Abstract
Genetically engineered mouse (GEM) models of lung tumorigenesis allow careful evaluation of lung tumor initiation, progression, and response to therapy. Using GEM models of oncogene-induced lung cancer, we demonstrate the striking similarity of the earliest stages of tumorigenesis induced by KRASG12D or BRAFV600E. Cre-mediated expression of KRASG12D or BRAFV600E in the lung epithelium of adult mice initially elicited benign lung tumors comprising cuboidal epithelial cells expressing markers of alveolar pneumocytes. Strikingly, in a head-to-head comparison, oncogenic BRAFV600E elicited many more such benign tumors and did so more rapidly than KRASG12D. However, despite differences in the efficiency of benign tumor induction, only mice with lung epithelium expression of KRASG12D developed malignant non-small-cell lung adenocarcinomas. Pharmacologic inhibition of MEK1/2 combined with in vivo imaging demonstrated that initiation and maintenance of both BRAFV600E- or KRASG12D-induced lung tumors was dependent on MEK→ERK signaling. Although the tumors dramatically regressed in response to MEK1/2 inhibition, they re-grew following cessation of drug treatment. Together, our findings demonstrate that RAF→MEK→ERK signaling is both necessary and sufficient for KRASG12D-induced benign lung tumorigenesis in GEM models. The data also emphasize the ability of KRASG12D to promote malignant lung cancer progression compared with oncogenic BRAFV600E.
INTRODUCTION
Lung cancer is the most prevalent malignancy in the industrialized world and was responsible for ~160,000 deaths in the USA in 2007 (1). Despite its prevalence and strikingly high mortality rates, the cellular origins of lung cancer remain obscure and therapeutic approaches to treat the disease have proven disappointingly ineffective (2). Consequently, the 5-year survival rate for patients with advanced lung cancer remains low, emphasizing the need for new therapeutic approaches to treat this disease.
The genetic heterogeneity of lung cancer has been revealed in more detail and in a manner that has direct implications for therapy (2, 3). For example, mutational activation of ERBB1, encoding the EGF receptor (EGFR), predicts for a clinical response to EGFR inhibitors such as Tarceva (4, 5). Similarly, lung cancers expressing an oncogenic EML4-ALK fusion protein respond well to Crizotinib, an inhibitor of ALK and MET protein kinases (6). However, lung cancers expressing mutationally activated KRAS do not respond to either EGFR or ALK inhibitors and, at least with Tarceva, the use of EGFR inhibitors is contra-indicated (7–9).
Mutationally activated KRAS binds to a multiplicity of effector proteins including (but not limited to) RAF protein kinases, PI3′-lipid kinases and RAL-GDS (10, 11). Consistent with an important role for RAS pathway signaling in lung cancer, mutational activation of BRAF or PIK3CA or silencing of PTEN are detected in a small percentage of lung cancers (12–15). Since mutationally activated KRAS remains an intractable pharmacological target, defining relevant RAS effector pathway(s) in lung cancer is of critical importance since potent and specific inhibitors of RAS effector kinases are being clinically tested for a number of different cancers (11).
Genetically engineered mouse (GEM) models of KRASG12D- or BRAFV600E-induced lung cancer have been described (16–19). In particular, mice carrying conditionally activated alleles of KRas (KRasLSL) or BRaf (BRafCA), in which oncogene expression is initiated by Cre recombinase at the normal chromosomal locus, have allowed exploration of the earliest events following oncogene activation as well as events involved in cancer progression and metastatic spread (17, 18, 20–22). Here we employ KRasLSL and BRafCA mice to directly compare the effects of oncogenic KRASG12D or BRAFV600E on benign lung tumorigenesis, malignant cancer progression and the importance of MEK1/2 signaling in tumor maintenance. KRASG12D and BRAFV600E-induced benign lung tumors share similar morphologic and histological characteristics and express markers of alveolar pneumocytes but not Clara cells. Despite the fact that BRAFV600E tumors formed faster and at higher multiplicity, they failed to display malignant progression. By contrast, KRASG12D-induced lung tumors routinely progressed to higher-grade adenocarcinomas. However, both KRASG12D- and BRAFV600E-induced lung tumors were sensitive to the anti-tumor effects of MEK1/2 inhibition. Consistent with this, tumor derived cell lines were growth arrested in vitro following MEK inhibitor treatment suggesting that MEK1/2 inhibition, either alone or in combination chemotherapy, might represent a viable strategy for targeting KRAS mutant lung cancers in humans.
MATERIALS & METHODS
Mice and Adenovirus delivery
All experiments involving mice were conducted in accordance with protocols approved by the UCSF Institutional Animal Care and Use Committee (IACUC). BRafCA (Braftm1Mmcm), KRasLSL (Krastm4Tyj), Aqp5−/− and LucRep (Tg(Actb-GFP-Luc) mice were bred and genotyped as previously described (17, 18, 23, 24). A stock of Adenovirus encoding Cre recombinase (Ad-Cre) was purchased from Viraquest (North Liberty, IA) and intranasal instillation for infection of the mouse lung epithelium was performed as previously described (25). BrdU (Sigma) was administered at ~30mg/kg by intra-peritoneal injection 20 hours prior to euthanasia.
Histology and quantification of lung tumor burden
Lungs were removed and fixed in zinc buffered formalin and stored in 70%(v/v) ethanol prior to paraffin embedding. 5μm sections were cut and slides were stained with Hematoxylin and Eosin (H&E). H&E stained slides were scanned using an Aperio ScanScope scanner. Quantification was performed using Aperio Spectrum ImageScope viewing software. Tumor number and size was measured per lung section and overall tumor burden was calculated as (area of lung section occupied by tumor)/(total area of section) in μm2. Differences in lung tumor grade were based on criteria established by Nikitin et al (26). Statistical significance of differences in lung tumor grade was assessed using the Exact Wilcoxon Rank sum test.
For effects of MEK inhibition on tumor prevention, 10 lung lobes from 3 vehicle treated and 13 lung lobes from 4 PD325901 treated KRasLSL mice were evaluated. For effects of MEK inhibition on tumor regression, 7 lung lobes from 2 vehicle treated and 8 lung lobes from 3 PD325901 treated KRasLSL mice were evaluated.
Drug treatments and bioluminescent imaging
PD0325901 (Hansun Trading Co.) was formulated in 0.5%(w/v) Hydroxy-Propyl-Methylcellulose (HPMT, Sigma) and administered by oral gavage at 12.5 mg/kg per mouse once per day for 5 days/week. Mice carrying the LucRep transgene were injected with Firefly D-Luciferin (Gold Biotechnology) intra-peritoneally and were imaged 10 minutes later using the Xenogen IVIS 100 bioluminescent imaging system. Bioluminescent signal measured in photons/second (p/s) was quantified using Live Image software (Caliper Life Sciences).
Immunostaining of mouse lung tissue and immunoblotting
Mouse lungs were fixed in formaldehyde overnight, processed, embedded in paraffin cut into 5μm sections and mounted on glass slides. Citrate-mediated antigen retrieval was performed and then the following antibodies were used for detection: anti-SP-C; anti-RAGE, anti-gp38 (Santa Cruz); anti-AQP5 (Calbiochem); anti-BrdU (Roche), anti-Ki67 (Abcam), anti-phospho-ERK1/2, and anti-phospho-S6 (Ser235/236) (Cell Signaling Technology).
Cell proliferation was assessed by counting the percentage of SP-C positive tumor cells that were also either BrdU positive by double label immunofluorescence. In BRafCA mice, 6 tumors were analyzed with 9 grids each for a total of 4112 SP-C positive cells evaluated. In KRasLSL mice 5 tumors were analyzed with 9 grids each for a total of 2831 SP-C positive cells evaluated. Similar numbers of cells were evaluated for the presence of SP-C/Ki67 double positive cells in BRAFV600E- versus KRASG12D-induced lung tumors.
