Abstract
The allosteric enzyme aspartate transcarbamoylase (ATCase) from Escherichia coli has been the subject of investigations for approximately 50 years. This enzyme controls the rate of pyrimidine nucleotide biosynthesis by feedback inhibition, and helps to balance the pyrimidine and purine pools by competitive allosteric activation by ATP. The catalytic and regulatory components of the dodecameric enzyme can be separated and studied independently. Many of the properties of the enzyme follow the Monod, Wyman Changeux model of allosteric control thus E. coli ATCase has become the textbook example. This review will highlight kinetic, biophysical, and structural studies which have provided a molecular level understanding of how the allosteric nature of this enzyme regulates pyrimidine nucleotide biosynthesis.
Keywords: allosteric enzyme, pyrimidine nucleotide biosynthesis, domain motions, concerted allosteric transition, feedback inhibition
Introduction
The first studies on the regulation of pyrimidine nucleotide metabolism were reported in 1956 by Yates and Pardee [1]. According to this study an end-product of pyrimidine nucleotide biosynthesis is capable of inhibiting the entire pathway by a mechanism that they called “feedback inhibition.” They also determined that the inhibition occurred at the formation of ureidosuccinic acid (now known as N-carbamoyl-L-aspartate, CA). However, in 1956 the enzyme responsible for the formation of ureidosuccinic acid was unknown.
After the reaction between carbamoyl phosphate (CP) and L-aspartate (Asp) to form N-carbamoyl-L-aspartate (ureidosuccinic acid) and inorganic phosphate (Pi) was established [2–4] (Fig. 1), the enzyme responsible for the reaction, aspartate transcarbamoylase (E.C.2.1.3.2, aspartate carbamoyltransferase, ATCase), was isolated and purified from Escherichia coli in 1960 [5]. Using analytical ultracentrifugation, Shepherdson and Pardee determined that the sedimentation coefficient of the enzyme was 11.5 ± 0.5 S. Using this value and estimates of the partial specific volume and asymmetric ratio, they calculated the molecular weight of the ATCase holoenzyme to be 220,000 ± 40,000 Da. Using purified enzyme, the mode of feedback inhibition by ATCase was investigated in detail by Gerhart and Pardee [6]. They determined that ATCase was inhibited by a variety of pyrimidine nucleotides with the extent of the inhibition increasing with the number of phosphates, with CTP and dCTP inhibiting the enzyme by 86% and 88% respectively at a concentration of 2 mM [6]. They also found weaker inhibition by GTP, dTTP and UTP. However, ATP or dATP, the products of purine nucleotide biosynthesis were able to activate the enzyme by 180% and 162% respectively at a concentration of 2 mM. Other important observations from this work included: (1) the addition of CTP or ATP shifted the cooperative aspartate saturation curve of ATCase to the right and left respectively, without altering the maximal velocity of the enzyme (Fig. 2), (2) the feedback inhibition by CTP and the activation by ATP could be eliminated without loss of enzyme activity by treatment of the enzyme with mercurial or silver compounds, 0.8 M urea, or by heating the enzyme for 4 min at 60° C, and (3) the activation of the enzyme by ATP could be reversed and completely eliminated by addition of CTP, suggesting that the two nucleotides compete for the same site on the enzyme [6]. Based upon these discoveries Gerhart and Pardee concluded that E. coli ATCase had site(s) on the enzyme involved in feedback inhibition by CTP and stimulation by ATP that were distinct from the active site(s), and the interactions between these sites could be uncoupled [6].
Fig. 1.
Aspartate transcarbamoylase catalyzes the reaction between carbamoyl phosphate (CP) and L-aspartate to form N-carbamoyl-L-aspartate (CA) and inorganic phosphate (Pi). Presumably, the reaction proceeds via a tetrahedral intermediate. The bisubstrate analog N-phosphonacetyl-L-aspartate (PALA), possessing many of the binding loci of the tetrahedral intermediate, is a potent inhibitor of the enzyme.
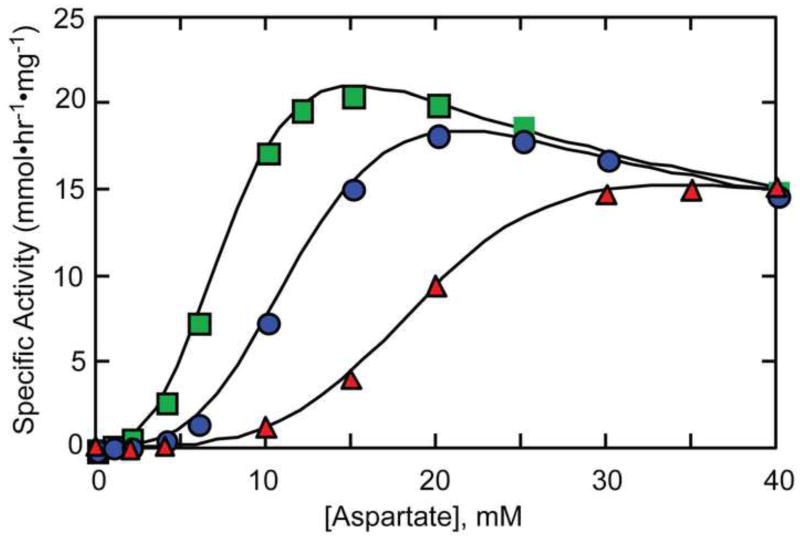
Fig. 2.
Aspartate saturation curves of ATCase in the absence of allosteric effectors (circles), in the presence of 4 mM ATP (squares) and in the presence of 2 mM CTP (triangles). The reactions were performed at 25°C in 0.05 M Tris-acetate buffer pH 8.3. At this pH substrate inhibition is observed at high concentrations of Asp.
In 1965 Gerhart and Schachman [7] showed that the ATCase holoenzyme was composed of two distinct types of subunits. They found that the treatment of the ATCase holoenzyme with mercurials or by heating to 60°C for 4 min was sufficient to breakdown the structure of the enzyme forming two types of subunits. One subunit was able to catalyze the reaction between aspartate and carbamoyl phosphate, not influenced by CTP or ATP (the catalytic subunit), and the other subunit was able to bind the regulatory nucleotides but was devoid of catalytic activity (the regulatory subunit). The dissociation of the enzyme into two distinct subunits provided an explanation for the observed uncoupling of feedback inhibition and stimulation of catalytic activity [7]. This work also confirmed that the catalytic subunit did not exhibit cooperativity as did the ATCase holoenzyme. The exact number of catalytic and regulatory subunits and the number of catalytic and regulatory chains within the enzyme was in question for some time. The quaternary structure was established, by 1968, by combining data on the primary sequence of the regulatory chains with crystallography data indicated that the molecule had both three-fold and two-fold axes of symmetry [8–10]. These data indicated that the E. coli holoenzyme is composed of six catalytic chains arranged into two trimers and six regulatory chains arranged into three dimers. Based upon the amino acid sequence of the catalytic and regulatory chains, the molecular weights of the holoenzyme, catalytic and regulatory chains are 308508, 34296 and 17122 Da respectively.
