Abstract
Inhibitors of HIV protease have proven to be important drugs in combination anti-HIV therapy. These inhibitors were designed to target mature protease and prevent viral particle maturation by blocking Gag and Gag-Pol processing by mature protease. Currently there are few data assessing the ability of these protease inhibitors to block the initial step in autoproteolytic processing of Gag-Pol. This unique step involves the dimerization of two Gag-Pol polyproteins and autocleavage of the Gag-Pol polyprotein by the embedded dimeric protease. We developed a plasmid encoding a modified form of Gag-Pol that can undergo autoprocessing only at the initial cleavage site between p2 and nucleocapsid. Using an in vitro transcription/translation system, we assessed the ability of six different approved protease inhibitors (darunavir, indinavir, nelfinavir, ritonavir, saquinavir, and tipranavir) to block this initial autocleavage step. Of these inhibitors, darunavir and saquinavir were the most effective. Darunavir and saquinavir were also the most effective at blocking the initial autoprocessing of full-length Gag-Pol in HIV-1-infected T cells. Thus, we have identified at least two HIV-1 protease inhibitors that have activity against the primary autocatalytic step of the embedded HIV-1 protease in Gag-Pol at concentrations that may be attained in HIV-1-infected patients. Due to unique aspects of the initial processing step, it may be possible to develop inhibitors with greater potency against this step, thus halting viral maturation at the earliest stages. The transcription/translation assay could be used to develop more potent inhibitors of this essential first step in viral maturation.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) protease is responsible for processing Gag and Gag-Pol polyproteins, leading to viral maturation. Thus, HIV-1 protease is an important therapeutic target for the treatment of HIV infection and AIDS, since protease inhibitors can prevent the production of infectious virions. Currently, a number of potent protease inhibitors are employed as part of drug combinations used to treat HIV/AIDS patients (for a review, see references 12 and 16). Although these combination regimens are quite effective in the treatment of AIDS, mutations in the protease can develop over time, leading to resistant and multidrug-resistant (MDR) strains of virus that lead to a new challenge in efforts to maintain effective treatment strategies. Viral resistance to protease inhibitors develops primarily in two ways: either directly through mutation of two or more amino acids within the mature, highly active protease (20) or indirectly through mutations of the protease cleavage sites within the Gag and Gag-Pol polyprotein (3, 25, 26).
HIV protease is encoded as part of a Gag-Pol polyprotein. This polyprotein forms a dimer primarily through binding of the embedded protease monomers, and this embedded protease then carries out a series of self-cleavage events (endogenous or cis protease activity). It is not known exactly how this autoprocessing is regulated in vivo, although we have previously shown that in vitro the activity of the initial autocatalytic step can be controlled by reversible oxidation of cysteines 67 and 95 of HIV-1 protease (6). Following a series of cleavage events by the embedded protease, the mature protease is released and can act in trans on Gag and Gag-Pol polyproteins, thus accelerating the maturation process. Due to constraints on the configuration of the embedded protease, its dimeric structure is expected to differ to some degree from that of mature protease. Precursor forms of the protease have lower dimerization affinities than mature protease, further indicating differences between precursor forms of protease and mature protease (14). This initial autocleavage is required for release of the mature protease and production of the mature virions. Moreover, because the structure of the protease in the Gag-Pol polyprotein is somewhat different from that of the mature protease, because the target site is in cis rather than in trans, and because dimerization of the Gag-Pol polyprotein is required before the first-cut autocleavage can take place, this step is a unique target for therapy, separate from the action of the mature protease dimer. Also, for the reasons listed above, one can hypothesize that certain mutations affecting protease inhibitors may not necessarily affect this step and vice versa.
HIV-1 protease inhibitors were initially developed to specifically target the active site of the mature protease. Limited research has been carried out to determine the effectiveness of protease inhibitors on the initial processing step within Gag-Pol. One study has demonstrated that the initial autoprocessing by the embedded protease is 10,000-fold less sensitive (when corrected for enzyme concentration) than mature protease to the inhibitor ritonavir (18). It was also found that the initial autoprocessing step was substantially less sensitive to ritonavir than subsequent processing steps (18), suggesting that the initial autocleavage differs most from cleavage by mature protease. In this study, we used a recently developed in vitro assay to assess the abilities of various protease inhibitors to block the initial intramolecular (cis processing) step of Gag-Pol processing between p2 and nucleocapsid (NC) within the Gag-Pol polyprotein, and we compared this to their abilities to block the processing by exogenous protease (trans processing) at this same site. We then examined the effect of these inhibitors on Gag and Gag-Pol processing in HIV-1-infected cells in culture and compared this to their effects in vitro.
MATERIALS AND METHODS
Plasmid construction and in vitro transcription/translation.
The plasmids pGPfs-wild type (pGPfs-WTHXB2), pGPfs-one cut (pGPfs-1C), and pGPfs-one cut encoding an inactive D25A protease (pGPfs-1Cpr−) were constructed as described previously (6). The pGPfs-1C construct was used to assess the activity of endogenous protease in Gag-Pol toward the p2/NC site in Gag-Pol, while pGPfs-1Cpr− was used to assess the activity of exogenously added protease toward the p2/NC cleavage site. The pGPfs-1C and pGPfs-1Cpr− Gag-Pol-encoding plasmids were constructed by introducing mutations to abolish all but the first processing cut site in pGPfs by site-directed mutagenesis using a Stratagene Quik-Change multisite-directed mutagenesis kit (Stratagene, Santa Clara, CA). Saquinavir (SQV) and ritonavir (RTV) were kindly provided by Roche Products, Ltd. (Welwyn Garden City, United Kingdom) and Abbott Laboratories (Abbott Park, IL), respectively. Nelfinavir (NFV) and indinavir (IDV) were kindly provided by Japan Energy, Inc., Tokyo, Japan. Tipranavir (TPV) was obtained from the NIH AIDS Reagent Program. Darunavir (DRV) was synthesized as previously described (8) and obtained from Arun K. Ghosh (Purdue University, West Lafayette, IN).