50μg aliquots of cell extract were analyzed by standard immunoblotting using antisera against the following proteins: phospho-MEK1/2 (pMEK1/2); phospho-ERK1/2 (pERK1/2), total ERK1/2 (tERK1/2), BIM (Epitomics), Cleaved Caspase 3 (CC3, Cell Signaling Technology), Cyclin D1 (Cell Signaling technology). Immunoblots were visualized using the Odyssey FC system (Li-Cor) and Image Studio software.
Electron Microscopy of BRAFV600E-induced lung tumors
Tumor bearing lungs from initiated BRafCA/+ mice were removed 11 weeks after Ad-Cre infection and fixed in 2%(v/v) glutaraldehyde, 1%(v/v) paraformaldehyde in 0.1M Sodium Cacodylate buffer at pH 7.4. Following fixation, samples were incubated in 2%(v/v) Osmium Tetroxide in the same buffer. Samples were then stained in 2%(v/v) aqueous Uranyl Acetate, dehydrated in acetone, infiltrated and then embedded in LX-112 resin (Ladd Research Industries, Burlington, VT). Toluidine blue stained semi-thin sections were made to locate the areas of interest. Samples were ultrathin sectioned on a Reichert Ultracut S ultramicrotome and counter stained with 0.8% lead citrate. Grids were examined on a JEOL JEM-1230 transmission electron microscope (JEOL USA, Inc., Peabody, MA) and photographed with a Gatan Ultrascan 1000 digital camera and Digital Micrograph software (Gatan Inc., Warrendale, PA).
Lung tumor cell isolation and culture
Lungs of tumor bearing BRafCA mice were perfused with Dispase (BD) and individual lobes were minced and further incubated with Dispase and Collagenase (Roche). Cell suspensions were then filtered and cultured in DMEM with 10%(v/v) normal calf serum. Recombination of the conditional BRafCA allele was verified using established genotyping protocols (18). Cell suspensions from lung cancer bearing KRasLSL mice were obtained in a similar manner and then passaged twice through orthotopic implantation into nude mice before single cell suspensions were generated and established in culture in DMEM with 10%(v/v) normal calf serum. The cell lines described here have not been separately authenticated using SNP or microsatellite DNA markers. Cell proliferation assays were performed using an Alamar Blue cell viability assay (Invitrogen). PD0325901 was dissolved in DMSO and applied to cells at a final concentration of 1μM. Similarly treated cells were permeabilized, stained with propidium iodide and analyzed by FACscan for DNA content.
RESULTS
BRafCA mice display an earlier onset and higher multiplicity of lung tumors than KRasLSL mice
Genetically engineered KRasLSL or BRafCA mice carry knock-in alleles of KRas or BRaf respectively at the normal chromosomal loci that allow for conditional expression of either oncogenic KRASG12D or BRAFV600E in response to Cre recombinase (17, 18). Intranasal instillation of Cre expressing Adenovirus (Ad-Cre) elicits oncogene expression in the lung epithelium in a manner in which the timing and frequency of oncogene expressing cells can be investigator controlled. Lung epithelium expression of KRASG12D is reported to lead initially to atypical adenomatous hyperplasia (AAH) and then to development of benign lung adenomas and ultimately to emergence of malignant non-small cell lung cancers (17). Lung epithelium expression of BRAFV600E also leads initially to AAH and then to benign lung adenomas but progression to frank malignancy is rare and appears to be constrained by cellular senescence (18). Despite their similarities, a thorough side-by-side comparison between these two models of lung tumorigenesis has not yet been reported.
To minimize the influence of genetic modifiers of lung tumorigenesis on our experiments, BRafCA/+ and KRasLSL/+ mice were bred into an FVB/N genetic background for a minimum of four generations and were then bred together to generate compound BRafCA/+; KRasLSL/+ mice. These mice were further intercrossed for at least three generations. Experimental mice were generated by breeding compound BRafCA/+; KRasLSL/+ heterozygous mice with FVB/N mice resulting in progeny heterozygous for either BRafCA or KRasLSL alleles on a predominantly FVB/N genetic background.
The lung epithelium of BRafCA/+ or KRasLSL/+ littermates was infected with 107 pfu of Ad-Cre to initiate expression of BRAFV600E or KRASG12D oncoproteins respectively. Mice were euthanized at 6 or 17 weeks following initiation of oncogene expression at which time lungs were processed for tumor analysis. Average tumor number and size was quantified and percentage tumor burden was calculated as the area occupied by tumors/total section area as described in Materials and Methods.
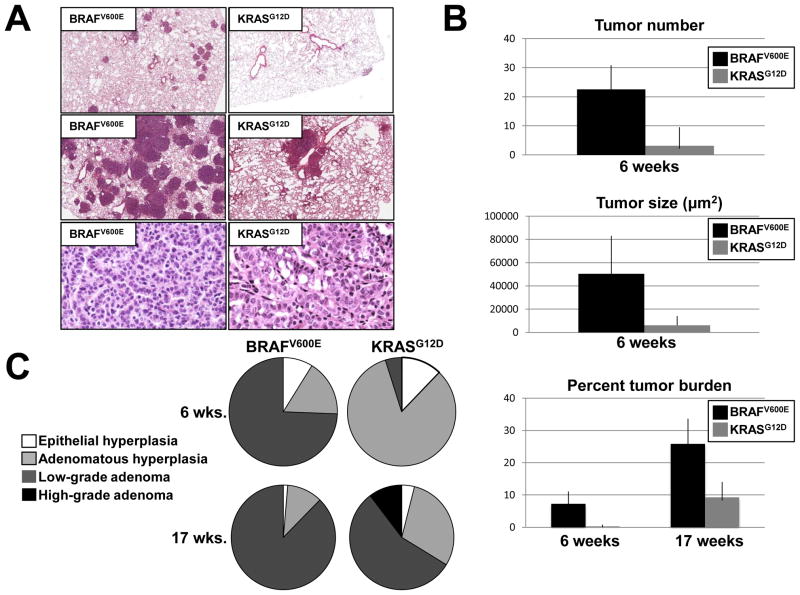
6 weeks post-initiation, BRAFV600E expression led to numerous detectable lung tumors and extensive tumor burden whereas very few lung tumors were detected in mice with KRASG12D expression in lung epithelium. 6 weeks after Ad-Cre infection, BRafCA mice had an average of ~23 lesions/lung section, whereas similarly treated KRasLSL mice had an average of ~3 lesions/section (p =0.0017, Fig. 1A top panels & Fig. 1B). In addition, individual tumors in the lungs of initiated BRafCA mice were approximately 8 times larger than lesions initiated in KRasLSL mice (50,370μm2 versus 6,231μm2, Figs. 1A & 1B) (p<0.0001). At 6 weeks overall lung tumor burden was significantly higher in BRafCA mice compared to their KRasLSL counterparts (7.3% versus 0.28%, p<0.0006, Fig. 1B)
Figure 1. Comparison of BRAFV600E and KRASG12D initiated lung tumorigenesis.
A: H&E staining of lung sections from BRafCA/+ or KRasLSL/+ mice 6 (top panels) or 17 (middle panels) weeks after intranasal infection with 107 pfu Ad-Cre. Scale bar represents 500μm. H&E staining of lung tumors observed in BRafCA/+ and KRasLSL/+ illustrate divergent tumor phenotypes at 17 weeks after Ad-Cre initiation (bottom panels). Bar represents 50μm. Images are 40× magnification.
