Abstract
A total of 403 nonduplicate isolates of Clostridium difficile were collected at three major teaching hospitals representing northern, central, and southern Taiwan from January 2005 to December 2010. Of these 403 isolates, 170 (42.2%) were presumed to be nontoxigenic due to the absence of genes for toxins A or B or binary toxin. The remaining 233 (57.8%) isolates carried toxin A and B genes, and 39 (16.7%) of these also had binary toxin genes. The MIC90 of all isolates for fidaxomicin and rifaximin was 0.5 μg/ml (range, ≤0.015 to 0.5 μg/ml) and >128 μg/ml (range, ≤0.015 to >128 μg/ml), respectively. All isolates were susceptible to metronidazole (MIC90 of 0.5 μg/ml; range, ≤0.03 to 4 μg/ml). Two isolates had reduced susceptibility to vancomycin (MICs, 4 μg/ml). Only 13.6% of isolates were susceptible to clindamycin (MIC of ≤2 μg/ml). Nonsusceptibility to moxifloxacin (n = 81, 20.1%) was accompanied by single or multiple mutations in gyrA and gyrB genes in all but eight moxifloxacin-nonsusceptible isolates. Two previously unreported gyrB mutations might independently confer resistance (MIC, 16 μg/ml), Ser416 to Ala and Glu466 to Lys. Moxifloxacin-resistant isolates were cross-resistant to ciprofloxacin and levofloxacin, but some moxifloxacin-nonsusceptible isolates remained susceptible to gemifloxacin or nemonoxacin at 0.5 μg/ml. This study found the diversity of toxigenic and nontoxigenic strains of C. difficile in the health care setting in Taiwan. All isolates tested were susceptible to metronidazole and vancomycin. Fidaxomicin exhibited potent in vitro activity against all isolates tested, while the more than 10% of Taiwanese isolates with rifaximin MICs of ≥128 μg/ml raises concerns.
INTRODUCTION
Clostridium difficile infection (CDI) is a major nosocomial threat and may surpass methicillin-resistant Staphylococcus aureus in some settings (28). Although the two most common therapies for CDI, metronidazole and vancomycin, are effective in resolving most cases (4, 7), there is concern that efficacy of metronidazole is declining in recent outbreaks and that overuse of vancomycin can lead to selection of vancomycin-resistant enterococci (2, 3, 7, 30, 40). Approximately 20 to 30% of patients have recurrence of CDI after successful treatment with metronidazole or vancomycin. In patients with multiple recurrences, tapered doses of vancomycin or use of a rifaximin “chaser” are sometimes effective (4, 7, 14, 15).
Not all C. difficile strains are pathogenic. Toxigenic strains harbor genes carried by the pathogenicity locus (PaLoc), including cdtA encoding enterotoxin A and cdtB encoding enterotoxin B as well as a negative regulator of their expression, cdtC (9). Emergence of a particularly virulent strain since 2000 has accounted for increased mortality in outbreaks in Europe, Canada, and the United States (24, 27, 29, 32, 39). This strain, restriction endonuclease analysis group type BI/pulsed-field gel electrophoresis type 1/PCR ribotype 027 (BI/NAP1/027), is characterized by its resistance to fluoroquinolones, mutations in the cdtC gene, and expression of an ADP-ribosylating binary toxin, encoded outside the PaLoc locus and not expressed in most toxigenic strains (31). Furthermore, the link between toxin profiles, antibiotypes (including clindamycin and quinolones), and epidemicity is important given the emergence and epidemic spread of pathogenic strains of C. difficile (33).
To date, BI/NAP1/027 has not been documented in Taiwan (5, 20, 25, 26). However, C. difficile clinical isolates resistant to fluoroquinolones have been found (26). Greater awareness in Taiwan in the last decade has prompted retrospective and prospective surveillance studies in some hospitals. Hsu et al. reported an incidence of 8 cases per 1,000 patient-days in Northern Taiwan during a 3-month period in 2003 (20). The same hospital conducted a 5-month prospective surveillance in high-risk units of the same hospital during 2010 and found a much lower incidence of 0.45 cases per 1,000 patient-days after initiating an aggressive hand-washing program (5, 25). In a teaching hospital in Southern Taiwan over a 15-month period during 2007 to 2008, a very similar rate of 0.43 cases per 1,000 patient-days was recorded, with a higher rate of 1.1 cases per 1,000 patient-days in the intensive care unit (5).
We recently reported the antibiotic susceptibility profiles and molecular epidemiology of 113 C. difficile isolates from two major teaching hospitals in Northern and Southern Taiwan (26). In the current study, we extend these results to the molecular and microbiological characterization of 403 isolates from three hospitals representing northern, central, and southern Taiwan. Susceptibility to clindamycin and major fluoroquinolones, a nonfluorinated quinolone (nemonoxacin), and antibiotics used clinically against CDI are reported and compared to genotypes for PaLoc toxins A and B and binary toxin and mutations in the DNA gyrase A and B genes. We also included fidaxomicin, a macrocyclic antibiotic with high specificity for C. difficile and inhibitory activity toward C. difficile RNA polymerase, and another RNA polymerase inhibitor, rifaximin, in this study.