Transcription and translation reactions were performed similarly to those described previously using a TnT T7 coupled reticulocyte lysate system (Promega, Madison, WI) except as noted below (6). The reactions (25 μl) were carried out with Easy-Tag [l-35S]methionine (>1,000 Ci/mmol) (Perkin Elmer, San Jose, CA). A master mix was prepared using the TnT rabbit reticulocyte lysate, reaction buffer, T7 polymerase, [35S]methionine, and amino acid mixture (minus methionine) provided with the system at ratios suggested by the manufacturer. Additionally, RNasin RNase inhibitor (Promega, Madison, WI) was added to the master mix at 1 μl per 25-μl reaction mixture. Nuclease-free water was used to bring the final volume of each reaction to 25 μl. The master mix was incubated for 15 min at 30°C, and the DNA plasmid constructs were then added to the master mix at a final concentration of 0.02 μg/μl. The master mix was distributed to individual tubes containing 1 μl of protease inhibitor in dimethyl sulfoxide (DMSO) to provide the desired final concentration. The reaction mixtures were incubated at 30°C for 45 min. For the exogenous protease assay, the reactions with and without protease inhibitors were first carried out for 45 min to produce full-length Gag-Pol. Then, the HIV-1 protease (250 nM final concentration; note that the final active concentration present within the in vitro translation reaction is indeterminate due to potential inhibitory effects of the lysate, refolding, and pH changes in the system) was added as described previously (6), and the reaction mixtures were incubated for an additional 10 min. Following incubation, each reaction was stopped with an equal volume of 3× lithium dodecyl sulfate-polyacrylamide gel electrophoresis (LDS-PAGE) loading buffer containing 150 mM dithiothreitol (DTT) (Invitrogen, Carlsbad, CA) and heated at 70°C for 15 min. The samples were cooled at room temperature for 30 min and loaded on a 1-mm 10-well 4 to 12% Bis-Tris NuPAGE gel (Invitrogen, Carlsbad, CA). The gels were run at 100 V for 10 min and then at 200 V for 70 min. The gels were washed in the following sequence: distilled water for 10 min, 10% acetic acid for 30 min, 10 quick washes with distilled water, and 10% glycerol for 15 min. Following the washing procedure, the gel was placed under a conventional dryer/vacuum for at least 90 min at 54°C and exposed to film for autoradiography using Kodak scientific imaging film (Kodak, Rochester, NY). Films were scanned and Gag-Pol-related bands were quantified via densitometry using Un-Scan-It software (Silk Scientific Corporation, Orem, UT).
Ratios of final product formation to the initial Gag-Pol protein substrate for each sample were determined as described previously (6). The percent processing was calculated as [(P1 + P2)/S1 + P1 + P2] × 100, where P1 and P2 are products and S1 is the substrate. Then, the percent inhibition for samples containing inhibitors was calculated by normalizing the percent processing in each sample containing inhibitor to the control sample with no inhibitor; specifically, percent inhibition was calculated as [(% processing without inhibitor − % processing with inhibitor)/(% processing without inhibitor)] × 100. The mean inhibition and standard deviation were calculated using data from three experiments for the concentrations tested, and a dose-response curve was calculated using Prism 4 (GraphPad Software, La Jolla, CA). Also, 50% inhibitory concentrations (IC50s) were calculated for each experiment, and these individual values were used to compare different drugs using the Student t test. The Student t test was performed on log-transformed data because the drug concentrations spanned several orders of magnitude. All P values are two-sided. A P value of less than or equal to 0.05 was considered significant, while a P value of less than or equal to 0.10 was considered a trend. No adjustment was made for multiple comparisons.
Effect of HIV-1 protease inhibitors on HIV-1 chronically infected H9 cells.
H9 cells chronically infected with HIV-1AO1 (7) were washed twice then plated (Costar, Corning, NY) at 250,000 cells per ml, 3 ml per well, in RPMI medium with Pen/Strep glutamine and 15% fetal calf serum. At least 30 min after plating, protease inhibitors were added at a final concentration of 3.3 or 10 μM (0.05% DMSO was added as a vehicle control to control wells). After 3 days, virus was collected and prepared for Western blotting. Cell suspensions were pelleted by centrifugation at 1,500 × g for 10 min. The supernatant from each treatment was collected and ultracentrifuged at 45,000 × g for 1 h to pellet virus. Viral pellets were washed twice with phosphate-buffered saline (PBS) containing 10 μM darunavir (added postcollection to prevent processing during the wash procedures) and ultracentrifuged at 45,000 × g for 30 min. Pellets were resuspended in 30 μl of 2× LDS-PAGE loading buffer containing 150 mM dithiothreitol (Invitrogen, Carlsbad, CA). Samples were boiled at 95°C for 5 min, and 10 μl of each sample were electrophoresed on a 4 to 12% bis-Tris polyacrylamide gel with MES (morpholineethanesulfonic acid) buffer using the NuPAGE system (Invitrogen, Carlsbad, CA). Proteins were transferred to nitrocellulose using the iBlot system (Invitrogen, Carlsbad, CA) and blocked in 5% dried milk/Tris-buffered saline (TBS) with 0.2% Tween 20 for 1 h at room temperature. After blocking, the membrane was probed for Gag and Gag-Pol processing products using a monoclonal antibody to HIV-1 p24 capsid protein (Advanced Biotechnologies, Columbia, MD), a monoclonal antibody to HIV-1 reverse transcriptase (RT) (Advanced Biotechnologies, Columbia, MD), or a monoclonal antibody to HIV-1 integrase (Int) (Novus, Littleton, CO). Membranes were then washed and incubated for 30 min with anti-mouse secondary antibody conjugated to either alkaline phosphatase or horseradish peroxidase. Bands were detected with either Western Blue stabilized alkaline phosphatase substrate (Promega, Madison, WI) or SuperSignal West Femto substrate (Thermo Scientific, Rockville, IL) as indicated in the figure legends. Minor contrast adjustments were made to the radiograms in PowerPoint (2004), version 11.5.5 (Macintosh).