B: Tumor number and size were quantified for BRAFV600E or KRASG12D-induced lung tumors at 6 weeks. Percent tumor burden was calculated as tumor area per total lung section area at 6 or 17 weeks.
C: Classification of tumor types observed in BRafCA/+ and KRasLSL/+ mice at 17 weeks post infection with Ad-Cre.
At 17 weeks post-initiation, lung tumors in BRafCA mice were confluent in many areas of the lung making individual tumor number and size difficult to quantify (Fig. 1A, middle panel). For this reason, only tumor burden was quantified. Lungs of BRafCA mice displayed approximately 26% tumor burden while KRasLSL mice had a burden of approximately 9% (p = 0.0009, Fig. 1A top panels & Fig. 1B). It was not possible to compare lung tumor burden between Ad-Cre treated BRafCA or KRasLSL mice at later time points due to the extensive benign tumor burden in the BRafCA mice, which led to respiratory distress and weight loss such that these mice had to be euthanized. By contrast, at a time when all of the Ad-Cre treated BRafCA had been euthanized, all of the similarly treated KRasLSL mice appeared healthy and maintained a normal weight despite the obvious lung tumor burden that they presented at euthanasia.
BRAFV600E-induced lung tumors fail to progress beyond low-grade, benign adenomas
To better characterize KRASG12D- or BRAFV600E-induced lung tumors, lesions were classified as epithelial hyperplasia (EH), adenomatous hyperplasia (AH), low- or high-grade adenomas (Fig. 1C) using criteria established by Nikitin et al (26). EH was classified as regions of hyperplasia originating from or directly associated with airways. Adenomatous hyperplasia was defined as lesions originating from the alveoli and measuring <100μm in diameter. Adenomas (lesions >100μm) were classified as being low- or high-grade depending on the degree of cellular atypia. Percentage of each tumor type per lung section was calculated as area of specific lesion type/area of total lesion area. The statistical significance of differences in lung tumorigenesis between BRafCA versus KRasLSL mice was assessed using the Exact Wilcoxon Rank sum test (27).
At six weeks, the lungs of Ad-Cre treated BRafCA or KRasLSL mice had a similar percentage of EH (9% and 12.5%, respectively, p = 0.23, Fig. 1C). The majority of lesions in BRafCA mice (~74%) were fully formed, low-grade adenomas, compared to only 5% of such lesions in the KRasLSL mice (p =0.002). Indeed, at six weeks, the vast majority of the lesions in the KRasLSL mice (85%) were adenomatous hyperplasias compared to only 17% of such lesions in BRafCA mice (p =0.04).
At 17 weeks, there was no significant difference in the percentage of epithelial hyperplasias between BRafCA and KRasLSL mice, (1.3% versus 4% respectively, p= 0.52). Adenomatous hyperplasias were more prevalent in KRasLSL mice compared to BRafCA mice (30% versus 11% respectively, p = .0005). The percentage of low-grade adenomas remained significantly higher in BRafCA mice compared to KRasLSL mice (88% versus 56% respectively, p= 0.002). Importantly, high-grade adenomas were only observed in KRasLSL mice, where they comprised 10% of the total observable lung lesions (p=0.02, Fig. 1C).
The majority of BRAFV600E-induced lung tumors appeared benign and adenomatous with a structured papillary pattern made up of seemingly well-differentiated, cuboidal epithelial cells and lacking characteristics of high-grade tumors (Fig. 1A, bottom panel). These observations are in accord with our previous studies demonstrating the benign nature of BRAFV600E-induced lung tumors and their low rate of spontaneous malignant progression (18). By contrast, despite the overall lower tumor burden, ~10% of lung tumors in KRasLSL mice appeared to be of higher grade, as evidenced by nuclear atypia such as prominent nucleoli and an increased nucleus to cytoplasm ratio (Fig. 1A, bottom panel). Finally, we observed no evidence of either local invasion, pleural effusion or distant metastatic spread in lung tumors arising in either BRafCA or KRasLSL mice, which is consistent with our previous observations and those of others (17, 18, 20).
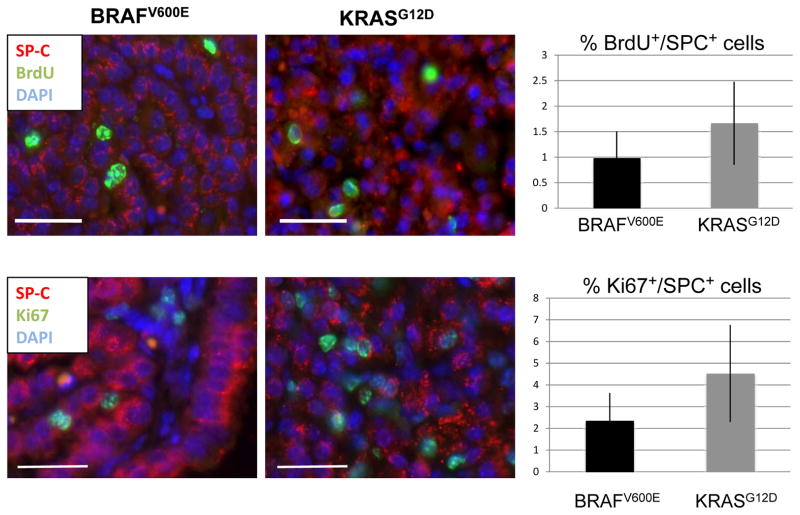
To determine if there was a difference in proliferation rate of KRASG12D- versus BRAFV600E-induced lung tumors 17 weeks after initiation, appropriate mice were injected with BrdU ~20 hours prior to euthanasia. Lung sections were stained with antibodies against BrdU and Surfactant Protein (SP-C), a marker for alveolar type 2 pneumocytes expressed in KRASG12D- and BRAFV600E-induced lung tumor cells (17, 18). Tumor cell specific proliferation was assessed by measuring the percentage of SP-C positive cells that also stained for BrdU incorporation by double label immunofluorescence. By these criteria, 0.98% of BRAFV600E expressing and 1.67% of KRASG12D expressing tumor cells were BrdU positive respectively (p = 0.12, Fig. 2). These data were confirmed by double label immunofluorescence for Ki67/SP-C double positive cells. By these criteria, 2.35% of BRAFV600E expressing and 4.53% of KRASG12D expressing tumor cells were Ki67/SP-C double positive (p = 0.08, Fig. 2B). Hence, at 17 weeks KRASG12D- and BRAFV600E-expressing tumors display a relatively low proliferative index and, although there was a trend to a higher proliferative index in KRASG12D-induced tumors, this did not reach statistical significance.
Figure 2. Quantification of proliferating cells in BRAFV600E and KRASG12D driven lung tumors.
A: BrdU (green) and SP-C (red) staining of BRAFV600E and KRASG12D driven tumors at 17 weeks. Double positive cells within fully formed adenomas were classified as proliferating. Bar represents 25μm. Images are 40× magnification.
B: Ki67 (green) and SP-C (red) staining of BRAFV600E and KRASG12D driven tumors at 17 weeks. Bar represents 25 μm. Images are 40× magnification.
