK, recombinant Rad53 or Rad53-KD (K227A) c, Flow cytometry of wild-type, SLD3-m25, dbf4-m25, SLD3-m25 dbf4-m25, or rad53Δ cells synchronized in G1 with α-factor and released into 0.033% MMS at 30°C. d, The 60 and 90 minute profiles from c, displayed together.
Intra-S-phase checkpoint defective mutants, such as rad53Δ, fail to prevent late origin firing in the presence of DNA damage and consequently proceed through S-phase faster than wild-type cells5,6. Cells expressing SLD3-m25 in the presence of methyl methanesulfonate (MMS) proceeded through S-phase faster than wild-type cells (Fig. 2c and d), although they progressed slower than rad53Δ cells. Like Sld3, Dbf4 functions to promote firing of all origins of replication10,11 and is phosphorylated in response to checkpoint activation in a Rad53-dependent manner12,13 (Fig. 1b and 2a). Dbf4 is a periodically expressed activator of the Cdc7 kinase, known to phosphorylate the replication helicases (MCMs)14–17. We reasoned that Dbf4 may also be inhibited by Rad53 to block late origin firing. Using a similar mass-spectrometry approach, six phosphorylation sites were identified on Dbf4 (Supplemental Table 1b). The six phosphorylation sites were mutated to generate a dbf4-m6 allele, however the Dbf4 damage-induced phosphorylation shift was intact when dbf4-m6 cells were treated with MMS (data not shown). Previous studies have shown that a Dbf4 mutant lacking the non-essential amino-terminus (1–109) is not phosphorylated in damage-treated cells18. For this reason, we mutated all serine and threonine residues in this non-essential domain of Dbf4 (except those matching the CDK consensus sequence), in addition to the six sites identified by mass spectrometry, to generate the dbf4-m25 allele. When expressed in cells, damage-induced Dbf4-m25 phosphorylation was reduced, but not eliminated (Fig. 2a), suggesting that not all damage-induced phosphorylation sites were identified or that Dbf4 is also phosphorylated by another damage-responsive kinase(s).
Cells expressing SLD3-m25, dbf4-m25, or both mutant alleles were viable and had no growth defects at any temperature tested (Supplemental Figs. 3 and 4a). Cells expressing dbf4-m25 alone appear to be intra-S-phase checkpoint competent (Fig. 2c and d). However, cells expressing SLD3-m25 and dbf4-m25 progressed faster through S-phase in the presence of MMS than cells expressing either allele alone (Fig. 2c and d and Supplemental Fig. 5), arguing that Sld3 and Dbf4 are both important for the block to late origin firing. The finding that SLD3-m25 dbf4-m25 cells progressed through S-phase slower than rad53Δ cells can best be explained by the fact the Dbf4-m25 is still partially phosphorylated in vivo (Fig. 2a). Damage-induced Rad53 phosphorylation and Sml1 degradation19 are intact in SLD3-m25 dbf4-m25 cells (Fig. 2a), suggesting that these mutations do not compromise checkpoint activation.
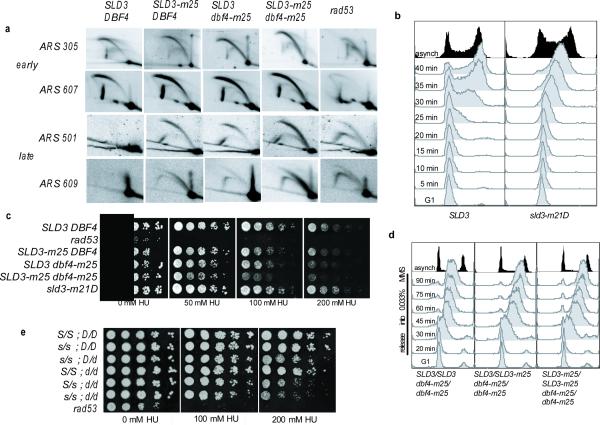
To unambiguously determine whether the fast S-phase observed in SLD3-m25 dbf4-m25 cells, in the presence of MMS, is due to the inappropriate firing of late origins of replication, firing was directly analyzed by two-dimensional gel electrophoresis20. In HU-treated cells, early origins ARS305 and ARS607 fired efficiently and with normal kinetics in SLD3-m25, dbf4-m25 and SLD3-m25 dbf4-m25 mutants compared to wild-type cells (Fig. 3a and Supplemental Fig. 6b). This finding is consistent with early origins being refractory to intra-S-phase checkpoint regulation5,6. We next looked at several late origins; ARS501, ARS609, ARS1007 and ARS1212 (Fig. 3a, Supplemental Fig. 6c and data not shown). In HU-treated cells, late origin firing, was not detected in wild-type cells, but was evident in rad53Δ cells. Inappropriate late origin firing was also observed in SLD3-m25, dbf4-m25 and, most strongly in the SLD3-m25 dbf4-m25 double mutant. In these mutant cells, late origins fired after early origins, confirming the finding that the temporal regulation of origin firing is not subject to intra-S-phase checkpoint regulation (Supplemental Fig. 6b and 6c). These results suggest that the fast S-phase observed in SLD3-m25 dbf4-m25 cells is due to the inappropriate firing of late replicating origins.