The importance of the control of a metabolic pathway by a metabolite not resembling the substrate and binding at a distinct site, the allosteric site, had been proposed by Monod, Changeux and Jacob in 1963 [11]. Molecules such as ATP and CTP become allosteric effectors capable of binding to the allosteric site(s) on the enzyme. The mechanism by which the allosteric effectors function is to induce a conformational change in the enzyme (the allosteric transition), which alter the active sites resulting in the observed changes in enzymatic activity.
In the 1965, Monod, Wyman and Changeux [12] proposed a model (the MWC model) to explain not only the cooperative aspartate saturation curve of E. coli ATCase, but also the inhibition and activation of the enzyme by CTP and ATP, respectively. The MWC model proposed that allosteric enzymes exist in an equilibrium between two states, the “tense” or T state and the “relaxed” or R state. The T and R states differ in substrate affinity and/or catalytic activity. Allosteric inhibitors shift the equilibrium towards T state, while allosteric activators shift the equilibrium towards the R state. For ATCase in the absence of ligands, the T to R equilibrium dramatically favors the T state by a factor of 250 or more [13].
ATCase follows an ordered Bi Bi reaction mechanism in which carbamoyl phosphate must bind before L-aspartate and the product N-carbamoyl-L-aspartate leaves the active site before inorganic phosphate [14]. In the presence of CP, the T to R equilibrium is significantly R shifted, although the T state is still in the majority. A shift of a majority of the enzyme to the R state can be induced by the binding of certain combinations of substrates and substrate analogs, the two natural substrates themselves, as well as bisubstrate analogs such as N-phosphonacetyl-L-aspartate (PALA) (see Fig 1).
At low concentrations of Asp and a saturating concentration of CP, PALA can activate ATCase [15]. However, if the concentration of PALA or Asp is increased, inhibition rather than activation is observed. The activation induced by low concentrations of PALA can be explained by the MWC model. At low concentrations of Asp the enzyme remains mainly in the low-activity, low-affinity T state, since the Asp concentration is insufficient to induce the transition to the R state for a majority of the enzyme population. However, the binding of PALA is able to stabilize the enzyme in the high-activity, high-affinity R state because its structure mimics the reaction’s transition state structure. The concerted transition to the R state allows a majority of active sites free to react with substrates and release products while a minority of active sites bound with PALA are inactive but stabilize the enzyme in the R state. Therefore, at low concentrations of PALA the activity increases; however, as the concentration of PALA is increased more and more of the active sites are filled by the non-hydrolyzable bisubstrate analog and the activity drops. At high concentrations of Asp and a saturating concentration of CP, no PALA activation is observed. Under these conditions, the enzyme population is already in the R state so the binding of PALA simply blocks the active site to which it binds and reduces the activity. The allosteric effectors directly alter the affinity of PALA for the enzyme. In the absence of allosteric effectors the average KD of PALA is 110 nM, decreasing to 65 nM in the presence of ATP and increasing to 266 in the presence of CTP [16].
The binding of the allosteric effectors
The earliest studies of the binding of the allosteric effectors to ATCase were performed by Gerhart and Schachman [7], Changeux et al. [17] and Winlund and Chamberlin [18]. The first two reports will not be discussed as the wrong number of subunits and molecular weights were used. In the latter work, the binding of CTP to the E. coli ATCase holoenzyme was measured by equilibrium dialysis. Winlund and Chamberlin found six CTP sites grouped into two classes, differing in affinity by about 50-fold. These experiments were extended to ATP and in the absence and presence of PALA [19]. In the presence of a saturating concentration of PALA, the binding of CTP was reduce by approximately 2 fold and the low affinity sites were barely detectable. ATP bound more weakly to the enzyme than CTP and the difference in affinity between the high and low affinity sites was reduced. However, the binding of ATP was not influenced by the presence of a saturating concentration of PALA. Numerous additional studies of ATP and CTP binding to the enzyme have been performed [20–25]. These studies, under a variety of conditions and using different experimental techniques, are consistent in the asymmetric binding of both CTP and ATP to the allosteric site; furthermore, this asymmetry is also observed for the binding of the allosteric effectors to the isolated regulatory subunits.
In 1989 Wild et al. [26] reported the discovery of a new allosteric inhibitor of E. coli ATCase, UTP. However, UTP only acted as an allosteric inhibitor in the presence of CTP and this synergistic effect was found to be most pronounced at pH 7. In the presence of CTP and UTP, E. coli ATCase was inhibited more than either nucleotide alone. This dual-mode inhibition was metabolically reasonable, since under normal cellular conditions there should not be an excess of UTP relative to CTP, as UTP is converted to CTP in the pyrimidine pathway. These authors state that “the synergistic inhibition of ATCase by both CTP and UTP provides a satisfying logic for ensuring a balance of endogenous pyrimidine nucleotide pools”.
Since the discovery that UTP can inhibit ATCase in the presence of CTP, additional binding studies have been performed [27–29]. There is a reciprocal relationship between the binding of CTP and UTP, CTP enhances the binding affinity for UTP, and UTP enhances the binding affinity for CTP. Furthermore, in the presence of UTP only the three high-affinity CTP sites are observed. In 1991, Zhang and Kantrowitz [27] proposed each regulatory dimer contains one binding site for CTP and one binding site for UTP. In the absence of UTP, the UTP sites exhibited a low affinity for CTP. UTP may be able to bind to the CTP sites but UTP binding to these sites has no influence on activity. The molecular basis for differentiation of these two sites could be due to a pre-existing asymmetry in the allosteric sites on the regulatory dimer or the asymmetry could be induced upon the binding of the first allosteric effector to the regulatory dimer. Details from the X-ray structures in the absence and presence of the nucleotides have provided support for a pre-existing asymmetry in the regulatory dimer (see below).
Quaternary structural changes during the allosteric transition
Upon addition of PALA to the enzyme there are a number of measurable alterations that occur. For example, the sedimentation coefficient decreases by 3.1% [30] as determined by analytical ultracentrifugation, while the radius of gyration increases from 45.8 ± 0.5 Å to 48.4 ± 1 Å [31] as determined by small-angle X-ray scattering (SAXS) in solution. Structural studies of ATCase have been performed by both X-ray crystallography and by SAXS. The structure of ATCase was first solved in 1972 at 5.5 Å by the Lipscomb group [32]. Today there are numerous structures of wild-type E. coli ATCase in the Protein Data Bank (see Table 1). The highest resolution structure of the T-state enzyme is in the presence of CTP (PDB entry 1ZA1, 2.1 Å), while the highest resolution structure of the R-state enzyme is in the presence of PALA (PDB entry 1D09, 2.1 Å).
Table 1.