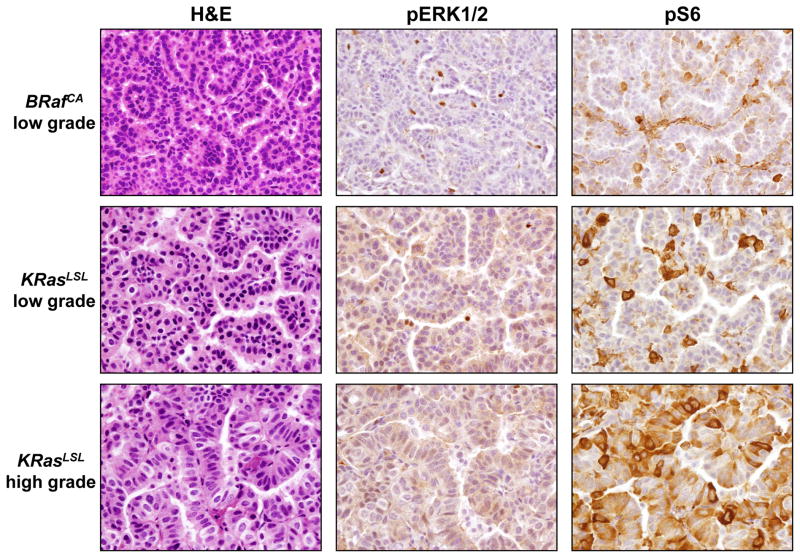
We next assessed signal pathway activation downstream of oncogenic KRASG12D or BRAFV600E by immunohistochemical staining for phospho-ERK1/2 (pERK1/2) or phospho-S6 (pS6) in low- or high-grade adenomas. In general, the levels of pERK1/2 and pS6 were low but detectable in both BRAFV600E and KRASG12D-induced low-grade adenomas. However, we did not detect a substantial elevation in pERK1/2 in KRASG12D-induced high-grade adenomas although elevated pS6 was detected in these tumors (Fig. 3). These data are consistent with the previous observations that neither KRASG12D nor BRAFV600E induce high-level signal pathway activation during the early stages of lung tumorigenesis (22).
Figure 3. Immunohistochemical staining for ERK1/2 activation in BRAFV600E and KRASG12D driven lung tumors.
Antibodies against phospho-ERK1/2 or phospho-S6 were used to stain low-grade adenomas from either BRafCA or KRasLSL mice and high-grade adenomas from KRasLSL mice.
BRAFV600E- and KRASG12D- induced lung tumors express markers of Alveolar type 1 and 2 pneumocytes
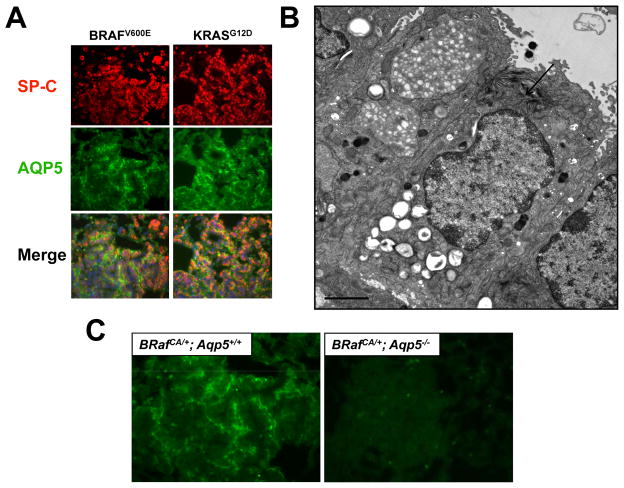
To further characterize lung tumors arising in Ad-Cre treated BRafCA or KRasLSL mice we stained for expression of markers of epithelial cells of the terminal bronchioles and alveoli: Clara Cell Antigen (CCA); Surfactant Protein-C (SP-C) and Aquaporin 5 (AQP5). Clara cells are CCA positive but negative for SP-C and AQP5. Alveolar type 2 (AT2) pneumocytes are SP-C positive but negative for CCA and AQP5. Alveolar type 1 (AT1) pneumocytes are AQP5 positive but negative for CCA and SP-C. As previously reported, adenomas arising in both the BRafCA and KRasLSL mice were largely CCA negative (data not shown). Interestingly, both BRAFV600E- and KRASG12D-induced lung tumors were positive for both SP-C and AQP5, a combination of marker expression not readily detected in the normal mouse lung epithelium. SP-C was localized in a punctate manner inside tumor cells consistent with its sequestration into lamellar bodies that are characteristic of AT2 cells (28). AQP5, an integral membrane water channel, appeared membranous and expressed asymmetrically on tumor cells consistent with its normal expression on the apical surface of AT1 cells (Fig. 4A) (29).
Figure 4. BRAFV600Eand KRASG12Ddriven lung tumors express the Alveolar Type 1 pneumocyte marker Aquaporin 5 (AQP5).
A: Immunofluorescent staining of BRAFV600E and KRASG12D driven lung tumors using antibodies against Surfactant Protein C (SP-C, red) and Aquaporin 5 (AQP5, green).
B: Electron microscopy image of a BRAFV600E-driven lung tumor demonstrating AT2-like cells with prominent lamellar body (red arrow). Bar represents 2μm.
C: BRAFV600E- driven tumors from mice either wild type or null for AQP5 stained with anti-AQP5 antiserum.
To rule out the possibility that the apparent double positivity of lung tumor cells for both SP-C and AQP5 was not simply due to the close juxtaposition of two different cell types, we analyzed BRAFV600E-induced lung tumors by electron microscopy. This analysis revealed that BRAFV600E-induced tumors were comprised of cuboidal AT2-like cells with prominent lamellar bodies (Fig. 4B, arrowed) and were not infiltrated by normal AT1-like cells. To further analyze the extent of tumor cell expression of AT1 markers, we stained for RAGE (Receptor for Advanced Glycosylation End Products) and Podoplanin. However, neither BRAFV600E- nor KRASG12D-induced lung tumors expressed these proteins (data not shown). To test whether the combination of SP-C and AQP5 expression is observed in human lung cancer, we stained sections of eight lung cancers with documented BRAF mutation with antisera against CCA, SP-C or AQP5 (30). However, human lung cancer cells in these sections were uniformly negative for the expression of these proteins (data not shown).
To allay concerns regarding the specificity of the anti-AQP5 antibody and to test for a possible role of AQP5 in lung tumorigenesis, we generated BRafCA mice homozygous for a null allele of Aqp5 (23). BRAFV600E expression was initiated in the lung epithelium of BRafCA; Aqp5+/+ or BRafCA; Aqp5−/− mice and these mice were analyzed for lung tumor formation 11 weeks later. Lack of AQP5 expression had no discernable effect on tumor frequency or the extent of tumor burden initiated by BRAFV600E. Indeed, BRafCA; Aqp5−/− mice developed benign adenomas that were indistinguishable from those that formed in control BRafCA; Aqp5+/+ mice. As expected, lung tumors arising in BRafCA; Aqp5+/+ mice were double positive for both SP-C and AQP5 expression, whereas lung tumors arising in BRafCA; Aqp5−/− mice were SP-C positive but negative for AQP5 (Fig. 4C). These data confirm the specificity of the AQP5 antiserum and indicate that BRAFV600E- (and by extension KRASG12D-) induced lung tumors are indeed positive for at least one marker of AT1 cells. However, based on their cuboidal morphology and the presence of lamellar bodies, tumor cells appear most similar to AT2 cells. Finally, these data unequivocally demonstrated that AQP5 is dispensable for BRAFV600E-induced lung tumorigenesis.
BRAFV600E- initiated lung tumors regress upon MEK1/2 inhibition
BRAFV600E-initiated lung tumorigenesis is prevented by the potent and specific MEK1/2 inhibitor PD0325901 (18, 31, 32). However, others have reported that inhibition of KRASG12D-initiated lung tumorigenesis requires blockade of both RAF→MEK→ERK and PI3′-kinase→AKT→mTorc signaling (33). Consequently, we sought to test the ability of single agent MEK1/2 inhibition (with PD0325901) to promote regression of established BRAFV600E- or KRASG12D-induced lung tumors. To facilitate this analysis we generated compound BRafCA; LucRep mice carrying the conditional BRafCA allele in combination with a Cre-activated Luciferase transgene (24). Ad-Cre infection of the lung epithelium of these mice initiates oncogenic BRAFV600E expression and the expression of Luciferase, which can be used to image tumorigenesis and response to pathway-targeted therapy using a Xenogen In Vivo Imaging System (IVIS).