Figure 3. Inappropriate firing of late origins in the presence of replication inhibitors.
a, Two-dimensional gel analysis of replication intermediates. Wild-type, SLD3-m25, dbf4-m25, SLD3-m25 dbf4-m25 or rad53Δ cells were synchronized with α-factor and released into 200 mM HU. Cells were collected at 30, 40, 50, 60 and 70 minutes into the HU release and pooled. 20 μg aliquots of DNA were analyzed by two-dimensional electrophoresis as previously described20. early origins; ARS305 and ARS607. late origins; ARS501 and ARS609. b, Flow cytometry of cells expressing SLD3-3Flag or a phosphorylation-mimetic allele sld3-m21D-3Flag. Cells were arrested in G1 with α-factor and released into media without α-factor at 30°C. c, 5-fold serial dilutions of wild-type or mutant strains grown on 0 mM, 50 mM, 100 mM, or 200 mM HU. d, Flow cytometry of diploid MAT a/a cells treated as in 2c, from SLD3/SLD3 dbf4-m25/dbf4-m25, SLD3/SLD3-m25 dbf4-m25/dbf4-m25, and SLD3-m25/SLD3-m25 dbf4-m25/dbf4-m25 cells. e, 5-fold serial dilutions, as in c, of diploid strains to assess dominance. Capital S and D represent wild-type SLD3 and DBF4, whereas lowercase s and d represent SLD3-m25 and dbf4-m25 respectively.
If Rad53 phosphorylation of Sld3 slows S-phase by blocking late origin firing, then constitutive Rad53 phosphorylation of Sld3 would constitutively slow S-phase, even in the absence of DNA damaging agents. To test this prediction, we created a phospho-mimetic allele, where twenty-one residues targeted by Rad53 were mutated to aspartic acids to mimic constitutive Sld3 phosphorylation (sld3-m21D). Cells expressing the sld3-m21D allele revealed a slow S-phase in the absence or presence of DNA-damaging agents (Fig. 3b and Supplemental Fig. 7a). The slow S-phase is not a result of checkpoint activation, as sld3-m21D cells do not elicit Rad53 phosphorylation in the absence of damage (Supplemental Fig. 7b). In addition, sld3-m21D cells grow with wild-type rates, are not HU-sensitive, and fire early origins of replication with efficiencies that are indistinguishable from those observed in wild-type cells (Fig. 3c and Supplemental Fig. 7c). This suggests that Sld3 phosphorylation is sufficient to inhibit origin firing, although effects on other aspects of replication may also contribute (Supplemental Fig. 1).
An Sld3-Dpb11 fusion has previously been shown to bypass the requirement for S-CDK activity for origin firing9. If Rad53 phosphorylation of Sld3 functions solely to inhibit an Sld3-Dpb11 interaction, then cells expressing an Sld3-Dpb11 fusion should be intra-S-phase checkpoint defective, akin to the SLD3-m25 allele. However, cells expressing the Sld3-Dpb11 fusion were neither intra-S-phase checkpoint defective (Supplemental Fig. 8) nor sensitive to growth on DNA damaging agents (data not shown). This suggests that, while Rad53 inhibits the Sld3-Dpb11 interaction, this inhibition is not sufficient to block late origin firing.
To test if growth of SLD3-m25 dbf4-m25 cells is impaired by DNA damaging agents, cells were spotted on HU and MMS plates and sensitivity was compared to wild-type and rad53Δ cells. SLD3-m25 dbf4-m25 cells are mildly sensitive to the replication inhibitor HU in a dosage-dependent manner (Fig 3c and Supplemental Fig. 4b), but were not sensitive to MMS (data not shown). Interestingly, mec1-100 cells, which are defective in the block to late origin firing, are also mildly HU-sensitive, but not MMS-sensitive21,22. It is possible that SLD3-m25 dbf4-m25 cells are sensitive to HU simply because these mutant alleles are hypomorphic. Two results suggested that this was not the case. First, the checkpoint-defective rad53-R70,R605A allele is epistatic to SLD3-m25 dbf4-m25: SLD3-m25 dbf4-m25 rad53-R70,R605A cells were not more HU-sensitive than rad53-R70,R605A cells (Supplemental Fig. 9a and 9b). Second, SLD3-m25 is dominant to wild-type for HU-sensitivity. In HU-sensitivity experiments using heterozygous diploid cells (Fig. 3e), homozygous SLD3/SLD3 dbf4-m25/dbf4-m25 diploid cells are resistant to HU. In contrast, heterozygous SLD3/SLD3-m25 dbf4-m25/dbf4-m25 diploid cells are mildly HU-sensitive, similarly to homozygous SLD3-m25/SLD3-m25 dbf4-m25/dbf4-m25 diploid cells (Fig. 3e). Similar results were obtained in flow cytometry experiments using heterozygous diploid cells (Fig. 3d). These results strongly suggest that the HU sensitivity seen in the SLD3-m25 dbf4-m25 double mutant is not due to insufficient Sld3 activity in the SLD3-m25 allele, but instead is due to its being refractory to Rad53 inhibition.
