Abstract
Chronic Hepatitis C virus (HCV) infection has been linked with B cell lymphoproliferative disorders and several autoimmune-related diseases. The mechanisms of how chronic viral infection affects B cell development and predisposes the patients to autoimmune manifestations are poorly understood. In this study, we established an experimental system to probe the B cell responses and characterize the antibodies from chronic-HCV-infected individuals. We identified an unusual polyclonal expansion of the IgM memory B cell subset in some patients. This B cell subset is known to be tightly regulated, and autoreactive cells are eliminated by tolerance mechanisms. Genetic analysis of the immunoglobulin (Ig) heavy chain variable gene (VH) sequences of the expanded cell population showed that the levels of somatic hypermutation (SHM) correlate with the extent of cell expansion in the patients and that the VH genes exhibit signs of antigen-mediated selection. Functional analysis of the cloned B cell receptors demonstrated autoreactivity in some of the expanded IgM memory B cells in the patients which is not found in healthy donors. In summary, this study demonstrated that, in some patients, chronic HCV infection disrupts the tolerance mechanism that normally deletes autoreactive B cells, therefore increasing the risk of developing autoimmune antibodies. Long-term follow-up of this expanded B cell subset within the infected individuals will help determine whether these cells are predictors of more-serious clinical manifestations.
INTRODUCTION
Hepatitis C virus (HCV) is a single-stranded RNA virus that is transmitted predominantly through direct blood contact, such as accidental needle sticks in the health care workplace, blood transfusions, and needle sharing among intravenous drug users (reviewed in reference 33). About 70% of those infected cannot clear the virus and succumb to persistent infection (reviewed in reference 22). Of those, more than half will develop some form of liver disease and ∼2 to 3% will develop liver cancer over a course of 20 to 40 years (4, 17). HCV has also been associated with B cell and antibody abnormalities (non-Hodgkin's lymphoma [NHL] [40]) and a number of autoimmune diseases (mixed cryoglobulinemia [MC] [2], celiac disease [56], Sjögren's syndrome [68], and systemic autoimmune diseases [53]). The contribution of the virus to these diseases is unclear. One hypothesis is that E2, a glycoprotein found on the surface of HCV, binds to CD81 instead of B cell receptors (BCRs) on B cells, stimulating cell proliferation (55) and hypermutation of immunoglobulin (Ig) genes (38). Prolonged stimulation by E2 during chronic infection can eventually lead to some form of B cell lymphoproliferative disorder. It is also thought that HCV itself may provide a chronic antigen stimulus that drives clonal expansion of B cells. In support of this, it was shown that Ig cloned from NHL biopsy specimens bound E2 (50). In addition, Charles et al. demonstrated an antigen-driven expansion of peripheral blood (PB) IgM+ CD27+ B cells that express Ig with rheumatoid factor activity in HCV individuals suffering from MC (12).
Studies investigating PB B cell responses in chronic HCV individuals without NHL or autoimmune diseases are conflicting and have not provided a clear overview on the impact of HCV on B cell development. We are interested in studying this patient cohort, as it may provide a clue to the predisposing factors for the development of disease. Racanelli et al. showed that there was a decrease in PB CD27+ memory B cells in patients with persistent HCV infection and that these cells differentiated into Ig-secreting cells independent of BCR engagement in vitro (51). Yet Fournillier et al. showed that the levels of naïve and memory B cells and their signaling via BCR stimulation were normal in HCV-infected individuals (19). Given the high frequency of detection of autoantibodies in chronic-HCV-infected patients (58), we considered it necessary to revisit this information to identify and define abnormalities in B cell regulation that can contribute to the understanding of how autoantibodies are elicited in chronic viral infection.
Newly generated B cells may undergo one of the two distinct developmental pathways: innate cell differentiation, including B-1 cells that undergo rearrangement of Ig genes in the absence of antigen stimulation and secrete natural antibodies, or adaptive cell differentiation, including B-2 cells that undergo antigen activation and secrete antigen-specific antibodies (21, 75). B-1 cells are primarily of the IgM phenotype of low affinity and have germ line or near-germ line sequences (10). Natural antibodies are known to be poly- and self-reactive and have a broad antibacterial activity in sera (76).
In contrast to B-1 cell development, B-2 cells are tightly regulated by the immune system, whereby the cells must traverse through tolerance checkpoints (42). Any disruption in these checkpoints can be indicated by abnormal levels of B cell subsets and aberrant antibody secretion in the sera. B-2 cell development begins with immature B cells in the bone marrow undergoing primary diversification of their antibodies via generation of different combinations of Ig V, D, and J gene segments [V(D)J recombination] (59). At this developmental checkpoint, a B cell with a self-reactive receptor is either deleted or rescued by receptor editing to acquire a new BCR (23). Selected cells exit into the periphery to become immature transitional (IT) B cells (CD19+ CD10+ CD27− CD21lo IgM+ IgD+ IgG−) (49). These cells comprise about 0.5 to 1% of total PB B cells in healthy individuals, and a proportion of those traverse through an additional tolerance checkpoint and are selected into the naïve compartment (9).
Naïve B cells (CD19+ CD27− IgM+ IgD+) comprise a large proportion, ∼40 to 60%, of the PB B cell compartment and continuously enter and exit secondary lymphoid organs to survey for cognate antigens necessary for activation. Once activated, a second diversification process occurs by generating point mutations in the Ig somatic hypermutation (SHM) to produce high-affinity antibodies (9). This activation can occur in a T-independent (TI) or T-dependent (TD) manner. TI antigen activation is further divided into two types: type I elicits a polyclonal stimulation of B cells via Toll-like receptor engagement, whereas type II elicits antigen-specific responses in which polysaccharides engage the BCRs (47). Marginal-zone (MZ) B cells (CD19+ CD27+ IgM+ IgD+), which reside at the junction of white and red pulp in the spleen and in the epithelium of mucosal lymphoid tissue, take part in type II activation (26). The circulating counterparts of these cells, MZ-like B cells, were initially considered to be IgM memory cells, but new studies clarify that they are a distinct B cell subset (30, 70) and comprise 5 to 10% of PB B cells (32). TD antigen activation occurs in germinal centers (GCs) in which B cells differentiate into either memory cells or long-lived plasma cells (27). Within the memory compartment, there are various subsets, including isotype-switched, IgM, and tissue-like memory. Whether these subsets are distinct lineages or stem from one pathway remains to be determined. Isotype-switched memory cells include cells that are CD27+, lack IgD, and express BCRs that are somatically mutated and switched to the IgG, IgA, or IgE isotype (41). IgM memory cells, first identified by Rajewsky et al. in 1997, also express CD27, lack IgD, and are somatically mutated, albeit to a lower frequency than the switched counterparts (29). The existence of IgM memory was later confirmed by Kuppers et al., who showed that these cells had mutations in the Bcl-6 gene indicating that they had traversed through a germinal center reaction (GCR) and are bona fide TD memory B cells (60). A distinction between marginal-zone-like and IgM memory B cells is highlighted by studies in hyper-IgM syndrome and X-linked lymphoproliferative disease patients who are unable to produce TD responses. These individuals retained somatically mutated MZ-like cells (CD19+ CD27+ IgM+ IgD+) but were unable to produce IgM memory (CD19+ CD27+ IgM+ IgD−) or isotype-switched memory cells (1, 71). IgM memory cells are in a relatively rare B cell subset that normally represents less than 1% of human PB B cells (70).
Finally, Ehrhardt and colleagues recently proposed a new memory B cell subset, tissue-like memory. These cells were first identified in human tonsil tissue, showed signs of antigen activation, did not express CD27, and were not responsive to stimuli (i.e., exhausted) (16). Since their discovery, exhausted B cells were found in the PB of HIV-infected individuals and this population of cells was enriched for HIV-specific responses (45). Taken together, it is thought that their lack of response to stimuli might contribute to the inefficient HIV-specific antibody production in chronically infected patients. The relative levels of this subset are estimated to be 1 to 3% of PB B cells (32).