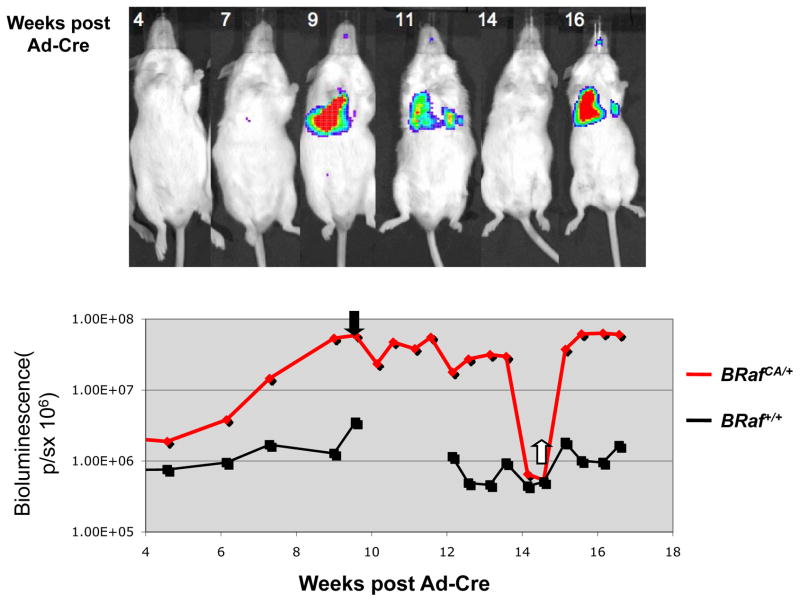
Bioluminescence (in photons/second, p/s) was measured in compound BRafCA; LucRep mice at different times after initiation with Ad-Cre (107 pfu). Baseline signal was determined to be 2×106 photons/second by measuring bioluminescence from Ad-Cre infected LucRep littermates lacking the Cre-activated BRafCA allele. Starting five weeks after Ad-Cre infection, bioluminescence in the lung was observed to increase in a representative BRafCA; LucRep mouse and at nine weeks had increased 30-fold to 6×107 p/s (Fig. 5). At this time PD0325901 was administered orally for five weeks on a 5 days/week dosing schedule. Over this period the Luciferase signal gradually decreased until it was below baseline (Fig. 5). In addition, at the time of drug administration, this mouse appeared hunched, had lost body weight and was displaying the signs of respiratory distress that typify the end-stage of confluent BRAFV600E-induced benign lung tumorigenesis. Following five weeks of drug treatment the mouse appeared healthy, had gained 8.7 grams in body weight (a ~40% increase) and no longer displayed signs of respiratory distress. Upon euthanasia of similarly treated BRafCA mice we could find little or no evidence of residual tumors following five weeks of MEK1/2 inhibition (Data not shown). This experiment was repeated with an additional group of 5 BRafCA; LucRep mice, all of which displayed robust tumor regression and health status improvements in response to MEK1/2 inhibition (Data not shown). Despite the profound tumor regression, cessation of MEK1/2 inhibitor administration led to rapid re-growth of lung tumors in all the mice. These data indicate that MEK→ERK signaling is required for both initiation and maintenance of BRAFV600E-induced lung tumors. However, the rapid re-growth of lung tumors following cessation of drug treatment indicates the presence of tumor initiating cells that are resistant to the cytotoxic effects of PD0325901.
Figure 5. BRAFV600E- driven tumors regress upon treatment with MEK inhibitor PD0325901.
Luciferase imaging of BRafCA/+ and BRaf+/+ Luciferase reporter mice following infection with 107 pfu Ad-Cre. Mice were administered 12.5 mg/kg PD32901 at nine weeks post infection and dosed five days a week for five weeks. Bioluminescent signal was measured in photons/second (p/s).
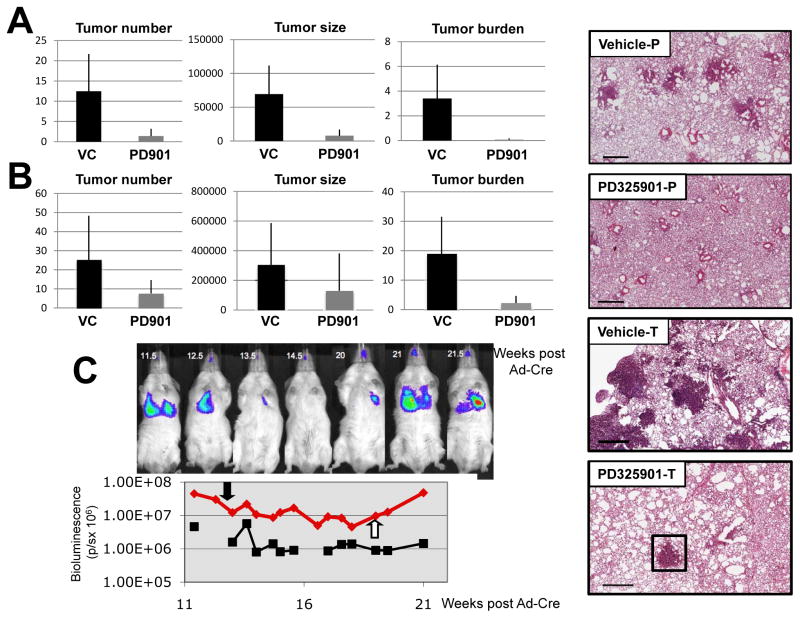
To assess the effect of MEK1/2 inhibition on KRASG12D-induced lung tumorigenesis a cohort of KRasLSL mice was initiated by infection with Ad-Cre and four weeks later, a time prior to the onset of tumorigenesis (Fig. 1), mice were treated with either vehicle or PD0325901 for six weeks at which time they were euthanized and lung tumor burden assessed (Fig. 6A, compare Vehicle-P versus PD325901-P). MEK1/2 inhibition with PD0325901 treatment was highly effective in the prevention of KRASG12D-driven lung tumors (Fig. 6A). Whereas vehicle treated mice had 12.5 tumors/lung section, PD0325901 treated mice had an average of 1.4 lung tumors/section – a 9-fold decrease in lung tumor incidence (p = .0012, Fig. 6A). In addition, we observed a 9-fold difference in the size of individual lesions when comparing control versus PD0325901 treated mice (69,354μm2 in vehicle treated mice compared to 7.837μm2 for drug treated mice, (p= .0002) (Fig. 6A). The differences in tumor number and size contributed to an overall 47-fold lower tumor burden in PD0325901 treated mice compared to vehicle treated littermates (3.4% versus 0.072%, p = .0005, Fig. 6A). These data indicate that RAF→MEK→ERK signaling is essential for the onset of KRASG12D-induced lung tumorigenesis.
Figure 6. MEK inhibition is effective in the prevention and regression of KRASG12D driven lung tumors.
A: KRasLSL/+ mice were initiated with 107 pfu Ad-Cre and then 4 weeks later administered 12.5 mg/kg PD0325901 dosed 5 times a week for six weeks. Lung tumor number, size and overall tumor burden were calculated as described previously. Representative H&E stained lung tumor sections from vehicle (Vehicle-P) or PD0325901 (PD325901-P) treated mice are presented. Bar represents 500μm.