X-ray structures of wild-type Escherichia coli asspartate transcarbamoylase holoenzyme and catalytic subunit in the Protein Data Bank
| PDB Code | Resolution (Å) | Space Group | Allosteric State | Active Site Ligandsa | Allosteric Site Ligands | Reference |
|---|---|---|---|---|---|---|
| Holoenzyme Structures | ||||||
| 1AT1 | 2.8 | P321 | R | PAM, MAL | [70] | |
| 1D09 | 2.1 | P321 | R | PALA | [35] | |
| 1Q95 | 2.5 | P212121 | R | PALA | [71] | |
| 1R0B | 2.9 | P212121 | R | FLC, Pi | [71] | |
| 1R0C | 2.4 | H3 | T | CA, Pi | [72] | |
| 1RAAc | 2.5 | P321 | T | CTP | [73] | |
| 1ZA1 | 2.2 | P321 | T | CTP | [74] | |
| 1ZA2 | 2.5 | P321 | T | CP | CTP | [74] |
| 2AIR | 2 | H3 | T | CP, AL0 | [75] | |
| 2AT1 | 2.8 | P321 | R | PAM, MAL | [70] | |
| 2ATC | 3 | H32 | T | [76] | ||
| 2FZC | 2.1 | P321 | T | EOP | [77] | |
| 2FZG | 2.3 | P321 | T | EOB | [77] | |
| 2FZK | 2.5 | P321 | T | EOZ | [77] | |
| 2H3E | 2.3 | P321 | R | 6PR | [78] | |
| 2IPO | 2.6 | P321 | R | 1IP | [79] | |
| 3AT1 | 2.8 | P321 | T | PAM | [70] | |
| 4AT1 | 2.6 | P321 | T | ATP | [70] | |
| 5AT1 | 2.6 | P321 | T | CTP | [70] | |
| 6AT1 | 2.5 | P321 | T | [80] | ||
| 7AT1 | 2.8 | P321 | R | PAM, MAL | ATP | [70] |
| 8AT1 | 2.8 | P321 | R | PAM, MAL | CTP | [70] |
| 8ATC | 2.5 | P321 | R | PALA | [33] | |
| Catalytic Subunit Structures | ||||||
| 1EKX | 1.95 | P212121 | PALA | [81] | ||
| 3CSU | 1.88 | P212121 | [82] | |||
Ligands in the active sites. Abbreviations used: PALA, N-phosphonacetyl-L- aspartate; PAM, phosphonoacetamide; MAL, malonate; CP, carbamoyl phosphate; Pi, inorganic phosphate; FLC, citrate; CA, carbamoyl aspartate; AL0, L-alanosine; EOP, (ethane-1,2-diylbis[imino(2-oxoethane-2,1-diyl)]) bis(phosphonic) acid; EOB, (1,3-phenylenebis[imino(2-oxoethane-2,1-diyl)]) bis(phosphonic) acid; EOZ, bis[(phosphonoacetyl)amino]benzoic acid; 6PR,(S)-4-amino-4-oxo-3-(2-phosphonoacetamido)butanoic acid; 1lP, (4S)-2-metyl-2,4-pentanediol.
A comparison of the structure of the ATCase holoenzyme in the absence and presence of PALA provide snapshots of the enzyme in the T- and R-structural states [33]. The changes in the molecular structure between the T and R states include an elongation of 11Å along the molecular three-fold axis. In the transformation between the T and R states there is a rotation of one catalytic trimer relative to the other catalytic trimer by 12°, and a rotation of each of the three regulatory dimers about their approximate two-fold axes by 15°. The allosteric transition of ATCase therefore corresponds to a molecular expansion along the three-fold axis with the above mentioned rotations of the catalytic and regulatory subunits (Fig. 3).
Fig. 3.
Quaternary structure of ATCase in the T (left) and R (right) states with the molecular 3-fold axis vertical (top) and viewed down the molecular 3-fold axis (bottom). The molecule expands 11Å along the 3-fold axis during the allosteric transition. During the T to R transition the regulatory dimers rotate ±6° around their respective 2-fold axes and the catalytic trimer rotate ±7.5° around the 3-fold axis. The catalytic chains are shown in shades of blue and the regulatory chains are shown in yellow and tan.
Interchain and intrachain structural changes during the allosteric transition
During the allosteric transition each of the catalytic chains undergoes domain motions that are coupled to the global quaternary conformation change. Each catalytic chain is composed of two folding domains; the Asp domain, primarily involved in the binding of aspartate and the CP domain, primarily involved in the binding of carbamoyl phosphate. Each regulatory chain is also composed of two folding domains: the Zn domain, primarily involved in the binding of the zinc cofactor, and the Al domain, primarily involved in the binding of allosteric effectors (see Fig. 4). The positions of the domains of the catalytic and regulatory chains undergo structural alterations relative to each other. The two domains of the catalytic chain close by 6.8° while the two domains of the regulatory chain open by 1.7° between the T and R states. The domain motions are also linked to more localized loop motions particularly in the catalytic chains. During the allosteric transition, the 50’s and 80’s loops of the CP domain, and the 240’s loop of the Asp domain undergo dramatic conformational changes helping to create the high-affinity, high-activity form of the enzyme (see Fig. 5). Thus, the allosteric transition involves not only global changes in the relative position of the subunits, but also local conformational changes within the individual chains which directly influence the structure, and therefore the catalytic efficiency of the active sites.
Fig 4.
The secondary structure of one catalytic and one regulatory chain of E. coli ATCase in the R-state. In the regulatory chain the β-sheets are shown in blue and the α-helices in maroon, while in the catalytic chain the β-sheets are shown in maroon and the α-helices in blue. The N- and C-termini of the catalytic and regulatory chains are indicated, as well as the Zn atom in the regulatory chain that is coordinated tetrahydrally to four cysteine residues. Each catalytic chain is composed of an aspartate (Asp) and a carbamoyl phosphate (CP) domain. Each regulatory chain is composed of an allosteric (Al) and a zinc (Zn) domain. CTP is shown as spheres bound to the allosteric site (Al site) in the regulatory chain, and phosphonacetamide and succinate, analogs of CP and Asp, respectively) are shown as spheres bound in the active site of the catalytic chain. This figure was drawn with PyMol [83] using data from PDB entry 8AT1 [84].
Fig. 5.
Structural changes of the α-carbon backbone associated with the binding of PALA to the unligated ATCase. Structure of one catalytic chain of E. coli ATCase along with the 80’s loop from the adjacent chain (80’s, c2). The structure of the unliganded enzyme is shown with blue highlights, while the structure of the ATCase•PALA complex is shown with red highlights, PALA is represented as spheres, and active site residues as sticks. The color gradient corresponds to 40 structures calculated linearly between the two determined X-ray structures, PDB entries 1ZA1 [74] and 1D09 [35].
The active site
The active site of ATCase exists in a number of conformations depending upon ligation. The unligated active site is observed in the T state. This form of the active site is the most open with an active site volume of 1898 Å3. The binding of CP induces only minor alterations in quaternary structure, but major conformational changes of the 50’s and 80’s loops reducing the active site volume to 945 Å3. The enzyme in the presence of PALA or CP and the aspartate analog succinate exhibit the elongated R-state structure. In this conformation the active sites are even more closed, with an active site volume of 541 Å3. The major conformational change that results in the loss of active site volume is the large movement of the 240’s loop (see Fig. 5). The structure of the ATCase•PALA complex has been used to model the structure of the proposed tetrahedral intermediate bound in the active site [34, 35]. The active site cavity in the ATCase•PALA complex is so closed that it shows no exit to solution. Presumably, PALA, and by inference the tetrahedral intermediate, holds the active site closed. Binding energy is lost upon the breakdown of the tetrahedral intermediate into the two products, allowing the active site to open for product release. Since the opening of the active site would first allow carbamoyl aspartate to leave, this also explains the observed ordered release of the products.