Since B cell development and activation are highly regulated by the immune system, we hypothesize that HCV infection disrupts these processes in such a way that the risk of the patients in developing autoimmune diseases is increased. To investigate this, we established an experimental system to probe the B cell response and characterize the antibodies from chronic-HCV-infected individuals. We identified an unusual polyclonal expansion of the IgM memory B cell subset in one-third of chronic-HCV-infected patients, and some of these cells broke the tolerance checkpoints and are reactive to self antigens. These cells may be precursors of autoimmune manifestations that resulted from chronic HCV infection.
MATERIALS AND METHODS
Study subjects.
Thirty-eight untreated chronic-HCV-infected patients (11 female, 27 male; mean age, 53.3 years; age range, 27 to 77 years) without B cell lymphoma, MC, or autoimmune disease were enrolled in the study during their routine doctor visit at The Scripps Clinic, La Jolla, CA. All of the HCV-infected individuals tested positive for HCV RNA. For our control group, 37 healthy human volunteers (18 female, 19 male; mean age, 40.1 years; age range, 25 to 66 years) were enrolled at the Normal Blood Donor Center at The Scripps Clinic. All of the healthy donors tested negative for hepatitis B virus, HCV, and HIV. Demographic information (age, sex, and race) for individual healthy donors can be found in Table S1 in the supplemental material. Demographic and clinical information for HCV-infected donors can be found in Table S2 in the supplemental material. This study and the human subject protocol have been approved by the institutional review boards at The Scripps Research Institute (TSRI) and The Scripps Clinic.
PBMC preparation.
Healthy and HCV-infected volunteers donated 50 ml of blood. Plasma and peripheral blood mononuclear cells (PBMCs) were immediately isolated by Histopaque (Sigma, St. Louis, MO) centrifugation. Plasma was harvested from the top layer and stored at −80°C until ready for analysis. PBMCs were harvested from the buffy coat layer, aliquoted at 1 × 108 cells/ml in 10% dimethyl sulfoxide (DMSO) in fetal bovine serum (FBS), and stored at −80°C until ready for analysis.
Flow cytometry analysis.
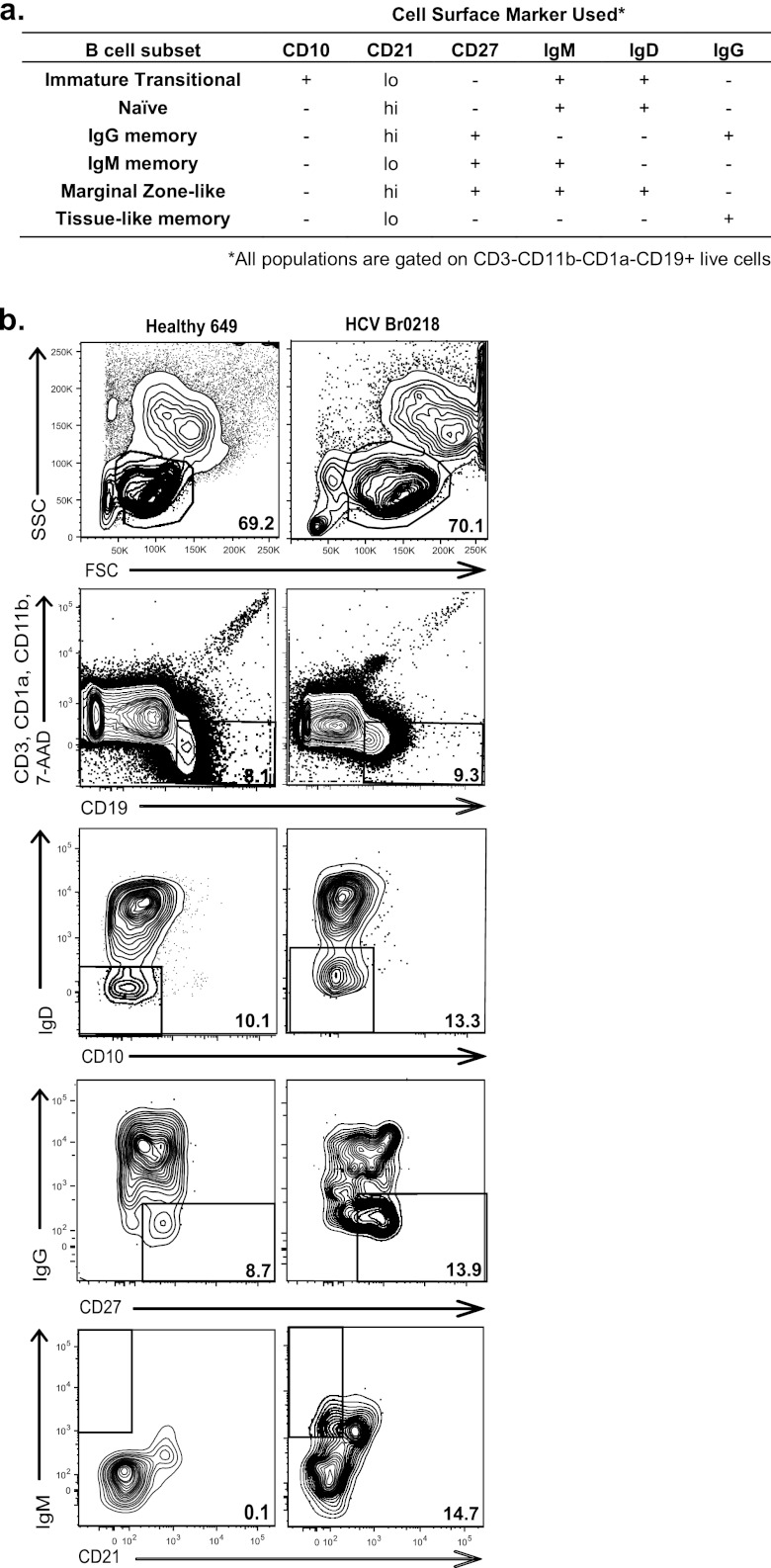
Cells were thawed slowly in prewarmed FBS (average of 75% recovery) and resuspended at 4 × 108 cells/ml in fluorescence-activated cell sorter (FACS) buffer (calcium- and magnesium-free phosphate-buffered saline [PBS], heat-inactivated 1% bovine serum albumin [BSA], 1 mM EDTA). The resuspended cells were aliquoted (25 μl/well, 1 × 107 cells) into a 96-well plate. Cells were blocked with anti-human FcγR antibody (eBioscience, San Diego, CA) for 20 min and then washed and resuspended in FACS buffer. For compensation controls, individual fluorochrome-conjugated antibody was added to each control well. For B cell subset staining, fluorochrome-conjugated antibodies were mixed and then added to each donor sample. The conjugated antibodies used to label surface markers are CD3, CD1a, and CD11b (phycoerythrin [PE]-Cy5), IgD (PE) (BD Biosciences, San Diego, CA), CD19 (Qdot605) (Invitrogen, Carlsbad, CA), CD21 (fluorescein isothiocyanate [FITC]), CD27 (PE-Cy7), IgM (Pacific Blue), CD10 (allophycocyanin [APC]-Cy7) (Biolegend, San Diego, CA), and IgG (Qdot655) (Molecular Probes, Carlsbad, CA). Cell/antibody mixtures were incubated for 15 min at 4°C in the dark. The cells were then washed twice and fixed with 4% paraformaldehyde (to inactivate any residual virus) for 20 min at 4°C in the dark. Cells were washed once and resuspended in 1 ml FACS buffer. Dead cells were eliminated from analysis by labeling with 0.1 μg 7-aminoactinomycin D (7-AAD) (BD Pharmingen, Carlsbad, CA). Samples were analyzed on a BD Digital LSR II flow cytometer or sorted on a FACSAria located at The Scripps Research Institute (TSRI) or the University of California, San Diego, flow cytometry core facility. These cytometers are equipped with a blue, red, violet, and near-UV diode laser base configuration capable of up to 14-color, 16-parameter analysis, 4-way sorting, and digital processing. Data were obtained using DIVA software and analyzed by FloJo software. Live cells were gated by forward (FSC) and side (SSC) scatter; doublets were excluded by FSC-W versus FSC-A and SSC-W versus SSC-A gates. T cells, dendritic cells, and macrophages were eliminated from analysis based on CD3, CD1a, and CD11b expression, respectively. CD19 was used to identify B cells. To further distinguish the B cell subsets, subsequent gating was applied using the presence or absence of the additional cell surface markers shown in Fig. 1 (26, 32, 43, 44, 63, 70).
