Abstract
Fluoroquinolone (FQ)-resistant extraintestinal pathogenic Escherichia coli (FQr ExPEC) strains from phylogenetic group B2 are undergoing epidemic spread. Isolates belonging to phylogenetic group B2 are generally more virulent than other E. coli isolates; therefore, resistance to FQs among group B2 isolates is concerning. Although clonal expansion of sequence type 131 (ST131) is a major factor, the contribution of additional clonal groups has not been quantified. Group B2 FQr ExPEC isolates from humans (n = 250) and dogs (n = 12) in Australia were screened for ST131, a recently recognized and rapidly emerging multidrug-resistant and virulent clonal group that is important in both human and companion animal medicine. Non-ST131 isolates underwent virulence genotyping, PCR-based O typing, partial multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), and FQ resistance mechanism analysis. Of 49 non-ST131 isolates (45 human, 4 canine), 49% (24 human, 2 canine) represented O-type O75 and exhibited conserved virulence genotypes (F10 papA allele, iha, fimH, sat, vat, fyuA, iutA, kpsMII, usp, ompT, malX, K1/K5 capsule) and MLST allele profiles corresponding with clonal complex CC14. Two clusters, each containing canine and human isolates, were identified by PFGE (differentiated by K1 and K5 capsules). Australian FQr O75 isolates exhibited commonality with an historical FQ-susceptible O75 urosepsis isolate (also CC14). The isolation from humans and dogs of highly similar FQr derivatives of the classic O75:K1/K5 (CC14) ExPEC lineage suggests recent acquisition of FQ resistance and potential cross-host-species transfer. This lineage should be targeted with ST131 in future epidemiological investigations of FQr ExPEC.
INTRODUCTION
Infections caused by extraintestinal pathogenic Escherichia coli (ExPEC) are very common among humans and dogs. ExPEC strains isolated from both host species, particularly those belonging to the more virulent phylogenetic group B2, have been shown to possess pathotypic and phylogenetic similarities (18, 24).
Treatment of ExPEC infections has become increasingly problematic due to the emergence and spread of clonally related multidrug-resistant (MDR) strains (12, 15, 32, 37). In particular, sequence type 131 (ST131), a newly recognized ExPEC clonal group from phylogenetic group B2, has shown recent and rapid global dissemination among humans (15, 25, 35). Multiple reports have documented the emergence of ST131 also in companion animals, including both as gastrointestinal carriage in healthy dogs (16) and as a cause of extraintestinal infection (8, 31, 33). ST131 strains typically possess a wide array of extraintestinal virulence genes and exhibit resistance to multiple antimicrobial agents of various classes, including fluoroquinolones and, frequently, extended-spectrum cephalosporins (30, 35).
Previously, among 262 group B2 fluoroquinolone-resistant (FQr) E. coli isolates from extraintestinal infections in humans and companion animals from the east coast of Australia, we found that ST131 accounted for 206 (82%) of 250 human isolates and 8 (69%) of 12 companion animal isolates (32). Here, to search for additional broad-host-range multiresistant clonal groups other than ST131, we assessed the 49 remaining non-ST131 group B2 isolates (45 from humans, 4 from dogs) for clonal structure, virulence genotypes, and evidence of interspecies transmission.
MATERIALS AND METHODS
Bacterial isolates.
In our previous study (32), 262 (250 human, 12 canine) phylogenetic group B2 FQr E. coli isolates were identified by PCR-based phylotyping among 702 FQr ExPEC clinical isolates obtained from humans and dogs in eastern Australia (2). The human isolates were obtained from two Brisbane private pathology laboratories processing samples from both hospitals (approximately 80 to 85% of submissions) and community clinics (approximately 15 to 20% of submissions) between October 2007 and October 2008, and the canine isolates were from four veterinary pathology laboratories (serving Brisbane, Sydney, and Melbourne) processing samples from both community clinics and referral hospitals between October 2007 and October 2009. Group B2 isolates underwent PCR-based screening for ST131-specific single-nucleotide polymorphisms (SNPs) in mdh and gyrB (15); this identified 213 (205 human, 8 canine) isolates as ST131 and 49 (45 human, 4 canine) as non-ST131 (32). For comparative purposes, four historical fluoroquinolone-susceptible phylogenetic group B2 O75 blood isolates from humans with urosepsis (strains 2H19, PM3, V21, and V8) (19) were included.