The allosteric site
The allosteric binding sites of ATCase are located in the allosteric domain of the regulatory chains (see Fig. 4). The allosteric domain has α/β topology and forms a modified Rossman fold [36]. The allosteric site is composed of four strands of anti-parallel β sheet with one helix bridging the first and three strands (see Fig. 4). The binding of ATP and CTP to the allosteric site involves both ionic and non-polar interactions. Many of the crystal structures of complexes of ATCase with ATP or CTP are in the P321 space group, which provides two independent catalytic and regulatory chains in the asymmetric unit. The interactions between ATP or CTP and the enzyme are not exactly the same in the two independently determined allosteric sites, often referred to as the r1 and r6 sites. Although many of the same residues are involved in the binding of ATP and CTP there are differences as well. Prominent ionic interactions involve the backbone of Ile12, Tyr89, and the side chains of Asp19, Lys60 and Lys94, while Ala11, Val17 and Ile86 are involved in non-polar interactions with the nucleotides (see Fig. 6). Shown in Fig. 7 are the conformational changes that occur locally in the allosteric site when the nucleotides bind. Fig. 7A and B show the molecular surface of the r1 chain in the T state with CTP or ATP bound, respectively, with the atoms of residues involved in allosteric-effector binding used to locally color the surface. The mesh corresponds to the surface as observed in the absence of nucleotides. Clearly, the allosteric site expands near the nucleoside and tightens near the phosphates. This local structural change is manifest in alterations of the interface between the regulatory dimers, which may propagate to changes at the interface between the regulatory and catalytic chains and to slight alterations to the quaternary structure of the enzyme (see below) [37]. Fig. 7C and D also show the surface of the allosteric site with CTP or ATP bound, respectively. However, in these two panels the mesh corresponds to the surface observed with the opposite nucleotide bound. Thus, the alterations in the allosteric site between the CTP and ATP bound forms can easily be discerned.
Fig. 6.
Stereoview of the allosteric site of ATCase in the T state. Superposition of the B chain of the ATCase structures with the allosteric inhibitor CTP (PDB entry 5AT1, green) and the allosteric activator ATP (PDB entry 4AT1, magenta) bound [80]. Interactions involving residues Val9, Ile12 and Tyr89 are from the protein backbone. Not shown are non-polar interactions with Ala11 and Ile86 with CTP and Glu10 and Ala11 with ATP. This figure was drawn using Molscript [85].
Fig. 7.
Surface representation of the allosteric site of ATCase colored by the atoms of the residues which interact with the allosteric effectors CTP and ATP (red, blue and white) showing changes in the surface upon the binding of the allosteric effectors. The binding of CTP (A) or ATP (B) causes the conformational changes from the unliganded surface (mesh) to the allosteric effector bound surface (solid). (C) The ATP-bound surface (mesh) changes to the solid surface when CTP binds. (D) The CTP bound surface (mesh) changes to the solid surface when ATP binds. This figure was drawn using the PDB entries 6AT1 (unliganded), 5AT1 (CTP bound) and 4AT1 (ATP bound) [80] using Chimera [86].
The nucleotides do alter the quaternary conformation of the enzyme, but neither CTP binding to the R-state enzyme nor ATP binding to the T-state enzyme is sufficient to shift the population that crystallizes to the opposite quaternary state. The binding of CTP to the R state does decrease the vertical separation between the catalytic trimers by 0.5 Å, while the binding of ATP to the T state increases the vertical separation by 0.4 Å [37]. Stevens et al. [37] have proposed that the local changes in the allosteric site upon nucleotide binding induce alterations in the regulatory dimer interface that are propagated to the regulatory-catalytic interface resulting in changes in the free energy between the T and R states, and thereby altering the T to R equilibrium [37].
Disruption of allosteric effects
Numerous modifications to the ATCase holoenzyme result in loss of allosteric effects. A few of the most interesting will be mentioned here. Enns and Chan [38] found that if the enzyme was cross-linked with tartaryl diazide in the presence of CP and the aspartate analog succinate, a fully functional enzyme resulted which had no cooperativity, high affinity for aspartate and no allosteric effector response. These authors suggested that the cross-linking locks the enzyme in the R state preventing any allosteric response. This result also agrees with the fact that if the T to R equilibrium is shifted to the R state, by addition of saturating concentrations of substrates, no allosteric response is observed. More recently, the holoenzyme was locked in the R state by creating a disulfide bridge between Cys241 in the upper catalytic chain (c1) with the symmetry related Cys241 in the lower catalytic chain (c4). For this experiment the wild-type residue Ala241 was replaced by cysteine via site-specific mutagenesis [39]. The ATCase holoenyzme with disulfide bridges exhibited no homotropic cooperativity and no response to the allosteric effectors, but exhibited full activity and enhanced substrate affinity. Both SAXS [39] and X-ray crystallography [40] have verified that this disulfide-linked holoenzyme is locked in the R-quaternary structure. Thus the ability of the enzyme to undergo the allosteric transition from the T to the R state is required for the allosteric response.
The regulatory subunits of ATCase, however, are not an absolute requirement for cooperativity. The disulfide-linked holoenzyme, discussed above, can be treated with ρ-hydroxymercuribenzoate to release the regulatory subunits and leave the two catalytic subunits still cross-linked (the c6 enzyme) [41]. The elimination of the structural constraints imposed by the regulatory subunits within the holoenzyme provides increased flexibility to the c6 enzyme, enhancing its activity over the wild-type holoenzyme and catalytic subunit. The covalent linkage between upper and lower catalytic trimers restores homotropic cooperativity so that substrate binding at one or so active sites stimulates binding at the other sites. Reduction of the disulfide bonds in the c6 enzyme resulted in catalytic subunits that displayed kinetic parameters similar to those of wild-type catalytic subunits [41]. The mutation of Arg105 to Ala in the active site of the enzyme produces a catalytic subunit that exhibits cooperativity. In this case the mutation reduces the affinity of the active site for substrates, but as substrates fill one active site, the activity of the other active sites is stimulated [42].
Modifications to the ATCase holoenzyme have also resulted in enzymes with no detectable homotropic cooperativity but still retained allosteric regulation. For example, catalytic chain residue Glu50 (CP domain) interacts with Arg234 (Asp domain) and Arg167 (Asp domain). When Glu50 is mutated to Ala, the resultant enzyme exhibits a 15-fold loss of activity, no cooperativity, but retains both activation by ATP and inhibition by CTP [43]. Furthermore, the allosteric effectors now function both at subsaturating and saturating concentrations of aspartate converting the enzyme from a K to a V type allosteric system [12]. The switch from K to a V type allosteric system suggests that without the domain stabilizing interactions between Glu50 and both Arg167 and Arg234 the mutant enzyme is unable to fully stabilize the R state of the enzyme.