Fig 1.
Representative scheme of the flow cytometry analysis used in this study. (a) Cell surface markers used to distinguish PB B cell subsets. (b) Flow cytometry analysis of healthy donor 649 (left) and HCV donor Br0218 (right) cells to quantify the percentage of IgM memory B cells in the total live B cells. Numbers are percentages of the indicated gates. Live lymphocytes (FSC versus SSC) per total PBMCs are gated. Total B cells (CD19+ CD3− CD1a− CD11b−) are gated per total live lymphocytes. To further distinguish the IgM memory B cell subset, subsequent gating was applied using the presence or absence of additional cell surface markers as shown.
Ig gene reverse transcription-PCR.
Single CD19+ CD3− CD27+ CD21lo IgM+ IgD− IgG− cells were sorted directly into 96-well plates containing 20 μl/well of catch buffer [4 U of RNasin (Promega, Madison, WI), 5 μl of 5× first-strand buffer (Invitrogen), 1.25 μl of 0.1 M dithiothreitol (DTT) (Invitrogen), and 0.06 ml of 1× Igepal (Sigma)] and immediately frozen on dry ice. Samples were stored at −80°C. cDNA was synthesized by adding 6 μl of master mix (3 μl of random hexamer primers [150 μg/ml] [Genelink, Longwood, FL], 2 μl of deoxynucleoside triphosphate [dNTP] mix, and 50 U of SuperScript III [Invitrogen]) to each well at 42°C for 10 min, 25°C for 10 min, 50°C for 60 min, and 94°C for 5 min. Individual IgH and IgL chain genes were amplified in two successive rounds of PCR (50 cycles each) using primers as described previously (73, 74). Ig genes were amplified from 3.5 μl of cDNA in 40 μl of the total reaction volume with 0.2 μM 5′ and 3′ primers or primer mixes, 312.5 μM concentrations of each dNTP, and 1 U of HotStar Taq DNA polymerase (Qiagen, Valencia, CA). All PCR products were purified (DNA Clean & Concentrator-5 kit; Zymo Research, Irvine, CA) and sequenced by Retrogen, Inc. (San Diego, CA). To identify V, D, and J gene segments and somatic mutations of the genes, the International ImMunoGeneTics information system (www.IMGT.org) was used (35). The net charge in the CDRH3 region was calculated by subtracting the number of negatively charged residues (aspartic acid or glutamic acid) from the number of positively charged residues (arginine, lysine, or histidine).
Antibody production and purification.
Recombinant antibodies were produced as described previously (73). In brief, 293T human embryonic kidney cells were cultured at 37°C in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% ultralow IgG fetal calf serum (FCS) (Invitrogen) and cotransfected with 12.5 μg of the IgH and IgL expression vectors. The transfected cell supernatant was changed after 4 h of incubation, harvested after 3 days of culture, and stored at −20°C until ready for use.
HCV binding assay.
Huh 7.5.1 clone 2 cells were infected with the JFH1 D193 clone (20) at a multiplicity of infection of 5 and harvested 3 days later for experiments. Cells (5 × 106) were fixed with 4% paraformaldehyde, permeabilized with 1× BD Perm/Wash buffer (BD Biosciences) for 20 min at 4°C in the dark, washed 1 time with FACS buffer, and blocked with 50 μl of BSA. Sample antibody or a positive control was added to each well, and cells were incubated for 20 min on ice in the dark. Cells were washed, and Alexa 488-conjugated anti-human IgG antibody (Invitrogen) was added to each well. Cells were incubated for 20 min on ice in the dark. Samples were run on a BD Digital LSR II flow cytometer located at TSRI flow cytometry core facility. Data were obtained using DIVA software. FloJo software was used for analysis. Live cells were gated by forward and side scatter; doublets were excluded by FSC-W versus FSC-A and SSC-W versus SSC-A gates. Alexa 488-positive cells were gated and quantified. A monoclonal HCV-specific antibody, AR3A, was used for a positive control (34).
Double-stranded DNA (dsDNA) enzyme-linked immunosorbent assay (ELISA).
Healthy and HCV-infected human plasma was heat inactivated at 56°C for 30 min and stored at −20°C at a 1:50 dilution in 1% BSA until ready for use. Plates were coated with 50 μl of 2.5-μg/ml salmon sperm overnight at 4°C. Plates were washed with wash buffer (0.02% Tween 20/PBS) twice and then blocked with 50 μl blocking butter (3% BSA/PBS) and incubated. The samples were added to wells (50 μl/well) in triplicate and incubated. Plates were washed three times. Alkaline phosphatase (AP)-conjugated anti-human IgG Fc (diluted 1:1,000) was added to wells and incubated. AP substrate (5 ml AP buffer/tablet; Sigma) was added to each well. Plates were read at an optical density at 405 nm (OD405) before maximal signals of the IgG standard reached 1.5. All incubations were performed for 45 min in a 37°C incubator.
ANA assay.
Anti-nuclear antigen (ANA) test kits were used according to the manufacturer's instructions (Cortez Diagnostics, Calabasas, CA). Samples that produce a binding signal at a 1/100 dilution were considered positive.
ALT assay.
Alanine transaminase (ALT) assays were performed by The Scripps Clinic with a routine hepatic panel assay.
VL determination.
Viral load (VL) in the samples collected for this study was determined using reverse transcription and real-time PCR assays as described previously (7, 28). Briefly, RNA was purified with a QIAamp MinElute virus spin column (Qiagen) and then treated with DNase I (Invitrogen), and cDNA was generated using the SuperScript III first-strand synthesis kit (Invitrogen) according to the manufacturer's recommendations. For the real-time PCR assay, reactions were carried out in 25-μl volumes with 5 μl cDNA and 20 μl RT-PCR master mix (250 nM TaqMan probe [6-FAM-CACCCTATCAGGCAGTACCACAAGGCC-TAMRA], 900 nM forward primer [T-149-F; 5′-TGCGGAACCGGTGAGTACA], 900 nM reverse primer [T149-R; 5′-AGGTTTAGGATTCGTGCTCAT]). A full-genome HCV plasmid (pUC-vJFH) (69) and plasma from healthy donors were used for positive and negative controls, respectively. To quantitate HCV transcript levels in human plasma, dilutions of the HCV plasmid (pUC-vJF) were used in a standard curve (dilutions ranged from 108 to 100 copies of each plasmid). All real-time PCRs were performed on a Bio-Rad iCycler. The protocol was as follows: (i) one cycle of 3 min at 95°C and (ii) 55 cycles of 15 s at 95°C and 1 min at 60°C. The conversion of genome equivalents/ml to IU/ml is 2.5:1.
Statistics.
P values for the flow cytometry B cell subset analysis were calculated using a Mann-Whitney U test. P values for CDRH3 length and positive charges were calculated by a two-tailed Student t test. Statistical analyses were performed using the analysis software within the GraphPad Prism package 5.01.
RESULTS
Expansion of the IgM memory B cell subset in chronic HCV infection.