B: KRasLSL/+ mice were initiated with 107 pfu Ad-Cre and then 6 weeks later administered 12.5 mg/kg PD0325901 dosed 5 times a week for four weeks. Lung tumor number, size and overall tumor burden were calculated as described previously. Representative H&E stained lung tumor sections from vehicle (Vehicle-T) or PD0325901 (PD325901-T) treated mice are presented. Residual lung tumor after PD325901 treatment is boxed. Bar represents 500μm.
C: Luciferase imaging of KRasLSL/+ and KRas+/+ Luciferase reporter mice following infection with 108 pfu Ad-Cre. Mice were administered 12.5 mg/kg PD032901 at 11.5 weeks post infection and dosed five days a week for 7.5 weeks. Bioluminescent signal was measured in photons/second (p/s).
Next we assessed the efficacy of PD0325901 in promoting regression of pre-existing KRASG12D-initiated lung tumors. Ten weeks after initiating KRASG12D expression, mice were treated with either vehicle or PD0325901 for a further four weeks at which time they were euthanized and lung tumor burden assessed (Fig. 6B, compare Vehicle-T versus PD325901-T). Mice treated with PD0325901 showed significantly fewer tumors per lung section compared to vehicle treated littermates (7.6 versus 25.3, p = 0.01). Moreover, whereas vehicle treated mice displayed mostly high-grade lung adenomas, lesions in PD0325901 treated mice were predominantly epithelial or adenomatous hyperplasias and thus generally smaller than tumors observed in vehicle treated mice. However the difference in overall lesion size between PD0325901 and vehicle treated mice was not significant (129679 μm2 versus 305584 μm2, p = 0.08). This is likely due to the presence of residual large adenomas (Fig. 6B, PD325901-T boxed) remaining after the full course of drug treatment, which were presumably resistant to the effects of MEK inhibition. Importantly, overall tumor burden was decreased approximately eight-fold in PD0325901 treated KRasLSL mice compared to control (19% compared to 2.2%, p = 0.0002, Fig. 6B).
To unequivocally demonstrate regression of pre-existing KRASG12D-induced lung tumors, we initiated tumorigenesis in a representative KRasLSL; LucRep mouse and monitored tumor growth by bioluminescent imaging as described above (Fig. 6C). Baseline signal (2.6 × 106 p/s) was determined as the average signal of KRas+/+; LucRep mice treated with the same dose (108 pfu) of Ad-Cre. 11.5 weeks after initiation of KRASG12D expression, this KRasLSL/+; LucRep mouse displayed a consistent bioluminescent signal ~17-fold above baseline at which time PD0325901 was administered. After 8.5 weeks of PD0325901 treatment, the bioluminescent signal from lung tumor burden was reduced ~6-fold (4.5×107 to 8×106 p/s), consistent with the reduction in tumor burden assessed by histochemical analysis of lung sections (Fig. 6B). However, unlike the situation with BRAFV600E-induced lung tumors, MEK1/2 inhibition did not result in the reduction of the bioluminescent signal to baseline. Moreover, as with BRAFV600E-induced lung tumors, cessation of PD0325901 administration led to rapid re-growth of KRASG12D expressing lung tumors requiring that this mouse be euthanized two weeks later (Fig. 6C). Cumulatively, these data provide compelling evidence that inhibition of RAF→MEK→ERK signaling both prevents the growth and promotes the regression of pre-existing of KRASG12D-induced lung tumors. However, the presence of adenomas at the end of the drug treatment period may explain the incomplete tumor regression observed in KRasLSL mice and thus why KRASG12D-driven lung tumors appear less sensitive to MEK1/2 inhibition than their BRAFV600E expressing counterparts.
BRAFV600E or KRASG12D expressing lung tumor derived cell lines are sensitive to MEK inhibition in vitro
The anti-tumor effects of PD0325901 against BRAFV600E- or KRASG12D-induced tumors in vivo could be mediated either by tumor cell autonomous or non-autonomous mechanisms or some combination thereof. Hence, to determine if there is a tumor cell autonomous response to MEK inhibition, we utilized cell lines isolated from either BRAFV600E or KRASG12D-induced lung tumors. Whereas lung tumor lines can be generated from lung cancer bearing KRasLSL mice, this is not the case for similarly treated BRafCA mice. This likely reflects the fact that BRAFV600E-induced lung tumors are benign and tumor cells are thought to be irreversibly arrested in the cell division cycle due to senescence (18). Consequently, we infected compound BRafCA/+; Trp53lox/lox mice with Ad-Cre to initiate BRAFV600E and silence TP53 expression. These mice develop abundant high-grade lung tumors with a propensity for malignant lung cancer progression (18). 10 weeks later mice were euthanized and independent BRAFV600E/TP53null lung cancer-derived cell lines were established as described in Materials and Methods. Cre-mediated recombination of the BRafCA and Trp53lox alleles in the various cell lines was confirmed by PCR. Independent KRASG12D expressing lung tumor derived cell lines were derived from Ad-Cre infected KRasLSL/+ mice in which spontaneous progression to NSCLC was observed. Immunoblot analysis revealed that KRASG12D expressing cell lines express detectable p53 (Shai & McMahon, Unpublished).
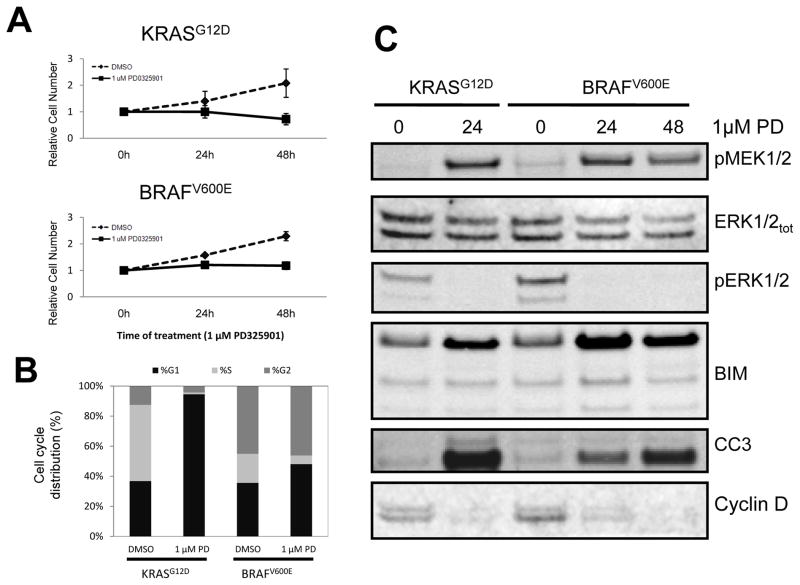
Treatment of BRAFV600E- or KRASG12D-expressing lung tumor cells with 1μM PD0325901 for 48 hours led to almost complete inhibition of proliferation compared to vehicle (DMSO) control (Fig. 7A) with comparable effects of PD0325901 observed on both BRAFV600E- and KRASG12D-expressing lung tumor cells. Analysis of the cell cycle profile of control versus PD0325901 treated cells revealed an accumulation of both KRASG12D and BRAFV600E expressing lung tumor cells in G1 and a decrease of cells in S phase (Fig. 7B), consistent with the observed inhibition of cell proliferation. In parallel, vehicle versus PD0325901 treated cells were stained for a marker of apoptosis (Annexin V) and cell death (PI). Whereas there was a modest increase of apoptotic and dead cells in PD0325901 treated cultures, the majority of cells remained Annexin V and PI negative (Data not shown).
Figure 7. Cells isolated from BRAFV600E- or KRASG12D- driven lung tumors are sensitive to MEK inhibition in vitro.
A. Lung tumor derived cell lines isolated from mice bearing KRASG12D- or BRAFV600E/TP53Null lung tumors were cultured in media containing 10%(v/v) fetal bovine serum and then treated with PD0325901 or vehicle control for 1, 24, and 48 hours. Relative cell number was assessed using an Alamar Blue cell viability assay.