RESULTS
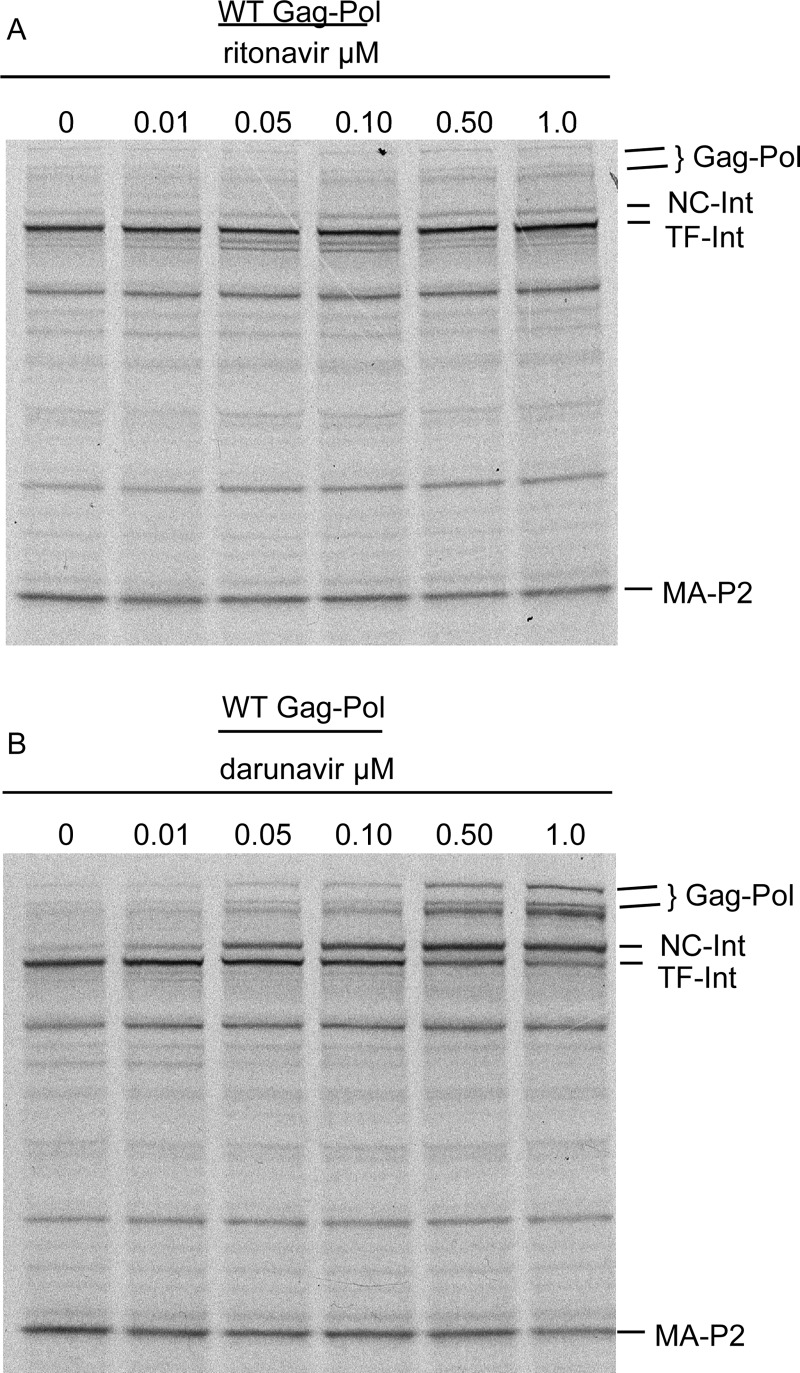
Previous studies demonstrated that the initial step in Gag-Pol processing is unique in that it is very insensitive (5 orders of magnitude difference) to inhibition by the classic protease inhibitor, ritonavir, compared to the mature protease dimer (18). Darunavir, one of the newest approved protease inhibitors for the treatment of HIV-1 infection, is a very potent active site inhibitor (11), and there is evidence to suggest that it can inhibit protease dimerization at higher drug concentrations (9). We were interested in testing the ability of darunavir to block Gag-Pol processing compared to ritonavir. To this end, we tested the ability of both drugs to block wild-type (WT) Gag-Pol processing in an in vitro translation system. A dose-response experiment (0.01 to 1.0 μM) utilizing a WT Gag-Pol-encoding plasmid demonstrated the expected weak activity for ritonavir (19) at preventing the first two processing steps by the embedded protease (cleavage of Gag-Pol at p2/NC and subsequent cleavage within the transframe protein, also known as p6*), resulting in an accumulation of TF-Int (Fig. 1A). In contrast, darunavir at the same concentrations showed a dose-dependent inhibition of both steps, leading to an accumulation of NC-Int and full-length Gag-Pol (Fig. 1B). The second step in processing was considerably more sensitive to darunavir than the first step as evidenced by the accumulation of NC-Int, demonstrating the particular insensitivity of the first step to inhibition by inhibitors in general (Fig. 1).
Fig 1.
Effect of ritonavir and darunavir on WT Gag-Pol processing. In vitro transcription/translation was carried out in the presence of [35S]methionine with pGPfs WT plasmid. The reaction was run in the presence of increasing concentrations of each drug at 0, 0.01, 0.05, 0.10, 0.50, and 1.0 μM as described in Materials and Methods. Radiograms of Gag-Pol processing in the presence of increasing concentrations of ritonavir (A) or darunavir (B). The location for Gag-Pol (Gag-Pol and Gag-Pol internal initiation band), the products of the first step of Gag-Pol processing (NC-Int and MA-p2), and the product of the second step of Gag-Pol processing (TF-Int) are indicated to the right. Shown is a representative of two experiments with similar results.
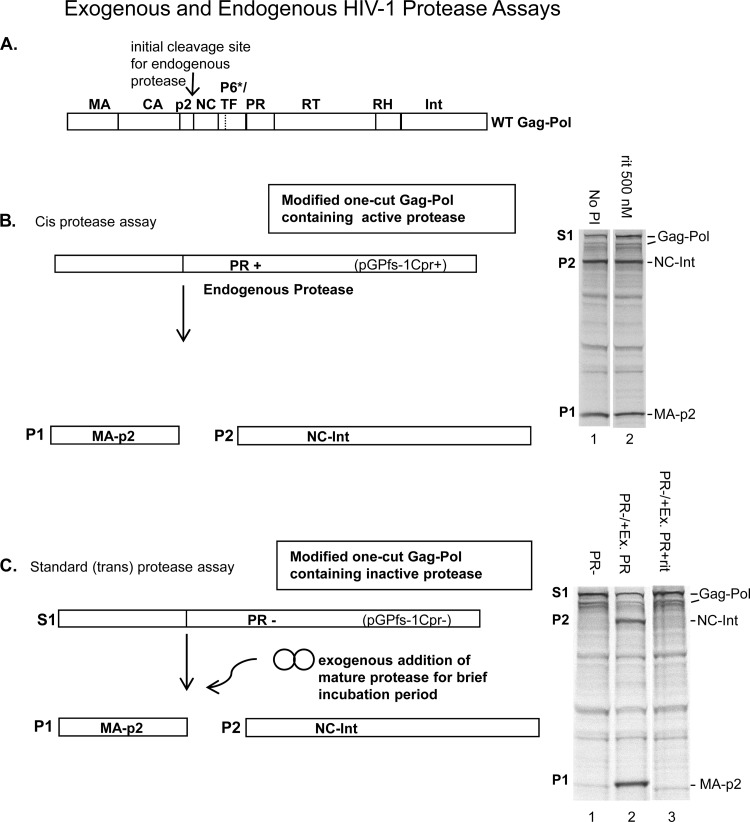
Because of the unique features of the first step in Gag-Pol polyprotein processing, we wanted to examine the ability of darunavir and other approved protease inhibitors to block this step. Six protease inhibitors (darunavir, indinavir, nelfinavir, ritonavir, saquinavir, and tipranavir) (see Fig. S1 in the supplemental material for structures) were screened for their ability to block the initial (autocatalytic) step in polyprotein processing, which involves dimerization of the two Gag-Pol monomers and cleavage between the p2 and NC protein of HIV-1 Gag-Pol (Fig. 2A) (19). To do this, we utilized a recently developed assay that allows measurement of the initial cleavage step by the embedded protease at the p2/NC cleavage site using a full-length mutated Gag-Pol plasmid in an in vitro transcription/translation system (6). HIV-1 Gag-Pol contains multiple cleavage sites (Fig. 2A), and therefore modification of the amino acid sequence of the Gag-Pol polyprotein generated in this assay was important to prevent subsequent cleavages and release of mature protease which could otherwise cleave in trans. In particular, we introduced mutations surrounding the other known polyprotein processing cleavage sites as described previously (6) (Fig. 2B). As expected from previous studies, ritonavir at 500 nM did not appreciably inhibit Gag-Pol processing at the first cut site by the embedded protease (Fig. 2B, compare lane 1 and lane 2 in radiogram) (18). To test processing at the initial cut site by exogenous protease as a comparison, we used a protease-negative (PR−) plasmid encoding Gag-Pol that also contained an active site mutation (D25A) that prevents autocleavage by endogenous protease (Fig. 2C), as described previously (6). As seen in the radiogram in Fig. 2C, no autoprocessing of Gag-Pol takes place with this PR− plasmid (lane 1). However, with the addition of exogenous protease, substantial processing takes place (lane 2) and, as expected (18), ritonavir (500 nM) is very effective at blocking Gag-Pol processing by the exogenously added mature protease (lane 3).