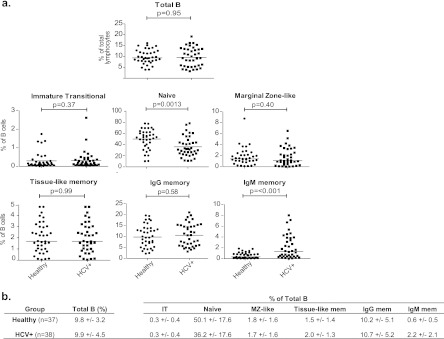
We hypothesized that chronic HCV infection can disrupt B cell regulation and predispose individuals to autoimmune diseases and that the disrupted homeostasis can be detected in PB cells. To investigate this, we first quantified the B cell subsets in healthy and chronic HCV donors by flow cytometry. B cells were distinguished based on expression of the pan-B cell marker CD19, while T cells, dendritic cells, and macrophages were excluded based on expression of CD3, CD1a, or CD11b. Subsequently, the presence or absence of multiple cell surface markers was used to identify different B cell subsets (Fig. 1a) (26, 32, 43, 44, 63, 70). In each individual, the percentage of the parent gate was multiplied to estimate the frequency of each subset in the total B cells. A representative flow cytometry schematic is shown in Fig. 1b. The frequencies of each B cell subset in the healthy (n = 37) and HCV-infected (n = 38) donors were determined and compared by a Mann-Whitney U test. We observed an increase in the IgM memory B cell subset in about one-third of the HCV-infected cohort that was not seen in the healthy donors (Fig. 2a; see also Tables S3 and S4 in the supplemental material). Furthermore, we observed a marked decrease in the naïve B cell population in the HCV-infected cohort. No difference in the frequencies of total B, IT, MZ-like, tissue-like, or IgG memory cells between the two groups was detected (Fig. 2a; see also Tables S3 and S4 in the supplemental material). The mean frequencies of each B cell subset in the two cohorts were summarized in Fig. 2b. The results suggest that chronic HCV infection can disrupt normal B cell homeostasis, specifically in the IgM memory and naïve B cell subsets.
Fig 2.
Frequencies of PB B cell subsets in healthy and HCV-infected cohorts. (a) Mann-Whitney U tests were employed to examine differences in percentages of total B cells of immature transitional (IT), naïve, marginal zone-like (MZ-like), tissue-like memory, IgG memory, and IgM memory B cells in 37 healthy and 38 HCV-infected donors. (b) Summary of mean percentages and standard deviations of B cell subsets. HCV+, HCV positive.
IgM memory B cell antibodies are self-reactive.
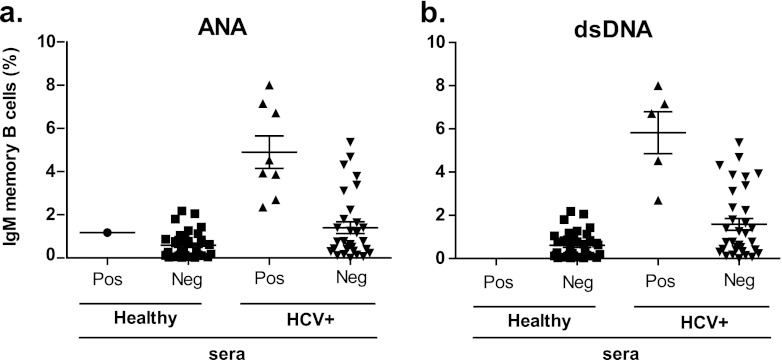
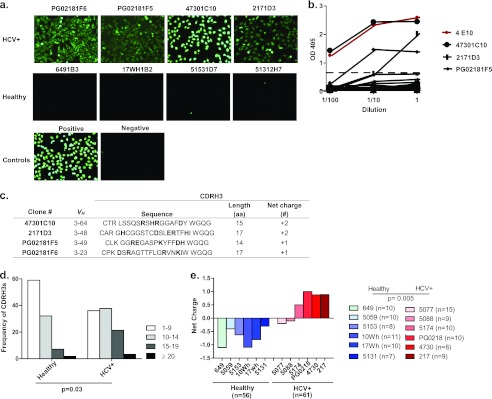
The above-described analysis presented evidence of B cell dysregulation in the IgM memory B compartment in a proportion of chronic-HCV-infected patients. Previous work by Tsuiji et al. showed that the development of this B cell subset is tightly regulated by tolerance mechanisms (65). Therefore, we asked whether this atypical expansion is associated with a break in tolerance allowing autoreactive cells to survive and escape into circulation. To investigate this, we examined the capacity of plasma antibodies of the healthy and HCV-infected cohorts to bind nuclear antigens by performing an immunofluorescence assay (IFA) on fixed Hep-2 cells (31) and dsDNA ELISAs (61). We found the presence of plasma ANAs (Fig. 3a; see also Table S4 in the supplemental material) and anti-dsDNA (Fig. 3b; see also Table S4 in the supplemental material) antibodies in HCV-infected patients with an expansion of this subset. However, there were also some patients with an expansion that did not bind to ANAs or dsDNA (Fig. 3; see also Table S4 in the supplemental material). All healthy individuals except one did not bind ANA or dsDNA (Fig. 3a and 3b; see also Table S3 in the supplemental material). To determine if the anti-ANA and anti-dsDNA antibodies found in the sera could be derived from the IgM memory B cell subset, the BCRs of singly sorted IgM memory B cells from six healthy and six HCV-infected donors were cloned and expressed as soluble antibodies. Four of the six HCV-infected donors (5174, PG0218, 4730, and 217) who had an expansion of the subset (2.4, 2.7, 4.5, and 6.7% of the total B cells, respectively) and who also tested positive for plasma ANA and dsDNA were used for this analysis. The remaining two HCV-infected individuals (5077 and 5088) displayed IgM memory B cell frequencies (0.5 and 0.6% of the total B cells, respectively) similar to those of healthy donors and did not test positive for plasma ANAs or dsDNAs. First, we assayed the antibodies for their capacity to bind nuclear antigens and found that 4/25 (16%) of the antibodies from the infected individuals tested positive whereas none of the antibodies cloned from healthy individuals did (0/25, 0%) (Fig. 4a). We also defined the subcellular staining patterns of these antibodies on the Hep-2 cells. We found that the antibodies predominantly recognized nuclear antigens staining with a homogeneous, speckled, and diffuse grainy pattern (31). This pattern is typically found with antibodies from patients with autoimmune diseases, such as systemic lupus erythematosus and rheumatoid arthritis (RA), indicating that the antibodies may be specific for DNA (3). To investigate this, we tested for reactivity in dsDNA ELISAs and found that 3 out of the 4 antibodies that tested positive for nuclear antigens also tested positive for dsDNA (Fig. 4b). Since autoreactive antibodies possess common genetic features (i.e., long [≥15 residues] and with a positive net charge) in their heavy chain complementarity-determining region 3 (CDRH3) (61, 72), we analyzed this region in the four antibodies that reacted to autoantigens. Indeed, these antibodies possessed long CDRH3s with a net positive charge, confirming that these antibodies possess autoreactive genetic features (Fig. 4c).
Fig 3.
Autoantibodies in plasma of healthy and HCV-infected donors. Plasma from healthy (n = 37) and HCV-infected (n = 38) donors were tested for the presence of ANA antibodies in a Hep-2 IFA stain (a) and anti-dsDNA antibodies in an ELISA (b).
Fig 4.
Autoantibodies in IgM memory B cell subsets. (a) Antibodies cloned from IgM memory B cells in HCV-infected donors that tested positive for nuclear antigens in the Hep-2 IFA stain. Four representative stains from antibodies isolated from healthy individuals are shown. (b) dsDNA ELISA results on IgM memory B cell antibodies from healthy (n = 25) and HCV-infected (n = 25) donors. The ANA and dsDNA reactive antibody 4E10 (red) is used for the positive control. The horizontal dotted line shows the cutoff OD405 value for positive reactivity as determined by a comparison to negative-control plasma from healthy and HCV-infected donors. (c) VH gene usage and CDRH3 characteristics, length (number of amino acids) and net charge (number of positively charged residues minus the number of negatively charged residues) of autoantibodies. CDRH3 regions from IgM memory B cells in healthy (n = 54) and HCV-infected (n = 61) donors were analyzed for length (d) and net charge (e) (healthy, blue shades; HCV+, red shades).
