Abstract
Porcine hemagglutinating encephalomyelitis (PHE) is caused by the coronavirus hemagglutinating encephalomyelitis virus (PHE-CoV), and the recent, rapid spread of PHE-CoV in piglets from many countries emphasizes the urgent need for a PHE-CoV vaccine. Here we use a murine model for evaluation of the induction of humoral and cellular immune responses by inactivated and PHE-CoV DNA vaccines in order to define the immune correlates for protection against PHE-CoV. The inactivated vaccine was composed of purified PHE-CoV and aluminum hydroxide gel (alum), which was chosen as an adjuvant because of its long history of safety for human use. The PHE-CoV DNA vaccine was constructed by subcloning the S1 gene of PHE-CoV into the pVAX1 vector to create the recombinant plasmid pV-S1. Our results showed that the inactivated PHE-CoV vaccine (IPV) elicited a high level of humoral immunity, resulting in good protection efficacy against PHE-CoV challenge. The IPV induced the IgG1 subclass of serum antibodies and expression of the cytokine interleukin-4 (IL-4), suggesting that the IPV generated a predominantly Th2-type immune response. The DNA vaccine was found to mediate primarily a cellular immune response with high levels of IgG2a and the cytokines IL-2 and gamma interferon (IFN-γ). However, mice that were vaccinated twice with the DNA vaccine and boosted with the IPV could mount a sufficient neutralizing antibody response against live PHE-CoV, with little variation in IgG1 and IgG2a levels, and showed high levels of IL-2 and IL-4. This response may activate both B and T cells to mount a specific humoral and cellular immune response that could, in turn, elicit a phagocyte-mediated defense against PHE-CoV infections to achieve viral clearance.
INTRODUCTION
Porcine hemagglutinating encephalomyelitis (PHE) is an acute, highly contagious disease in piglets that is caused by the coronavirus hemagglutinating encephalomyelitis virus (PHE-CoV), which is a member of the Coronaviridae family (6). PHE-CoV infects mainly piglets under the age of 3 weeks and causes vomiting, exhaustion, and obvious neurological symptoms. The mortality rate ranges from 20 to 100% (11). In 1962, the pathogen was isolated for the first time in vivo from breastfeeding pigs suffering from encephalomyelitis in Canada (12). In 1969, an antigenically identical virus was isolated in England from suckling pigs showing anorexia, depression, and vomiting but no clear signs of encephalomyelitis (10). Animals that did not die had stunted growth, and thus, the condition was called “vomiting and wasting disease” (VWD). Mengeling and Cutlip (22) were later able to reproduce both forms of the disease experimentally using the same field isolates. PHE has been reported in all of the major pig-producing countries of Europe, Asia, and North America, where it appears to be endemic with no clinical outbreaks (5, 22). PHE-CoV was first reported in China in 1986; eventually, it occurred both on the mainland and in Taiwan Province (6).
Studies of the chemical composition of PHE-CoV (4, 23) have revealed that it is an RNA virus with five polypeptides, four of which—the nucleocapsid (N), membrane (M), spike (S), and hemagglutinin-esterase (HE) proteins—are glycosylated. The coronavirus S glycoprotein is a major determinant of neurovirulence (16, 34) and is responsible for viral attachment to the cellular receptor and for fusion of the viral and cellular membranes, resulting in virus entry. The S glycoprotein can also induce neutralizing antibodies in vivo or in vitro, as well as cell-mediated immunity (1). Many viral antigens (19), including S1 from transgenic plants, have been demonstrated to be effective at inducing mucosal and serum immune responses in animals. The natural host of PHE-CoV is the pig, but the virus has been adapted experimentally for replication in mice and Wistar rats (36). The virus is neurotropic in mice, but susceptibility was found to be influenced by age and the route of inoculation (37). Under experimental conditions, the disease has been reproduced in most instances following oronasal exposure of nonimmune pigs to PHE-CoV during the first few weeks of life (2). Clinical signs may vary, however. In a study in which the virulence of several PHE-CoV field isolates was compared, symptom severity was related to differences in host susceptibility and the apparent virulence of each isolate (22). In contrast, older pigs and neonatal pigs that had received antibodies in colostrum were usually clinically unaffected when they were exposed to PHE-CoV under otherwise similar conditions (2). Thus, neonatal pigs are usually protected by passively acquired colostral antibodies, and they subsequently develop an age-related resistance to the potential clinical effects of the virus.
Recently, the incidence of PHE among pigs in many countries was found to be on the rise, resulting in great economic losses to the pig industry (33). In August 2006, some of the pig farms in Argentina experienced outbreaks of PHE, leading to 1,226 deaths, morbidity rates as high as 52.6%, and a mortality rate of 16.9% (28). In 2007, PHE outbreaks occurred twice on pig farms in Jilin Province, with incidence rates among 20-day-old piglets as high as 100% and reported mortality rates of 48% and 100% (6). Remarkably, this recent virus isolate showed a high degree of genetic and antigenic homology to the 1962 reference strain PHE-CoV-67N (32). Thus, the pig industry cannot ignore the potential hazards of PHE, and the development of a PHE-CoV vaccine to control PHE-CoV infection will be economically and medically significant.
Following an established vaccine protocol is one of the best ways to shorten the time and minimize the costs of new vaccine development. Currently, developmental methods for inactivated vaccines are well established, and such vaccines exhibit good immunogenicity, high stability, high security, ease of storage, and other benefits (27). DNA vaccine technologies, however, are relatively new and seek to use gene therapy to express protective immunity by inserting original genes into cells to produce antigens through endogenous protein expression; expressed proteins are presented to the immune system, causing long-term and sustained antibody responses and cytotoxic T-lymphocyte (CTL)-based cell immunity (18). In this study, the cDNA of the PHE-CoV S1 gene was directionally subcloned into vector pVAX1 downstream of the cytomegalovirus (CMV) promoter to create a recombinant S1 gene DNA vaccine plasmid. At the same time, PHE-CoVs were inactivated using formaldehyde, and an appropriate amount of alum was added to produce the inactivated PHE-CoV vaccine. To determine the immunogenicities of the inactivated PHE-CoV and PHE-CoV DNA vaccine candidates, we immunized mice intramuscularly with the candidate vaccines and assessed the efficacies of the humoral and cellular immune responses elicited against PHE-CoV infection in the murine model.