MATERIALS AND METHODS
Bacterial isolates.
A total of 403 nonduplicate isolates of C. difficile, including 332 isolates from National Taiwan University Hospital (NTUH), 40 from National Cheng Kung University Hospital (NCKUH), and 31 from China Medical University from January 2005 to December 2010, were obtained for analysis. These isolates were recovered from stool specimens of patients with unexplained fever or concurrent gastrointestinal symptoms, such as diarrhea, abdominal discomfort, or ileus. Not all patients were confirmed as having CDI by toxin assays.
Antimicrobial susceptibility testing.
MICs of the 403 isolates to 12 antimicrobial agents were determined using the agar dilution method recommended by the Clinical and Laboratory Standards Institute (CLSI) (6), with the exception of daptomycin. An inoculum of 105 CFU of bacteria was applied to each plate of supplemented Brucella blood agar (BBL Microbiology Systems, Cockeysville, MD) using a Steers replicator. The plates were incubated in an anaerobic chamber for 48 h at 35°C. For daptomycin susceptibility assays, the broth microdilution method using Brucella broth with hemin (5 μg/ml), vitamin K1 (1 μg/ml), lysed horse blood (5%), and calcium (50 μg/ml) was used (6). The 12 antimicrobial agents used for susceptibility testing were obtained from their corresponding manufacturers: fidaxomicin (Optimer Pharmaceuticals Inc., San Diego, CA); rifaximin, vancomycin, and metronidazole (Sigma, St. Louis, MO); ciprofloxacin and moxifloxacin (Bayer Co., West Haven, CT); levofloxacin (Daiichi Pharmaceuticals, Tokyo, Japan); gemifloxacin (LG Chem Investments, Seoul, South Korea); nemonoxacin (TaiGen Biotechnology, Co. Ltd., Taipei, Taiwan); daptomycin (Cubist Pharmaceuticals, Lexington, MA); and tigecycline (Pfizer Inc., New York, NY).
The MIC was defined as the lowest concentration of each antimicrobial agent that inhibited the growth of the tested isolate. C. difficile ATCC 700057 and Bacteroides fragilis ATCC 25285 were used for quality control for each run of susceptibility testing. The MIC interpretive breakpoints for metronidazole, clindamycin, and moxifloxacin followed the guidelines recommended by the CLSI (6), and breakpoints for vancomycin (susceptible, MIC of ≤2 μg/ml; and resistant, MIC of >2 μg/ml) were those recommended by The European Committee on Antimicrobial Susceptibility Testing (EUCAST) (13) (Table 1). Breakpoints are not established for rifaximin and fidaxomicin.
Table 1.
In vitro susceptibilities of 403 isolates of C. difficile to fidaxomicin, rifaximin, and 10 other antimicrobial agents
| Agent | MIC (μg/ml)a |
No. (%) of isolates |
|||
|---|---|---|---|---|---|
| Range | 50% | 90% | Susceptible | Resistant | |
| Fidaxomicin | ≤0.015–0.5 | 0.12 | 0.25 | ||
| Rifaximin | ≤0.015–>128 | ≤0.015 | >128 | ||
| Metronidazoleb | ≤0.03–4 | 0.5 | 0.5 | 403 (100) | 0 (0) |
| Vancomycinc | 0.06–4 | 0.5 | 1 | 401 (99.5) | 2 (0.5) |
| Clindamycinb | 0.06–>256 | 8 | >256 | 55 (13.6) | 296 (73.5) |
| Ciprofloxacin | 0.5–128 | 16 | 64 | ||
| Moxifloxacinb | 0.06–32 | 2 | 16 | 322 (79.9) | 72 (17.9) |
| Levofloxacin | 1–>128 | 4 | 128 | ||
| Gemifloxacin | 0.25–>32 | 2 | 32 | ||
| Nemonoxacin | 0.25–>32 | 1 | 8 | ||
| Tigecycline | ≤0.03–1 | 0.06 | 0.06 | ||
| Daptomycin | 0.06–8 | 1 | 1 | ||
MICs were determined by the agar dilution method with the exception of daptomycin, for which the broth microdilution method was used.
MIC breakpoints applied were those recommended for anaerobes by the Clinical and Laboratory Standards Institute (CLSI-2007, M11-A7) (6). For metronidazole, susceptible, ≤8 μg/ml; resistant, ≥ 32 μg/ml. For clindamycin, susceptible, ≤2 μg/ml; resistant, ≥ 8 μg/ml. For moxifloxacin, susceptible, ≤2 μg/ml; resistant, ≥ 8 μg/ml.
For vancomycin there are no CLSI-recommended MIC breakpoints. Breakpoints are those recommended by The European Committee on Antimicrobial Susceptibility Testing (EUCAST) (susceptible, ≤2 μg/ml; resistant, >2 μg/ml) (13). The two isolates resistant to vancomycin both had vancomycin MICs of 4 μg/ml.
Genotyping and sequencing.
Presence of tcdA, tcdB, cdtA, and cdtB genes were determined by multiplex PCR as described previously (13). Moxifloxacin-nonsusceptible isolates (moxifloxacin MICs of ≥4 μg/ml) were subjected to partial sequencing of gyrA and gyrB genes after PCR amplification of 390-bp fragments of each (13).