Fig 2.
Schematic diagram for Gag-Pol polyprotein precursor (WT), one-cut Gag-Pol polyprotein precursor containing active endogenous protease, and Gag-Pol precursor containing inactive protease. (A) The WT Gag-Pol contains nine major cleavage sites as shown. The initial processing site is indicated with an arrow at the p2/NC junction, and the processing site within the transframe protein (the second cleavage site) is indicated with a dashed line since it is located within the transframe protein). (B) The one-cut Gag-Pol substrate (S1) retains the initial p2/NC cleavage site and an active embedded HIV-1 protease. Cleavage by the embedded protease yields two major products, P1 and P2 (MA-p2 and NC-Int, respectively) as shown. The substrate and products are indicated in the radiogram to the right (lane 1). As shown, in the presence of 500 nM ritonavir there is little effect on the embedded protease activity as shown previously (lane 2). (C) Schematic diagram of the one-cut Gag-Pol substrate (S1) that retains the initial p2/NC cleavage site but with a mutation (D25A) rendering the embedded HIV-1 protease inactive (PR−). In the absence of protease only unprocessed Gag-Pol is produced by the PR− plasmid construct (lane 1). Cleavage by exogenously added (trans) mature protease yields the same two major products, P1 and P2 (MA-p2 and NC-Int, respectively) (lane 2), as shown in panel B. In the presence of 500 nM ritonavir the activity of the exogenous protease is substantially inhibited (lane 3). The other minor bands in both radiograms are inherent in the in vitro translation system.
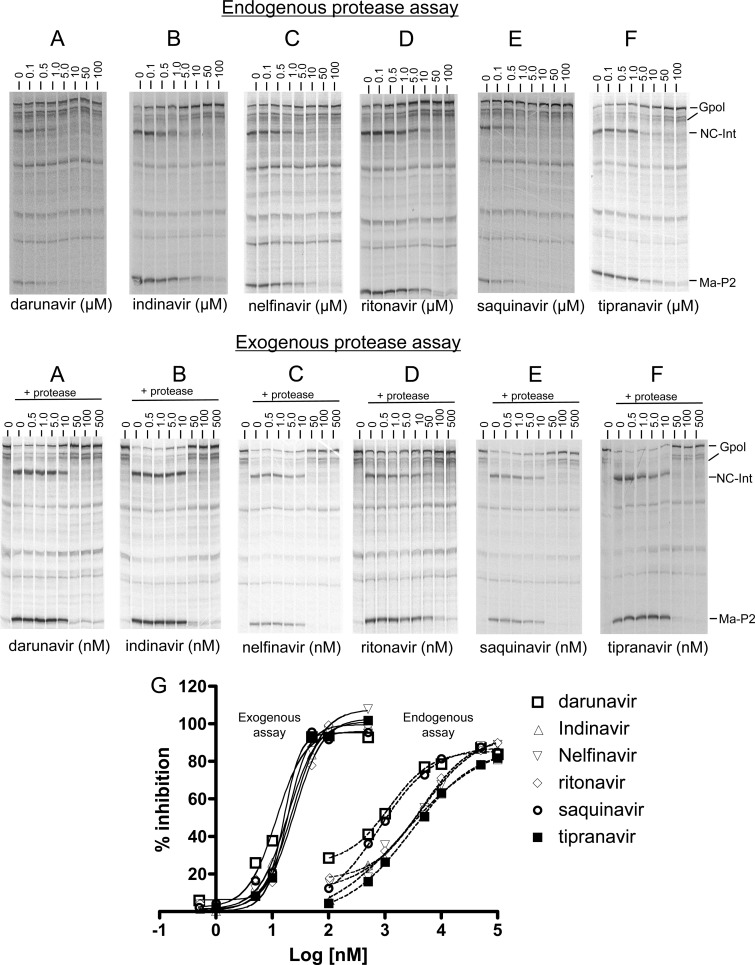
All drugs were tested in the endogenous protease assay at concentrations ranging from 0.01 μM to 100 μM (Fig. 3, top panels). The individual dose-response curves for each drug tested in the endogenous assay and the corresponding error bars at each concentration tested can be found in Fig. S2A in the supplemental material. All drugs had the ability to inhibit the initial processing step in a dose-dependent manner (Fig. 3A to F). At the highest concentration tested (100 μM), all drugs significantly blocked processing of Gag-Pol at the initial cut site by 80% or more (Fig. 2A). However, the dose-response curves for darunavir and saquinavir appeared to be shifted to the left in the endogenous assay (Fig. 3G) compared to those for the other drugs, indicating a relatively greater potency of these drugs in blocking the initial endogenous autocleavage. Darunavir had a calculated IC50 of 1.2 μM (P = 0.03 compared to the IC50 of 4.6 μM for ritonavir) and saquanavir had a calculated IC50 of 1.0 μM (P = 0.08, a trend toward a difference compared to ritonavir) for the initial autocleavage of Gag-Pol in this assay, whereas all the other drugs tested had IC50s of 3.2 μM or more (P > 0.1, neither a significant difference nor a trend compared to ritonavir) (Table 1).
Fig 3.
