Abstract
The fmtA gene is a member of the Staphylococcus aureus core cell wall stimulon. The FmtA protein interacts with β-lactams through formation of covalent species. Here, we show that FmtA has weak d-Ala-d-Ala-carboxypeptidase activity and is capable of covalently incorporating C14-Gly into cell walls. The fluorescence microscopy study showed that the protein is localized to the cell division septum. Furthermore, we show that wall teichoic acids interact specifically with FmtA and mediate recruitment of FmtA to the S. aureus cell wall. Subjection of S. aureus to FmtA concentrations of 0.1 μM or less induces autolysis and biofilm production. This effect requires the presence of wall teichoic acids. At FmtA concentrations greater than 0.2 μM, autolysis and biofilm formation in S. aureus are repressed and growth is enhanced. Our findings indicate dual roles of FmtA in S. aureus growth, whereby at low concentrations, FmtA may modulate the activity of the major autolysin (AtlA) of S. aureus and, at high concentrations, may participate in synthesis of cell wall peptidoglycan. These two roles of FmtA may reflect dual functions of FmtA in the absence and presence of cell wall stress, respectively.
INTRODUCTION
The fmtA gene was identified as a methicillin resistance factor in Staphylococcus aureus by Komatsuzawa et al. (23). Inactivation of fmtA leads to increased sensitivity of methicillin-resistant S. aureus strains (MRSA) to Triton X-100 and β-lactams and decreases the level of highly cross-linked peptidoglycan (PG) (22, 23). Cell wall inhibitors such as oxacillin, methicillin, cefoxitin, phosfomycin, and bacitracin cause upregulation of the transcription levels of fmtA (22).
Several studies on the genome-wide S. aureus response to cell wall inhibitors have demonstrated that their biological activity causes upregulation of fmtA (3, 26, 27, 41, 46). In addition, inactivation of genes involved with cell wall biosynthesis, such as murF, leads to higher levels of fmtA (41). Hence, fmtA is considered part of the core cell wall stimulon (26).
The function of fmtA is not known. The fmtA gene has been linked to biofilm formation and autolysis, two interconnected processes in bacteria whereby the ability of S. aureus to produce biofilms is linked to its ability to undergo a controlled autolysis process (2, 25). Inactivation of fmtA diminishes the ability of S. aureus to form biofilms (4, 44) and enhances autolysis (4, 23). Boles et al. showed the cell wall of the S. aureus fmtA mutant to lack wall teichoic acids (WTAs) (4). Teichoic acids are phosphate-rich glycopolymers referred to as WTAs (Fig. 1) when connected to the peptidoglycan (PG) or as lipoteichoic acids (LTAs) when connected to the cytoplasmic membrane (51, 54). They make up as much as 60% of the total cell mass in Gram-positive bacteria and have been implicated in a variety of processes, including biofilm production and autolysis (13, 21, 33, 39, 49). Recent reports have implicated WTAs in spatial and temporal regulation of the S. aureus major autolysin (AtlA) and penicillin-binding protein 4 (PBP4) (1, 40).
Fig 1.

Schematic illustration of S. aureus cell envelope. PG, peptidoglycan, WTA, wall teichoic acid; LTA, lipoteichoic acid; D-Ala, d-alanine; GlcNAc, N-acetylglucosamine; ManNAc, N-acetylmannosamine; P, phosphate.
The primary structure of FmtA harbors two of the three conserved motifs of PBPs, SXXK and S(Y)XN (where X indicates any amino acid) while lacking the KTG motif. The protein forms covalent species with β-lactam compounds; however, this interaction is weak, suggesting that FmtA is intrinsically resistant to β-lactam inactivation (10). The other four native PBPs of S. aureus are sensitive to β-lactam. The methicillin-resistant S. aureus strains have acquired an additional PBP, PBP2a, which also interacts poorly with β-lactams (36).
S. aureus remains a major concern for public health due to resistance to a wide range of antibiotics. Understanding how it copes with cell wall stress would provide insights toward new treatment strategies. Further, genome-wide screening studies have shown that inhibition of nonessential genes could be a successful strategy in sensitizing S. aureus to antibiotics that otherwise have failed in clinical settings due to resistance (5, 20, 28, 43).
Here we investigate the relationship of fmtA to cell wall biosynthesis. Our new findings shed light on the role of fmtA in S. aureus methicillin resistance, autolysis, and biofilm formation.
MATERIALS AND METHODS
Materials and chemical reagents.
Growth media were purchased from EMD Bioscience. Enzymes and chemicals were purchased from Sigma and New England BioLabs. Lipoteichoic acids were purchased from InvivoGen. Amplex Red (AR) was purchased from Molecular Probes, Inc. 14C-Gly was purchased from Perkin Elmer. Monoclonal anti-FmtA serum was purchased from GeneScript Corporation. Sterile 96-well tissue culture plates were purchased from Corning Incorporated. Safranin was purchased from bioMérieux Canada.
Construction of FmtAΔ27S127A mutant.
FmtAΔ27 refers to the mature protein sequence. The signal peptide sequence (27 amino acids at the N terminus) was not included in the cloning process of fmtA (10). This facilitated the isolation of the protein from the cytoplasm. The mature protein lacks any cysteine residues in the primary structure. Hence, isolation of this otherwise periplamismic protein from the cytoplasm should not have any effect on the folding of the protein (10).
Mutation of Ser127 to Ala was carried out using a QuikChange site-directed mutagenesis kit (Stratagene). The pET24a (+)::fmtA27 vector (10) was amplified using Pfu Turbo DNA polymerase and a pair of mutagenic primers, DirS127A (5′-CGATGTTTTTAATAGGTGCAGCTCAAAAATTTTC-3′) and RevS127A (5′-GAAAATTTTTGAGCTGCACCTATTAAAAACATCG-3′) (the mutated nucleotides are italicized). The PCR product was digested with the DpnI restriction endonuclease and used to transform Escherichia coli BL21(DE3). The nucleotide sequence of the mutant variants was verified by DNA sequencing. The purifications of FmtAΔ27 and the S12A mutant were carried as described by Fan et al. (10).
dd-Carboxypeptidase assay.