B: Lung tumor derived cell lines described in A were treated for 24 hours with either PD0325901 or vehicle control at which time permeabilized cells were stained with propidium iodide to assess the percentage of cells in the G0/G1, S or G2/M phases of the cell division cycle.
C: Lung tumor derived cell lines expressing KRASG12D or BRAFV600E were treated with PD0325901 for the indicated times at which time cell lysates were analyzed for the phosphorylation or expression of MEK1/2, ERK1/2, BIM, Cleaved Caspase 3 (CC3) or Cyclin D1 as indicated.
In parallel, cell extracts were analyzed to assess the effect of MEK inhibition on signal pathway activation. As expected, PD0325901 treatment led to increased phospho-MEK1/2 but decreased phospho-ERK1/2, consistent with the mechanism of action of this agent (Fig. 7C). Both cell lines also displayed decreased expression of cyclin D1 consistent with the known role of ERK1/2 signaling in Cyclin D1 regulation (34). We also detected modest induction of the pro-apoptotic BCL-2 family member BIM, consistent with its regulation by ERK signaling (35), and the presence of cleaved-Caspase-3 (CC3) in PD0325901 treated cells. Cumulatively, these data suggest that the anti-tumor effect of PD0325901 in mouse models of KRASG12D- or BRAFV600E-induced lung tumorigenesis results, at least in part, from direct anti-proliferative effects of the agent on tumor cells mediated by cell cycle arrest and accompanied by a modest induction of apoptosis.
DISCUSSION
Mutationally activated KRAS is a feature common to ~25% of all cancers and is particularly prevalent in pancreas, colon and lung cancer (2, 3, 36). Consistent with its documented importance in human lung cancer, expression of oncogenic KRASG12D in the mouse lung epithelium leads to the development of non-small cell adenocarcinomas, some of which have a propensity for metastasis (17, 20, 21). However, it remains unclear which of its various effector pathways are critical for KRASG12D-induced lung tumor initiation and maintenance. Here we demonstrate RAF→MEK→ERK signaling is both necessary and sufficient for the early stages of KRASG12D-induced tumorigenesis up to the formation of low-grade adenomas. Consistent with this, expression of oncogenic BRAFV600E in the lung epithelium leads to development of benign lung tumors with most of the same histological and morphological features but with strikingly enhanced efficiency compared to KRASG12D. One explanation for this difference may be the efficiency of Cre-mediated recombination of the conditional BRafCA and KRasLSL alleles, which is not readily measurable in the lung epithelium. Another possibility is that there may be more cells with tumorigenic potential which express BRAF compared to KRAS. Finally, the efficiency of lung tumorigenesis may reflect a better ability of BRAFV600E to elevate MEK→ERK signaling compared to KRASG12D. Although we did not see a difference in pERK between KRASG12D or BRAFV600E-induced lung tumors, we were not able to assess pERK levels in single initiated cells to determine if there may be differences between the two oncogenes in signal pathway engagement. Although this highly reproducible difference in the efficiency of BRAFV600E versus KRASG12D-induced lung tumorigenesis was surprising, it is not without precedent. For example, whereas BRAFV600E elicits papillary thyroid cancer when expressed in mouse thyrocytes, KRASG12D expression had no effect in the thyroid unless combined with PTEN silencing (37, 38).
Although BRAFV600E elicits abundant lung tumors at early time points they remain low-grade, benign and rarely progress to lung cancer (18). By contrast, despite the overall reduced efficiency of lung tumorigenesis, KRasLSL mice invariably develop high-grade tumors with a strong propensity for further malignant progression (17). Whereas evidence suggests that high-grade lesions emerge from more benign tumors, it remains possible that these lesions emerge from a different cell of origin (17, 22). To explore the possibility that KRASG12D- or BRAFV600E-induced lung tumors may display phenotypic differences reflective of their cell of origin, we stained tumors for markers of cells of the distal lung epithelium. Whereas both KRASG12D- and BRAFV600E-induced lung tumors types lacked CCA expression, they were uniformly double positive for SP-C and AQP5, markers of AT-2 and AT-1 pneumocytes respectively and a combination of markers not detected in normal lung. However, by most other criteria, KRASG12D and BRAFV600E-induced lung tumor cells resembled AT-2 cells. Consequently, it is unclear why the AT2-like cells in KRASG12D or BRAFV600E driven lung tumors express an AT1 marker like AQP5, but increased signaling through the ERK MAP kinase pathway may influence expression of this protein in AT2 cells. Overall, the data stresses the similarity of the cellular phenotype of both KRASG12D- or BRAFV600E-induced lung tumors and suggests that the ability of KRASG12D to promote malignant progression at late time points may be a result of differences in signaling caused by this oncogene perhaps reflected by the elevated phospho-S6 detected in high-grade KRASG12D-induced tumors. Finally, It has been proposed that the cell of origin of KRASG12D-driven lung tumors is a brochioalveolar stem cell (BASC), which co-expresses both CCA and SP-C (39). Although we detected small numbers of double CCA/SP-C positive cells in BRAFV600E driven tumors, not all tumors contained such cells and their frequency was less than 1% of the total tumor cells (Not shown).
To explore the importance of MEK→ERK signaling in BRAFV600E or KRASG12D driven tumors we employed PD0325901, a highly selective and potent inhibitor of MEK1/2 (31, 32). In vivo bioluminescent imaging demonstrated the ability of this agent, not only to prevent the onset of BRAFV600E-induced tumorigenesis (18), but to promote dramatic regression of pre-existing lung tumors. PD0325901 treatment also effectively prevented the formation of KRASG12D-driven tumors and promoted the regression of pre-existing lung tumors, consistent with the work of others (33, 40). However, the magnitude of tumor regression in the KRasLSL mice was less dramatic than that observed in BRafCA mice and, at euthanasia, we detected fully formed adenomas in the lungs of PD0325901 treated KRasLSL mice, which were presumably resistant to the cytotoxic effects of MEK inhibition. In addition, despite the striking effects of MEK1/2 inhibition on BRAFV600E-induced lung tumorigenesis, we were unable to eradicate tumor-initiating cells from the lungs of these animals, which rapidly re-grew lung tumors upon cessation of drug treatment. The reasons for these observations are the subject of ongoing investigation.
Tumor cell autonomous effects of MEK1/2 inhibition were established using cultured cells isolated from BRAFV600E or KRASG12D driven lung tumors. Interestingly, although such cell lines were sensitive to the anti-proliferative effects of MEK inhibition and displayed some evidence of apoptosis, we did not observe substantial involution of these cell cultures. This leaves open the possibility that MEK1/2 inhibition has both tumor cell autonomous and non-autonomous effects that contribute to the regression of BRAFV600E- and KRASG12D-induced lung tumors in vivo. However, these data emphasize the central importance of RAF→MEK→ERK signaling in KRASG12D-induced lung tumorigenesis in the mouse. Since mutationally activated KRAS remains an intractable pharmacological target, it is possible that targeting MEK1/2, either alone or in combination chemotherapy, may represent a viable approach for targeting KRAS-induced lung cancer (11).
Acknowledgments
We thank past and present members of the McMahon Lab who helped with this work in particular Drs. David Dankort, Eric Collisson, Anny Shai and Anthony Karnezis. In addition, we thank Shon Greenberg for critical review of the manuscript. We thank Allan Balmain for advice on genetic modifiers of mouse lung cancer and Byron Hann, Scott Kogan and Adam Olshen of the Helen Diller Family Comprehensive Cancer Center Preclinical Therapeutics, Mouse Pathology and Biostatistics cores respectively for their advice and guidance in the analysis of GEM models of lung tumorigenesis. Finally, we thank Samuel Yousem (University of Pittsburgh) for human lung cancer specimens, Jinny Wong (Gladstone Electron Microscopy Core) for microscopy assistance, the UCSF RTOG Biospecimen Resource for access to the Aperio scanner, Alan Verkman (UCSF) and Tyler Jacks (MIT) for providing Aqp5 knockout and KRas(LSL) mice respectively.