Inhibition of Gag-Pol cleavage (cis and trans) by HIV-1 protease inhibitors. (Top) For the endogenous protease assay, in vitro transcription/translation was carried out in the presence of [35S]methionine with the one-cut Gag-Pol plasmid encoding active endogenous protease as described in Materials and Methods. The reactions were run in the presence of increasing concentrations of each drug at 0, 0.1, 0.5, 1.0, 5.0, 10, 50, and 100 μM as described in Materials and Methods. (Bottom) For the exogenous assay, in vitro transcription/translation was carried out for 1.5 h in the presence of [35S]methionine with the one-cut Gag-Pol plasmid encoding inactive endogenous protease. HIV-1 protease was then added to each reaction and terminated 10 min later. The reactions were run in the presence of increasing concentrations of each drug at 0, 0.5, 1.0, 5.0, 10, 50, 100, and 500 nM. (G) Graphical overlay for the dose-response curves for each drug in the two assays. The percentage of inhibition for endogenous and exogenous assays was calculated as [(percentage of processing without drug − percentage of processing with drug)/(percentage of processing without drug)] × 100. The percentage of processing for each lane was calculated as [(sum of products)/(substrate + sum of products)] × 100. Each value represents the mean of results of three separate experiments except for that for tipranavir (exogenous), which represents the mean of results of two separate experiments. The individual dose-response curves are shown in Fig. S2 in the supplemental material. The resultant curves were obtained using the sigmoidal nonvariable slope dose-response equation in Prism 4 and represent the mean ± the standard deviation of results of three separate experiments. For tipranavir (exogenous), the mean of results of two experiments and the range are shown.
Table 1.
Plasma levels and activity comparison for protease inhibitors in the endogenous and exogenous Gag-Pol protease assays
| Drug | Cmin–Cmax (μM)a | IC50b |
Relative potency (IC50 end//IC50 ex ratio)c | |
|---|---|---|---|---|
| Endogenous (μM) | Exogenous (nM) | |||
| Darunavir | 4.3–10.7 | 1.2 ± 0.8 | 13.2 +/− 5.1 | 91 |
| Indinavir | 0.67–10.1 | 3.4 ± 1.5 | 21.8 ± 6.4 | 156 |
| Nelfinavir | 2.4–6.6 | 5.3 ± 3.3 | 28 ± 18.8 | 189 |
| Ritonavir | 0.24–1.3 | 4.6 ± 1.4 | 24.5 +/− 6.8 | 188 |
| Saquinavir | 0.11–4.2 | 1.0 ± 0.5 | 18.9 ± 4.4 | 53 |
| Tipranavir | 0.59–30 | 3.2 ± 1.2 | 21.8d | 146 |
Plasma levels for darunavir, indinavir, and saquinavir were obtained when coadministered (boosted) with ritonavir. Plasma levels for darunavir, indinavir, nelfinavir, ritonavir, saquinavir, and tipranavir were from references 23, 4, 10, 23, 1, and 5, respectively, and represent the range of Cmin to Cmax values.
IC50s are expressed as the mean ± standard deviation from results of three separate experiments. The IC50s shown were obtained using the sigmoidal nonvariable slope dose-response equation in Prism 4.
IC50 end, endogenous IC50; IC50 ex, exogenous IC50.
The value for tipranavir in the exogenous assay is the averaged result from two separate experiments (IC50s of 25.2 nM and 18.3 nM).
The six drugs were also screened in the in vitro system utilizing exogenously added mature protease. In this case, a lower range of concentrations (from 0.5 to 500 nM) were used to assess the inhibition of the initial cut site cleavage carried out by exogenous protease (exogenous protease assay). As noted, the construct used in this assay encodes the one-cut Gag-Pol with an inactive D25A embedded protease, so the p2/NC site is available for cleavage only by the exogenously added mature HIV-1 protease. As expected, all the protease inhibitors were highly active in this assay and were significantly more effective at blocking cleavage of the initial cut site by mature dimeric protease than by the embedded protease (Fig. 3A to F). The individual dose-response curves for each drug tested in the exogenous assay and the corresponding error bars at each concentration can be found in Fig. S2B in the supplemental material. Inspection of the radiograms for each drug revealed a significant level of inhibition at concentrations higher than 10 nM (Fig. 3A to F). While IC50s for the drugs in the endogenous protease assay were in the low micromolar range (1.0 to 5.3 μM), the IC50s in this exogenous protease assay clustered in the low nanomolar range (Table 1). An overlay of the dose-response curves did not reveal any striking differences between the drugs in the exogenous assay (Fig. 3G). The relative potency of each drug in the exogenous protease assay is shown in Table 1. Darunavir was the most active in this assay (IC50 = 13.2 nM; P = 0.04 compared to ritonavir, which had an IC50 of 24.5 nM), followed by saquinavir (IC50 = 18.9 nM; P = 0.05 compared to ritonavir, a trend), while none of the other drugs were significantly different from ritonavir. The drugs were all substantially more potent in the exogenous assay compared to the endogenous assay (P < 0.01 for each drug). Comparing the ratio of activities in the endogenous assay to that for exogenous protease (Table 1), the relatively most effective endogenous inhibitor appeared to be saquinavir (IC50 ratio of 1:53), followed by darunavir (IC50 ratio of 1:91); all the other drugs had ratios of more than 1:140. The lower ratio for saquinavir was due to its relatively greater potency in the endogenous assay.