Carboxypeptidase activity of FmtAΔ27 against the substrate surrogate Nα,Nε-diacetyl-l-Lys-d-Ala-d-Ala (the surrogate for the peptidoglycan donor strand) was monitored with a fluorescence-based assay (17). A typical 100-μl assay mixture consisted of 6 mM tripeptide–10 μM FmtAΔ27–0.1 M Tris (pH 8.5) buffer. The surrogate for the peptidoglycan acceptor strand, Gly-Gly peptide, was added to one set of reaction mixtures at a 1:10 ratio (donor to acceptor).
Transpeptidase activity assays.
Transpeptidase activity of FmtAΔ27 was investigated using radioactive 14C-labeled glycine (14C-Gly) as the acceptor. Cross-linking experiments were carried out using cell wall isolated from S. aureus RN4220. A typical 100-μl reaction mixture consisted of 50 μg of cell wall, FmtAΔ27 at different concentrations (0 to 30 μM), and 1 mM 14C-Gly, all in 0.1 M Tris (pH 8.5) buffer. In a control reaction, albumin (15 μM) or ovalbumin (15 μM) was incubated with cell wall (50 μg) and 14C-Gly (1 mM).
Reaction mixtures were incubated for 2 h at 37°C. Following incubation, samples were spun down at 25,000 × g for 30 min at 4°C. Pellets containing cell walls were washed twice with 0.1 M Tris (pH 8.5) buffer to remove any nonspecifically bound 14C-Gly. The incorporated radioactivity was recorded by a Tri-carb liquid scintillation analyzer (Packard). The same experiments were carried out with FmtAΔ27S127A at 30 μM.
Preparation of S. aureus cell walls with and without wall teichoic acid.
Cell walls from S. aureus RN4220 were isolated as previously reported (7). Removal of WTAs from cell walls was carried out using hydrofluoric acid (HF) based on a protocol by Vialle et al. (47). The cell wall preparation resulting in peptidoglycan with no WTAs attached is referred to as peptidoglycan (PG). Binding of FmtAΔ27 to cell walls was performed as previously described (10).
Investigation of the FmtAΔ27 hydrolase activity.
FmtAΔ27 (2.5 μM) was incubated with PG (1 mg/ml) in 50 mM sodium phosphate (pH 7.0) buffer (1 ml) at 37°C for different times (0.5, 1, 2, 3, 4, and 5 h). The change in absorbance was monitored at 450 nm. Mutanolysin from Streptomyces globisporus was used as a positive control.
In another experiment, PG (1 mg/ml)–50 mM phosphate (pH 7.0) buffer was incubated with 10 μM FmtAΔ27 in a 20-μl reaction mixture. In control experiments, PG was digested with mutanolysin and undigested PG. The reaction mixture was loaded into a C18 reverse-phase high-performance liquid chromatography (HPLC) column (Persuit; Varian) (5 μ; 250 by 4.6 mm). A linear gradient of 0% to 30% methanol in a 100 mM phosphate (pH 2.8) buffer was used to elute any peptidoglycan fragments.
Investigation of FmtAΔ27 interaction with WTA by the use of acidic nPAGE/H+.
Interaction of FmtAΔ27 with WTAs was investigated with native polyacrylamide gel electrophoresis (nPAGE)/H+. A typical reaction mixture (15 μl) consisted of 10 μg of FmtAΔ27 (15 μM final concentration) and 5 μg of WTA in the presence of NaCl at different concentrations (100, 300, or 500 mM), all in 50 mM sodium phosphate (pH 7.0) buffers. Another reaction mixture containing 8 μg of PBP2a (15 μM final concentration) was also prepared using a method similar to that described above. Bovine serum albumin (BSA), ovalbumin, RNaseA, and carbonic anhydrase (10 μg each) were used as negative controls. The PAGE gels were stained with Coomassie blue.
Investigation of FmtAΔ27 interaction with WTA by trypsin digestion.
FmtAΔ27 (40 μM) was incubated either with or without WTAs (1.25 μg in 12.5 mM sodium phosphate [pH 5.5] buffer and 0.2% sodium azide; 80-μl reaction volume) for 1 h at 37°C. The mixture was subjected to trypsin digestion based on a protocol from a ProteoExtract kit (CalBiochem) (trypsin assay concentration = 0.2 μg/μl) and further incubated at 37°C. Aliquots of 10 μl were removed at defined time intervals, and the digestion was terminated with 10 μl of sodium dodecyl sulfate (SDS) loading dye, followed by heating at 60°C for 5 min. Samples were analyzed by 15% Tris-Tricine SDS-PAGE. Each experiment was repeated three times.
In the case of limited trypsin digestion, the samples were subjected to trypsin only for 15 min and the protein samples were analyzed by matrix-assisted laser desorption ionization—time of flight (MALDI-TOF) mass spectroscopy (MS) at the Advanced Protein Technology Center, Hospital for Sick Children, Toronto, Canada.
CD spectroscopy studies of FmtAΔ27 in the presence of WTAs.
Far-UV (200 to 260 nm) circular dichroism (CD) spectra of FmtAΔ27 (14 μM in 50 mM sodium phosphate [pH 7.0] buffer), in the presence of WTAs, were recorded at 22°C using a Jasco J-810 instrument and a cuvette with a 1.0-mm path length. The average of three scans was recorded and corrected for the signals from the buffer, PG fragment, and WTAs.
Autolysis assays.
S. aureus RN4220 cells were grown in tryptic soy broth (TSB) overnight at 37°C. Cells were diluted 200-fold in fresh TSB medium and grown with shaking at 37°C to an optical density at 620 nm (OD620) of 0.8. Cells were harvested by centrifugation (6,300 × g, 10 min) and washed twice with ice-cold water. The pellet was resuspended in 1× phosphate-buffered saline (PBS) (pH 7.0) to an OD620 of 1.00 and dispensed into a 96-well plate (200 μl per well). Cells were incubated at 37°C in the presence of various concentrations of FmtAΔ27, with agitation at 200 rpm. Autolysis was measured as a decrease in OD620 over time. In the case of the S. aureus ΔtarO mutant and its parental strain (EBII16), Mueller-Hinton broth (MHB) medium was used and FmtAΔ27 was kept at 0.2 μM.
Biofilm assay.