To investigate if these autoreactive genetic features are unique properties of the cloned antibodies or represent the general characteristics of the expanded IgM memory B cell population, we performed analogous CDRH3 analysis of sorted single IgM memory B cells from the healthy and HCV-infected donors. We found a trend of long and positively charged CDRH3s in the expanded IgM memory B cell compartment of the HCV-infected individuals but not in the healthy controls (Fig. 4d; see also Tables S5 and S6 in the supplemental material). Moreover, the HCV-infected individuals without IgM memory B cell expansion did not possess positively charged CDRH3s (Fig. 4e; see also Tables S5 and S6 in the supplemental material). Overall, functional and genetic characterizations of the expanded IgM memory B cells point to the same conclusion that some cells in this subset are autoreactive, resulting from a breakdown of immune tolerance due to chronic HCV infection.
Expanded IgM memory cells are driven by antigen activation.
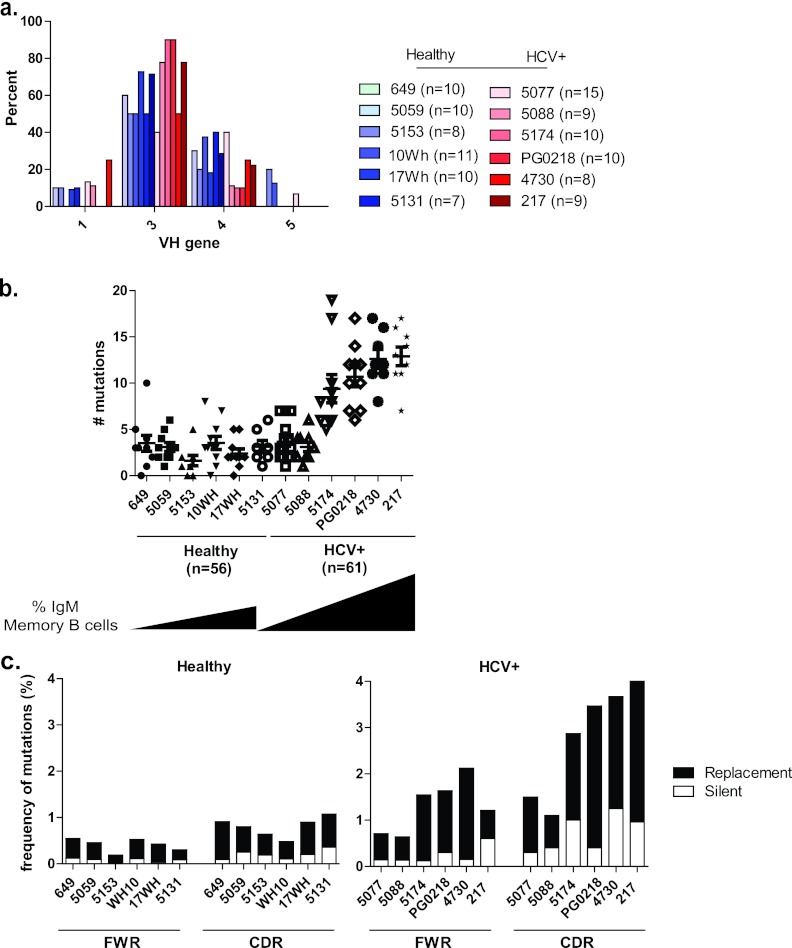
We have found an expanded IgM memory B cell subset that displays properties of autoreactivity in persistently infected individuals. To test if the expansion was due to monoclonal B cell expansion, which may lead to malignancy, or polyclonal expansion due to antigen stimulation, we performed an in-depth genetic analysis of the antibody variable heavy chains (VH) from sorted single IgM memory B cells of six healthy and six HCV-infected donors. The VH regions were sequenced, and the sequences were aligned to their closest germ line counterparts using the online tool of the International ImMunoGeneTics information system (35) to determine VH gene usage and SHM frequency. We found that the responses were polyclonal, utilizing VH gene families equivalent to those of healthy individuals (Fig. 5a; see also Tables S5 and S6 in the supplemental material). We also found that VH genes of the IgM memory B cells in the HCV-infected patients had undergone SHM (Fig. 5b; see also Tables S5 and S6 in the supplemental material). Furthermore, the extent of SHM correlated directly with the magnitude of the IgM memory B cell response. In infected individuals with an expansion of this subset, the number of mutations ranged from 5 to 19 (average, 11.4), compared to a range of 0 to 10 mutations (average, 2.7) in healthy individuals. The two HCV-infected individuals without an expanded IgM memory cell subset displayed slightly higher numbers of mutations (average, 3.4) than did healthy individuals, although the increase is statistically insignificant (P = 0.3) (Fig. 5b; see also Tables S5 and S6 in the supplemental material). Thus, the expanded cells have undergone SHM without isotype switching. Since the expanded population is polyclonal and somatically mutated, this suggests that the expansion is being driven by antigens. On the other hand, monoclonal or oligoclonal expansion suggests proliferative disorder in this subset (12). To address if this is the case, the patterns of mutations in the framework regions (FWRs) and complementarity-determining regions (CDRs) of the VH genes were analyzed. In SHM, although nucleotide substitutions occur in both FWRs and CDRs, an antigen-driven SHM favors mutations in the CDRs (52). Therefore, one would expect mutations to accumulate in the CDRs rather than the FWRs if the expansion of the IgM memory subset was antigen driven. Indeed, a higher frequency of mutations was found in the CDRs than in the FWRs in individuals with an expanded IgM memory B subset (Fig. 5c). In comparison, fewer mutations and a lower CDR-to-FWR ratio were found in the IgM memory B cells of healthy donors (Fig. 5c), confirming an antigen-activated “memory” phenotype for the expanded IgM subset in the HCV-infected individuals. Moreover, in antigen-selected Ig, replacement (R) mutations are overrepresented in CDRs, whereas the opposite is true for silent (S) mutations (6, 52, 62). Typically, a replacement-to-silent (R/S) ratio greater than 3 in the CDRs is consistent with an antigen-driven selection mechanism (62). The R/S ratio in the CDR regions of the expanded IgM memory B cells was greater than 3, consistent with these cells having undergone positive selection (Table 1). Collectively, the expanded populations of IgM memory B cells in the HCV-infected patients display a molecular signature of having undergone SHM through an antigen-mediated selection process.
Fig 5.
IgH gene features of IgM memory B cells. (a) Analysis of antibodies from IgM memory B cells, including the VH gene repertoire in 6 healthy (n = 54, shades of blue) and 6 HCV-infected (n = 61, shades of red) donors. (b) Number of mutations in the VH gene in 6 healthy (n = 54) and 6 HCV-infected (n = 61) donors. The black triangles correspond to the magnitude of the IgM memory B cell response. (c) Frequency of mutations of VH genes as calculated by the number of replacement (R; black bars) and silent (S; white bars) nucleotide exchanges per base pair in FWRs and CDRs in healthy (n = 6) and HCV-infected (n = 6) donors.
Table 1.
Analysis of positive antigen selection of the expanded IgM memory B cell compartment
| HCV-infected donor | Mean R/S ratioa |
|
|---|---|---|
| FWR | CDR | |
| 5174 | 1.6 | 4.7 |
| PG0218 | 4.3 | 4.1 |
| 4730 | 2.0 | 6.1 |
| 217 | 3.7 | 3.9 |
| Avg | 2.9 | 4.7 |
R/S ratios were calculated by dividing the number of R mutations by the number of S mutations in either the FWR1, 2, and 3 or CDR1 and 2 regions. For sequences where the denominator was 0, 1 was used to obtain a ratio value.