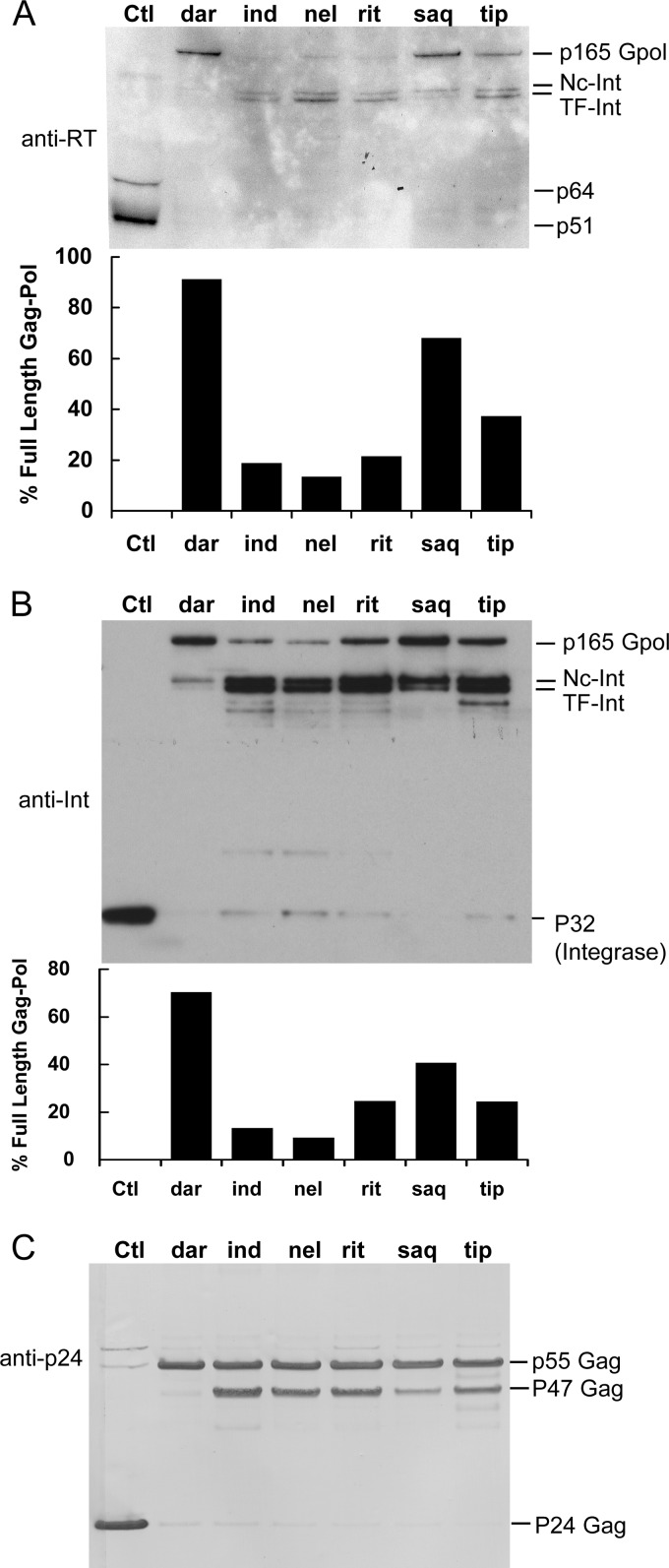
Since these data provided evidence that darunavir and saquinavir were more active at blocking the initial step in Gag-Pol processing than the other drugs tested, we were interested in determining if these two drugs also contributed to relatively greater inhibition of polyprotein processing in HIV-1-infected cells. Therefore, we assessed the ability of the six drugs to block processing of the full-length Gag-Pol polyproteins in H9 T cells chronically infected with HIV-1. These cells were treated for 3 days with the drugs at 10 μm, a concentration that is capable of blocking the endogenous activity of each drug in vitro by more than 50% and also completely blocking mature protease activity (Fig. 3G). Virus released in the supernatant was collected by ultracentrifugation and analyzed by Western blotting with antibodies to reverse transcriptase (RT), integrase (Int), and p24. RT and Int are two proteins found within the Gag-Pol polyprotein but not within the Gag polyprotein. As shown in Fig. 4A, full-length Gag-Pol was readily detected (migration distance equal to that obtained for Gag-Pol produced in the in vitro translation system) with antibody to RT in samples treated with darunavir and saquinavir. Smaller amounts were also detected in the presence of tipranavir (Fig. 4A). The main bands detected on the blot in the presence of the other drugs were bands migrating at locations consistent with the initial processing of Gag-Pol. The measured size (in kDa) of the migrated first cut products of Gag-Pol (doublet labeled NC-Int and TF-Int [Fig. 3]) and their detection with both integrase and RT antibodies (Fig. 4) provide additional evidence that these are products of the first two processing steps of Gag-Pol, formed from cleavage at the p2/NC site and within transframe protein. We performed densitometry on this blot to assess the percentage of full-length Gag-Pol to processed products. The results show that darunavir and saquinavir had the highest percentage of full-length Gag-Pol and were the more effective drugs (Fig. 4A, bar graph).
Fig 4.
Inhibition of HIV-1 p165 Gag-Pol polyprotein processing and HIV-1 p55 Gag processing in the presence of HIV-1 protease inhibitors. H9 cells chronically infected with HIV-1 were treated for 3 days with control (Ctl) vehicle (0.05% DMSO) or 10 μM the indicated inhibitor. The supernatant was collected and virus pelleted by ultracentrifugation. The viral pellets were analyzed by Western blotting with the following monoclonal antibodies. (A) Monoclonal antibody to HIV-1 reverse transcriptase (RT); shown is one representative blot from two experiments. (B) Monoclonal antibody to HIV-1 integrase (Int); shown is a representative blot from three experiments. (C) Monoclonal antibody to HIV-1 p24 capsid protein; shown is a representative blot from three experiments with similar results. The locations for p165 Gag-Pol, Nc-Int, TF-Int, RT subunits (p64 and p51), p55 Gag, p47 Gag, and p24 Gag and Int (integrase) are indicated. Below the blots in panels A and B is the quantification of the percent Gag-Pol over the total bands detected [(pixels for Gag-Pol/total pixels) × 100].
The same samples were also probed with an integrase antibody (Fig. 4B). Overall, Gag-Pol bands were more readily detected with this antibody. Virus treated with vehicle (DMSO) alone contained the expected band for integrase, p32 (Fig. 4B). Interestingly, mature integrase was not detected in the presence of darunavir or saquinavir but small amounts were detected with the other four drugs (Fig. 4B, lanes 3, 4, 5, and 7), indicating that some processing is occurring at the C-terminal end of Gag-Pol. Again, we performed densitometry on the blot to assess the percentage of full-length Gag-Pol to total products. The results were similar to those obtained with antibody to RT, indicating that darunavir and saquinavir were more effective than the other four drugs at blocking full-length Gag-Pol processing, with darunavir being the most potent. Again, accumulation of immature viral proteins was observed for the four drugs that were ineffective at blocking the first step of full-length Gag-Pol processing, providing additional evidence that these drugs could still substantially block subsequent processing of Gag-Pol (doublet in Fig. 4A and 4B). This indicates a clear difference in sensitivity of the first processing step relative to subsequent steps to inhibition by the drugs utilized. These data parallel those seen for the same drugs in the endogenous in vitro translation assay and therefore provide further evidence that saquinavir and darunavir can prevent the initial (cis) autocatalytic step of Gag-Pol in HIV-1-infected cells. We also assessed the effect of the drugs on virus processing using a lower dose (3 μM) of the drugs (see Fig. S3 in the supplemental material). Again, darunavir and saquinavir were clearly the most effective at blocking the processing of Gag-Pol to its mature products compared to all the other drugs (Fig. S3). Overall, these data suggest that the results of the endogenous assay correlate with the activity of these drugs in blocking Gag-Pol polyprotein processing in chronically infected cells.
We also assessed the ability of the drugs to block p55 Gag processing in chronically infected H9 cells using an antibody to the p24 capsid protein of HIV-1. Processing of the Gag polyprotein is effected in trans by the mature protease but does not involve endogenous autocleavage. As the Gag polypeptide is synthesized approximately 20 times more than Gag-Pol during virus production, Western blots probed with the p24 antibody primarily assess differences in the extent of Gag polyprotein processing by mature protease. Virus treated with vehicle only (DMSO) contained the expected band for mature capsid protein, p24 (Fig. 4C). Among the different drugs tested, darunavir and saquinavir appeared to be the most effective at blocking p55 processing (Fig. 4C). In particular, darunavir completely blocked the processing of p55 Gag. This greater effect of darunavir and saquinavir at blocking p55 processing was consistently seen in multiple experiments. This result is consistent with the greater activity of these two drugs against mature protease (Table 1). However, it is also possible that the activity of these drugs in blocking endogenous Gag-Pol autoprocessing (Fig. 3 and Table 1, endogenous results), resulting in less mature protease, contributes to this increased level of inhibition of polyprotein processing.