Cultures of S. aureus RN4220 in TSB, or of the ΔtarO mutant and EBII16 strain in MHB, were grown overnight and normalized by optical density. The cell cultures were next diluted 50-fold into fresh TSB supplemented with 0.2% glucose. Wells of a sterile, 96-well flat-bottomed tissue culture plate (Corning Incorporated) were filled with 200-μl aliquots of the diluted culture. FmtAΔ27 was added to the assay at various final concentrations (0 to 6 μM). Cultures were incubated for 24 h at 37°C in the closed plate and assayed for biofilm formation according to the method of Christensen et al. (6). Adherent cells were fixed with Bouin fixative (Sigma) and stained with 0.1% Safranin (bioMérieux, Canada) for 10 min. Wells were then washed with running tap water. Images of the inverted plates were taken using a digital camera. Biofilms were dissolved using 30% glacial acetic acid for 15 min, and relative biofilm formation was also assayed by reading absorbance at 562 nm using a plate reader.
Construction of gfp-fmtAΔ27 fusion.
The fmtAΔ27 amplicon was ligated into a pcDNA3.1/NT-GFP-TOPO vector (Invitrogen). To facilitate subcloning of gfp-fmtA chimera into the pET24a vector, a silent mutation was introduced by QuikChange site-directed mutagenesis (Stratagene) to remove the NdeI restriction site in the gfp gene. The following mutagenic primers were used: Dir- (5′-CGTTATCCGGATCACATGAAACGGCATGAC-3′) and Rev (5′-GTCATGCCGTTTCATGTGATCCGGATAAG-3′). Introduction of the mutation was confirmed by DNA sequencing analysis. Another set of primers was designed to amplify the gfp-fmtAΔ27 fusion gene for the final cloning to the pET24a vector: Dir (5′-AGCCATATGGCCAGCAAAGGAGAAGAAC-3′) and Rev- (5′-AGCGAATTCTTATTATTGAACAATAACACCCTTCG-3′) (NdeI and EcoRI restriction sites are italicized). The successful cloning of the fusion gene into pET24a was also confirmed by DNA sequencing analysis.
The final construct, gfp-fmtAΔ27::pET24a, was used to transform E. coli BL21(DE3) cells. Expression and purification of green fluorescent protein-FmtAΔ27 (GFP-FmtAΔ27) was carried out as described before (10). In addition, the activity of the chimeric FmtAΔ27 was investigated using fluorescent penicillin (Bocillin) and binding to cell walls isolated from S. aureus EBII16 and ΔtarO mutant, as previously described (10).
Investigation of GFP-FmtAΔ27 localization to S. aureus by fluorescence microscopy.
S. aureus EBII16 and ΔtarO mutant strains were grown overnight in MHB growth media. Cells were diluted 200-fold in fresh MHB and grown to an OD600 of ∼0.6. Cells were harvested by centrifugation at 9,300 × g for 10 min. The cells were washed with 1× phosphate-buffered saline (PBS) and resuspended in 1× PBS buffer to give a final OD600 of 1.2.
In a typical binding assay, 100 μl of 5 μM GFP-FmtAΔ27 protein solution was mixed with 400 μl of washed S. aureus cells to yield a final concentration of 1 μM GFP-FmtAΔ27. Reaction mixtures were incubated for 10 min at room temperature. Staphylococcal cells with bound protein were sedimented by centrifugation at 16,000 × g for 3 min. The cells were washed with PBS and resuspended in PBS. A drop of bacterial suspension was placed on a polylysine-coated glass slide and immediately analyzed using epifluorescence microscopy performed with a laser-scanning confocal microscope (Fluoview FV300). Similar experiments were carried out at GFP-FmtAΔ27 concentrations of 2, 5, and 10 μM. Prior to analysis by microscopy, mixtures were incubated with 1 μM protein for 0.5, 1, and 2 h to measure binding time of GFP-FmtAΔ27 to S. aureus cells.
To assess the effect of oxacillin and vancomycin on the localization of FmtA, S. aureus EBII16 and ΔtarO mutant strains were exponentially exposed to either oxacillin (final concentration, 4 μg/ml) or vancomycin (final concentration, 20 μg/ml) for 1 h. Aliquots were sampled at the final dose. Sequential dilutions were developed on MHB agar plates to determine the number of viable cells. The rest of the cell cultures were processed as described above.
RESULTS
FmtAΔ27 displays dd-carboxypeptidase activity.
In this study, we worked with the mature FmtA protein, FmtAΔ27, which lacks the signal peptide sequence (10). FmtAΔ27 catalyzed the removal of the d-Ala from the diacetyl tripeptide substrate (a surrogate for the PG donor strand). The rate of d-Ala removal increased in the presence of Gly-Gly, which is a surrogate for the PG acceptor strand. Different ratios of donor to acceptor were investigated, and the optimum ratio was 1:10. Under these conditions, the catalytic efficiency of dd-carboxypeptidase activity of FmtA was measured to be 0.745 × 10−6 M−1 s−1. The FmtAΔ27S127A mutant was completely devoid of this activity.
Potential transpeptidase activity of FmtAΔ27 was investigated by monitoring the catalytic incorporation of 14C-Gly into S. aureus cell walls. Albumin and ovalbumin, proteins unrelated to PG biosynthesis, served as negative controls in these experiments (Fig. 2A). FmtAΔ27 covalently incorporated 14C-Gly into the cell walls, but albumin and ovalbumin did not. The 14C labeling of cell walls by FmtAΔ27 depended on the concentration of the protein (Fig. 2A). The level of 14C-Gly incorporation into the cell walls by the FmtAΔ27S127A mutant was the same as the background levels (in the absence of FmtAΔ27) (Fig. 2B).
Fig 2.

Investigation of the 14C-Gly incorporation into cell wall isolated from S. aureus RN4220 by FmtAΔ27. (A) FmtAΔ27 at different protein concentrations and albumin and ovalbumin (15 μM) were incubated with cell wall (50 μg [100-μl final volume]) isolated from S. aureus RN220 in the presence of 14C-Gly (1 mM). (B) Comparison of the activity of FmtAΔ27 to the activity of the FmtAΔ27S127A mutant. The concentration of each of the two proteins was 15 μM. Error bars represent standard deviations calculated from at least three independent experiments.
FmtAΔ27 interacts with S. aureus WTAs.
FmtAΔ27 binds to cell walls (10). To identify the cell wall component essential for FmtAΔ27 recruitment, we investigated the binding of FmtAΔ27 to cell walls digested with either lysostaphin or mutanolysin. Lysostaphin cleaves the pentaglycine bridge in PG and decreases the degree of PG cross-linking (glycan strands are less cross-linked). Mutanolysin cleaves the glycan strands after N-acetylmuramic acid and produces different oligomers of muropeptides.