The allosteric transition
Numerous studies on E. coli ATCase have been performed to provide support that the enzyme follows the MWC model. Howlett et al. [13] evaluated the kinetic and physical properties of the enzyme and concluded that they were internally consistent with the MWC model. From these calculations they determined that the [T]/[R] ratio was 250 and that the T state was 3.3 kcal/mol more stable than the R state. The binding of the first substrate in the ordered-binding mechanism, CP, reduced the [T]/[R] ratio to 7, and the combination of CP and ATP reduced the [T]/[R] ratio even further to 2. In the presence of CTP alone, the [T]/[R] ratio was 1250, while the combination of CTP and CP shifted the [T]/[R] ratio to 35 [13]. Small-angle X-ray scattering experiments in solution suggested that CTP and ATP influence activity by changing the affinity of the active sites for the substrates, and only to a small extent by modifying the [T]/[R] ratio [44]. However, neither of these studies have provided formal thermodynamic or structural proof for the existence of an equilibrium between the T and R states of the enzyme. Fetler et al. [45] were able to use a mutant version of ATCase in the absence of ligands to provide direct evidence for a thermodynamic equilibrium between different quaternary structures. The mutation of Asp236 to Ala (D236A) in the catalytic chains of the enzyme weakens a catalytic-regulatory interface interaction destabilizing the T state, and thereby shifting the [T]/[R] ratio to approximately 1. For the D236A enzyme, CTP and Mg-ATP shift the T to R equilibrium towards the T state and the R state respectively. The SAXS curve for the D236A enzyme is temperature dependent, in a reversible manner, indicating an alteration in the [T]/[R] ratio and therefore a dynamic equilibrium between the two states. [45].
Importance of intradomain and intrachain interactions
Site-specific mutagenesis has been used extensively on ATCase to determine the specific function of many of the residues implicated as functionally important in the various X-ray crystal structures of the enzyme [46]. Many of these studies have been directed at determining which of the interactions between subunits and between domains are important for the stabilization of the T and R allosteric states of the enzyme. Based upon these mutagenesis experiments, many of the interactions that stabilize the T and R states have been established. In the more compact T state, stabilizing interactions exist at the c1:c4 interface (and by symmetry c2:c5 and c3:c6) and at the c1:r4 interface (and by symmetry between c2:r5, c3:r6, c4:r1, c5:r2, and c6:r3) (see Fig. 8). The c1:c4 and the c1:r4 interfaces will be used here as representatives. In the expanded R state, all interactions at the c1:r4 and c1:r4 interfaces are lost, although a number of interactions switch between interchain to intrachain.
Fig. 8.
A schematic representation of the interactions that have been identified as important for the allosteric transition in ATCase by site-specific mutagenesis in the T (A) and R (B) states. For clarity, only one catalytic chain from each of the upper (c1) and lower (c4) catalytic subunits is shown. Because of the molecular 3-fold axis, the various interactions shown here are repeated in the c2/c5, and c3/c6 pairs. In the T state stabilizing interactions exist between the c1 and c4 catalytic chains, and between the c1 catalytic and the r1 regulatory chains. The 240’s loops of c1 and c4 undergo a large alteration in position and change from being side by side in the T state to almost one on top of the other in the R state. In the R state, the interchain interactions between c1 and c4 and between c1 and r4 are lost. All of the interactions indicated have been shown experimentally to stabilize the T or R states of the enzyme. Interactions between chains not involved in allosteric-state stabilization are not shown.
Three sets of interactions have been studied that are particularly important for the stability of the T state: (1) the Glu239c1 interaction with both Lys164c4 and Tyr165c4, (2) the Asp236c1 interaction with Lys143r4, and (3) the Ser238c1 interaction with Asn111r4 (see Fig. 8A). None of these interactions exist in the R state. (see Fig. 8B) However, in the R state, Glu239c1 forms new interchain interactions with Lys164c1 and Tyr165c1, while Asn111r4 forms a new interaction with Glu109c4. In this fashion, T-state interactions are replaced by approximately equally energetic R-state interactions. The D236Ac [47], K143Ar [48], E239Qc [49] and N111Ar [48] mutations all substantially destabilize the T state of the enzyme. Based upon sedimentation velocity [48] and SAXS [50, 51] experiments, these mutant enzymes either shift the T to R equilibrium towards the R state or the mutations result in a new T′ state, which has an intermediate structure between the wild-type T and R states. In the case of the D236A mutant enzyme, where there is direct evidence that the mutation alters the [T]/[R] ratio [45], the binding of CP or phosphonacetamide, an analog of CP, alone is sufficient to convert the enzyme into the R structure [45, 52].
Besides the interchain interactions shown in Fig. 8, and mentioned above, there are also interdomain bridging interactions that stabilize the T and R states of ATCase. These interactions are between the Asp and CP domains of the catalytic chains. As one example, consider Glu50 in the CP domain. In the T state Glu50 forms a salt link with Arg105, holding Arg105 out of the active site. In the R state the link between these two residues is lost and a new interdomain interactions are observed between Glu50 with both Arg167 and Arg234 of the Asp domain. These new interactions are made possible by the reorientation of the 50’s and 240’s loops that occurs during the T to R transition, as well as, a closure of the CP and Asp domains by ~7° [53]. The interactions involving Glu50 not only stabilize the active site in the domain-closed high-activity, high-affinity conformation, but also position the side chains of Arg105 and Arg167 to directly interact with the substrates. The interdomain interactions are more important for R-state than T-state stabilization.
A concerted allosteric transition
There have been numerous studies on the nature of the allosteric transition in ATCase and as to whether or not the transition is concerted [54–57], however, the determination of the actual number of molecules required to induce the allosteric transition has been more elusive. As mentioned above, the E. coli ATCase holoenzyme can be dissociated into its component catalytic and regulatory subunits. Furthermore, purified catalytic and regulatory subunits can be mixed to reconstitute the holoenzyme. The ability of E. coli ATCase to be reconstituted has provided the opportunity to create hybrid versions of the enzyme containing two different catalytic subunits as well as more complex hybrid molecules [58–62]. By addition of a chromatographic handle to the catalytic chains, it has been possible to create hybrids with varying numbers of wild-type and modified chains [63, 64]. Using this technique, Macol et al. [65] constructed a series of modified ATCase holoenzymes that had varying ratios of catalytic chains unable to bind PALA and normal catalytic chains (see Fig. 9). Small-angle X-ray scattering was then used to determine if the binding of PALA to hybrids that had 1 or 2 functional active sites was sufficient to cause the ATCase holoenzyme to shift from the T to the R state [65]. As a control two holoenzymes were prepared in an identical fashion; one with all wild-type catalytic chains and one with all mutant catalytic chains. The wild-type holoenzyme displayed the characteristic scattering pattern of the T state when unligated, and the characteristic scattering pattern of the R state upon the addition of PALA (Fig. 9A). The mutant holoenzyme did not show any change in scattering in the absence or presence of 10 mM PALA (Fig. 9B), confirming the inability of the mutant enzyme to undergo the allosteric transition. Analysis showed that even the hybrid holoenzyme with 1 wild-type and 5 mutant catalytic chains was capable of undergoing the allosteric transition upon PALA binding (Fig. 9C). The SAXS data showed that the binding of one PALA molecule per enzyme (six catalytic sites) was sufficient to cause the T to R transition and agrees with the kinetic experiments of Foote and Schachman [66]. Thus, one molecule of PALA is sufficient to cause the entire ATCase holoenzyme to switch from the T to R the state, a true concerted allosteric transition. Since the binding affinity of PALA is substantially higher than the natural substrates, this experiment does not conclusively prove that one molecule of aspartate in the presence of carbamoyl phosphate can induce the same transition.