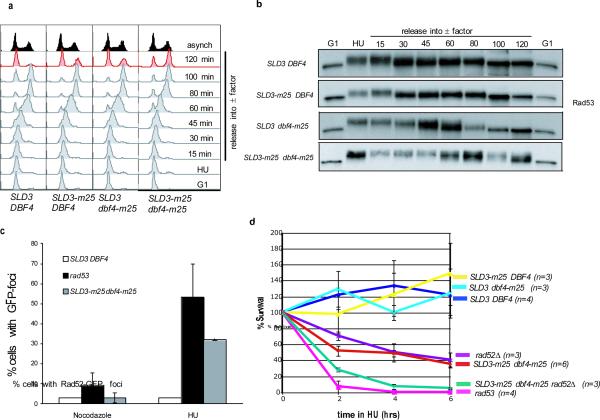
Previous studies have shown that wild-type cells can efficiently recover from HU arrest, whereas rad53Δ cells cannot23,24. We asked if SLD3-m25 dbf4-m25 cells could recover from transient blocks to DNA replication. Wild-type, SLD3-m25 and dbf4-m25 single mutant cells recovered from an HU arrest and divided, entering the next cell cycle by 120 minutes (Fig. 4a). However, only a fraction of SLD3-m25 dbf4-m25 cells appeared to recover immediately from an HU arrest by 120 minutes; the majority remained arrested in mitosis, suggesting that these cells harbored DNA damage (Fig. 4a). We then examined Rad53 de-phosphorylation kinetics. In wild-type, SLD3-m25, and dbf4-m25 cells, Rad53 became phosphorylated during the HU arrest and was de-phosphorylated after the HU release. In contrast, SLD3-m25 dbf4-m25 cells retained Rad53 phosphorylation for 120 minutes after HU release (Fig. 4b). While it is likely that inappropriate firing of late origins leads to a proportional increase in fork collapse, Sld3 phosphorylation likely provides additional HU resistance through other uncharacterized means.
Figure 4. Inappropriate firing of late origins elicits DNA damage.
a, Flow cytometry of wild-type or mutant strains synchronized in G1 with α-factor, released into a 200 mM HU block for 2 hours, then released into medium with α-factor. b, Immunoblots of samples from a, taken at indicated time points and probed for Rad53. c, Quantification of cells with one or more Rad52-GFP foci after incubating for 90 minutes in the presence of nocodazole or 200 mM HU. Error bars represent s.e.m.; n = 3. d, Survival assay of strains synchronized in G1 and released into 200 mM HU for 2, 4 or 6 hours. Error bars represent s.e.m; n is indicated on graph.
Rad52 functions in recombinational repair and can potentially repair or restart broken or stalled replication forks. Rad52-GFP foci have been shown to form in response to replication fork collapse during S-phase in several checkpoint mutants25,26. In wild-type cells, however, foci were not observed when cells are treated with either HU or MMS25,27. Consistently, we observed Rad52-GFP foci in 3% of wild-type and 53% of rad53Δ cells treated with HU. Interestingly, 32% of SLD3-m25 dbf4-m25 cells treated with HU formed Rad52-GFP foci (Fig. 4c) with some cells containing multiple foci (Supplemental Figs 10, 11, and 12). We also observed that SLD3-m25 dbf4-m25 double mutants cells exhibited a 50% viability loss after a 4 hour HU treatment (Fig. 4d). Consistent with the observed increase in Rad52 foci, the mild viability loss seen in SLD3-m25 dbf4-m25 cells after transient HU treatment is exacerbated when RAD52 is also deleted. This suggests that in HU, SLD3-m25 dbf4-m25 cells repair by homologous recombination to survive. As expected, rad53Δ cells show a severe viability loss after HU treatment due the additional roles of Rad53 in fork stabilization23,24. Taken together, these data suggest that failure to block late origins in the presence of replication inhibitors results in an increased number of stalled replication forks, and perhaps increases the propensity of replication fork collapse. Rad52-dependent pathways may function to resolve or restart these stalled forks and its loss likely generates problems at the fork that ultimately contribute to cell lethality.