Digestion of cell walls with mutanolysin resulted in as much as an 80% decrease of FmtAΔ27 binding to cell walls (Fig. 3), whereas digestion of cell walls with lysostaphin resulted in a 40% decrease of FmtAΔ27 binding (Fig. 3). The persistence of FmtAΔ27 binding to cell walls digested by lysostaphin was investigated further. We looked at the components of cell wall present in our cell wall preparations and took note that WTAs were not removed from the cell wall preparations. WTAs were removed from the cell wall preparations by HF. The newly prepared cell walls are referred to as peptidoglycan (PG) here.
Fig 3.
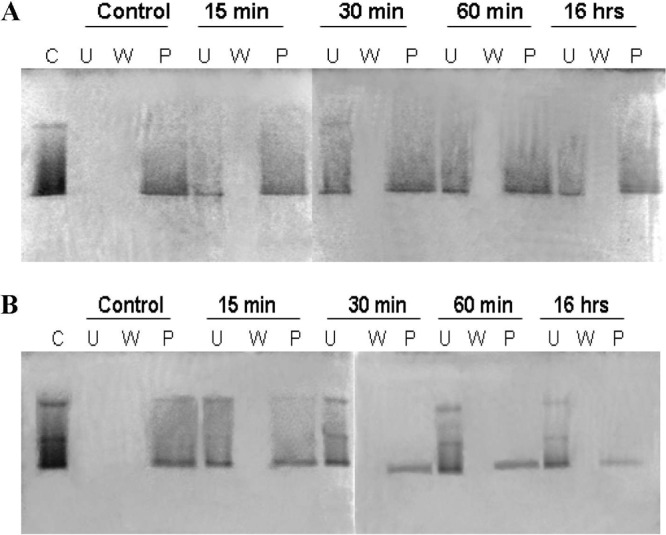
FmtAΔ27 binding to cell wall. Cell wall (40 μg) was digested with lysostaphin (A) or mutanolysin (B) at different time intervals. Binding of FmtAΔ27 (2.5 μM) to cell wall products was investigated with 12.5% SDS-PAGE. Samples represent the control (C), unbound (U), wash (W), and pellet (P) fractions. The SDS-PAGE gels were stained with Coomassie blue.
FmtAΔ27 failed to bind to PG (Fig. 4). This experiment was repeated with cell walls isolated from the ΔtarO mutant. The tarO gene is involved in the first step of WTA biosynthesis, and, as such, S. aureus tarO mutant cell walls lack WTAs (8). The in vitro binding assays with cell walls isolated from the ΔtarO mutant showed that these cell walls could not recruit FmtAΔ27; the protein remained predominately in the unbound fraction (Fig. 4).
Fig 4.
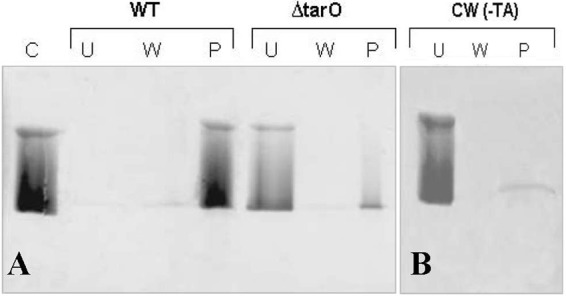
The effect of removal of WTAs on binding of FmtAΔ27 to cell wall. (A) Cell walls (100 μg) of S. aureus RN4220 (WT) and the tarO deletion strain (ΔtarO) were incubated with FmtAΔ27 at 2.5 μM for 2 h at 4°C and analyzed by 12.5% SDS-PAGE. (B) Cell walls with removed WTA [CW (-TA)] (100 μg) isolated from S. aureus RN4220 were incubated with FmtAΔ27 (15 μM). Experimental conditions were the same as described for panel A. Samples represent control (C), unbound (U), wash (W), and pellet (P) fractions. The SDS-PAGE gels was stained with Coomassie blue.
The WTA-binding properties of FmtAΔ27 were further studied using WTAs isolated from S. aureus RN4220. Incubation of FmtAΔ27 with WTAs resulted in a lack of electrophoretic resolution of the protein by the use of an nPAGE/H+ gel (Fig. 5). We concluded that FmtAΔ27 could not enter the gel in the presence of WTAs; however, addition of NaCl to the reaction mixture facilitated resolution of FmtAΔ27 by the nPAGE/H+ procedure. At 500 mM NaCl, the protein was resolved completely (Fig. 5A). The presence of salt alone had no effect on the electrophoretic resolution of FmtAΔ27 by nPAGE/H+ (Fig. 5B). These results strongly indicate that FmtAΔ27 interacts with WTAs. Similar experiments were carried out with PBP2a, a penicillin-binding protein that has transpeptidase activity. PBP2a displayed an affinity for the WTAs and behaved in the same way as FmtA (Fig. 5C). Proteins unrelated to cell wall metabolism, including RNaseA, and carbonic anhydrase, did not show any binding to WTAs, while BSA showed a weak affinity for teichoic acids (Fig. 6).
Fig 5.
FmtAΔ27 interaction with WTAs analyzed by nPAGE/H+. (A) FmtAΔ27 (15 μM) was incubated with 5 μg of WTAs in the presence of different concentrations of NaCl in a 15-μl reaction volume. Samples were analyzed with nPAGE/H+. (B) FmtA (15 μM) was incubated with different concentrations of NaCl, and samples were analyzed with nPAGE/H+. (C) PBP2a (15 μM) was incubated with 5 μg of WTA in a 15-μl reaction volume in the absence or in the presence of 500 mM NaCl. The nPAGE gels were stained with Coomassie blue.
Fig 6.

Investigation of the interaction between TAs and protein unrelated to peptidoglycan biosynthesis. Lipoteichoic acids (LTAs; 5 μg) were incubated with bovine serum albumin (BSA; pI 4.6), ovalbumin (pI 4.5 to 4.6), RNaseA (pI 9.63), or carbonic anhydrase (pI 5.88 to 6.55) at 10 μg each in a 15-μl reaction volume. Samples were analyzed by nPAGE/H+. Protein bands were stained with Coomassie blue.
WTAs induce a conformational change in FmtAΔ27 structure.