MATERIALS AND METHODS
Virus strain.
The virus strain used in this study was PHE-CoV-67N (GenBank accession no. AY078417). The viruses were propagated by passage in porcine kidney epithelial cells (PK-15 cells) (9), purified by sucrose density gradient centrifugation, and stored at −80°C until use.
Preparation of the inactivated PHE-CoV vaccine.
Purified PHE-CoV was inactivated with formaldehyde as described previously (7). The inactivated virus was then diluted in phosphate-buffered saline (PBS) and was mixed with Alhydrogel diluted 9:1 according to the reference protocol in “The Veterinary Biological Products Quality Standard in China” (http://www.moa.gov.cn/zwllm/zcfg) to yield a final concentration of 250 μg/ml virus and 1.7 mg/ml alum. The inactivated PHE-CoV vaccines (IPV) were stored at 4°C until use.
A patent application (K. Chen, F. Gao, W. He, H. Lu, D. Song, W. Gao, C. Zhao, Z. Li, Y. Lan, and D. Zang, 8 May 2012, Chinese patent application 201010165332.7) has been made for the inactivated PHE-CoV vaccines and the method of their preparation.
Preparation of the PHE-CoV DNA vaccine.
The S1 gene was amplified from PHE-CoV genomic RNA and was subcloned into the pVAX1 vector (Invitrogen, Carlsbad, CA) to construct recombinant plasmid pVAX1-S1. PHE-CoV was purified by sucrose density gradient centrifugation, and viral RNA was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA). The S1 gene was amplified using the following oligonucleotide primers: forward, 5′-CGGAATTCGTGCCATCTATTAGCTCTGAAGT-3′; reverse, 5′-TTGCGGCCGCAAGTATGCCCTGGCCTGTAAT G-3′.
Reverse transcription-PCR (RT-PCR) was performed in a total reaction volume of 25 μl using the One-Step RT-PCR kit (TaKaRa, Dalian, China). Reverse transcription was performed at 42°C for 1 h. PCR was performed with an initial denaturation at 96°C for 5 min, followed by 30 cycles at 95°C for 50 s, 50°C for 50 s, and 72°C for 2 min, with a final extension at 72°C for 10 min. The PCR products were then purified and cloned into the pMD18-T vector to create plasmid pTS1. The gene fragment was recovered from pTS1 by double-enzyme cleavage using EcoRI/NotI and was directionally subcloned into pVAX1 downstream of the CMV promoter to create the pV-S1 construct. The accuracy of the constructs was confirmed by restriction enzyme digestion and sequencing. Plasmids were purified using the Gene JET Plasmid Miniprep kit (K0503; MBI Fermentas) and were suspended in Tris-EDTA (TE) buffer to a final concentration of 1 mg/ml. The recombinant strains were screened by PCR, and the expression products were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis (Fig. 1). The results demonstrated the presence of a 73-kDa protein, confirming successful construction of the pVAX1-S1 recombinant plasmid. The PHE-CoV DNA vaccine (pV-S1) was then stored at −80°C until use.
Fig 1.
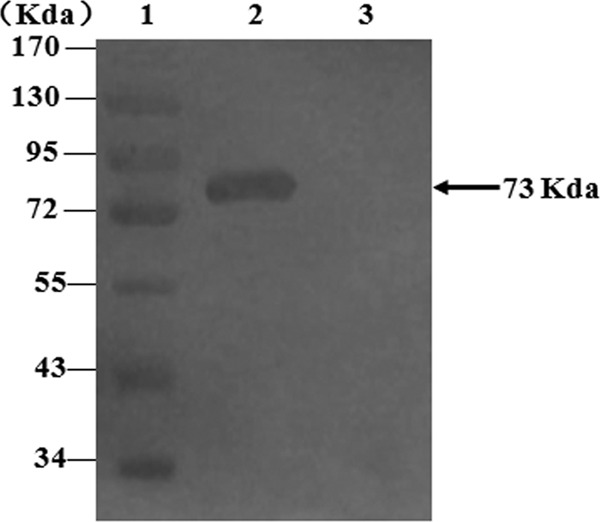
The S1 gene was amplified from PHE-CoV genomic RNA and was subcloned into pVAX1 to construct recombinant plasmid pVAX1-S1. The expression product was analyzed by Western blot analysis. Lane 1, PageRuler prestained protein ladder; lane 2, recombinant plasmid pVAX1-S1; lane 3, plasmid pVAX1.
Mice and immunization.
Female BALB/c mice, 6 to 8 weeks old, were purchased from the Department of Laboratory Animals, General Hospital of Shenyang Military Region. Animals were maintained under pathogen-free conditions and were randomly divided into two groups. Group 1 consisted of mice (10 per vaccine group) that were immunized at 0, 2, and 4 weeks, sacrificed at 6 weeks, and examined for both humoral and cell-mediated immune responses resulting from immunization with the different PHE-CoV vaccines. The mice were immunized intramuscularly either (i) with 0.2 ml of inactivated PHE-CoV (IP), (ii) with 0.2 ml of IPV, (iii) with 0.2 ml of PBS as a negative control, (iv) with 50 μg of pV-S1 (S1), (v) with 0.2 ml of IPV at 0 weeks and with 50 μg of pV-S1 at 2 and 4 weeks (IPV + 2S1), (vi) with 50 μg of pV-S1 at 0 and 2 weeks and with IPV at week 4 (2S1 + IPV), or (vii) with 50 μg of pVAX1. Group 2 consisted of mice (10 per vaccine group) that were immunized as described above but were subsequently challenged with live PHE-CoV at 6 weeks.
VN assay.
The virus neutralization (VN) assay is a more reliable measure than the hemagglutination inhibition (HI) test for the detection of PHE-CoV infection (31). The serum for the VN assay was inactivated at 56°C for 30 min and was diluted with minimum essential medium (MEM). Twofold dilutions of heat-inactivated serum were tested for the presence of antibodies that would neutralize the infectivity of 100 50% tissue culture infective doses (TCID50) of PHE-CoV in PK-15 cell monolayers. The serum dilution that completely inhibited cytopathic effect (CPE) in 50% of the wells was calculated as described previously (29).
Determination of the antigen-specific antibody subclasses using an ELISA.