RESULTS
Antimicrobial susceptibilities.
Susceptibilities to 16 antimicrobial agents, including 4 used clinically to treat CDI (metronidazole, vancomycin, rifaximin, and fidaxomicin), were determined for 403 clinical isolates of C. difficile. Susceptibilities of the test strain C. difficile ATCC 700057 to fidaxomicin, rifaximin, vancomycin, and metronidazole were with the CLSI standard ranges, and susceptibility of B. fragilis ATCC 25285 to metronidazole was also within the established range. Table 1 presents the ranges of MIC values of individual isolates and MIC50 and MIC90 values for each agent. Distribution of isolates according to susceptibility is presented for four antibiotics for which MIC breakpoints have been established.
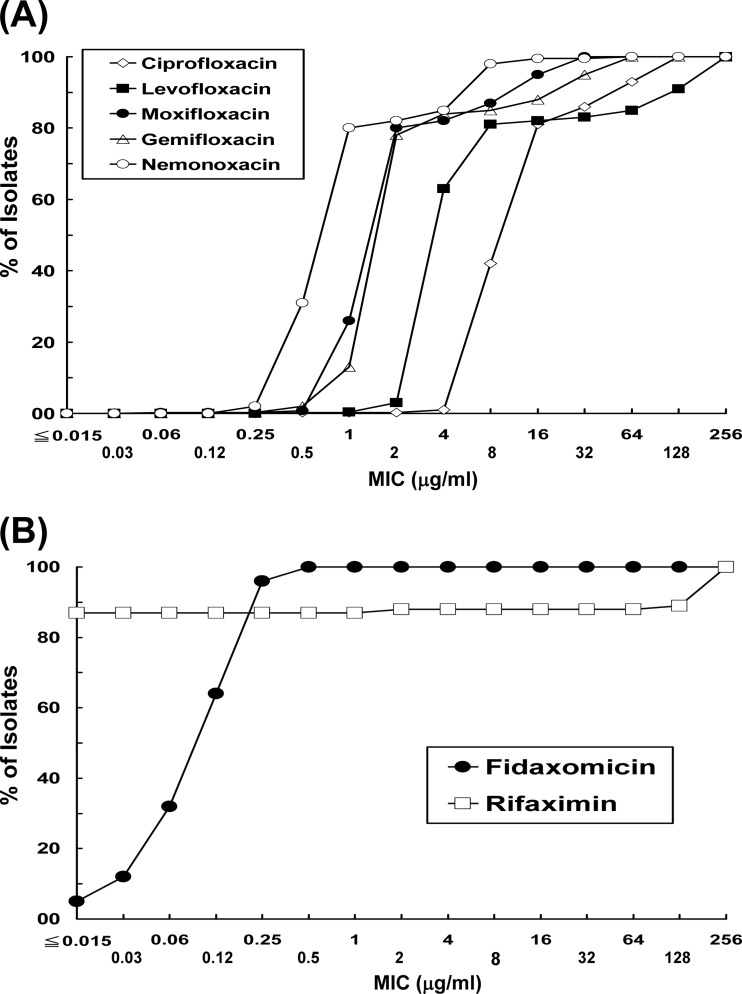
Only 13.6% of isolates were fully susceptible to clindamycin, and the MIC90 value was >256 μg/ml. Among the fluoroquinolones, susceptibility was lowest for levofloxacin (MIC90, 128 μg/ml), followed by ciprofloxacin (MIC90, 64 μg/ml), gemifloxacin (MIC90, 2 μg/ml), and moxifloxacin (MIC90, 16 μg/ml); 80% of isolates were fully susceptible to moxifloxacin. Isolates were more susceptible to the nonfluorinated quinolone, nemonoxacin (MIC90, 8 μg/ml; range 0.25 to >32 μg/ml). The majority of isolates were inhibited by tigecycline (MIC90, 0.06 μg/ml) and daptomycin (MIC90, 1 μg/ml), although the range of MIC values was 0.06 to 8 μg/ml for daptomycin. Distribution of isolates by MIC value is plotted in Fig. 1A for the quinolones. Susceptibility can be ranked in order as nemonoxacin (most susceptible), moxifloxacin/gemifloxacin, levofloxacin, and ciprofloxacin (least susceptible).
Fig 1.
Distribution of MICs among clinical isolates of C. difficile to five quinolones (A) and fidaxomicin and rifaximin (B).
Two isolates (0.5%) were resistant to vancomycin (MIC, 4 μg/ml) by EUCAST criteria, and none were resistant to metronidazole. Ninety percent of isolates were susceptible to metronidazole at 0.5 μg/ml (range, ≤0.03 to 4 μg/ml), and 90% were inhibited by vancomycin at 1 μg/ml (range, 0.06 to 4 μg/ml). Fidaxomicin exhibited potent in vitro activities against these isolates; the range of MIC values was ≤0.015 to 0.5 μg/ml, and 90% of isolates were inhibited at 0.25 μg/ml. In contrast, there was a wide range of MICs to rifaximin (≤0.015 to >128 μg/ml; MIC90 of >128 μg/ml): 352 (87.3%) isolates with MICs of ≤0.25 μg/ml, four (1.0%) with MICs of 4 μg/ml, three (0.7%) with MICs of 128 μg/ml, and 44 (10.9%) with MICs of >128 μg/ml (Fig. 1B).