The CD experiments showed that FmtAΔ27 undergoes a dose-dependent conformational change in the presence of teichoic acids (Fig. 7). The conformational change is associated with a reduction in FmtAΔ27 α-helix content, as interpreted from a decrease in the CD signal at 222 nm. The reduction in α-helix content was calculated to be 6% in the presence of WTAs at 6.25 μg/ml, 14% in the presence of WTAs at 12.5 μg/ml, and 21% in the presence of WTAs at 25 μg/ml (Fig. 7).
Fig 7.

Investigation of FmtAΔ27 interaction with WTAs by CD. The CD spectra of FmtAΔ27 (14 μM) were collected in the absence and in the presence of different concentrations of WTAs. The CD spectra were recorded using 50 mM sodium phosphate buffer (pH 7.0) at 22°C. The data points represent averages of the results of three scans and were corrected for buffer contribution.
The interpretation of the effect of WTAs on FmtAΔ27 secondary structure was supported by trypsin digestion experiments (Fig. 8). The digestion profiles of FmtAΔ27 differed in the presence and absence of WTAs. Three major fragments were released upon incubation of FmtAΔ27 with trypsin; however, when WTAs (6.25 μg/ml) were present, six fragments were produced (Fig. 8). These observations provide further evidence that WTAs interact with FmtAΔ27.
Fig 8.

Investigation of FmtAΔ27 interaction with WTAs by trypsin digestion. FmtAΔ27 (40 μM) was digested with trypsin in the absence (−TA) and presence (+TA) of WTAs (1.25 μg, 80 μl). Aliquots (10 μl) were removed at different time intervals and quenched by addition of loading buffer and heating at 60°C. Samples were analyzed with 15% Tris-Tricine SDS-PAGE.
To identify the specific region in the protein involved with binding to WTAs, FmtAΔ27 was subjected to a 15-min trypsin digestion in the absence and presence of WTAs. Analysis of the released peptides by MALDI-TOF MS (see Fig. S1S in the supplemental material) showed a notable difference: a peptide with a mass of 1,369 Da was not detected when the digestion was carried out in the presence of WTA. Instead, a peptide with a mass of 853 Da exhibited more abundance. The peptide with a mass of 1,369 Da mapped to amino acids 312 to 323 in the C-terminal portion of FmtAΔ27, and the peptide with a mass of 853 Da mapped to amino acids 155 to 163 of FmtAΔ27.
FmtAΔ27 induces autolysis of S. aureus.
Binding of FmtAΔ27 to WTA suggests that it could affect processes regulated by WTA. One of the important processes controlled by WTA and LTA is autolysis. The activity of AtlA, the major autolysin of S. aureus, has been shown to be controlled by teichoic acids (14, 40, 50, 51). We investigated the potential effect of FmtAΔ27 on autolysis of S. aureus RN4220 by incubating cells with different concentrations of FmtAΔ27. We discovered that FmtAΔ27 exogenously added to S. aureus cells affected autolysis of cells in a dual mode. At concentrations of up to 0.1 μM, FmtAΔ27 induced autolysis over 1 h, whereas at concentrations higher than 0.2 μM, an enhanced growth of bacteria was observed instead (Fig. 9A). A similar observation was also made for the ΔtarO mutant, which lacks WTAs (8) (Fig. 9B).
Fig 9.

FmtAΔ27 effect on autolysis of S. aureus RN4220. (A) Autolysis of S. aureus RN4220 cells in the presence of different concentrations of FmtAΔ27 and 1× PBS buffer, monitored for 1 h (empty circles) and 2 h (filled circles). Error bars indicate standard deviations. (B) The effect of 0.2 μM FmtAΔ27 on autolysis of S. aureus tarO parental strain (WT) and tarO-deleted mutant strain (ΔtarO).
FmtAΔ27 induces biofilm formation in S. aureus.
Controlled autolysis is essential for formation of biofilms (15, 19); therefore, FmtAΔ27-induced autolysis suggests that FmtAΔ27 might be involved in biofilm production in S. aureus. Indeed, recent reports show that inactivation of FmtA leads to cell biofilm formation deficiency (4, 44). Our study showed that biofilm production in the S. aureus RN4220 strain increases with the FmtAΔ27 concentration up to 0.1 μM and that it decreases at FmtA concentrations above 0.2 μM (Fig. 10), much like the dual effect observed for FmtA-induced autolysis.
Fig 10.

Stimulation of biofilm formation by FmtAΔ27 in S. aureus RN4220. (A) Detection of biofilm formation of S. aureus RN4220 plates in the presence of various concentrations of FmtAΔ27 following 24 h of growth in polystyrene flat-bottomed tissue culture plates. (B) Quantification (A562) of biofilm growth with various concentrations of FmtAΔ27. Data represent the averages of the results of at least three independent experiments. Error bars indicate standard deviations. (C) Detection of biofilm formation of S. aureus wild-type strain (WT) and ΔtarO mutant strain (ΔtarO) in the presence of 0.2 μM FmtAΔ27 after 24 h of growth in polystyrene flat-bottomed tissue culture plates.
We investigated the WTA effect on the ability of FmtAΔ27 to induce biofilm formation by comparing biofilm formation of the ΔtarO mutant with that of the wild-type strain in the presence or absence of 0.2 μM FmtAΔ27. These studies showed that, although FmtAΔ27 induced maximum biofilm formation in the S. aureus wild-type strain, it was entirely unable to induce this process in the ΔtarO mutant strain despite similar conditions (Fig. 10C).
FmtAΔ27 localizes in the division septum.
To investigate localization of FmtA to S. aureus, we fused FmtAΔ27 to the C terminus of GFP. GFP-FmtAΔ27 expressed well in E. coli (25 mg/liter), and the protein was purified using the protocol for FmtAΔ27 purification (10). The GFP tag showed no effect on the interaction of FmtA with β-lactams or cell walls (see Fig. S2S in the supplemental material). We therefore concluded that GFP-fused FmtAΔ27 is indistinguishable from wild-type FmtAΔ27.
Gründling and Schneewind reported that the GFP protein alone does not bind to S. aureus cells (16). Our fluorescence microscopy studies showed that GFP-FmtAΔ27 bound at the division septum in the dividing S. aureus RN4220 cells, where cell wall synthesis is reported to occur in S. aureus (33). In nondividing cells, GFP-FmtAΔ27 was localized as two dots corresponding to the ring of the future division plane (Fig. 11). A localization pattern similar to that of GFP-FmtAΔ27 has been reported for PBP4 of S. aureus (1). In the ΔtarO mutant, GFP-FmtAΔ27 was observed all over the cell wall but with no specific accumulation at the division septum (Fig. 11).