References
- 1.Hayat MJ, Howlader N, Reichman ME, Edwards BK. Cancer Statistics, Trends, and Multiple Primary Cancer Analyses from the Surveillance, Epidemiology, and End Results (SEER) Program. Oncologist. 2007 Jan;12(1):20–37. doi: 10.1634/theoncologist.12-1-20. [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008 Sep 25;359(13):1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herbst RS, Lippman SM. Molecular signatures of lung cancer--toward personalized therapy. N Engl J Med. 2007 Jan 4;356(1):76–8. doi: 10.1056/NEJMe068218. [DOI] [PubMed] [Google Scholar]
- 4.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004 May 20;350(21):2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 5.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004 Sep 7;101(36):13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007 Aug 2;448(7153):561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 7.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005 Jul 14;353(2):123–32. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 8.Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med. 2005 Jul 14;353(2):133–44. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 9.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005 Jan;2(1):e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007 Apr;7(4):295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 11.Gysin S, Salt M, Young A, McCormick F. Therapeutic strategies for targeting ras proteins. Genes Cancer. 2011 Mar;2(3):359–72. doi: 10.1177/1947601911412376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002 Dec 1;62(23):6997–7000. [PubMed] [Google Scholar]
- 13.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004 Apr 23;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 14.Kohno T, Takahashi M, Manda R, Yokota J. Inactivation of the PTEN/MMAC1/TEP1 gene in human lung cancers. Genes Chromosomes Cancer. 1998 Jun;22(2):152–6. doi: 10.1002/(sici)1098-2264(199806)22:2<152::aid-gcc10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 15.Forgacs E, Biesterveld EJ, Sekido Y, Fong K, Muneer S, Wistuba, et al. Mutation analysis of the PTEN/MMAC1 gene in lung cancer. Oncogene. 1998 Sep 24;17(12):1557–65. doi: 10.1038/sj.onc.1202070. [DOI] [PubMed] [Google Scholar]
- 16.Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, Lizak MJ, et al. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001 Dec 15;15(24):3249–62. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001 Dec 15;15(24):3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007 Feb 15;21(4):379–84. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim CF, Jackson EL, Kirsch DG, Grimm J, Shaw AT, Lane K, et al. Mouse models of human non-small-cell lung cancer: raising the bar. Cold Spring Harb Symp Quant Biol. 2005;70:241–50. doi: 10.1101/sqb.2005.70.037. [DOI] [PubMed] [Google Scholar]
- 20.Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM, et al. Suppression of lung adenocarcinoma progression by Nkx2–1. Nature. 2011 May 5;473(7345):101–4. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005 Nov 15;65(22):10280–8. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 22.Feldser DM, Kostova KK, Winslow MM, Taylor SE, Cashman C, Whittaker CA, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010 Nov 25;468(7323):572–5. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma T, Song Y, Gillespie A, Carlson EJ, Epstein CJ, Verkman AS. Defective secretion of saliva in transgenic mice lacking aquaporin-5 water channels. J Biol Chem. 1999 Jul 16;274(29):20071–4. doi: 10.1074/jbc.274.29.20071. [DOI] [PubMed] [Google Scholar]
- 24.Lyons SK, Meuwissen R, Krimpenfort P, Berns A. The generation of a conditional reporter that enables bioluminescence imaging of Cre/loxP-dependent tumorigenesis in mice. Cancer Res. 2003 Nov 1;63(21):7042–6. [PubMed] [Google Scholar]
- 25.Fasbender A, Lee JH, Walters RW, Moninger TO, Zabner J, Welsh MJ. Incorporation of adenovirus in calcium phosphate precipitates enhances gene transfer to airway epithelia in vitro and in vivo. J Clin Invest. 1998 Jul 1;102(1):184–93. doi: 10.1172/JCI2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikitin AY, Alcaraz A, Anver MR, Bronson RT, Cardiff RD, Dixon D, et al. Classification of proliferative pulmonary lesions of the mouse: recommendations of the mouse models of human cancers consortium. Cancer Res. 2004 Apr 1;64(7):2307–16. doi: 10.1158/0008-5472.can-03-3376. [DOI] [PubMed] [Google Scholar]
- 27.Wilcoxon F. Individual comparisons by ranking methods. Biometrics Bulletin. 1945;1(6):80–3. [Google Scholar]
- 28.Rooney SA, Young SL, Mendelson CR. Molecular and cellular processing of lung surfactant. FASEB J. 1994 Sep;8(12):957–67. doi: 10.1096/fasebj.8.12.8088461. [DOI] [PubMed] [Google Scholar]
- 29.Verkman AS. Aquaporins at a glance. J Cell Sci. Jul 1;124(Pt 13):2107–12. doi: 10.1242/jcs.079467. [DOI] [PubMed] [Google Scholar]
- 30.Yousem SA, Nikiforova M, Nikiforov Y. The histopathology of BRAF-V600E-mutated lung adenocarcinoma. Am J Surg Pathol. 2008 Sep;32(9):1317–21. doi: 10.1097/PAS.0b013e31816597ca. [DOI] [PubMed] [Google Scholar]
- 31.Sebolt-Leopold JS. MEK inhibitors: a therapeutic approach to targeting the Ras-MAP kinase pathway in tumors. Curr Pharm Des. 2004;10(16):1907–14. doi: 10.2174/1381612043384439. [DOI] [PubMed] [Google Scholar]
- 32.Ohren JF, Chen H, Pavlovsky A, Whitehead C, Zhang E, Kuffa P, et al. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat Struct Mol Biol. 2004 Dec;11(12):1192–7. doi: 10.1038/nsmb859. [DOI] [PubMed] [Google Scholar]
- 33.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008 Dec;14(12):1351–6. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, et al. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995 Oct 6;270(40):23589–97. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 35.Gillings AS, Balmanno K, Wiggins CM, Johnson M, Cook SJ. Apoptosis and autophagy: BIM as a mediator of tumour cell death in response to oncogene-targeted therapeutics. FEBS J. 2009 Nov;276(21):6050–62. doi: 10.1111/j.1742-4658.2009.07329.x. [DOI] [PubMed] [Google Scholar]
- 36.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49(17):4682–9. [PubMed] [Google Scholar]
- 37.Miller KA, Yeager N, Baker K, Liao XH, Refetoff S, Di Cristofano A. Oncogenic Kras requires simultaneous PI3K signaling to induce ERK activation and transform thyroid epithelial cells in vivo. Cancer Res. 2009 Apr 15;69(8):3689–94. doi: 10.1158/0008-5472.CAN-09-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charles RP, Iezza G, Amendola E, Dankort D, McMahon M. Mutationally activated BRAF(V600E) elicits papillary thyroid cancer in the adult mouse. Cancer Res. 2011 Jun 1;71(11):3863–71. doi: 10.1158/0008-5472.CAN-10-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005 Jun 17;121(6):823–35. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 40.Ji H, Wang Z, Perera SA, Li D, Liang MC, Zaghlul S, et al. Mutations in BRAF and KRAS converge on activation of the mitogen-activated protein kinase pathway in lung cancer mouse models. Cancer Res. 2007 May 15;67(10):4933–9. doi: 10.1158/0008-5472.CAN-06-4592. [DOI] [PubMed] [Google Scholar]