In addition to self antigens, we also investigated whether these B cells had been activated by viral antigens. Molecular mimicry of HCV antigens to self antigens may also lead to the production of antiviral B cells that cross-react with self antigens (48). We assayed the cloned BCRs for their capacity to bind to HCV antigens expressed in an infected human liver cell line (Huh 7.5.1). The results show that the expanded cells are not HCV specific or at least are not binding to HCV antigens at a high enough affinity for detection (see Fig. S1 in the supplemental material).
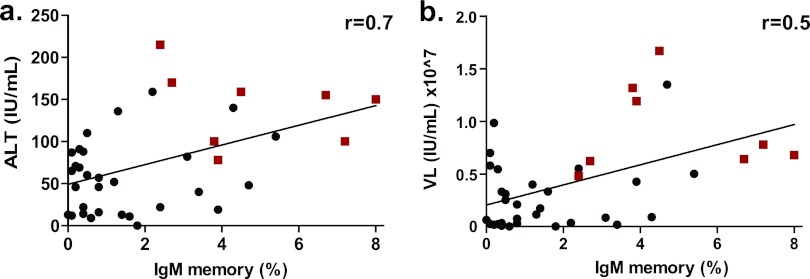
Liver damage and viral load correlation with IgM memory B cell expansion and production of autoantibodies.
The expanded IgM memory B subset in the HCV-infected patients appeared to be antigen activated, with genetic features consistent with autoreactivity, and some of their BCRs bind nuclear antigens. Although none of the patients were suffering from autoimmune-related diseases, some have hepatitis and signs of liver damage (see Table S2 in the supplemental material). We speculated that the extent of this atypical cell expansion was a function of liver damage in the patients. Therefore, we examined the correlation between liver damage, viral load (VL), and the extent of IgM memory B cell expansion and development of autoantibodies. Liver damage was assessed by an ALT test that was given to HCV-infected individuals to monitor their health during routine doctor visits. Plasma viral load was determined by a limiting-dilution real-time RT-PCR. Indeed, an increase in ALT levels (i.e., liver damage) and VL correlated directly with the extent of IgM memory B cell expansion (Fig. 6a and b). Moreover, all the individuals having autoreactive plasma (which included the individuals with expressed autoreactive IgM memory B cells) had an increase in ALT or VL coupled with an expanded IgM memory B cell subset (Fig. 6a and b).
Fig 6.
Correlation analysis of liver damage. IgM memory B cell expansion (%) versus the ALT level (a) and viral load (b) (in-house quantification) are indicated for the HCV-infected cohort (n = 38). Red squares and black circles represent individuals whose plasma tested positive and negative for ANA reactivity, respectively. The correlation coefficient (r) was calculated using Spearman's method.
DISCUSSION
In this study, we have shown that B cell homeostasis and tolerance are disrupted in chronic-HCV-infected individuals, posing a potential risk for the development of autoimmune diseases. Specifically, there is an expansion of IgM memory B cells producing autoreactive antibodies and a decrease in the naïve B cell population that can be detected in PB. While autoreactive antibodies are frequently detected in HCV-infected patients (up to 70% of patients), in most patients, their levels are not high enough to be of significant clinical concern (18). In this study, we focused on the IgM memory B cell subset because previous work by Tsuiji et al. showed that the development of this subset is tightly regulated by tolerance mechanisms such that autoreactive cells are eliminated (65). We investigated the expanded IgM memory cells, and a genetic analysis of their Ig genes showed that these cells are somatically mutated and antigen selected even though they had not undergone isotype switching. This raises the interesting possibility that these cells may have participated in a short-lived GCR given the evidence that the GCR is a dynamic process whereby SHM precedes isotype switching (37). These cells might not have participated in the GCR long enough to switch isotypes. It has been shown that antibodies from memory B cells that are generated from a short-lived GCR display reduced antigen affinity (25). Consistent with this, we found that Igs from the IgM memory B cell subset had relatively low numbers of mutations. Antibodies with low to moderate mutation levels are predominantly of low affinity to the target antigens (52).
Since the cloned BCRs from the IgM memory B cell subset demonstrated autoreactive features, another possibility is that they are derived from ectopic GCRs outside the lymphatic system. These reactions were first identified in patients with autoimmune diseases, including multiple sclerosis (15), Sjögren's syndrome (57), RA (54), and Hashimoto's thyroiditis (5), and they have recently been identified in allografts (13) and cancer (14). They have been associated with the production of autoantibodies (39), and therefore, we speculate that they are also present in chronic HCV infection. Ectopic GCRs arise when normal immune responses fail to clear antigens under chronic stimulation. This can occur if there are insufficient numbers of scavenger cells to clear out large amounts of antigens or the antigen is an integral part of the target tissue (e.g., self antigen). The dynamics of this process have yet to be determined and most likely vary in different clinical states (66). Ectopic GCs are architecturally similar to conventional GCs in that they have clearly defined B and T cell zones and high endothelial venules such that the cells may circulate throughout the periphery (8). Moreover, recent evidence has shown the development of ectopic GCs, consisting of stromal cells and reticular conduits that are essential for the organization of B, T, and antigen-presenting cells, during inflammation in human liver, pancreatic, and kidney tissue (36). It appears that the tolerance checkpoints are not tightly regulated in ectopic GCRs because several studies report the presence of autoantibodies in these structures (24, 57, 77). Since liver damage and viral load correlate with IgM memory B cell expansion and autoantibody production, ectopic GCRs might have developed in the infected individuals due to the continuous activation of B cells driven by chronic liver damage. We propose that the autoreactive IgM memory B cells are generated by ectopic GCRs. In some chronic-HCV-infected patients, there may be an excessive expansion of these cells in ectopic GCRs to a level that can be monitored in peripheral blood samples.
The expanded population of IgM memory cells has a CD21lo phenotype. This raises the possibility that the cells may be anergic, since two groups have independently reported recently that CD27+ CD21lo IgM+ IgD− PB B cells in HCV-infected patients with MC were not responsive to stimuli (11, 64). Notably, a different group reported recently that the identical B cell subset succumbs to B cell anergy regardless of CD21 phenotype (67), suggesting that CD21 may not be an appropriate marker for anergic B cells. Therefore, functional assays to determine the proliferative capability of the expanded subset are needed to determine if these cells are anergic. However, we were not able to determine conclusively the proliferative status of these cells due to the low number of IgM memory B cells obtained per blood sample/individual (∼500 cells). An alternative scenario of low CD21 expression is that these cells could be in a transitional stage differentiating into plasma cells. Moir and colleagues used CD21 as a marker to designate an activated memory B cell that is transitioning to a plasma cell when characterizing B cell perturbations in HIV-infected individuals (43). These CD21lo B cells did not proliferate but did secrete antibody and had plasma-like morphology, suggesting that they were transitioning to plasma cells (46).
Our results differ from previous work showing a decrease in the memory population in HCV-infected individuals (51). The discrepancy may be due to the improvement of B cell subset characterization in this study. On the other hand, Fournillier et al. (19) demonstrated that there was no difference in the frequencies of different B cell subsets in chronic-HCV-infected individuals; however, their cohort size consisted of only nine individuals. Due to variations between individuals, this cohort size might not be adequate to observe a significant difference in this study. With a cohort size quadruple their number (n = 38) and a comparable cohort of healthy donors, we were able to identify statistical differences in the frequencies of B cell subsets.
The direct correlation between the expansion of IgM memory and the extent of SHM frequency suggests that this is an antigen-driven process. A follow-up longitudinal study will help decipher if the linked expansion, SHM, and generation of autoreactive cells are ongoing and associated with the development of autoimmune diseases. This is important because if true, the expansion of this B cell subset may provide a predictive marker for tracking the development of autoimmune diseases, allowing intervention therapies ahead of time.