Fig 11.

Fluorescence microscopy imaging of FmtAΔ27 localization in S. aureus. Pictures show localization of GFP-FmtAΔ27 at the cell septum in S. aureus RN4220 (WT strain) and loss of septal localization in the ΔtarO strain. S. aureus RN4220 cultures and ΔtarO mutant strains were incubated with 1 μM GFP-FmtAΔ27 at room temperature (RT) for 10 min. Pictures were taken with an epifluorescence microscope after removing unbound GFP-FmtAΔ27. Fluorescence (GFP), differential interference contrast (DIC), and overlap of GFP and DIC images are shown. Red arrows identify the GFP-FmtAΔ27 protein in S. aureus cells. Bar, 1 μm. (B) A conceptual model of FmtA location in S. aureus based on GFP-FmtA localization studies.
Exposure of S. aureus to either oxacillin or vancomycin (each of which inhibits the cross-linking step in peptidoglycan biosynthesis by binding, respectively, to the transpeptidase domain of PBPs or the d-Ala-d-Ala portion of the peptidoglycan) did not have any effect on the localization of GFP-FmtAΔ27. A similar observation was made for PBP4 (1). In the case of PBP2, however, the presence of these antibiotics caused delocalization of the PBP2 from the division site, suggesting that PBP2 requires substrate binding for its correct localization (35).
DISCUSSION
The fmtA gene was identified as a methicillin resistance factor (23), and it is present in all S. aureus strains. It is part of the core cell wall stimulon (26). Its expression level is elevated by the presence of cell wall inhibitors and knockout of genes that are directly involved with peptidoglycan biosynthesis (3, 23, 26, 27, 42, 46). Furthermore, inactivation of fmtA results in a lack of WTAs on S. aureus cell walls and deficiency in biofilm production (4, 44).
The function of FmtA is not known. In an earlier study, we showed that FmtA is capable of forming covalent species with β-lactams involving the serine-active site residue of the SXXK conserved motif (10). The β-lactam binding activity of FmtA suggests that it is a PBP. Of the four well-known PBPs of S. aureus, PBP1 is a monofunctional transpeptidase that is essential for viability of S. aureus (48). PBP2 is a bifunctional enzyme with both transglycosylase and transpeptidase activity, and it is essential for viability of S. aureus (37). PBP3 and PBP4 are believed to be monofunctional and dispensable transpeptidases. Furthermore, PBP2 and PBP4 were shown to work together to synthesize a highly cross-linked peptidoglycan (24).
In this report, we show that FmtA has dd-carboxypeptidase activity; it catalyzes the removal of the last d-Ala residue from the Nα,Nε-diacetyl-l-Lys-d-Ala-d-Ala peptide (a surrogate for the PG donor strand). However, it shows low enzymatic activity. In the presence of Gly-Gly (a surrogate for the PG acceptor strand), the catalytic efficiency of FmtAΔ27 dd-carboxypeptidase activity was determined to be 0.745 × 10−6 M−1 s−1. In comparison, the catalytic efficiency of the dd-carboxypeptidase activity of PBP4 of S. aureus was determined to be 15.7 M−1 s−1 (30). The activity of other well-studied PBPs has also been reported to be higher, ranging from 10 M−1 s−1 for PBP5 of E. coli to 2.9 × 104 M−1 s−1 for PBP3 of N. gonorrhoeae, according to a study using the same substrate as used in our study (42).
The low enzymatic activity of FmtA could be due to three main reasons: (i) it lacks the third conserved motif, the KTG motif (this motif has been shown to be involved in substrate binding and catalysis) (9, 29, 52); (ii) the l-Lys-d-Ala-d-Ala is not the FmtA native substrate; (iii) the DD-carboxypeptidase activity is reminiscent of that of a PBP from which FmtA had evolved; and (iv) the enzyme may require interaction with a ligand in the periplasmic space to become active. Several proteins involved with either peptidoglycan biosynthesis or its remodeling have been shown to become active in the presence of either the native substrate or a particular ligand. For example, PBP2a of S. aureus becomes active upon binding to PG (12), E. coli PBP1a and PBP1b (two lipoproteins) are activated through interactions with membrane-bound proteins LpoA and LpoB (32, 45) and PBP5 of E. coli requires binding to the membrane to become active (38).
In spite of the poor activity of the enzyme observed using PG surrogates, we discovered that FmtAΔ27 is capable of incorporating 14C-Gly into isolated S. aureus cell walls. This observation suggests that FmtA may have transpeptidase activity, which agrees with previous reports that inactivation of fmtA is associated with a decrease in the level of the highly cross-linked peptidoglycan and an increase in the sensitivity of S. aureus to oxacillin (22, 23).
The reported link between FmtA and the level of cross-linked PG should be noted, as the cross-linked sites in PG are reported to be targets of teichoic acid attachments in Streptococcus pneumoniae and Bacillus licheniformis (11, 53). Interestingly, inactivation of fmtA leads to a lack of teichoic acid attachment in S. aureus (4). In light of these reports, it is possible that the native function of FmtA might be involved with teichoic acid attachment through mediation of cross-linked PG synthesis or marking the teichoic acid attachment sites in PG. In this study, we demonstrated that FmtA interacts specifically with WTAs. Proteins unrelated to cell wall biosynthesis (BSA, ovalbumin, RNaseA, and carbonic anhydrase) showed no interactions with WTAs, suggesting that the negative charge on WTAs does not enable indiscriminate protein binding.
WTAs are proposed to be temporal and spatial regulators of peptidoglycan cross-linking that operate by mediating localization of proteins associated with PG biosynthesis and remodeling, such as PBP4 and AtlA (1, 40). Although a direct interaction between these two proteins and WTAs has not been demonstrated, our study shows that interaction of WTAs with PG-associated proteins is not limited to FmtA. PBP2a, the major source of resistance to β-lactams in S. aureus, interacts with WTAs in the same way as FmtA. This finding suggests that interaction of PG-associated proteins, such as PBPs, with WTAs could be more common than previously thought.