Genotypes and antimicrobial susceptibilities.
Of the 403 isolates, 57.8% (233/403) were potentially toxigenic by genotype, carrying both tcdA and tcdB genes. Of those, 16.7% (39/233) also possessed the genes for the binary toxin, cdtA and cdtB; this represents 9.6% (39/403) of all isolates (Tables 2 and 3). The remaining 42.2% (170/403) of isolates did not carry the genes encoding toxins A and B and are presumably not toxigenic. There was no clear pattern linking antibiotic susceptibility to genotype according to the toxin genes, with the exception that many tcdA+B+ cdtA−B− isolates were resistant to rifaximin, and the MIC90 value was >128 μg/ml. Susceptibilities to fidaxomicin were identical across all three genotypes, and susceptibilities to vancomycin and metronidazole differed by no more than 2-fold between genotypes. The 39 isolates carrying the binary toxin genes tended to be more susceptible to clindamycin (MIC90, 32 μg/ml) than those without cdtA and cdtB genes (MIC90, >256 μg/ml). There was no more than a 2-fold difference in MIC90 values between genotypes for the remaining eight agents.
Table 2.
Pathogenicity locus and binary toxin genotypes and in vitro susceptibilities of C. difficile isolates to fidaxomicin, rifaximin, and other antimicrobial agents
| Agent | MIC (μg/ml)a |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
tcdA+
tcdB+
cdtA+
cdtB+ (n = 39 isolates) |
tcdA+
tcdB+
cdtA−
cdtB− (n = 194 isolates) |
tcdA−
tcdB−
cdtA−
cdtB− (n = 170 isolates) |
|||||||
| Range | 50% | 90% | Range | 50% | 90% | Range | 50% | 90% | |
| Fidaxomicin | ≤0.015–0.5 | 0.12 | 0.25 | ≤0.015–0.5 | 0.12 | 0.25 | ≤0.015–0.5 | 0.12 | 0.25 |
| Rifaximin | ≤0.015–2 | 0.015 | 0.015 | ≤0.015–>128 | 0.015 | >128 | ≤0.015–>128 | 0.015 | 0.06 |
| Metronidazoleb | 0.12–1 | 0.25 | 1 | ≤0.03–1 | 0.5 | 0.5 | 0.06–4 | 0.5 | 1 |
| Vancomycinc | 0.25–2 | 0.5 | 0.5 | 0.06–4 | 0.5 | 1 | 0.25–4 | 0.5 | 1 |
| Clindamycinb | 0.12–>256 | 8 | 32 | 0.5–>256 | 16 | >256 | 0.06–>256 | 8 | >256 |
| Ciprofloxacin | 8–128 | 8 | 64 | 4–128 | 16 | 64 | 0.5–128 | 16 | 64 |
| Moxifloxacinb | 1–32 | 2 | 16 | 0.06–32 | 2 | 16 | 0.5–32 | 2 | 16 |
| Levofloxacin | 2–>128 | 4 | >128 | 1–>128 | 4 | 128 | 1–>128 | 4 | 128 |
| Gemifloxacin | 1–>32 | 2 | >32 | 0.5–>32 | 2 | 16 | 0.25–>32 | 2 | 32 |
| Nemonoxacin | 0.25–16 | 1 | 8 | 0.25–>32 | 1 | 8 | 0.25–>32 | 1 | 8 |
| Tigecycline | ≤0.03–0.12 | 0.06 | 0.12 | ≤0.03–0.5 | 0.06 | 0.06 | ≤0.03–1 | 0.06 | 0.06 |
| Daptomycin | 0.25–4 | 1 | 2 | 0.06–4 | 0.5 | 1 | 0.06–8 | 0.5 | 1 |
MICs were determined by the agar dilution method with the exception of daptomycin, for which the broth microdilution method was used.
MIC breakpoints applied were those recommended for anaerobes by the Clinical and Laboratory Standards Institute (CLSI-2007, M11-A7) (6). For metronidazole, susceptible, ≤8 μg/ml; resistant, ≥ 32 μg/ml. For clindamycin, susceptible, ≤2 μg/ml; resistant, ≥ 8 μg/ml. For moxifloxacin, susceptible, ≤2 μg/ml; resistant, ≥ 8 μg/ml.
For vancomycin there are no CLSI-recommended MIC breakpoints. Breakpoints are those recommended by The European Committee on Antimicrobial Susceptibility Testing (EUCAST) (susceptible, ≤2 μg/ml; resistant, >2 μg/ml) (13). The two isolates resistant to vancomycin both had vancomycin MICs of 4 μg/ml.
Table 3.