An enzyme-linked immunosorbent assay (ELISA) was used to determine the subclasses of serum anti-PHE-CoV antibodies at 6 weeks by using previously described methods with some modifications (6). Briefly, the purified PHE-CoV was diluted in carbonate buffer (0.05 M; pH 9.6) to a final concentration of 40 μg/ml, and a volume of 100 μl/well was used to coat 96-well polystyrene microtiter plates, which were then incubated overnight (16 to 18 h) at 4°C. The plates were washed three times with PBS-Tween 20 (PBST), and nonspecific binding sites were blocked with 200 μl/well of 3% (wt/vol) bovine serum albumin (BSA) at 37°C for 2 h. The plates were washed three times; 10 μl of serum and 90 μl of PBS (containing 1% BSA) were added to each well; and the plates were incubated for 1.5 h at 37°C. To detect the serum subclasses, anti-mouse IgG1, IgG2a, IgG2b, IgG3, and IgA antibodies (1:1,000; Sigma) were added to the wells, and the plates were incubated for 1 h at 37°C. After three washes with PBST, the plates were incubated with a peroxidase-conjugated anti-mouse IgG antibody (1:4,000; Sigma). After four washes with PBST, 100 μl/well of o-phenylenediamine (OPD) was added for color development, and then the plates were incubated in the dark at 37°C. The reaction was stopped by the addition of 2 M H2SO4 solution, and the absorbance of each well at 490 nm was measured using an ELX800 Universal microplate reader.
Cellular immune responses of the two PHE-CoV candidate vaccines.
A lymphocyte transformation assay was used to detect the cellular immune responses of the immunized mice that were elicited by the different PHE-CoV vaccines. Single-cell suspensions of splenocytes from the immunized mice were prepared. The mice were killed by cervical dislocation; the spleens were harvested and disrupted; and the splenocytes were added to 10 ml of RPMI 1640 (Gibco, Invitrogen Corporation) and were centrifuged at 1,000 × g for 10 min. The cells were suspended in RPMI 1640 supplemented with 10% fetal calf serum (FCS) at a concentration of 2.5 × 106 cells/ml. Cell suspensions (100 μl) were added to cell culture plates (American, Costar) and were treated with concanavalin A (ConA; 2.5 μg/ml) and inactivated PHE-CoV (final concentration, 1 μg/ml); 100 μl of RPMI 1640 was used as a control. Assays were repeated 3 times for each serum. Plates were incubated at 37°C under 5% CO2 for 48 h. Then 10 μl of 3-(4,5-dimethyl-2-thazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT; 5 mg/ml) was added to each well, and the plates were incubated for another 4 h. Finally, 100 μl of dimethyl sulfoxide (DMSO) was added to each well, and the plates were incubated in the dark at room temperature for 10 min before the optical density at 570 nm (OD570) was measured using an ELX800 Universal microplate reader.
Regional T-cell responses to the two PHE-CoV candidate vaccines.
Splenocyte suspensions were prepared as described above. Cells were diluted with PBS to a concentration of 1 × 107/ml, and then 100-μl aliquots was added to fluorescein isothiocyanate (FITC)-conjugated anti-mouse CD3+, phycoerythrin (PE)/Cy5-conjugated anti-mouse CD8+, and PE-conjugated anti-mouse CD4+ antibodies (BioLegend, San Diego, CA). Cell mixtures were incubated on ice for 30 min, washed twice with PBS (0.01 M; pH 7.4), centrifuged at 1,000 × g for 10 min, and then suspended in 600 μl of PBS, followed by detection using flow cytometry.
Enzyme-linked immunosorbent spot (ELISPOT) assay.
Three mice per treatment were sacrificed; the spleens were collected; single-cell suspensions stimulated with inactivated PHE-CoV were prepared; and levels of the cytokines interleukin-2 (IL-2), IL-4, tumor necrosis factor alpha (TNF-α), and gamma interferon (IFN-γ) were detected by using an ELISA kit (R&D) according to the manufacturer's instructions. Cell culture supernatants were diluted 1:50 in microtiter plates. Then 50 μl of a biotin-labeled antibody was added, and the plates were incubated at 37°C for 90 min. A wash solution (300 μl/well) was added for 1 min, and the plates were washed 4 times. An avidin-horseradish peroxidase (HRP)-labeled antibody (100 μl/well) was added, and the plates were first incubated at 37°C for 30 min and then washed 4 times. Finally, 100 μl/well of the color reagent tetramethyl benzidine (TMB) was added, and the plates were incubated in the dark for 10 min, after which the reaction was stopped by the addition of the stop solution, and the absorbance of each well at 450 nm was measured using an ELX800 Universal microplate reader.
Statistical analysis.
All data are presented as means for the immunized mice in each group ± standard deviations (SD). SPSS, version 13.0 for Windows, was used for statistical analysis. Differences in humoral or cellular immune responses between groups were analyzed using one-way analysis of variance. The least-significant-difference t test was used for between-group comparisons. P values of <0.05 and <0.01 were considered statistically significant.
RESULTS
Neutralizing-antibody responses to different PHE-CoV vaccines.
Blood samples were obtained from the tail veins of the group 1 mice at 2, 4, and 6 weeks, and the sera were collected by centrifugation. The sera were then used for VN detection (Fig. 2). In contrast to the PBS and pVAX1 controls, the group 1 mice vaccinated with different PHE-CoV vaccines demonstrated detectable serum neutralizing antibody titers at 2 weeks. The VN titer was highest, at 29, after three immunizations with IPV, and the addition of alum to the IPV preparation increased the serum neutralizing antibody levels significantly over those with other immunization protocols (P < 0.05). Mice immunized with pV-S1 or IPV + 2S1 showed similar serum neutralizing antibody titers (P > 0.05), but the titers for 2S1 + IPV were higher than those for S1 and IPV + 2S1. No serum neutralizing antibody responses could be detected within the 6-week period in mice immunized with either the PBS or the pVAX1 control.
Fig 2.