Fig. 9.
Scattering curves for the reconstituted ATCase holoenzymes, (A) all six catalytic chains are wild-type (yellow), (B) all six catalytic chains are mutant unable to bind PALA (purple), and (C) a hybrid with only one catalytic chain capable of binding PALA. All holoenzyme species were at a final concentration of 37 mg/ml. Scattering was performed at 25 °C in 40 mM KH2PO4 buffer, pH 7.0 in the absence (●) and presence (○) of 1 mM PALA. The scattering curves are expressed as the scattering vector s (s = (2 sin θ/λ), where 2θ and λ are the scattering angle and the wavelength of the X-ray beam, respectively) [65].
Kinetics of the allosteric transition
The measurable change in the shape of ATCase during the allosteric transition may be the largest of any allosteric enzyme. This shape change is easily measured by SAXS (see Fig. 9A). As synchrotrons have become more powerful and X-ray detectors have become more sensitive, SAXS measurements that previously required minutes have been reduced to milliseconds. These improvements have allowed SAXS to be used as the detector for stopped-flow kinetics equipment (time-resolved SAXS) [67, 68]. Using time-resolved SAXS it has been possible to measure directly the kinetics of the allosteric transition of ATCase with the natural substrates. The first set of experiments were performed at −5°C in buffer with 30% ethylene glycol [56] and later, with more advanced equipment, at temperatures between 4° and 25°C without ethylene glycol [69]. Since the SAXS pattern of the R state of ATCase has a substantially larger integrated intensity than then the SAXS pattern of the T state, the integrated intensity of the SAXS pattern was used to monitor the allosteric transition. When Asp is mixed with the ATCase•CP complex there is a fast change from the T state to the R state within about 50 msec at 4°C (see Fig. 10). Following the conversion of the enzyme to the R state, the majority of the enzyme population remains in the high-activity, high-affinity R state until the substrates start to be depleted (~1.5 sec in Fig. 10), upon which the enzyme population starts to shift back to the T state. During this period, each enzyme molecule could have different combinations of reactants and products in each active site. When the binding energy of the mixture of reactants and products in the active sites of a particular enzyme molecule is insufficient to stabilize the R-state structure, the enzyme relaxes back to the more stable T-state structure (see Fig. 10). The allosteric effectors directly alter the kinetic profile shown in Fig. 10 [69]. An activation energy of 13.0 ± 1.4 kcal/mol was also measured for the allosteric transition from the T to the R states [69]. Analysis of the data for the first 50 msec after mixing the ATCase•CP complex with Asp revealed a biphasic reaction, with the faster phase accounting for about 75% of the intensity change. These results suggest that there may be a metastable intermediate formed during the T to R transition or the structural change may represent a composite of T-state species with different ligation states, each with its own particular rate of transition to the R state.
Fig. 10.
Time evolution of the ATCase structural change at 5°C as determined by time-resolved SAXS. Individual scattering patterns were recorded at 17 millisecond intervals. Scattering intensities were integrated over the s range 0.012–0.024 Å−1 and plotted as a function of time. The integrated intensity of the T-state was normalized to zero. The reaction can be divided into three phases. The initial transition from the T to the R state is observed upon with mixing of Asp with the ATCase•CP complex at time zero (red). The steady-state structural phase (green) is observed when the majority of the enzyme population is in the R state and substrates are in excess, and the relaxation phase (blue) is observed when the enzyme population is shifting back to the T state as substrates are exhausted.
Conclusions
In the T state, E. coli aspartate transcarbamoylase can be considered constrained in its compressed quaternary structure, with an open active site with low catalytic activity and low affinity for substrates. Salt-links between the upper and lower catalytic chains (c1:c4) and between upper catalytic chains and lower regulatory chains (c1:r4) provide stabilization for the T-state structure. In the T state the two domains of the active site are in the open conformation with many of the catalytically important sidechains pointing away from the active site. The first substrate to bind in the ordered binding mechanism, CP, alone or in combination with the allosteric activator destabilizes the T state and shifts the [T]/[R] ratio towards the R state. However, the binding of Asp (in the presence of CP) is required to convert most of the population to the R state. Asp binding causes a compression of the CP and Asp domains of the catalytic chains. However, the final formation of the active site must await the repositioning of the 80’s and 240’s loops, which can only occur after the expansion of the enzyme along its three-fold axis, with the resulting formation of the R-state structure. At this stage most of the active sites are in their high-activity, high-affinity conformation ready to catalyze the reaction with the characteristic properties of the R state. As long as sufficient substrates are available, the enzyme remains in the R state, but when the substrates are exhausted there is no longer sufficient binding stabilization to keep the enzyme in the R state and it collapses back to the T state. Although E. coli ATCase is extremely complex, numerous studies over the last 50 years have provided a molecular level understanding of how the allosteric nature of this enzyme regulates pyrimidine nucleotide biosynthesis.
Supplementary Material
Acknowledgments
The work on ATCase by the author was supported by Grant GM26237 from the National Institutes of Health and Grant CHE-0923264 from the National Science Foundation. The author would also like to dedicate this paper to the memory of Prof. William N. Lipscomb and Dr. Hiro Tsuruta who pioneered the studies of ATCase by X-ray crystallography and time-resolved SAXS.