Susceptibility distribution of 403 clinical isolates of C. difficile by genotypes to four agents with MIC interpretive breakpoints by the Clinical and Laboratory Standards Institute (6)
| Agent | No. (%) of isolates for each genotype |
|||||
|---|---|---|---|---|---|---|
|
tcdA+
tcdB+
cdtA+
cdtB+ (n = 39 isolates) |
tcdA+
tcdB+
cdtA−
cdtB− (n = 194 isolates) |
tcdA−
tcdB−
cdtA−
cdtB− (n = 170 isolates) |
||||
| Susceptible | Resistant | Susceptible | Resistant | Susceptible | Resistant | |
| Metronidazolea | 39 (100) | 0 (0) | 194 (100) | 0 (0) | 170 (100) | 0 (0) |
| Vancomycinb | 39 (100) | 0 (0) | 193 (99) | 1 (1) | 169 (99) | 1 (1) |
| Clindamycina | 8 (21) | 24 (62) | 14 (7) | 151 (78) | 33 (19) | 121 (71) |
| Moxifloxacina | 29 (74) | 10 (26) | 157 (81) | 33 (17) | 136 (80) | 29 (17) |
MIC breakpoints applied were those recommended for anaerobes by the Clinical and Laboratory Standards Institute (CLSI-2007, M11-A7) (6).
For vancomycin there are no CLSI-recommended MIC breakpoints. Breakpoints are those recommended by The European Committee on Antimicrobial Susceptibility Testing (EUCAST) (13).
For those agents with resistance breakpoints defined, results are presented by distribution of isolates among susceptible, intermediate, and resistant classes according to genotype. There was no pattern linking resistance to genotype around the toxins A and B and binary toxin genes, although neither of the two vancomycin-resistant isolates carried the binary toxin genes. Isolates with reduced susceptibility to moxifloxacin (MIC ≥ 4 μg/ml), as well as the other fluoroquinolones, were found in all genotype classes: tcdA+B+ cdtA+B+ (10/39, 25.6%); tcdA+B+ cdtA−B− (37/194, 19.1%); and tcdA−B− cdtA−B− (34/170, 20.0%).
Gyrase mutations.
Of the 403 isolates, 81 (20.1%) had reduced susceptibility to moxifloxacin (MIC ≥ 4 μg/ml) and 72 (88.9%) of those were fully resistant (MIC ≥ 8 μg/ml) (Table 4). All but 8 isolates with reduced susceptibility to moxifloxacin harbored amino acid substitutions in gyrA alone (52 isolates), gyrB alone (16 isolates), or both (5 isolates). One had multiple gyrA mutations (at Asp81, Arg90, Asp103, and Glu123) and a single gyrB substitution (at Asp426), and another had 6 gyrB amino acid substitutions as well as a Thr82-to-Ile substitution in gyrA. The most common substitution in gyrA was Thr82 to Ile (52/57 isolates with gyrA mutations), and the most common gyrB substitution was Asp426 to Asn (9/21 isolates with gyrB mutations). There was a high level of cross-resistance to ciprofloxacin and levofloxacin, but some isolates remained susceptible to gemifloxacin or nemonoxacin at 0.5 μg/ml.
Table 4.
MICs of quinolones and substitutions in GyrA and GyrB for 81 isolates of C. difficile with reduced susceptibility to moxifloxacin (MIC ≥ 4 μg/ml)
| No. of isolates with indicated MICs | MIC (μg/ml)a |
No. of isolates with indicated amino acid substitutions | Amino acid substitutionsb |
|||||
|---|---|---|---|---|---|---|---|---|
| Moxi | Cipro | Levo | Gemi | Nemo | GyrA | GyrB | ||
| 9 | 4 | 8–32 | 4–>64 | 2–>32 | 0.5–8 | 2 | Thr82 to Ile | NF |
| 1 | Asp71 to Gly | NF | ||||||
| 1 | Thr82 to Ala | NF | ||||||
| 5 | NF | NF | ||||||
| 19 | 8 | 16–128 | 32–>128 | 0.5–32 | 4–>32 | 2 | Thr82 to Ile | NF |
| 1 | Asp71 to Val | NF | ||||||
| 1 | Asp81 to Asn | NF | ||||||
| 7 | NF | Asp426 to Asn | ||||||
| 3 | NF | Asp426 to Val | ||||||
| 1 | NF | Glu466 to Lys | ||||||
| 1 | NF | Ser416 to Ala | ||||||
| 1 | Thr82 to Ile | Asp426 to Val | ||||||
| 1 | Thr82 to Ile | Ser416 to Ala | ||||||
| 1 | Asp81 to Asn | Asp426 to Val | ||||||
| Arg90 to Lys | ||||||||
| Asp103 to Asn | ||||||||
| Glu123 to Lys | ||||||||
| 33 | 16 | 16–128 | 4–>128 | 0.5–>32 | 0.5–16 | 27 | Thr82 to Ile | NF |
| 2 | NF | Asp426 to Asn | ||||||
| 1 | NF | Ser416 to Ala | ||||||
| 1 | NF | Arg377 to Gly | ||||||
| 1 | Thr82 to Ile | Ser416 to Ala | ||||||
| 1 | Thr82 to Ile | Arg389 to Pro | ||||||
| Glu399 to Lys | ||||||||
| Asp409 to Asn | ||||||||
| Val423 to Phe | ||||||||
| Arg457 to Thr | ||||||||
| Asp465 to Tyr | ||||||||
| 20 | 32 | 16–128 | 64–>128 | 2–>32 | 0.5–16 | 17 | Thr82 to Ile | NF |
| 3 | NF | NF | ||||||
Moxi, moxifloxacin; Cipro, ciprofloxacin; Levo, levofloxacin; Gemi, gemifloxacin; Nemo, nemonoxacin.