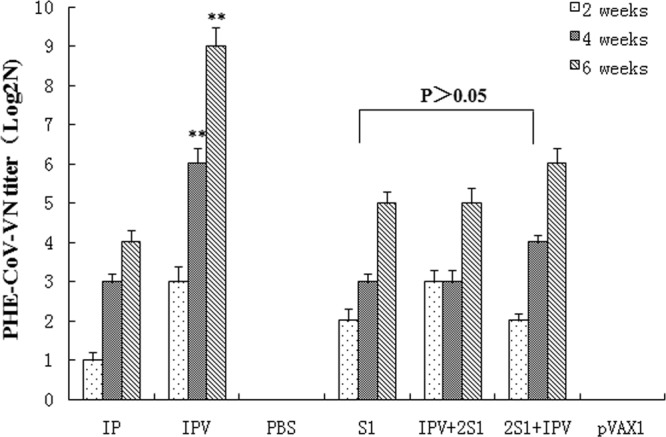
Neutralizing antibody responses to different PHE-CoV vaccines. Blood was obtained from the tail veins of group 1 mice at 2, 4, and 6 weeks, and the sera were used to detect VN antibodies; titers are expressed as log2N and were calculated using the method of Reed and Muench. The VN titers determined at 6 weeks postimmunization indicate no significant differences among the S1, IPV + 2S1, and 2S1 + IPV groups (P > 0.05). However, among the immune vaccine groups, the VN titers were significantly higher in the IPV group than in the other groups (IP, S1, IPV + 2S1, and 2S1 + IPV) (**, P < 0.01).
Subclasses of serum antibodies elicited after immunization with different PHE-CoV vaccines.
The subclasses of serum anti-PHE-CoV antibodies in individual sera collected at 6 weeks were detected by ELISA using anti-mouse IgG1, IgG2a, IgG2b, IgG3, and IgA secondary antibodies (Fig. 3). Interestingly, the level of anti-PHE-CoV IgG2a in mice immunized with IPV was comparable to that in mice immunized with the virion alone, whereas the level of anti-PHE-CoV IgG1 was higher in mice immunized with IPV than in mice immunized with IP without alum (P < 0.05). In contrast, the levels of IgG2a were significantly higher in the pV-S1 and IPV + 2S1 groups, but the 2S1 + IPV group showed little variation in the high levels of IgG1 and IgG2a detected. Notably, the levels of IgG2b, IgG3, and IgA antibodies were low in the sera of mice immunized with the different PHE-CoV vaccines.
Fig 3.
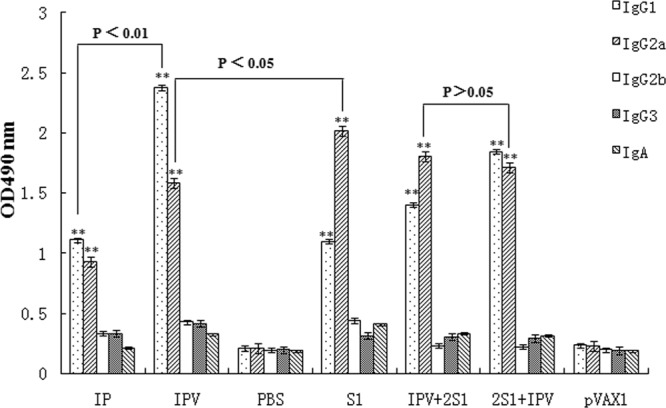
Subclasses of serum antibodies following immunization with different PHE-CoV vaccines. PHE-CoV-specific IgG titers (means ± SD for 10 mice) were detected using ELISA and secondary antibodies (anti-mouse IgG1, IgG2a, IgG2b, IgG3, and IgA) on individual serum samples at 6 weeks. The levels of anti-PHE-CoV IgG2a and IgG1 in mice immunized with any of the five vaccines (IP, IPV, S1, 2S1 + IPV, or IPV + 2S1) were significantly different from those in the control groups (PBS and pVAX1) (**, P < 0.01).
Protective efficacies of PHE-CoV vaccines against challenge with PHE-CoV.
To assess the efficacies of different PHE-CoV vaccines, group 2 mice were immunized and were subsequently challenged with live PHE-CoV at 6 weeks. From the vaccination period to the time of PHE-CoV challenge, the mice appeared normal in activity and appetite and showed no signs of adverse reaction to any of the vaccines at the administration site. Brain samples were collected from mice that were deeply anesthetized and killed 1 week after challenge. The collected tissues were weighed and homogenized in 10 volumes of PBS, and then the tissue suspensions were centrifuged at 7,000 rpm for 10 min. The supernatants were used for RNA extraction and RT-PCR (9), and the results are shown in Table 1. Viral RNA was undetectable in the brain homogenates of animals vaccinated with IPV or 2S1 + IPV, indicating that these two vaccines could effectively reduce infection and death rates for mice. Interestingly, PHE-CoV RNA was still detectable in the brain homogenates of mice vaccinated with S1 or IPV + 2S1, even though the protective effect was the same as that with IPV. For both the PBS and pVAX1 control groups, PHE-CoV RNA was detectable in the brain homogenates of the vaccinated mice, which died 4 days after intranasal challenge with live PHE-CoV.
Table 1.
Protective efficacies of different immunizations against challenge with PHE-CoV
| Vaccine or control | No. (%) of samples positive for virus by RT-PCR | No. of live mice | Protection rate (%) |
|---|---|---|---|
| IP | 6 (60) | 4 | 40 |
| IPV | 0 (0) | 10 | 100 |
| PBS | 10 (100) | 0 | 0 |
| S1 | 2 (20) | 10 | 100 |
| IPV + 2S1 | 1 (10) | 10 | 100 |
| 2S1+IPV | 0 (0) | 10 | 100 |
| pVAX1 | 10 (100) | 0 | 0 |
Cellular immune responses to different PHE-CoV vaccines.