DISCUSSION
The initial step in Gag-Pol polyprotein processing is unique in that (i) it requires the initial dimerization of full-length Gag-Pol, (ii) it consists of processing at p2/NC via a cis intramolecular cleavage event, and (iii) it was found by Pettit et al. to be approximately 10,000-fold more resistant than fully mature dimeric protease to an active site inhibitor, ritonavir (18). Pettit et al. also found that following the initial step in processing, there was a substantial (27-fold) increase in sensitivity to ritonavir for the second cis cleavage step, which takes place within the transframe protein. This initial cis autocleavage of Gag-Pol is essential for the release of mature protease, as well as other essential HIV proteins, and blocking it will thus prevent viral maturation. Moreover, it is likely that blocking this step will result in a reduction of active protease, and inhibition of this step would be expected to work in tandem with inhibition of mature protease to reduce overall protease activity.
Because the protease embedded in the Gag-Pol polyprotein is constrained by being part of a larger polyprotein, its tertiary structure is most likely different from that of mature protease, and drugs optimized against mature protease may not necessarily target this step. Indeed, all the drugs had substantially less activity against the endogenous protease activity than against the activity of the mature protease added exogenously to the Gag-Pol polyprotein. However, darunavir and saquinavir appeared to be the most active in blocking the initial endogenous Gag-Pol processing step in in vitro translation experiments. These two drugs were also more potent at blocking the activity of exogenously added protease, but they had relatively better activity in the endogenous assay, compared to the exogenous assay, than the other drugs tested. Darunavir and saquinavir were also clearly better than the other four drugs at blocking the initial step of Gag-Pol processing when tested in HIV-1-infected cells. Cursory inspection of the chemical structures of these two drugs (see Fig. S1 in the supplemental material) did not reveal any obvious chemical similarities to explain their unique behavior.
When these drugs were tested on HIV-1-infected cells, all inhibitors were able to block the later steps of Gag-Pol processing to a substantial degree as evidenced by the accumulation of partially processed forms of Gag-Pol detectable with Int and RT antibodies. This further demonstrates the unique insensitivity of the initial step in processing to most active site inhibitors. This difference suggests that a structural change may take place in the embedded protease following the initial cleavage of Gag-Pol that allows much greater binding by the active site protease inhibitors. It should be noted that interpretation of the results with infected cells is complicated because the initial cleavage can occur either in cis by embedded protease or in trans by mature protease, once it forms. At the same time, it is expected that detection of uncleaved Gag-Pol represents inhibition of the initial autocleavage, especially as the drugs tested are highly active toward mature protease. These results are in contrast, to some degree, with the recent work by Louis et al., who demonstrated that one of the late cleavage events, as assessed in a transframe (p6*) precursor form of protease (TF/PR), remains highly resistant to active site inhibitors (13). However, they did find darunavir and saquinavir to be more active at blocking TF/PR precursor processing than the other tested inhibitors in their Escherichia coli expression assay system. In their system, which utilizes an artificially truncated Gag-Pol, the late transframe cleavage event is thought to take place in cis and therefore may have certain properties that resemble the initial cleavage of Gag-Pol. It is possible that these drugs can at least partially inhibit Gag-Pol processing when administered to infected patients. The IC50 for inhibition of Gag-Pol processing by darunavir falls well within the plasma levels reported in patients (23). Also, the IC50 for inhibition of Gag-Pol processing by saquinavir falls within the plasma levels obtained for saquinavir (1). However, studies have shown that inhibitor concentrations required to achieve 95% protease inhibition in the presence of human serum can be up to 10 times higher than those required in the absence of serum (22). Although this suggests that the levels achieved in plasma may not be sufficient to substantially affect the first cut, it is important to note that our assays on the first cut and in infected cells are done in the presence of 60% rabbit reticulocyte or 15% serum, respectively, which could also lead to significant protein binding.
In previous studies, our group found evidence to suggest that darunavir, as well as tipranavir, blocked protease dimerization as measured by a cellular fluorescence resonance energy transfer (FRET) assay (9). These results suggest that the activity of darunavir in the endogenous assay may in part be related to an ability of darunavir to block Gag-Pol dimerization at the protease region. However, tipranavir was not as effective as darunavir or saquinavir in either the endogenous assay or infected cells, and in turn, saquinavir had little activity in blocking the cellular FRET assay, suggesting that factors other than dimerization inhibition may be affecting the relative activities of the drugs in the endogenous assay.
Darunavir has a binding constant in the low picomolar range for the mature HIV protease and induces a different set of resistance mutations than other drugs in its class (15). It is arguably the most potent of the HIV-1 protease inhibitors, and previous studies have indicated that it is more difficult to generate protease-resistant HIV-1 with darunavir than the other existing protease inhibitors (11). Saquinavir is unique in that it induces primarily the L90M and G48V resistance mutations in protease (2), which differ from those induced for other drugs such as indinavir and ritonavir (mutations commonly occur at 46, 63, and 82) (21, 24). It is conceivable that these different mutation patterns are in part related to their activity on the endogenous Gag-Pol protease activity, although additional studies will be needed to sort this out. Also, resistance to darunavir occurs more slowly than to other protease inhibitors, and it is possible that its activity against endogenous Gag-Pol protease activity, as well as against the active protease, may provide constraints on the development of resistance mutations. Our one-cut Gag-Pol assay system could provide a means to determine if patients receiving darunavir or saquinavir develop resistance mutations that directly affect the ability of the embedded protease to undergo initial processing in the presence of these drugs.
The results of the current study show that certain active site protease inhibitors and especially darunavir and saquinavir have some limited activity against endogenous autocleavage by embedded Gag-Pol, and this may contribute to the activity of these drugs in patients. However, the activity of these drugs is relatively weak compared to their activity against mature protease, and it may be worthwhile to develop new agents that preferentially target this initial step. The tertiary structure of embedded Gag-Pol is likely to be different from that of mature protease because it is part of a larger structure and is tethered to its substrate. In addition, its activity is initially focused on a single first-cut site, which may further affect the target structure for drug inhibition. Also, dimerization of Gag-Pol needs to take place before the embedded protease can begin autoprocessing; therefore, Gag-Pol may be targeted by dimerization inhibitors. In contrast, once the mature protease dimer forms, the strong affinity for dimeric protease may make it difficult to reverse the dimerization. Thus, it may be possible to develop novel drugs whose activity is targeted against this initial autocleavage step, possibly by blocking Gag-Pol dimerization, as suggested by others (13), or through binding to the Gag-Pol active site. Because Gag-Pol autocleavage is essential to produce a mature protease, such a drug may work cooperatively with mature protease inhibitors to block viral maturation. Another potential advantage of such drugs is that they would more completely prevent the release of the essential viral enzymes RT and Int from Gag-Pol than mature protease inhibitors.