In conclusion, we find an unusual expansion of IgM memory B cells in HCV-infected individuals and that some of these cells are autoreactive. The data suggest that these cells escape tolerance mechanisms (65) and exit into the peripheral circulation in chronic HCV infection. Long-term follow-up of this expanded B cell subset within the infected individuals will help determine whether these are precursors of more-serious clinical manifestations.
Supplementary Material
ACKNOWLEDGMENTS
We thank all individuals for blood donations. We are grateful to David A. Thorley-Lawson of Tufts University for data analysis and critical reading of the manuscript and Michael Huber of the University of Zurich for help with data analysis and useful discussions. We thank Patrick Wilson of the University of Chicago for the IgG, Igκ, and Igλ expression vectors. We also thank Gregg Silverman of New York University for comments on the manuscript, Takaji Wakita for the pUC-vJFH plasmid, and the following individuals from TSRI for helpful discussions, reagents, and support: Dennis Burton (research resources), Francis Chisari (Huh 7.5.1 clone 2 cells and JFH1 virus clone D193), Michael Zwick (4E10 antibody), Stefan Wieland (for help optimizing the quantitative RT-PCR assay), Tony Cooper and Yuxing Li (for help with the single-cell PCR assay), Colleen Doyle-Cooper and Yi Ting Koh (for help with the dsDNA and ANA assays), and Arthur Kim (for help in PBMC preparation). We thank TSRI and the UC San Diego Center for AIDS Research flow cytometry core facility (supported by NIH grant AI36214) for cell sorting and Mary-Lou Souders and Priscilla Crisler of The Scripps Clinic for blood draws.
This work was supported by TSRI Immunology Training Grant NIH/NIAID T32 A1007244-28 (J.E.R.), NIH grants AI80916 and AI79031 (M.L.), and NIH grant AI71084 (to Dennis Burton, supporting early experiments of this study).
This is TSRI manuscript number 21674.
Footnotes
Published ahead of print 23 May 2012
Supplemental material for this article may be found at http://cvi.asm.org/.
REFERENCES
- 1.Agematsu K, et al. 1998. Absence of IgD-CD27(+) memory B cell population in X-linked hyper-IgM syndrome. J. Exp. Med. 102:853–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agnello V, Chung RT, Kaplan LM. 1992. A role for hepatitis C virus infection in type II cryoglobulinemia. N. Engl. J. Med. 327:1490–1495 [DOI] [PubMed] [Google Scholar]
- 3.Almqvist N, Winkler TH, Martensson IL. 2011. Autoantibodies: focus on anti-DNA antibodies. Self Nonself 2:11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alter HJ, Seeff LB. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17–35 [DOI] [PubMed] [Google Scholar]
- 5.Armengol MP, et al. 2001. Thyroid autoimmune disease: demonstration of thyroid antigen-specific B cells and recombination-activating gene expression in chemokine-containing active intrathyroidal germinal centers. Am. J. Pathol. 159:861–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahler DW, Levy R. 1992. Clonal evolution of a follicular lymphoma: evidence for antigen selection. Proc. Natl. Acad. Sci. U. S. A. 89:6770–6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bissig KD, et al. 2010. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J. Clin. Invest. 120:924–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carragher DM, Rangel-Moreno J, Randall TD. 2008. Ectopic lymphoid tissues and local immunity. Semin. Immunol. 20:26–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carsetti R, Rosado MM, Wardmann H. 2004. Peripheral development of B cells in mouse and man. Immunol. Rev. 197:179–191 [DOI] [PubMed] [Google Scholar]
- 10.Casali P, Notkins AL. 1989. Probing the human B-cell repertoire with EBV: polyreactive antibodies and CD5+ B lymphocytes. Annu. Rev. Immunol. 7:513–535 [DOI] [PubMed] [Google Scholar]
- 11.Charles ED, et al. 2011. Clonal B cells in patients with hepatitis C virus-associated mixed cryoglobulinemia contain an expanded anergic CD21low B-cell subset. Blood 117:5425–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charles ED, et al. 2008. Clonal expansion of immunoglobulin M+CD27+ B cells in HCV-associated mixed cryoglobulinemia. Blood 111:1344–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng J, et al. 2011. Ectopic B-cell clusters that infiltrate transplanted human kidneys are clonal. Proc. Natl. Acad. Sci. U. S. A. 108:5560–5565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coppola D, et al. 2011. Unique ectopic lymph node-like structures present in human primary colorectal carcinoma are identified by immune gene array profiling. Am. J. Pathol. 179:37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corcione A, et al. 2004. Recapitulation of B cell differentiation in the central nervous system of patients with multiple sclerosis. Proc. Natl. Acad. Sci. U. S. A. 101:11064–11069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrhardt GR, et al. 2005. Expression of the immunoregulatory molecule FcRH4 defines a distinctive tissue-based population of memory B cells. J. Exp. Med. 202:783–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fattovich G, Stroffolini T, Zagni I, Donato F. 2004. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology 127:S35–S50 [DOI] [PubMed] [Google Scholar]
- 18.Ferri S, et al. 2008. HCV and autoimmunity. Curr. Pharm. Des. 14:1678–1685 [DOI] [PubMed] [Google Scholar]
- 19.Fournillier A, et al. 2004. Analysis of B-lymphocyte differentiation in patients infected with hepatitis C virus. J. Med. Virol. 72:566–574 [DOI] [PubMed] [Google Scholar]
- 20.Gastaminza P, et al. 2010. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J. Virol. 84:10999–11009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosn EE, Sadate-Ngatchou P, Yang Y, Herzenberg LA. 2011. Distinct progenitors for B-1 and B-2 cells are present in adult mouse spleen. Proc. Natl. Acad. Sci. U. S. A. 108:2879–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guidotti LG, Chisari FV. 2006. Immunobiology and pathogenesis of viral hepatitis. Annu. Rev. Pathol. 1:23–61 [DOI] [PubMed] [Google Scholar]
- 23.Hertz M, Nemazee D. 1998. Receptor editing and commitment in B lymphocytes. Curr. Opin. Immunol. 10:208–213 [DOI] [PubMed] [Google Scholar]
- 24.Humby F, et al. 2009. Ectopic lymphoid structures support ongoing production of class-switched autoantibodies in rheumatoid synovium. PLoS Med. 6:e1 doi:10.1371/journal.pmed.0060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inamine A, et al. 2005. Two waves of memory B-cell generation in the primary immune response. Int. Immunol. 17:581–589 [DOI] [PubMed] [Google Scholar]
- 26.Jackson SM, Wilson PC, James JA, Capra JD. 2008. Human B cell subsets. Adv. Immunol. 98:151–224 [DOI] [PubMed] [Google Scholar]
- 27.Jacob J, Przylepa J, Miller C, Kelsoe G. 1993. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. III. The kinetics of V region mutation and selection in germinal center B cells. J. Exp. Med. 178:1293–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapadia SB, Brideau-Andersen A, Chisari FV. 2003. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. U. S. A. 100:2014–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein U, Kuppers R, Rajewsky K. 1997. Evidence for a large compartment of IgM-expressing memory B cells in humans. Blood 89:1288–1298 [PubMed] [Google Scholar]
- 30.Kruetzmann S, et al. 2003. Human immunoglobulin M memory B cells controlling Streptococcus pneumoniae infections are generated in the spleen. J. Exp. Med. 197:939–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar Y, Bhatia A, Minz RW. 2009. Antinuclear antibodies and their detection methods in diagnosis of connective tissue diseases: a journey revisited. Diagn. Pathol. 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lanzavecchia A, Sallusto F. 2009. Human B cell memory. Curr. Opin. Immunol. 21:298–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lauer GM, Walker BD. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41–52 [DOI] [PubMed] [Google Scholar]