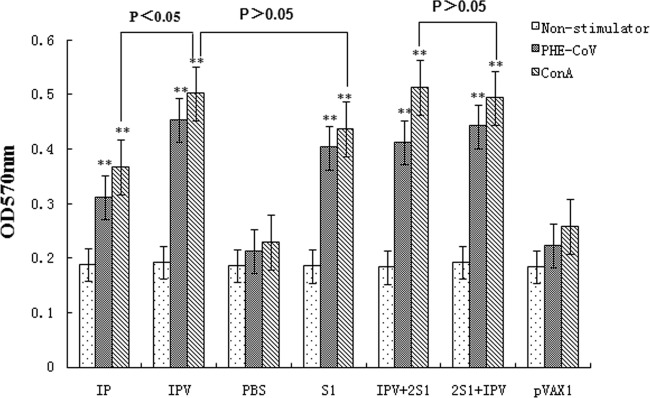
To examine whether the vaccinated mice mounted a T-cell response against PHE-CoV, MTT chronometry and lymphocyte proliferation assays were used. Splenocytes from immune mice were stimulated with PHE-CoV (specific antigen) and ConA (nonspecific antigen). Representative results of the proliferation study are shown in Fig. 4. Compared with the background OD (0.183 ± 0.005) and the OD of the positive control (0.508 ± 0.003), mice immunized with inactivated PHE-CoV plus alum (IPV group) mounted a stronger proliferation response than mice immunized with inactivated PHE-CoV only (IP group) (P < 0.05), indicating that the addition of alum enhances the cellular immune response. However, the OD value for the IPV + 2S1 group was slightly higher than that for the 2S1 + IPV group (P > 0.05), and there were no obvious differences between the OD values of the 2S1 + IPV group and the S1 group (P > 0.05).
Fig 4.
To examine whether vaccinated mice mounted a T-cell response against PHE-CoV, MTT chronometry and lymphocyte proliferation tests were used. Immunized mice were stimulated with PHE-CoV and ConA, and the OD570 of splenocyte proliferation was determined. Compared with the background OD (0.183 ± 0.005) and the OD of the positive control (0.508 ± 0.003). The OD values of the five vaccine groups (IP group, IPV group, S1 group, 2S1+IPV group, and IPV + 2S1 group) showed significant differences (**, P < 0.01). Mice immunized with inactivated PHE-CoV plus alum (the IPV group) induced a stronger proliferation response than mice immunized with inactivated PHE-CoV only (the IP group) (*, P < 0.05). However, the OD value of the 2S1 + IPV group increased slightly compared with that of the IPV + 2S1 group (P > 0.05), and the OD values showed no obvious differences between the IPV + 2S1 group and the S1 group (P > 0.05).
Subsets of specific memory T-cell responses.
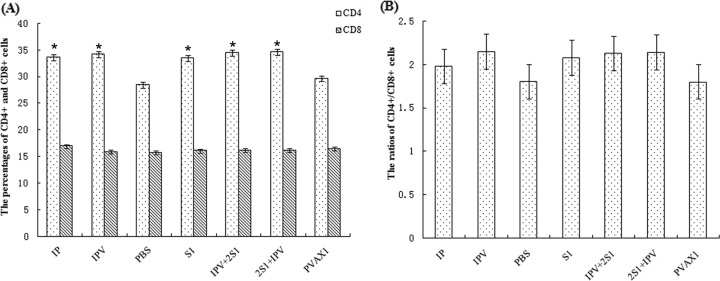
Activated CD4+ and CD8+ T lymphocytes are among the most crucial of the antiviral effectors. To characterize the T-cell response further, we used flow cytometry to examine the CD4+ and CD8+ splenocytes collected from immunized mice. The percentages of CD4+ cells in mice immunized with IPV (inactivated PHE-CoV plus alum), IP (inactivated PHE-CoV alone), S1 (pVAX1-S1), 2S1 + IPV (plasmid pVAX1-S1 and inactivated PHE-CoV plus alum), or IPV + 2S1 (inactivated PHE-CoV plus alum and plasmid pVAX1-S1) increased significantly over those in mice immunized with either the PBS or the pVAX1 control (P < 0.05); the percentages of CD8+ cells, however, were not obviously different for the different immunization groups (P > 0.05) (Fig. 5A). The percentage of activated CD4+ cells in the IPV group was slightly higher than that in the IP group (P > 0.05), indicating that the alum enhanced specific memory T-cell responses. The percentage of activated CD4+ cells in the 2S1 + IPV group was slightly higher than that in the IPV + 2S1 group (P > 0.05). The ratios of CD4+ to CD8+ cells (Fig. 5B) in mice immunized with the vaccines were higher than those in the control groups. These results indicate that the different immunization protocols were capable of inducing an active T-cell immune response in mice.
Fig 5.
The CD4+ and CD8+ splenocytes collected from immunized mice were analyzed using flow cytometry. (A) The percentages of CD4+ cells in mice immunized with IPV, IP, S1, 2S1 + IPV, or IPV + 2S1 were all significantly higher than those for mice immunized with the PBS or pVAX1 control (*, P < 0.05). (B) The CD4+/CD8+ cell ratios were higher in mice immunized with vaccines than in the control groups.
Antigen-specific T-cell cytokine responses.
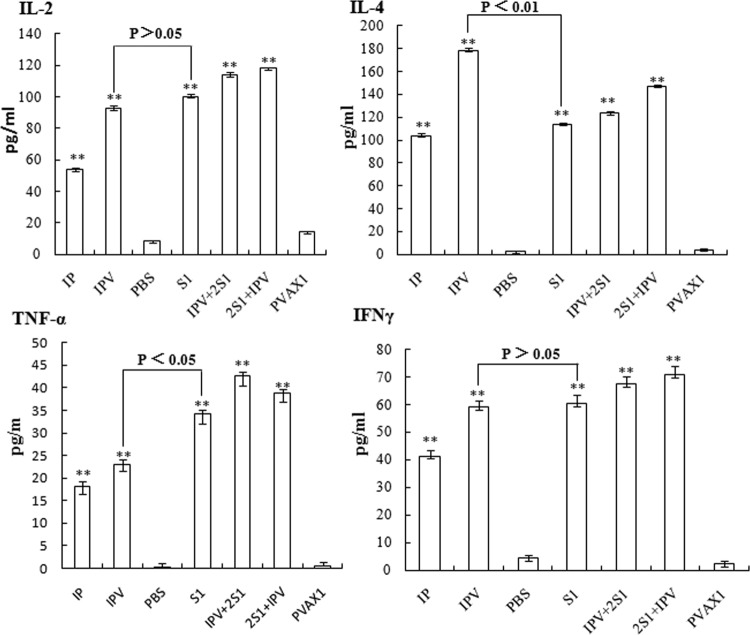
To assess the production of cytokines elicited by the two PHE-CoV vaccine candidates, the frequencies of cells producing IL-2, IL-4, TNF-α, and IFN-γ at the single-cell level were determined using an ELISPOT assay. As shown in Fig. 6, both IPV and S1 could induce the production of all four of the cytokines tested in mice, while the control groups exhibited only low numbers of nonspecific spots. However, the IPV group produced mainly IL-4, at levels higher than those for the IP group. The 2S1 + IPV group showed higher levels of IL-2 and IFN-γ than the other groups and had a high level of IL-4. The levels of TNF-α showed little difference between groups except for the control group, and the IPV + 2S1 group had higher TNF-α levels than the 2S1 + IPV group. IPV + 2S1 may produce Th1- and Th2-dominated immune responses and may elicit a phagocyte-mediated defense against PHE-CoV infections that is involved in the clearance of the virus.