The interaction of FmtA with teichoic acids led us to investigate two processes in S. aureus that are closely linked to teichoic acids, autolysis and biofilm production. We observed that exogenous FmtAΔ27 induced autolysis and biofilm production in S. aureus at concentrations lower than 0.1 μM. However, at FmtAΔ27concentrations higher than 0.2 μM, S. aureus exhibited enhanced growth and deficiency in biofilm production. These observations corroborate recent reports that S. aureus ΔfmtA mutants have lost their ability to precisely control the autolysis activity of autolysins (4) and their ability to effectively form biofilms (4, 44). The ability of S. aureus to form biofilms is intimately linked to its ability to undergo controlled autolysis (2, 25). We therefore propose that the effect FmtA has on biofilm formation is linked to its ability to induce controlled autolysis.
The FmtA role in autolysis and, hence, biofilm formation does not result from a direct role of FmtA in autolysis, as FmtA does not have autolysis activity (see Fig. S3S in the supplemental material). It is possible that binding of FmtA to WTAs may activate S. aureus autolysins. The major autolysin of S. aureus is AtlA (30), which is involved in the separation of daughter cells after cell division. Studies have shown that WTAs control the activity of AtlA (14, 40, 50, 51) and that modulation of the WTA negative charge affects AtlA activity (33, 39). Binding of FmtA to WTAs might affect the activity of AtlA by modulating the negative charge of WTAs; however, we cannot exclude the possibility of interactions between FmtA and AtlA.
The dual mode of FmtA activity for S. aureus, induction and inhibition of autolysis, suggests that FmtA might serve two different roles in S. aureus, both dependent of the FmtA concentration in the cell; at low concentrations (≤0.1 μM), FmtA may be involved in positively controlling the autolysis process, and at higher protein concentrations (≥0.2 μM), FmtA may repress the activity of autolysins and may be involved in PG biosynthesis.
In this study, we discovered that FmtA localizes at the cell division septum, most likely by binding to the cross wall (Fig. 11). PBP4 and AtlA also localize in the cell division septum (1, 40). The localization data clearly show that FmtA is initially localized at the division poles and then moves toward the center of the bacterium as the cross-wall is synthesized (Fig. 11B). The presence of cell wall inhibitors did not cause delocalization of the protein, demonstrating that PG does not mediate the FmtA recruitment to the division septum. Our finding that FmtA interacts with WTAs suggests that WTAs might recruit FmtA to the cell wall. However, no mature WTAs have been shown to be present in the cross-wall region (40). It is plausible that FmtA can be recruited either by a precursor of WTAs in the cross-wall region or by another target. Based on the effect that FmtA has on autolysis, we cannot exclude the possibility that FmtA interacts with AtlA; AtlA also localizes in the cross-wall region (40). Incidentally, the structural genes of atlA and fmtA are close to each other in S. aureus genomes and are separated by three structural gene loci, SA0906, SA0907, and SA0908 (gene numbering per the S. aureus N315 strain), with no known functions.
On a final note, it is intriguing that one autolysin and two PBPs (FmtA and PBP4) colocalize in the same region of the cell wall during the process of cell division. It is proposed that PG-synthesizing enzymes and autolysins form a complex, which would be significantly more efficient in PG biosynthesis (18). Our localization studies, and those performed with AtlA and PBP4, suggest that AtlA, FmtA, and/or PBP4 may work cooperatively to mediate PG remodeling during normal cell growth conditions. In the presence of cell wall inhibitors, FmtA could repress autolysis and mediate growth, probably by enhancing PG biosynthesis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant to D.G.-K. from the Canadian Institutes of Health Research.
The ΔtarO deletion strain (EBII44) and its parental strain (EBII16) were kindly provided by Eric D. Brown's laboratory at McMaster University, Hamilton, Ontario, Canada.
Footnotes
Published ahead of print 7 May 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Atilano ML, et al. 2010. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc. Nat. Acad. Sci. 107:18991–18996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bayles KW. 2007. The biological role of death and lysis in biofilm development. Nat. Rev. Microbiol. 5:721–726 [DOI] [PubMed] [Google Scholar]
- 3. Bernal P, et al. 2010. Insertion of epicatechin gallate into the cytoplasmic membrane of methicillin-resistant Staphylococcus aureus disrupts penicillin-binding protein (PBP) 2a-mediated beta-lactam resistance by delocalizing PBP2. J. Biol. Chem. 285:24055–24065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boles BR, Thoendel M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146 doi:10.1371/journal.pone.0010146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Campbell J, et al. 2011. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 6:106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Christensen GD, et al. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for adherence of Staphylococci to medical devices. J. Clin. Microbiol. 22:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. De Jonge BLM, Chang Y-S, Gage D, Tomasz A. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J. Biol. Chem. 267:11248–11254 [PubMed] [Google Scholar]
- 8. D'Elia MA, et al. 2006. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensible pathway. J. Bacteriol. 188:4183–4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dubus A, Wilkin JM, Raquet X, Normark S, Frere JM. 1994. Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the Lys-Thr(Ser)-Gly triad. Biochem. J. 301(Pt. 2):485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fan X, et al. 2007. Diversity of penicillin-binding proteins. Resistance factor FmtA of Staphylococcus aureus. J. Biol. Chem. 282:35143–35152 [DOI] [PubMed] [Google Scholar]
- 11. Fischer H, Tomasz A. 1985. Peptidoglycan cross-linking and teichoic acid attachment in Streptococcus pneumoniae. J. Bacteriol. 163:46–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fuda C, Suvorov M, Vakulenko SB, Mobashery S. 2004. The basis of resistance to β-lactam antibiotics by penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. J. Biol. Chem. 279:40802–40806 [DOI] [PubMed] [Google Scholar]
- 13. Garcia-Bustos JF, Tomasz A. 1987. Teichoic acid containing muropeptides from Streptococcus pneumoniae as substrate for the pneumococcal autolysin. J. Bacteriol. 169:447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giudicelli S, Tomasz A. 1984. Attachment of pneumococcal autolysin to wall teichoic acids, an essential step in enzymatic wall degradation. J. Bacteriol. 158:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Götz F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367–1378 [DOI] [PubMed] [Google Scholar]
- 16. Gründling A, Schneewind O. 2006. Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J. Bacteriol. 188:2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gutheil WG, Stefanova MS, Nicholas RA. 2000. Fluorescent coupled enzyme assays for D-alanine: application to penicillin-binding protein and vancomycin activity assays. Anal. Biochem. 287:196–202 [DOI] [PubMed] [Google Scholar]