NF, amino acid substitutions in gyrA or gyrB were not found.
DISCUSSION
We characterized antibiotic susceptibility patterns of 403 clinical isolates of C. difficile in Taiwan collected over a 6-year period (2005 to 2010); around 58% of the isolates were toxigenic by genotype. We found all 403 isolates to be fully susceptible to metronidazole with MICs of ≤4 μg/ml. All but two isolates were susceptible to vancomycin, and only one vancomycin-resistant isolate was a toxigenic strain; that particular isolate carried toxin A, toxin B, and binary toxin genes and had a MIC for vancomycin of 4 μg/ml, still orders of magnitude below fecal levels of vancomycin achieved during treatment (17). No vancomycin- or metronidazole-resistant clones were found among 100 clinical isolates from South Korea during 2006 to 2008 (23). Among 112 clinical isolates cultured in China in late 2008 to early 2009, none had reduced susceptibility to metronidazole, but two isolates had a vancomycin MIC of 4 μg/ml (22). Our earlier study found also that all 113 isolates of C. difficile collected in Taiwan during 2001 to 2009 were susceptible to metronidazole, but, as in this study, some had reduced susceptibility to vancomycin (MIC, 4 μg/ml).
In this study, fidaxomicin, which has recently been approved by the U.S. Federal Drug Administration and the European Medicines Agency for the treatment of CDI, had potent in vitro activity against all the isolates tested (18). The MIC50 (0.12 μg/ml) and MIC90 (0.25 μg/ml) values for fidaxomicin were identical to those found for 716 isolates from patients at enrollment in clinical trials of fidaxomicin in North America and Europe (16). Another RNA polymerase inhibitor, rifaximin, has been used against CDI following standard vancomycin therapy. We found that 10.9% of isolates in this study were not effectively inhibited by rifaximin at 128 μg/ml. All rifaximin-resistant isolates lacked the binary toxin genes, but some contained toxin A and toxin B genes and some were nontoxigenic. In the same study cited for fidaxomicin, rifaximin-resistant C. difficile clones were isolated in the United States, Germany, and Italy, but not in the United Kingdom, Belgium, France, Spain, Sweden, or Canada. That study and ours found no evidence of cross-resistance to fidaxomicin; this is not unexpected since the two transcriptional inhibitors interact with different regions of RNA polymerase (37).
One-fifth of the isolates had reduced susceptibility to moxifloxacin (MIC of ≥4 μg/ml), with cross-resistance to ciprofloxacin and levofloxacin. Some moxifloxacin-nonsusceptible isolates were susceptible to gemifloxacin and nemonoxacin. There was no correlation between fluoroquinolone resistance and the presence of the binary toxin gene, two identifying characteristics of the hypervirulent 027 strain. The first identified and most common mutation associated with fluoroquinolone resistance in C. difficile (1, 8, 11, 35, 36, 38) was the single most common gyrase mutation among these fluoroquinolone reduced susceptibility isolates, gyrA Thr82 to Ile, present in 52 isolates (64%). One other isolate had a Thr82-to-Ala mutation, not previously reported, although a Thr82-to-Val mutation was reported in an isolate from China (22). Some isolates with the gyrA Thr82-to-Ile substitution alone had low to intermediate resistance (MICs of 4 to 8 μg/ml) to moxifloxacin in contrast to other studies that found this substitution to be associated only with high-level resistance to moxifloxacin (MICs of ≥16 μg/ml) among isolates from France and Canada (10, 38). The single most common gyrB substitution was Asp426 to Asn in nine isolates, which as a single substitution was associated with moxifloxacin MICs from 4 to 16 μg/ml. Asp426 to Asn or Val substitutions have been associated with reduced fluoroquinolone susceptibility in several studies (10, 12, 21, 35, 38).
We also found previously unreported amino acid substitutions in the gyrB gene, two of which apparently can independently confer resistance to moxifloxacin and other fluoroquinolones: Ser416 to Ala and Glu466 to Lys were each found as the only gyrase amino acid substitution in one isolate each with a MIC of 8 μg/ml. Ser416 to Ala was also identified in two other isolates which carried the Thr82-to-Ile (gyrA) mutation as well. Six amino substitutions in GyrB in one fluoroquinolone-resistant isolate may be silent since they were accompanied by the GyrA Thr82-to-Ile substitution. Site-directed mutagenesis would be required to determine whether any of these substitutions independently or together reduce susceptibility to moxifloxacin; none have been reported in fluoroquinolone-resistant C. difficile isolates before.
Of the 81 moxifloxacin-nonsusceptible isolates, eight had no mutations in the regions of gyrA and gyrB established as being important for susceptibility to quinolones; three of those were resistant to moxifloxacin at 32 μg/ml, while the remaining 5 were intermediate in susceptibility to moxifloxacin (MICs, 4 μg/ml). We cannot rule out that other mutations outside these regions exist in the gyrase genes. Other mechanisms of quinoline resistance have been identified; a pentapeptide repeat protein encoded by qnrA acts in trans to protect DNA gyrase from quinolone activity (19). qnr genes are found on resistance plasmids as well as chromosomally in some Gram-positive species, including a toxigenic laboratory strain of C. difficile, ATCC 9689 (19, 34).