Fig 6.
To assess the production of cytokines elicited by the two PHE-CoV vaccine candidates, the frequencies of cells producing IL-2, IL-4, TNF-α, and IFN-γ at the single-cell level were determined using an ELISA kit. The levels of all of the cytokines (IL-2, IL-4, TNF-α, and IFN-γ) produced by the immunized mice show significant differences between the five vaccine groups (IP, IPV, S1, 2S1 + IPV, and IPV + 2S1) and the control groups (PBS and pVAX1) (**, P < 0.01). The IPV group produced mainly IL-4 and did so at a higher level than the S1 group (**, P < 0.01). However, the levels of IL-2 and IFN-γ were not obviously different for the IPV and S1 groups (P > 0.05).
DISCUSSION
Pigs are the only species known to be naturally susceptible to PHE-CoV infection. Most of the infections in this species are subclinical, and the overall economic impact of the disease is low. Serological surveys have revealed that infection of swine with PHE-CoV is very common and is probably a worldwide occurrence. In fattening pigs, 31% of the sera tested in Canada (2), 46% in Northern Ireland (20), 49% in England (5), 52 to 82% in Japan (14), and 0 to 89% in the United States (21) were positive, depending on the region surveyed. Conversely, Neuvonen et al. (24) found that 40 Finnish elite breeding pig herds were free of seropositive animals. Furthermore, most pigs receive protective antibodies in colostrum; however, piglets without PHE-CoV antibodies, especially specific-pathogen-free (SPF) pigs (30) from breeding farms in several countries, including China, are at greater risk of PHE-CoV infection. Moreover, with the recessive spread of PHE-CoV, the viral genes may mutate to reinforce virulence, and this is another potential threat of PHE-CoV. Animals infected with PHE-CoV appear wasted and stunted, which affects their performance and the quality of their meat. Thus, the potential hazard of this disease for the pig industry cannot be ignored. Currently, there are no reports of a PHE-CoV vaccine in China or elsewhere, although several countries use “subclinical infection” as a means of prevention. In “subclinical infection,” 1-month-antepartum sows are exposed, either by contact with sick pigs, by spray, or by intramuscular injection, to weak strains of PHE-CoV so that the sow can protect the piglet through colostrum antibodies (12, 22). However, this method of inducing immunity is dangerous, since PHE-CoV variants with different levels of virulence could undergo genetic recombination to increase the overall virulence of the virus. Therefore, the development of safe, high-performance vaccines for the prevention and treatment of PHE-CoV has important practical significance.
Here we describe the construction of inactivated and PHE-CoV DNA candidate vaccines and their use in different immunization protocols in a murine model. The immunization protocols were used to evaluate the ability of the vaccines to induce serum neutralizing antibodies and cellular immune responses in order to define correlates of immunity for protection against PHE-CoV. We found that vaccination with IPV induced levels of serum neutralizing antibodies much higher than those for other vaccination groups and that the levels of neutralizing antibodies (VN titers as high as 29) increased with the time of immunization and after three immunizations of the mice. Thus, the addition of alum to the IPV preparation was found to increase the serum VN activity significantly. The aluminum hydroxide adjuvant is still the only adjuvant approved by the FDA for use in human vaccines; its mechanism of action appears to be a storage effect (8). Notably, pV-S1 could induce VN antibodies, and boosters were found to increase the level of antigen presentation in vivo. The combined use of IPV with the DNA vaccine induced even higher levels of serum neutralizing antibodies, but the VN titer in the 2S1 + IPV group was higher than that in the IPV + 2S1 group. Thus, the addition of alum to inactivated PHE-CoV or the combination with S1 immunization increased the time over which the immune animals could effectively stimulate a humoral immune response.
Previous studies have shown that the protective effect of DNA vaccines is not dependent on higher doses but that immune boosters designed to improve the antigen expression level could induce a high level of antibody response and fully effective immune protection (18). In this study, the S1 gene of PHE-CoV was subcloned into the pVAX1 vector, the only eukaryotic expression vector approved for human use by the FDA, to construct the recombinant plasmid pV-S1, which was used as the PHE-CoV DNA vaccine. Mice were given three intramuscular injections of IPV or S1 alone or combined immunizations with IPV + 2S1 or 2S1 + IPV; these immunizations resulted in the induction of serum neutralizing antibodies and a protection efficacy of 100% against PHE-CoV challenge, but the mice did not receive full biological protection, because PHE-CoV could still be detected by RT-PCR in the S1 and IPV + 2S1 groups. The results of subclass serum antibody analysis showed that IPV mediated a primarily IgG1 immune response, whereas S1 mediated an IgG2a response, which suggests that the IPV may induce a predominantly Th2-type immune response, since alum is known to selectively stimulate an IgG1-dominant, type 2 immune response (15).
In addition to humoral immune responses, we also demonstrated the induction of T-cell immune responses in our study. The splenocytes of immune mice were stimulated with PHE-CoV and ConA. The spleen lymphocytes proliferated conspicuously in the S1 and IPV groups, but the proliferation rate was lower than those for the combined immunization groups (the IPV + 2S1 and 2S1 + IPV groups), indicating that IPV and S1 could systematically induce a sufficient cellular immune response following combined immunization to good effect. T lymphocytes are generally divided into cytotoxic T (Tc) cells, suppressor T (Ts) cells, delayed-type hypersensitivity T (TD) cells, and helper/inducer T (Th) cells. The distribution of CD4+ on the Th cell surface can increase the number of memory cells, which respond rapidly upon reexposure to pathogens and thus play a vital role in protection against viral challenge (3). CD8+ is distributed on the surfaces of Ts and Tc cells; following activation, naïve antigen-specific CD8+ T cells are able to proliferate quickly and differentiate into potent effector cells capable of rapid cytokine production and the cytolytic killing of target cells (13). We used flow cytometry to determine the subsets of specific memory T-cell responses found in the immunized mice in our study. The percentages of activated CD3+ CD4+ cells were significantly increased among all immunization groups except the control groups, whereas the 2S1 + IPV group had the highest ratios of CD3+ CD4+ to CD3+ CD8+ cells among all immunization groups. Thus, combined immunization with 2S1 + IPV was successful in inducing cellular immunity, although either IPV or S1 alone could also elicit high regional levels of CD4+ and CD8+ cells in spleen lymphocytes.