Abbreviations used
- ATCase
E. coli aspartate transcarbamoylase holoenzyme
- CP
carbamoyl phosphate
- Asp
L-aspartic acid
- CA
N-carbamoyl-L-aspartate
- PALA
N-phosphonacetyl-L-aspartate
- c1,c2, c3
the three catalytic chains of ATCase in the upper catalytic trimer
- c4,c5, c6
the three catalytic chains of ATCase in the lower catalytic trimer, with c4 below c1
- r1/r6, r2/r4 and r3/r5
the three regulatory dimers of ATCase
- SAXS
small-angle X-ray scattering
- 50’s loop
a loop in the catalytic chain of ATCase comprised of residues 49–58
- 80’s loop
a loop in the catalytic chain of ATCase comprised of residues 73–88
- 240’s loop
a loop in the catalytic chain of ATCase comprised of residues 230–245
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yates RA, Pardee AB. J Biol Chem. 1956;221:757–770. [PubMed] [Google Scholar]
- 2.Jones ME, Spector L, Lipmann F. J Am Chem Soc. 1955;77:819–820. [Google Scholar]
- 3.Lowenstein JM, Cohen PP. J Biol Chem. 1956;235:57–78. [PubMed] [Google Scholar]
- 4.Reichard P, Hanshoff G. Acta Chem Scand. 1956;10:548–560. [Google Scholar]
- 5.Shepherdson M, Pardee AB. J Biol Chem. 1960;235:3233–3237. [Google Scholar]
- 6.Gerhart JC, Pardee AB. J Biol Chem. 1962;237:891–896. [PubMed] [Google Scholar]
- 7.Gerhart JC, Schachman HK. Biochemistry. 1965;4:1054–1062. doi: 10.1021/bi00882a012. [DOI] [PubMed] [Google Scholar]
- 8.Wiley DC, Lipscomb WN. Nature. 1968;218:1119–1121. doi: 10.1038/2181119a0. [DOI] [PubMed] [Google Scholar]
- 9.Weber K. J Biol Chem. 1968;243:543–546. [PubMed] [Google Scholar]
- 10.Weber KK. Nature. 1968;218:1116–1119. doi: 10.1038/2181116a0. [DOI] [PubMed] [Google Scholar]
- 11.Monod J, Changeux JP, Jacob F. J Mol Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 12.Monod J, Wyman J, Changeux JP. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 13.Howlett GJ, Blackburn MN, Compton JG, Schachman HK. Biochemistry. 1977;16:5091–5099. doi: 10.1021/bi00642a023. [DOI] [PubMed] [Google Scholar]
- 14.Wedler FC, Gasser FJ. Arch Biochem Biophys. 1974;163:57–68. doi: 10.1016/0003-9861(74)90454-8. [DOI] [PubMed] [Google Scholar]
- 15.Collins KD, Stark GR. J Biol Chem. 1971;246:6599–6605. [PubMed] [Google Scholar]
- 16.Newell JA, Markby DW, Schachman HK. J Biol Chem. 1989;264:2476–2481. [PubMed] [Google Scholar]
- 17.Changeux J, Gerhart J, Schachman H. Biochemistry. 1968;7:531–538. doi: 10.1021/bi00842a007. [DOI] [PubMed] [Google Scholar]
- 18.Winlund CC, Chamberlin MJ. Biochem Biophys Res Commun. 1970;40:43–49. doi: 10.1016/0006-291x(70)91043-0. [DOI] [PubMed] [Google Scholar]
- 19.Gray CW, Chamberlin M, Gray D. J Biol Chem. 1973;248:6071–6079. [PubMed] [Google Scholar]
- 20.Suter P, Rosenbusch JP. J Biol Chem. 1976;251:5986–5991. [PubMed] [Google Scholar]
- 21.Suter P, Rosenbusch JP. Eur J Biochem. 1976;70:191–196. doi: 10.1111/j.1432-1033.1976.tb10969.x. [DOI] [PubMed] [Google Scholar]
- 22.Suter P, Rosenbusch JP. J Biol Chem. 1977;252:8136–8141. [PubMed] [Google Scholar]
- 23.Hammes GG, Porter RW, Wu CW. Biochemistry. 1970;9:2992–2994. doi: 10.1021/bi00817a009. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto S, Hammes GG. Biochemistry. 1973;12:1388–1394. doi: 10.1021/bi00731a019. [DOI] [PubMed] [Google Scholar]
- 25.Tondre C, Hammes GG. Biochemistry. 1974;13:3131–3136. doi: 10.1021/bi00712a020. [DOI] [PubMed] [Google Scholar]
- 26.Wild JR, Loughrey-Chen SJ, Corder TS. Proc Natl Acad Sci U S A. 1989;86:46–50. doi: 10.1073/pnas.86.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Kantrowitz ER. J Biol Chem. 1991;266:22154–22158. [PubMed] [Google Scholar]
- 28.England P, Hervé G. Biochemistry. 1992;31:9725–9732. doi: 10.1021/bi00155a028. [DOI] [PubMed] [Google Scholar]
- 29.Mendes KR, Martinez JA, Kantrowitz ER. ACS Chem Biol. 2010;5:499–506. doi: 10.1021/cb9003207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howlett GJ, Schachman HK. Biochemistry. 1977;16:5077–5083. doi: 10.1021/bi00642a021. [DOI] [PubMed] [Google Scholar]
- 31.Moody MF, Vachette P, Foote AM. J Mol Biol. 1979;133:517–532. doi: 10.1016/0022-2836(79)90405-4. [DOI] [PubMed] [Google Scholar]
- 32.Wiley DC, Evans DR, Warren SG, McMurray CH, Edwards BF, Franks WA, Lipscomb WN. Cold Spring Harb Symp Quant Biol. 1972;36:285–290. doi: 10.1101/sqb.1972.036.01.038. [DOI] [PubMed] [Google Scholar]
- 33.Ke HM, Lipscomb WN, Cho Y, Honzatko RB. J Mol Biol. 1988;204:725–747. doi: 10.1016/0022-2836(88)90365-8. [DOI] [PubMed] [Google Scholar]
- 34.Gouaux JE, Krause KL, Lipscomb WN. Biochem Biophys Res Commun. 1987;142:893–897. doi: 10.1016/0006-291x(87)91497-5. [DOI] [PubMed] [Google Scholar]
- 35.Jin L, Stec B, Lipscomb WN, Kantrowitz ER. Proteins: Struct Funct Genet. 1999;37:729–742. [PubMed] [Google Scholar]
- 36.Rao ST, Rossmann MG. Journal of Molecular Biology. 1973;76:241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- 37.Stevens RC, Lipscomb WN. Proc Natl Acad Sci U S A. 1992;89:5281–5285. doi: 10.1073/pnas.89.12.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enns CA, Chan WC. J Biol Chem. 1978;253:2511–2513. [PubMed] [Google Scholar]
- 39.West JM, Tsuruta H, Kantrowitz ER. J Biol Chem. 2002;277:47300–47304. doi: 10.1074/jbc.M209913200. [DOI] [PubMed] [Google Scholar]
- 40.Mendes KR, Kantrowitz ER. J Mol Biol. 2010;401:940–948. doi: 10.1016/j.jmb.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mendes KR, Kantrowitz ER. Biochemistry. 2010;49:7694–7703. doi: 10.1021/bi1010333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stebbins JW, Xu W, Kantrowitz ER. Biochemistry. 1989;28:2592–2600. doi: 10.1021/bi00432a037. [DOI] [PubMed] [Google Scholar]
- 43.Newton CJ, Kantrowitz ER. Biochemistry. 1990;29:1444–1451. doi: 10.1021/bi00458a015. [DOI] [PubMed] [Google Scholar]
- 44.Hervé G, Moody MF, Tauc P, Vachette P, Jones PT. J Mol Biol. 1985;185:189–199. doi: 10.1016/0022-2836(85)90190-1. [DOI] [PubMed] [Google Scholar]