DNA replication and the intra-S-phase checkpoint machinery are conserved in organisms ranging from yeast to humans. Recently several vertebrate candidate Sld3 orthologues have been identified, including Treslin/Ticcr, GEMC1 and DUE-B28. Of these candidates, Treslin/Ticcr shares significant domain conservation with Sld329. It remains to be seen if ATM/ATR or Chk2 also inhibit any of these new DNA replication factors upon DNA damage.
Supplementary Material
Acknowledgements
We would like to thank members of the Toczyski, Morgan, Li and O'Farrell labs for helpful discussions. A special thanks to David Morgan, Geeta Narlikar, and Joachim Li for intellectual contributions, Jennifer Benanti and Michael Downey for critical reading of this manuscript, Svetlana Makovets and Margaret Hoang for assistance with two-dimensional DNA gels. We thank Hiroyuki Araki for strains, Robert Sclafani for plasmids and John Diffley for communicating results prior to publication. A heartfelt thanks to Nayeli Lopez for help with GFP-foci and colony quantification. Funding was provided by a Ford Foundation Pre-Doctoral Diversity Fellowship and a National Institutes of Health grant GM059691.
Footnotes
Methods A full description of methods is provided in Supplementary Information as Supplementary Methods. Strains and plasmids are listed in Supplementary Tables S2 and S3.
Author Information The authors declare no competing financial interests.
References
- 1.Painter RB, Young BR. Radiosensitivity in ataxia-telangiectasia: a new explanation. Proc Natl Acad Sci U S A. 1980;77:7315–7317. doi: 10.1073/pnas.77.12.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young BR, Painter RB. Radioresistant DNA synthesis and human genetic diseases. Hum Genet. 1989;82:113–117. doi: 10.1007/BF00284040. [DOI] [PubMed] [Google Scholar]
- 3.Larner JM, Lee H, Hamlin JL. Radiation effects on DNA synthesis in a defined chromosomal replicon. Mol Cell Biol. 1994;14:1901–1908. doi: 10.1128/mcb.14.3.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 5.Santocanale C, Diffley JF. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 6.Shirahige K, et al. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 7.Kamimura Y, Tak YS, Sugino A, Araki H. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. Embo J. 2001;20:2097–2107. doi: 10.1093/emboj/20.8.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka S, et al. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 9.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 10.Donaldson AD, Fangman WL, Brewer BJ. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev. 1998;12:491–501. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bousset K, Diffley JF. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinreich M, Stillman B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. Embo J. 1999;18:5334–5346. doi: 10.1093/emboj/18.19.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncker BP, Shimada K, Tsai-Pflugfelder M, Pasero P, Gasser SM. An N-terminal domain of Dbf4p mediates interaction with both origin recognition complex (ORC) and Rad53p and can deregulate late origin firing. Proc Natl Acad Sci U S A. 2002;99:16087–16092. doi: 10.1073/pnas.252093999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lei M, et al. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang W, McDonald D, Hope TJ, Hunter T. Mammalian Cdc7-Dbf4 protein kinase complex is essential for initiation of DNA replication. Embo J. 1999;18:5703–5713. doi: 10.1093/emboj/18.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP. Incorporation into the prereplicative complex activates the Mcm2-7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev. 2009;23:643–654. doi: 10.1101/gad.1759609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463:113–117. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabrielse C, et al. A Dbf4p BRCA1 C-terminal-like domain required for the response to replication fork arrest in budding yeast. Genetics. 2006;173:541–555. doi: 10.1534/genetics.106.057521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X, Rothstein R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad Sci U S A. 2002;99:3746–3751. doi: 10.1073/pnas.062502299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman KL, Brewer BJ. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 21.Tercero JA, Longhese MP, Diffley JF. A central role for DNA replication forks in checkpoint activation and response. Mol Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 22.Paciotti V, Clerici M, Scotti M, Lucchini G, Longhese MP. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol Cell Biol. 2001;21:3913–3925. doi: 10.1128/MCB.21.12.3913-3925.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopes M, et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 24.Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barlow JH, Rothstein R. Rad52 recruitment is DNA replication independent and regulated by Cdc28 and the Mec1 kinase. Embo J. 2009;28:1121–1130. doi: 10.1038/emboj.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 27.Alabert C, Bianco JN, Pasero P. Differential regulation of homologous recombination at DNA breaks and replication forks by the Mrc1 branch of the S-phase checkpoint. Embo J. 2009;28:1131–1141. doi: 10.1038/emboj.2009.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labib K. How do Cdc7 and Cyclin-dependent Kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24:1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez-Pulido L, Diffley JFX, Ponting CP. Homology explains the functional similarities of Treslin/Ticrr and Sld3. Curr Biol. 2010;20:R509–510. doi: 10.1016/j.cub.2010.05.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.