Cytokines are generally micromolecular polypeptides that are secreted by the immune cells and that play an important role in the regulation of cell function with respect to cell growth and differentiation, the transmission of information between cells, and the immunological competency of cells involved in specific immune responses (25). Accordingly, the expression of different cytokines divides the T cells into Th1 and Th2 cell subsets. The major cytokines associated with the Th1 cell subsets are IL-2, TNF-α, IFN-γ, and others; the main role of these cytokines is to enhance Tc cellular cytotoxicity and cell-mediated immune responses (35). Th2 cell subsets can secrete IL-4, IL-5, and IL-10; the main role of these cytokines is to promote antibody production and mediate humoral immune responses (17). We determined the frequencies of cells producing IL-2, IL-4, TNF-α, and IFN-γ at the single-cell level by using an ELISPOT assay. The IPV group induced mainly IL-4 and did so at a higher level than the IP group, indicating that the IPV may tend to select T-cell subsets with a Th2 phenotype. S1 immunization and the combined immunizations, 2S1 + IPV and IPV + 2S1, induced mainly IL-2, TNF-α, and IFN-γ, suggesting that S1 selects for a strong Th1-biased cellular immune response and that pV-S1 may play an important role in stimulating Th1-dominated immune responses. Interestingly, the 2S1 + IPV group induced high levels of IL-2 and IL-4, which may produce Th1- and Th2-dominated immune responses that may, in turn, elicit a phagocyte-mediated defense against PHE-CoV infections that is important to viral clearance.
Currently, DNA vaccines are less able than traditional inactivated vaccines to overcome the effects of maternal antibodies so as to induce immune responses (38). Thus, after the birth of young animals, when the maternal antibody levels decrease enough to prevent resistance to viral infection, immunization with DNA vaccines may be an effective preventive measure. Furthermore, both humoral and cellular immune responses are known to contribute to protection against coronavirus infection (26), though T cells may also play a role in exacerbating the pathology of some animal coronavirus infections (35).
In summary, we report the first side-by-side description of two PHE-CoV vaccines evaluated for their abilities to induce immunogenicity. Our data indicate that IPV were more effective than DNA vaccines at inducing humoral immune responses and protection against PHE-CoV. However, mice that were vaccinated twice with DNA vaccines and were then boosted with IPV could activate both B and T cells to induce specific humoral and cellular immune responses with good efficacy against challenge with live PHE-CoV. Thus, combined immunization with DNA and inactivated vaccines could serve as a candidate protocol for the prevention of PHE-CoV. Such direct comparisons of PHE-CoV vaccines in animal models will not only determine which vaccine strategies are more effective but also shorten the time required for the best vaccine candidates to progress into the pig-testing phase.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (no. 31072134 and 31172291) and the Young Scientist Program of the Science and Technology Development Plan in Jilin Province (no. 20090154).
K.C. and K.Z. performed most of the experiments and wrote the manuscript. D.S. and W.H. participated in the project design. W.G., C.Z., L.W., and W.P. performed the MTT assay, ELISPOT assay, and statistical analysis. F.G. and C.W. conceived of the study and participated in its design and coordination. All authors read and approved the final manuscript.
Footnotes
Published ahead of print 18 April 2012
REFERENCES
- 1. Almazan F, DeDiego ML, Galan C, Alvarez E, Enjuanes L. 2006. Identification of essential genes as a strategy to select a SARS candidate vaccine using a SARS-CoV infectious cDNA. Adv. Exp. Med. Biol. 581: 579–583 [PubMed] [Google Scholar]
- 2. Appel M, Greig AS, Corner AH. 1965. Encephalomyelitis of swine caused by a haemagglutinating virus. IV. Transmission studies. Res. Vet. Sci. 6: 482–489 [PubMed] [Google Scholar]
- 3. Bourgeois C, Veiga-Fernandes H, Joret AM, Rocha B, Tanchot C. 2002. CD8 lethargy in the absence of CD4 help. Eur. J. Immunol. 32: 2199–2207 [DOI] [PubMed] [Google Scholar]
- 4. Callebaut PE, Pensaert MB. 1980. Characterization and isolation of structural polypeptides in haemagglutinating encephalomyelitis virus. J. Gen. Virol. 48: 193–204 [DOI] [PubMed] [Google Scholar]
- 5. Cartwright SF, Lucas M, Cavill JP, Gush AF, Blandford TB. 1969. Vomiting and wasting disease of piglets. Vet. Rec. 84: 175–176 [DOI] [PubMed] [Google Scholar]
- 6. Chen K, et al. 2011. Development of an immunochromatographic strip for serological diagnosis of porcine hemagglutinating encephalomyelitis virus. J. Vet. Diagn. Invest. 23: 288–296 [DOI] [PubMed] [Google Scholar]
- 7. Chen KY, et al. 2009. Comparison of different adjuvant inactive vaccines for hemagglutinating encephalomyelitis virus immunizing BALB/c mice. Anim. Husbandry Vet. Med. 6: 34–37 (In Chinese.) [Google Scholar]
- 8. Clements CJ, Griffiths E. 2002. The global impact of vaccines containing aluminium adjuvants. Vaccine 20(Suppl. 3): S24–S33 [DOI] [PubMed] [Google Scholar]
- 9. Gao W, et al. 2011. Vomiting and wasting disease associated with hemagglutinating encephalomyelitis viruses infection in piglets in Jilin, China. Virol. J. 8: 130 doi:10.1186/1743-422X-8-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Greig AS. 1969. Vomiting and wasting disease of piglets. Vet. Rec. 85: 99–100 [DOI] [PubMed] [Google Scholar]