This study describes the diversity of toxigenic and nontoxigenic strains of C. difficile found in the health care settings in Taiwan. There is no evidence of increasing resistance to the antibiotics commonly used to treat CDI, metronidazole, and vancomycin. In addition, fidaxomicin exhibited potent in vitro activity against all isolates, while there was concerning resistance to another transcription inhibitor, rifaximin.
ACKNOWLEDGMENTS
This study was supported in part by grants from Optimer Biotechnology, Inc., Taiwan, ROC.
We thank Sharon Dana, Ph.D., for assisting with preparation of the manuscript.
Footnotes
Published ahead of print 16 April 2012
REFERENCES
- 1. Ackermann G, et al. 2001. Resistance to moxifloxacin in toxigenic Clostridium difficile isolates is associated with mutations in gyrA. Antimicrob. Agents Chemother. 45:2348–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Al-Nassir WN, et al. 2008. Both oral metronidazole and oral vancomycin promote persistent overgrowth of vancomycin-resistant enterococci during treatment of Clostridium difficile-associated disease. Antimicrob. Agents Chemother. 52:2403–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baines SD, et al. 2008. Emergence of reduced susceptibility to metronidazole in Clostridium difficile. J. Antimicrob. Chemother. 62:1046–1052 [DOI] [PubMed] [Google Scholar]
- 4. Bauer MP, Kuijper EJ, van Dissel JT. 2009. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): treatment guidance document for Clostridium difficile infection (CDI). Clin. Microbiol. Infect. 15:1067–1079 [DOI] [PubMed] [Google Scholar]
- 5. Chung CH, et al. 2010. Clostridium difficile infection at a medical center in southern Taiwan: incidence, clinical features and prognosis. J. Microbiol. Immunol. Infect. 43:119–125 [DOI] [PubMed] [Google Scholar]
- 6. Clinical and Laboratory Standards Institute 2007. Methods for antimicrobial susceptibility testing of anaerobic bacteria. Approved standard M11-A7, 7th ed Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7. Cohen SH, et al. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect. Control Hosp. Epidemiol. 31:431–455 [DOI] [PubMed] [Google Scholar]
- 8. Deneve C, et al. 2009. Effects of subinhibitory concentrations of antibiotics on colonization factor expression by moxifloxacin-susceptible and moxifloxacin-resistant Clostridium difficile strains. Antimicrob. Agents Chemother. 53:5155–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dingle KE, et al. 2011. Clinical Clostridium difficile: clonality and pathogenicity locus diversity. PLoS One 6:e19993 doi:10.1371/journal.pone.0019993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dridi L, Tankovic J, Burghoffer B, Barbut F, Petit JC. 2002. gyrA and gyrB mutations are implicated in cross-resistance to ciprofloxacin and moxifloxacin in Clostridium difficile. Antimicrob. Agents Chemother. 46:3418–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drudy D, Kyne L, O'Mahony R, Fanning S. 2007. gyrA mutations in fluoroquinolone-resistant Clostridium difficile PCR-027. Emerg. Infect. Dis. 13:504–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drudy D, et al. 2006. High-level resistance to moxifloxacin and gatifloxacin associated with a novel mutation in gyrB in toxin-A-negative, toxin-B-positive Clostridium difficile. J. Antimicrob. Chemother. 58:1264–1267 [DOI] [PubMed] [Google Scholar]
- 13. European Committee on Antimicrobial Susceptibility Testing 2011. Clinical breakpoint tables, version 1.3. European Committee on Antimicrobial Susceptibility Testing, London, United Kingdom: http://www.eucast.org/eucast_susceptibility_testing/breakpoints/ Accessed 21 February 2012 [Google Scholar]
- 14. Garey KW, et al. 2011. A randomized, double-blind, placebo-controlled pilot study to assess the ability of rifaximin to prevent recurrent diarrhoea in patients with Clostridium difficile infection. J. Antimicrob. Chemother. 66:2850–2855 [DOI] [PubMed] [Google Scholar]
- 15. Garey KW, Jiang ZD, Bellard A, Dupont HL. 2009. Rifaximin in treatment of recurrent Clostridium difficile-associated diarrhea: an uncontrolled pilot study. J. Clin. Gastroenterol. 43:91–93 [DOI] [PubMed] [Google Scholar]
- 16. Goldstein EJ, et al. 2011. Comparative susceptibilities to fidaxomicin (OPT-80) of isolates collected at baseline, recurrence, and failure from patients in two phase III trials of fidaxomicin against Clostridium difficile infection. Antimicrob. Agents Chemother. 55:5194–5199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gonzales M, et al. 2010. Faecal pharmacokinetics of orally administered vancomycin in patients with suspected Clostridium difficile infection. BMC Infect. Dis. 10:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hardesty JS, Juang P. 2011. Fidaxomicin: a macrocyclic antibiotic for the treatment of Clostridium difficile infection. Pharmacotherapy 31:877–886 [DOI] [PubMed] [Google Scholar]