- 45.Fetler L, Kantrowitz ER, Vachette P. Proc Natl Acad Sci U S A. 2007;104:495–500. doi: 10.1073/pnas.0607641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevens RC, Chook YM, Cho CY, Lipscomb WN, Kantrowitz ER. Protein Eng. 1991;4:391–408. doi: 10.1093/protein/4.4.391. [DOI] [PubMed] [Google Scholar]
- 47.Newton CJ, Kantrowitz ER. Proc Natl Acad Sci U S A. 1990;87:2309–2313. doi: 10.1073/pnas.87.6.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eisenstein E, Markby DW, Schachman HK. Biochemistry. 1990;29:3724–3731. doi: 10.1021/bi00467a019. [DOI] [PubMed] [Google Scholar]
- 49.Ladjimi MM, Kantrowitz ER. Biochemistry. 1988;27:276–283. doi: 10.1021/bi00401a042. [DOI] [PubMed] [Google Scholar]
- 50.Tauc P, Vachette P, Middleton SA, Kantrowitz ER. J Mol Biol. 1990;214:327–335. doi: 10.1016/0022-2836(90)90164-H. [DOI] [PubMed] [Google Scholar]
- 51.Chan RS, Sakash JB, Macol CP, West JM, Tsuruta H, Kantrowitz ER. J Biol Chem. 2002;277:49755–49760. doi: 10.1074/jbc.M208919200. [DOI] [PubMed] [Google Scholar]
- 52.Stieglitz KA, Dusinberre KJ, Cardia JP, Tsuruta H, Kantrowitz ER. J Mol Biol. 2005;352:478–486. doi: 10.1016/j.jmb.2005.07.046. [DOI] [PubMed] [Google Scholar]
- 53.Stieglitz K, Stec B, Baker DP, Kantrowitz ER. J Mol Biol. 2004;341:853–868. doi: 10.1016/j.jmb.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 54.Fetler L, Tauc P, Vachette P. J Appl Cryst. 1997;30:781–786. [Google Scholar]
- 55.Tsuruta H, Kihara H, Sano T, Amemiya Y, Vachette P. J Mol Biol. 2005;348:195–204. doi: 10.1016/j.jmb.2005.02.041. [DOI] [PubMed] [Google Scholar]
- 56.Tsuruta H, Vachette P, Sano T, Moody MF, Tauc P, Amemiya Y, Wakabayashi K, Kihara H. Biochemistry. 1994;33:10007–10012. doi: 10.1021/bi00199a026. [DOI] [PubMed] [Google Scholar]
- 57.Werner WE, Schachman HK. J Mol Biol. 1989;206:221–230. doi: 10.1016/0022-2836(89)90535-4. [DOI] [PubMed] [Google Scholar]
- 58.Meighen EA, Pigiet V, Schachman HK. Proc Natl Acad Sci USA. 1970;65:234–241. doi: 10.1073/pnas.65.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang YR, Schachman HK. Proc Natl Acad Sci U S A. 1980;77:5187–5191. doi: 10.1073/pnas.77.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schachman HK, Pauza CD, Navre M, Karels MJ, Wu L, Yang YR. Proc Natl Acad Sci USA. 1984;81:115–119. doi: 10.1073/pnas.81.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robey EA, Schachman HK. Proc Natl Acad Sci USA. 1985;82:361–365. doi: 10.1073/pnas.82.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eisenstein E, Han MS, Woo TS, Ritchey JM, Gibbons I, Yang YR, Schachman HK. J Biol Chem. 1992;267:22148–22155. [PubMed] [Google Scholar]
- 63.Sakash J, Kantrowitz ER. J Biol Chem. 2000;275:28701–28707. doi: 10.1074/jbc.M005079200. [DOI] [PubMed] [Google Scholar]
- 64.Sakash JB, Chan RS, Tsuruta H, Kantrowitz ER. J Biol Chem. 2000;275:752–758. doi: 10.1074/jbc.275.2.752. [DOI] [PubMed] [Google Scholar]
- 65.Macol CP, Tsuruta H, Stec B, Kantrowitz ER. Nat Struc Biol. 2001;8:423–426. doi: 10.1038/87582. [DOI] [PubMed] [Google Scholar]
- 66.Foote J, Schachman HK. J Mol Biol. 1985;186:175–184. doi: 10.1016/0022-2836(85)90267-0. [DOI] [PubMed] [Google Scholar]
- 67.Tsuruta H, Nagamura T, Kimura K, Igarashi T, Kajita A, Wang ZX, Wakabayashi K, Amemiya Y, Kihara H. Rev Sci Instrum. 1989;60:2356–2358. [Google Scholar]
- 68.Kihara H. J Synchrotron Rad. 1994;1:74–77. doi: 10.1107/S0909049594006618. [DOI] [PubMed] [Google Scholar]
- 69.West JM, Xia J, Tsuruta H, Guo W, O’Day EM, Kantrowitz ER. J Mol Biol. 2008;384:206–218. doi: 10.1016/j.jmb.2008.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gouaux JE, Lipscomb WN. Biochemistry. 1990;29:389–402. doi: 10.1021/bi00454a013. [DOI] [PubMed] [Google Scholar]
- 71.Huang J, Lipscomb WN. Biochemistry. 2004;43:6415–6421. doi: 10.1021/bi030213b. [DOI] [PubMed] [Google Scholar]
- 72.Huang J, Lipscomb WN. Biochemistry. 2004;43:6422–6426. doi: 10.1021/bi0302144. [DOI] [PubMed] [Google Scholar]
- 73.Kosman RP, Gouaux JE, Lipscomb WN. Proteins: Struct Funct Genet. 1993;15:147–176. doi: 10.1002/prot.340150206. [DOI] [PubMed] [Google Scholar]
- 74.Wang J, Stieglitz KA, Cardia JP, Kantrowitz ER. Proc Natl Acad Sci U S A. 2005;102:8881–8886. doi: 10.1073/pnas.0503742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang J, Lipscomb WN. Biochemistry. 2006;45:346–352. doi: 10.1021/bi051543u. [DOI] [PubMed] [Google Scholar]
- 76.Honzatko RB, Crawford JL, Monaco HL, Ladner JE, Edwards BFP, Evans DR, Warren SG, Wiley DC, Ladner RC, Lipscomb WN. J Mol Biol. 1982;160:219–263. doi: 10.1016/0022-2836(82)90175-9. [DOI] [PubMed] [Google Scholar]
- 77.Heng S, Stieglitz KA, Eldo J, Xia J, Cardia JP, Kantrowitz ER. Biochemistry. 2006;45:10062–10071. doi: 10.1021/bi0601095. [DOI] [PubMed] [Google Scholar]
- 78.Eldo J, Cardia JP, O’Day EM, Xia J, Tsuruta H, Kantrowitz ER. J Med Chem. 2006;49:5932–5938. doi: 10.1021/jm0607294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cardia JP, Eldo J, O’Day EM, Kantrowitz ER. 2007 will be resubmitted. [Google Scholar]
- 80.Stevens RC, Gouaux JE, Lipscomb WN. Biochemistry. 1990;29:7691–7701. doi: 10.1021/bi00485a019. [DOI] [PubMed] [Google Scholar]
- 81.Endrizzi JA, Beernink PT, Alber T, Schachman HK. Proc Natl Acad Sci U S A. 2000;97:5077–5082. doi: 10.1073/pnas.090087197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beernink PT, Endrizzi JA, Alber T, Schachman HK. Proc Natl Acad Sci U S A. 1999;96:5388–5393. doi: 10.1073/pnas.96.10.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.DeLano WL. DeLano Scientific LLC. San Carlos, CA, USA: 2002. http://www.pymol.org. [Google Scholar]
- 84.Gouaux JE, Stevens RC, Lipscomb WN. Biochemistry. 1990;29:7702–7715. doi: 10.1021/bi00485a020. [DOI] [PubMed] [Google Scholar]
- 85.Kraulis PJ. J Appl Cryst. 1991;24:946–950. [Google Scholar]
- 86.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.