- 34.Law M, et al. 2008. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 14:25–27 [DOI] [PubMed] [Google Scholar]
- 35.Lefranc MP. 2001. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 29:207–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Link A, et al. 2011. Association of T-zone reticular networks and conduits with ectopic lymphoid tissues in mice and humans. Am. J. Pathol. 178:1662–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu YJ, et al. 1996. Within germinal centers, isotype switching of immunoglobulin genes occurs after the onset of somatic mutation. Immunity 4:241–250 [DOI] [PubMed] [Google Scholar]
- 38.Machida K, Cheng KT, Pavio N, Sung VM, Lai MM. 2005. Hepatitis C virus E2-CD81 interaction induces hypermutation of the immunoglobulin gene in B cells. J. Virol. 79:8079–8089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manzo A, Pitzalis C. 2007. Lymphoid tissue reactions in rheumatoid arthritis. Autoimmun. Rev. 7:30–34 [DOI] [PubMed] [Google Scholar]
- 40.Matsuo K, et al. 2004. Effect of hepatitis C virus infection on the risk of non-Hodgkin's lymphoma: a meta-analysis of epidemiological studies. Cancer Sci. 95:745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McHeyzer-Williams LJ, McHeyzer-Williams MG. 2005. Antigen-specific memory B cell development. Annu. Rev. Immunol. 23:487–513 [DOI] [PubMed] [Google Scholar]
- 42.Meffre E, Wardemann H. 2008. B-cell tolerance checkpoints in health and autoimmunity. Curr. Opin. Immunol. 20:632–638 [DOI] [PubMed] [Google Scholar]
- 43.Moir S, et al. 2010. B cells in early and chronic HIV infection: evidence for preservation of immune function associated with early initiation of antiretroviral therapy. Blood 116:5571–5579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moir S, Fauci AS. 2009. B cells in HIV infection and disease. Nat. Rev. Immunol. 9:235–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moir S, et al. 2008. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J. Exp. Med. 205:1797–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moir S, et al. 2001. HIV-1 induces phenotypic and functional perturbations of B cells in chronically infected individuals. Proc. Natl. Acad. Sci. U. S. A. 98:10362–10367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Obukhanych TV, Nussenzweig MC. 2006. T-independent type II immune responses generate memory B cells. J. Exp. Med. 203:305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oldstone MB. 2005. Molecular mimicry, microbial infection, and autoimmune disease: evolution of the concept. Curr. Top. Microbiol. Immunol. 296:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palanichamy A, et al. 2009. Novel human transitional B cell populations revealed by B cell depletion therapy. J. Immunol. 182:5982–5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quinn ER, et al. 2001. The B-cell receptor of a hepatitis C virus (HCV)-associated non-Hodgkin lymphoma binds the viral E2 envelope protein, implicating HCV in lymphomagenesis. Blood 98:3745–3749 [DOI] [PubMed] [Google Scholar]
- 51.Racanelli V, et al. 2006. Antibody production and in vitro behavior of CD27-defined B-cell subsets: persistent hepatitis C virus infection changes the rules. J. Virol. 80:3923–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajewsky K. 1996. Clonal selection and learning in the antibody system. Nature 381:751–758 [DOI] [PubMed] [Google Scholar]
- 53.Ramos-Casals M, et al. 2009. Systemic autoimmune diseases in patients with hepatitis C virus infection: characterization of 1020 cases (The HISPAMEC Registry). J. Rheumatol. 36:1442–1448 [DOI] [PubMed] [Google Scholar]
- 54.Randen I, Mellbye OJ, Forre O, Natvig JB. 1995. The identification of germinal centres and follicular dendritic cell networks in rheumatoid synovial tissue. Scand. J. Immunol. 41:481–486 [DOI] [PubMed] [Google Scholar]
- 55.Rosa D, et al. 2005. Activation of naive B lymphocytes via CD81, a pathogenetic mechanism for hepatitis C virus-associated B lymphocyte disorders. Proc. Natl. Acad. Sci. U. S. A. 102:18544–18549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruggeri C, et al. 2008. Celiac disease and non-organ-specific autoantibodies in patients with chronic hepatitis C virus infection. Dig. Dis. Sci. 53:2151–2155 [DOI] [PubMed] [Google Scholar]
- 57.Salomonsson S, et al. 2003. Cellular basis of ectopic germinal center formation and autoantibody production in the target organ of patients with Sjogren's syndrome. Arthritis Rheum. 48:3187–3201 [DOI] [PubMed] [Google Scholar]
- 58.Sansonno L, et al. 2009. B cells and HCV: an infection model of autoimmunity. Autoimmun. Rev. 9:93–94 [DOI] [PubMed] [Google Scholar]
- 59.Schatz DG. 2004. V(D)J recombination. Immunol. Rev. 200:5–11 [DOI] [PubMed] [Google Scholar]
- 60.Seifert M, Kuppers R. 2009. Molecular footprints of a germinal center derivation of human IgM+(IgD+)CD27+ B cells and the dynamics of memory B cell generation. J. Exp. Med. 206:2659–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shlomchik MJ, Aucoin AH, Pisetsky DS, Weigert MG. 1987. Structure and function of anti-DNA autoantibodies derived from a single autoimmune mouse. Proc. Natl. Acad. Sci. U. S. A. 84:9150–9154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shlomchik MJ, Marshak-Rothstein A, Wolfowicz CB, Rothstein TL, Weigert MG. 1987. The role of clonal selection and somatic mutation in autoimmunity. Nature 328:805–811 [DOI] [PubMed] [Google Scholar]
- 63.Tangye SG, Good KL. 2007. Human IgM+CD27+ B cells: memory B cells or “memory” B cells? J. Immunol. 179:13–19 [DOI] [PubMed] [Google Scholar]
- 64.Terrier B, et al. 2011. Expansion of functionally anergic CD21−/low marginal zone-like B cell clones in hepatitis C virus infection-related autoimmunity. J. Immunol. 187:6550–6563 [DOI] [PubMed] [Google Scholar]
- 65.Tsuiji M, et al. 2006. A checkpoint for autoreactivity in human IgM+ memory B cell development. J. Exp. Med. 203:393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vinuesa CG, Sanz I, Cook MC. 2009. Dysregulation of germinal centres in autoimmune disease. Nat. Rev. Immunol. 9:845–857 [DOI] [PubMed] [Google Scholar]
- 67.Visentini M, et al. 2011. The V(H)1-69-expressing marginal zone B cells expanded in HCV-associated mixed cryoglobulinemia display proliferative anergy irrespective of CD21low phenotype. Blood 118:3440–3442 [DOI] [PubMed] [Google Scholar]
- 68.Vitali C. 2011. Immunopathologic differences of Sjogren's syndrome versus sicca syndrome in HCV and HIV infection. Arthritis Res. Ther. 13:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wakita T, et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weller S, et al. 2004. Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood 104:3647–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weller S, et al. 2001. CD40-CD40L independent Ig gene hypermutation suggests a second B cell diversification pathway in humans. Proc. Natl. Acad. Sci. U. S. A. 98:1166–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winkler TH, Fehr H, Kalden JR. 1992. Analysis of immunoglobulin variable region genes from human IgG anti-DNA hybridomas. Eur. J. Immunol. 22:1719–1728 [DOI] [PubMed] [Google Scholar]
- 73.Wrammert J, et al. 2008. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453:667–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu X, et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoshimoto M, et al. 2011. Embryonic day 9 yolk sac and intra-embryonic hemogenic endothelium independently generate a B-1 and marginal zone progenitor lacking B-2 potential. Proc. Natl. Acad. Sci. U. S. A. 108:1468–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou ZH, Tzioufas AG, Notkins AL. 2007. Properties and function of polyreactive antibodies and polyreactive antigen-binding B cells. J. Autoimmun. 29:219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zuckerman NS, et al. 2010. Somatic hypermutation and antigen-driven selection of B cells are altered in autoimmune diseases. J. Autoimmun. 35:325–335 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.