- 19. Hernandez A, Sanchez MB, Martinez JL. 2011. Quinolone resistance: much more than predicted. Front. Microbiol. 2:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hsu MS, Wang JT, Huang WK, Liu YC, Chang SC. 2006. Prevalence and clinical features of Clostridium difficile-associated diarrhea in a tertiary hospital in northern Taiwan. J. Microbiol. Immunol. Infect. 39:242–248 [PubMed] [Google Scholar]
- 21. Huang H, Weintraub A, Fang H, Nord CE. 2009. Antimicrobial resistance in Clostridium difficile. Int. J. Antimicrob. Agents 34:516–522 [DOI] [PubMed] [Google Scholar]
- 22. Huang H, et al. 2010. Antimicrobial susceptibility and heteroresistance in Chinese Clostridium difficile strains. Anaerobe 16:633–645 [DOI] [PubMed] [Google Scholar]
- 23. Kim H, et al. 2010. Investigation of toxin gene diversity, molecular epidemiology, and antimicrobial resistance of Clostridium difficile isolated from 12 hospitals in South Korea. Korean J. Lab. Med. 30:491–497 [DOI] [PubMed] [Google Scholar]
- 24. Kuijper EJ, et al. 2007. Update of Clostridium difficile-associated disease due to PCR ribotype 027 in Europe. Euro Surveill. 12:E1–E2 [DOI] [PubMed] [Google Scholar]
- 25. Lee YC, et al. 28 December 2011. Changing incidence and clinical manifestations of Clostridium difficile-associated diarrhea detected by combination of glutamate dehydrogenase and toxin assay in Northern Taiwan. J. Microbiol. Immunol. Infect. [Epub ahead of print.] doi:10.1016/j.jmii.2011.12.001 [DOI] [PubMed] [Google Scholar]
- 26. Lin YC, et al. 2011. Antimicrobial susceptibilities and molecular epidemiology of clinical isolates of Clostridium difficile in Taiwan. Antimicrob. Agents Chemother. 55:1701170–1701175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McDonald LC, et al. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353:2433–2441 [DOI] [PubMed] [Google Scholar]
- 28. Miller BA, Chen LF, Sexton DJ, Anderson DJ. 2011. Comparison of the burdens of hospital-onset, healthcare facility-associated Clostridium difficile infection and of healthcare-associated infection due to methicillin-resistant Staphylococcus aureus in community hospitals. Infect. Control Hosp. Epidemiol. 32:387–390 [DOI] [PubMed] [Google Scholar]
- 29. Miller M, et al. 2010. Health care-associated Clostridium difficile infection in Canada: patient age and infecting strain type are highly predictive of severe outcome and mortality. Clin. Infect. Dis. 50:194–201 [DOI] [PubMed] [Google Scholar]
- 30. Musher DM, et al. 2005. Relatively poor outcome after treatment of Clostridium difficile colitis with metronidazole. Clin. Infect. Dis. 40:1586–1590 [DOI] [PubMed] [Google Scholar]
- 31. O'Connor JR, Johnson S, Gerding DN. 2009. Clostridium difficile infection caused by the epidemic BI/NAP1/027 strain. Gastroenterology 136:1913–1924 [DOI] [PubMed] [Google Scholar]
- 32. Pepin J, Valiquette L, Cossette B. 2005. Mortality attributable to nosocomial Clostridium difficile-associated disease during an epidemic caused by a hypervirulent strain in Quebec. CMAJ 173:1037–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pituch H, et al. 2007. Toxin profiles and resistances to macrolides and newer fluoroquinolones as epidemicity determinants of clinical isolates of Clostridium difficile from Warsaw, Poland. J. Clin. Microbiol. 45:1607–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rodriguez-Martinez JM, et al. 2008. Qnr-like pentapeptide repeat proteins in Gram-positive bacteria. J. Antimicrob. Chemother. 61:1240–1243 [DOI] [PubMed] [Google Scholar]
- 35. Spigaglia P, Barbanti F, Dionisi AM, Mastrantonio P. 2010. Clostridium difficile isolates resistant to fluoroquinolones in Italy: emergence of PCR ribotype 018. J. Clin. Microbiol. 48:2892–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Spigaglia P, et al. 2008. Fluoroquinolone resistance in Clostridium difficile isolates from a prospective study of C. difficile infections in Europe. J. Med. Microbiol. 57:784–789 [DOI] [PubMed] [Google Scholar]
- 37. Srivastava A, et al. 2011. New target for inhibition of bacterial RNA polymerase: ‘switch region.’ Curr. Opin. Microbiol. 14:532–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Walkty A, et al. 2010. Molecular characterization of moxifloxacin resistance from Canadian Clostridium difficile clinical isolates. Diagn. Microbiol. Infect. Dis. 66:419–424 [DOI] [PubMed] [Google Scholar]
- 39. Warny M, et al. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084 [DOI] [PubMed] [Google Scholar]
- 40. Zar FA, Bakkanagari SR, Moorthi KM, Davis MB. 2007. A comparison of vancomycin and metronidazole for the treatment of Clostridium difficile-associated diarrhea, stratified by disease severity. Clin. Infect. Dis. 45:302–307 [DOI] [PubMed] [Google Scholar]