- 11. Greig AS, Girard A. 1969. Serological comparison of hemagglutinating encephalomyelitis viruses isolated from different outbreaks. Can. J. Comp. Med. 33: 25–28 [PMC free article] [PubMed] [Google Scholar]
- 12. Greig AS, et al. 1962. A hemagglutinating virus producing encephalomyelitis in baby pigs. Can. J. Comp. Med. Vet. Sci. 26: 49–56 [PMC free article] [PubMed] [Google Scholar]
- 13. Harty JT, Tvinnereim AR, White DW. 2000. CD8+ T cell effector mechanisms in resistance to infection. Annu. Rev. Immunol. 18: 275–308 [DOI] [PubMed] [Google Scholar]
- 14. Hirai K, Chang CN, Shimakura S. 1974. A serological survey on hemagglutinating encephalomyelitis virus infection in pigs in Japan. Nihon Juigaku Zasshi 36: 375–380 [DOI] [PubMed] [Google Scholar]
- 15. HogenEsch H. 2002. Mechanisms of stimulation of the immune response by aluminum adjuvants. Vaccine 20(Suppl. 3): S34–S39 [DOI] [PubMed] [Google Scholar]
- 16. Iacono KT, Kazi L, Weiss SR. 2006. Both spike and background genes contribute to murine coronavirus neurovirulence. J. Virol. 80: 6834–6843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jordan MB, Mills DM, Kappler J, Marrack P, Cambier JC. 2004. Promotion of B cell immune responses via an alum-induced myeloid cell population. Science 304: 1808–1810 [DOI] [PubMed] [Google Scholar]
- 18. Kodihalli S, Kobasa DL, Webster RG. 2000. Strategies for inducing protection against avian influenza A virus subtypes with DNA vaccines. Vaccine 18: 2592–2599 [DOI] [PubMed] [Google Scholar]
- 19. Li W, et al. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426: 450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McFerran JB, Clarke JK, Connor TJ, Knox ER. 1971. Serological evidence of the presence of haemagglutinating-encephalitis virus in Northern Ireland. Vet. Rec. 88: 339–340 [DOI] [PubMed] [Google Scholar]
- 21. Mengeling WL. 1975. Incidence of antibody for hemagglutinating encephalomyelitis virus in serums from swine in the United States. Am. J. Vet. Res. 36: 821–823 [PubMed] [Google Scholar]
- 22. Mengeling WL, Cutlip RC. 1976. Pathogenicity of field isolants of hemagglutinating encephalomyelitis virus for neonatal pigs. J. Am. Vet. Med. Assoc. 168: 236–239 [PubMed] [Google Scholar]
- 23. Myung SJ, et al. 2006. 15-Hydroxyprostaglandin dehydrogenase is an in vivo suppressor of colon tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 103: 12098–12102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Neuvonen E, Ek-Kommonen C, Veijalainen P, Schulman A. 1982. Absence of hemagglutinating encephalomyelitis virus in Finnish elite breeding pig herds. Nord. Vet. Med. 34: 334–335 [PubMed] [Google Scholar]
- 25. Nicollier-Jamot B, Ogier A, Piroth L, Pothier P, Kohli E. 2004. Recombinant virus-like particles of a norovirus (genogroup II strain) administered intranasally and orally with mucosal adjuvants LT and LT(R192G) in BALB/c mice induce specific humoral and cellular Th1/Th2-like immune responses. Vaccine 22: 1079–1086 [DOI] [PubMed] [Google Scholar]
- 26. Pope M, et al. 1996. Resistance of naive mice to murine hepatitis virus strain 3 requires development of a Th1, but not a Th2, response, whereas pre-existing antibody partially protects against primary infection. J. Immunol. 156: 3342–3349 [PubMed] [Google Scholar]
- 27. Prince GA, et al. 2001. Monophosphoryl lipid A adjuvant reverses a principal histologic parameter of formalin-inactivated respiratory syncytial virus vaccine-induced disease. Vaccine 19: 2048–2054 [DOI] [PubMed] [Google Scholar]
- 28. Quiroga MA, et al. 2008. Hemagglutinating encephalomyelitis coronavirus infection in pigs, Argentina. Emerg. Infect. Dis. 14: 484–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reed L, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27: 493–497 [Google Scholar]
- 30. Ren ZW, Li M, Li JP. 2001. The production technology of SPF pigs: applications of modern pig raising in China. Anim. Sci. Vet. Med. 18: 56–58 (In Chinese.) [Google Scholar]
- 31. Sasaki I, Kazusa Y, Shirai J, Taniguchi T, Honda E. 2003. Neutralizing test of hemagglutinating encephalomyelitis virus (HEV) in FS-L3 cells cultured without serum. J. Vet. Med. Sci. 65: 381–383 [DOI] [PubMed] [Google Scholar]
- 32. Sasseville AM, Boutin M, Gelinas AM, Dea S. 2002. Sequence of the 3′-terminal end (8.1 kb) of the genome of porcine haemagglutinating encephalomyelitis virus: comparison with other haemagglutinating coronaviruses. J. Gen. Virol. 83: 2411–2416 [DOI] [PubMed] [Google Scholar]
- 33. Sasseville AM, Gelinas AM, Sawyer N, Boutin M, Dea S. 2001. Biological and molecular characteristics of an HEV isolate associated with recent acute outbreaks of encephalomyelitis in Quebec pig farms. Adv. Exp. Med. Biol. 494: 57–62 [DOI] [PubMed] [Google Scholar]
- 34. Woo PC, et al. 2005. SARS coronavirus spike polypeptide DNA vaccine priming with recombinant spike polypeptide from Escherichia coli as booster induces high titer of neutralizing antibody against SARS coronavirus. Vaccine 23: 4959–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu CY, et al. 2002. Distinct lineages of TH1 cells have differential capacities for memory cell generation in vivo. Nat. Immunol. 3: 852–858 [DOI] [PubMed] [Google Scholar]
- 36. Yagami K, Hirai K, Hirano N. 1986. Pathogenesis of haemagglutinating encephalomyelitis virus (HEV) in mice experimentally infected by different routes. J. Comp. Pathol. 96: 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yagami K, Izumi Y, Kajiwara N, Sugiyama F, Sugiyama Y. 1993. Neurotropism of mouse-adapted haemagglutinating encephalomyelitis virus. J. Comp. Pathol. 109: 21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang ZY, et al. 2004. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature 428: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]