Previous work by Kaplan et al. showed that the order of Gag-Pol processing by the wild-type protease is highly conserved (17) and that mutations in the protease alter the cleavage pattern due to changes in the ability to interact with Gag-Pol (17). Because the structure and constraints of this step may be different from that of mature protease, different mutations may confer resistance to this step than to that of mature protease, and drugs or drug combinations that inhibit both the first cut and mature protease may help thwart the development of resistant mutations. The assay described in this paper may have utility in helping to examine additional drugs for activity against this unique step in the viral replication cycle and to help in the development of novel agents acting at this step. These results may also lead to the development of additional novel assays to screen for inhibitors of this step.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
We thank Hiroaki Mitsuya for his interesting and helpful discussions.
Footnotes
Published ahead of print 16 April 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Bickel M, et al. 2009. Once-daily treatment with saquinavir mesylate (2000 mg) and ritonavir (100 mg) together with a fixed-dose combination of abacavir/lamivudine (600/300 mg) or tenofovir/emtricitabine (245/200 mg) in HIV-1-infected patients. J. Antimicrob. Chemother. 64:1260–1264 [DOI] [PubMed] [Google Scholar]
- 2. Bragman K. 1996. Saquinavir: an HIV proteinase inhibitor. Adv. Exp. Med. Biol. 394:305–317 [DOI] [PubMed] [Google Scholar]
- 3. Burlet S, et al. 2005. Prospects for the resistance to HIV protease inhibitors: current drug design approaches and perspectives. Curr. Pharm. Des. 11:3077–3090 [DOI] [PubMed] [Google Scholar]
- 4. Cressey TR, et al. 2005. Low-doses of indinavir boosted with ritonavir in HIV-infected Thai patients: pharmacokinetics, efficacy and tolerability. J. Antimicrob. Chemother. 55:1041–1044 [DOI] [PubMed] [Google Scholar]
- 5. Croom K. F., Keam. S. J. 2005. Tipranavir: a ritonavir-boosted protease inhibitor. Drugs 65:1669–1677; discussion, 1678–1679 [DOI] [PubMed] [Google Scholar]
- 6. Daniels SI, et al. 2010. The initial step in human immunodeficiency virus type 1 GagProPol processing can be regulated by reversible oxidation. PLoS One 5:e13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davis DA, et al. 2006. Inhibition of HIV-1 replication by a peptide dimerization inhibitor of HIV-1 protease. Antiviral Res. 72:89–99 [DOI] [PubMed] [Google Scholar]
- 8. Ghosh AK, Leshchenko S, Noetzel M. 2004. Stereoselective photochemical 1,3-dioxolane addition to 5-alkoxymethyl-2(5H)-furanone: synthesis of bis-tetrahydrofuranyl ligand for HIV protease inhibitor UIC-94017 (TMC-114). J. Org. Chem. 69:7822–7829 [DOI] [PubMed] [Google Scholar]
- 9. Koh Y, et al. 2007. Potent inhibition of HIV-1 replication by novel non-peptidyl small molecule inhibitors of protease dimerization. J. Biol. Chem. 282:28709–28720 [DOI] [PubMed] [Google Scholar]
- 10. Kruse G, et al. 2005. The steady-state pharmacokinetics of nelfinavir in combination with tenofovir in HIV-infected patients. Antivir. Ther. 10:349–355 [PubMed] [Google Scholar]
- 11. Lefebvre E, Schiffer CA. 2008. Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir. AIDS Rev. 10:131–142 [PMC free article] [PubMed] [Google Scholar]
- 12. Llibre JM. 2009. First-line boosted protease inhibitor-based regimens in treatment-naive HIV-1-infected patients—making a good thing better. AIDS Rev. 11:215–222 [PubMed] [Google Scholar]
- 13. Louis JM, Aniana A, Weber IT, Sayer JM. 2011. Inhibition of autoprocessing of natural variants and multidrug resistant mutant precursors of HIV-1 protease by clinical inhibitors. Proc. Natl. Acad. Sci. U. S. A. 108:9072–9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Louis JM, Clore GM, Gronenborn AM. 1999. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat. Struct. Biol. 6:868–875 [DOI] [PubMed] [Google Scholar]
- 15. Mitsuya Y, Liu TF, Rhee SY, Fessel WJ, Shafer RW. 2007. Prevalence of darunavir resistance-associated mutations: patterns of occurrence and association with past treatment. J. Infect. Dis. 196:1177–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Naggie S, Hicks C. 2010. Protease inhibitor-based antiretroviral therapy in treatment-naive HIV-1-infected patients: the evidence behind the options. J. Antimicrob. Chemother. 65:1094–1099 [DOI] [PubMed] [Google Scholar]
- 17. Pettit SC, Clemente JC, Jeung JA, Dunn BM, Kaplan AH. 2005. Ordered processing of the human immunodeficiency virus type 1 GagPol precursor is influenced by the context of the embedded viral protease. J. Virol. 79:10601–10607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pettit SC, Everitt LE, Choudhury S, Dunn BM, Kaplan AH. 2004. Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism. J. Virol. 78:8477–8485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pettit SC, Gulnik S, Everitt L, Kaplan AH. 2003. The dimer interfaces of protease and extra-protease domains influence the activation of protease and the specificity of GagPol cleavage. J. Virol. 77:366–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ridky T, Leis J. 1995. Development of drug resistance to HIV-1 protease inhibitors. J. Biol. Chem. 270:29621–29623 [DOI] [PubMed] [Google Scholar]
- 21. Roberts NA. 1995. Drug-resistance patterns of saquinavir and other HIV proteinase inhibitors. AIDS 9(Suppl. 2):S27–S32 [PubMed] [Google Scholar]
- 22. Schon A, del Mar Ingaramo M, Freire E. 2003. The binding of HIV-1 protease inhibitors to human serum proteins. Biophys. Chem. 105:221–230 [DOI] [PubMed] [Google Scholar]
- 23. Sekar V, et al. 2010. Pharmacokinetics of darunavir/ritonavir and rifabutin coadministered in HIV-negative healthy volunteers. Antimicrob. Agents Chemother. 54:4440–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Svicher V, et al. 2005. Novel human immunodeficiency virus type 1 protease mutations potentially involved in resistance to protease inhibitors. Antimicrob. Agents Chemother. 49:2015–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yusa K, Harada S. 2004. Acquisition of multi-PI (protease inhibitor) resistance in HIV-1 in vivo and in vitro. Curr. Pharm. Des. 10:4055–4064 [DOI] [PubMed] [Google Scholar]
- 26. Zhang YM, et al. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 71:6662–6670 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.