- 18. Höltje JV. 1995. From growth to autolysis: the murein hydrolases in Escherichia coli. Arch. Microbiol. 164:243–254 [DOI] [PubMed] [Google Scholar]
- 19. Houston P, Rowe SE, Pozzi C, Waters EM, O'Gara JP. 2011. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect. Immun. 79:1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jo DS, Montgomery CP, Yin S, Boyle-Vavra S, Daum RS. 2011. Improved oxacillin treatment outcomes in experimental skin and lung infection by a methicillin-resistant Staphylococcus aureus isolate with a vraSR operon deletion. Antimicrob. Agents Chemother. 55:2818–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koehl JL, et al. 2004. Cell wall composition and decreased autolytic activity and lysostaphin susceptibility of glycopeptide-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 48:3749–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Komatsuzawa H, Ohta K, Labischinski H, Sugai M, Suginaka H. 1999. Characterization of fmtA, a gene that modulates the expression of methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 43:2121–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Komatsuzawa H, et al. 1997. Cloning and characterization of the fmt gene which affects the methicillin resistance level and autolysis in the presence of Triton X-100 in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 41:2355–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Łeski TA, Tomasz A. 2005. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J. Bacteriol. 187:1815–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mann EE, et al. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One 4:e5822 doi:10.1371/journal.pone.0005822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McAleese F, et al. 2006. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S. aureus-type resistance to vancomycin. J. Bacteriol. 188:1120–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCallum N, Spehar G, Bischoff M, Berger-Bachi B. 2006. Strain dependence of the cell wall-damage induced stimulon in Staphylococcus aureus. Biochim. Biophys. Acta 1760:1475–1481 [DOI] [PubMed] [Google Scholar]
- 28. Memmi G, Filipe SR, Pinho MG, Fu Z, Cheung A. 2008. Staphylococcus aureus PBP4 is essential for beta-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob. Agents Chemother. 52:3955–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Monnaie D, et al. 1994. Role of residue Lys315 in the mechanism of action of the Enterobacter cloacae 908R beta-lactamase. Biochemistry 33:5193–5201 [DOI] [PubMed] [Google Scholar]
- 30. Navratna V, et al. 2010. Molecular basis for the role of Staphylococcus aureus penicillin binding protein 4 in antimicrobial resistance. J. Bacteriol. 192:134–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oshida T, et al. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. U. S. A. 92:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paradis-Bleau C, et al. 2010. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143:1110–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peschel A, Vuong C, Otto M, Götz F. 2000. The D-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob. Agents Chemother. 44:2845–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pinho MG, Errington J. 2003. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol. Microbiol. 50:871–881 [DOI] [PubMed] [Google Scholar]
- 35. Pinho MG, Errington J. 2005. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol. Microbiol. 55:799–807 [DOI] [PubMed] [Google Scholar]
- 36. Pinho MG, de Lencastre H, Tomasz A. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. U. S. A. 98:10886–10891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pinho MG, Filipe SR, de Lencastre H, Tomasz A. 2001. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J. Bacteriol. 183:6525–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Potluri L, et al. 2010. Sepetal and lateral wall localization of PBP5, the major D-D-carboxypeptidase of Escherichia coli, requires substrate recognition and membrane attachment. Mol. Microbiol. 77:300–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rice KC, Bayles KW. 2008. Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 72:85–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schlag M, et al. 2010. Role of Staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol. Microbiol. 75:864–873 [DOI] [PubMed] [Google Scholar]
- 41. Sobral RG, Jones AE, Des Etages SG. 2007. Extensive and genome-wide changes in the transcription profile of Staphylococcus aureus induced by modulating the transcription of the cell wall synthesis gene murF. J. Bacteriol. 189:2376–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stefanova ME, et al. 2003. Neisseria gonorrhoeae penicillin-binding protein 3 exhibits exceptionally high carboxypeptidase and beta-lactam binding activities. Biochemistry 42:14614–14625 [DOI] [PubMed] [Google Scholar]
- 43. Suzuki T, Campbell J, Swoboda JG, Walker S, Gilmore MS. 2011. Role of wall teichoic acids in Staphylococcus aureus endophthalmitis. Invest. Ophthalmol. Vis. Sci. 52:3187–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tu Quoc PH, et al. 2007. Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect. Immun. 75:1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Typas A, et al. 2010. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143:1097–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Utaida S, Dunman PM, Macapagal D. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719–2732 [DOI] [PubMed] [Google Scholar]
- 47. Vialle S, Sepulcri P, Dubayle J, Talaga P. 2005. The teichoic acid (C-polysaccharide) synthesized by Streptococcus pneumoniae serotype 5 has a specific structure. Carbohydr. Res. 340:91–96 [DOI] [PubMed] [Google Scholar]
- 48. Wada A, Watanabe H. 1998. Penicillin-binding protein 1 of Staphylococcus aureus is essential for growth. J. Bacteriol. 180:2759–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wecke J, Madela K, Fischer W. 1997. The absence of D-alanine from lipoteichoic acid and wall teichoic acid alters surface charge, enhances autolysis and increases susceptibility to methicillin in Bacillus subtilis. Microbiology 143:2953–2960 [DOI] [PubMed] [Google Scholar]
- 50. Weidenmaier C, et al. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomical infections. Nat. Med. 10:243–245 [DOI] [PubMed] [Google Scholar]
- 51. Weidenmaier CaP. 2008. Teichoic acids and related cell wall glycopolymers in gram positive physiology and host interactions. Nat. Rev. Microbiol. 6:276–287 [DOI] [PubMed] [Google Scholar]
- 52. Wilkin JM, Dubus A, Joris B, Frere JM. 1994. The mechanism of action of DD-peptidases: the role of threonine-299 and -301 in the Streptomyces R61 DD-peptidase. Biochem. J. 301(Pt. 2):477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wyke AW, Ward JB. 1975. The synthesis of covalently-linked teichoic acid and peptidoglycan by cell-free preparations of Bacillus licheniformis. Biochem. Biophys. Res. Commun. 65:877–885 [DOI] [PubMed] [Google Scholar]
- 54. Xia G, Kohler T, Peschel A. 2010. The wall teichoic acid and lipoteichoic acid polymers of Staphylococcus aureus. Int. J. Med. Microbiol. 300:148–154 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