O typing and further isolate selection.
As O types may be a primary indicator of clonal association, the 49 FQr non-ST131 group B2 isolates underwent PCR-based O typing to detect 13 known clonal group- and/or sepsis-associated rfb variants (O1, O2, O4, O6, O11, O12, O15, O16, O18, O25a, O25b, O75, and O157) (3, 4). Twenty-six isolates belonging to O75 (i.e., 24 human and 2 canine isolates) were further characterized, as detailed below. In addition, the remaining 2 non-O75 canine isolates and 6 (30%; randomly selected) of the 21 remaining non-O75 human isolates were also characterized to determine the level of diversity among the non-O75 isolates.
Susceptibility testing.
The 34 selected isolates underwent disk diffusion testing for susceptibility to 19 antimicrobials, including amikacin, amoxicillin-clavulanic acid, ampicillin, aztreonam, cefepime, cefoxitin, ceftazidime, cephalothin, chloramphenicol, ciprofloxacin, enrofloxacin, gentamicin, imipenem, nitrofurantoin, piperacillin, piperacillin-tazobactam, streptomycin, tetracycline, and trimethoprim-sulfamethoxazole (5, 6). Resistance was defined as nonsusceptibility (i.e., resistance or intermediate susceptibility) according to standardized interpretative criteria (5, 6). An isolate's resistance score was the number of antimicrobials to which the isolate demonstrated resistance, whereas the resistance profile was the combination of antimicrobials to which the isolate exhibited resistance. Resistance profiles were examined to provide information about the extent and specific patterns of resistance, plus provide some (albeit weak) indication regarding clonality.
Virulence gene profiling and lactose fermentation determination.
Isolates were screened for 52 ExPEC-associated virulence marker genes and their variants using established multiplex PCR assays (11, 13, 19, 21). Here, the term “virulence genes” is used to include proven or putative virulence-associated genes and associated variants. Virulence scores reflected the total number of virulence genes identified, with adjustment for multiple detection of certain operons (i.e., pap, sfa-foc, and kps). Virulence genotype similarity between isolates was calculated as the number of virulence genes concordantly detected in both isolates divided by the total number of virulence genes detected in either isolate. Virulence profiles provide some indication of the organism's virulence potential (as reflected in the total number and particular combination of constituent virulence genes), and they may have associations with specific clonal groups and thus provide another marker for clonality independently of their virulence implications. Lactose fermentation was determined by plating on MacConkey agar.
PFGE.
For a more discriminating assessment of within- and across-species genomic commonality, the 34 selected Australian isolates and the 4 historical isolates underwent comparative pulsed-field gel electrophoresis (PFGE) analysis according to a standardized protocol (34). Dice coefficient-based similarity dendrograms were constructed within BioNumerics software (Bio-Rad) according to the unweighted-pair group method with arithmetic mean. Isolates were considered to represent the same pulsotype if they exhibited ≥94% profile similarity, which approximates to a ≤3-band difference (39).
MLST.
To classify the study isolates as to clonal group and to enable interlaboratory comparisons, the 34 selected Australian isolates underwent partial multilocus sequence typing (MLST) (for fumC, gyrB, and recA) according to the Achtman system (http://mlst.ucc.ie/mlst/dbs/Ecoli). Subsequently, to give a full 7-locus MLST, the remaining 4 loci were analyzed for 9 of these isolates (4 canine, 5 human). The 4 historical O75 isolates also underwent full 7-locus MLST.
Determination of MICs.
Twenty-three randomly selected non-ST131 FQr isolates, including half of the O75 isolates (13 human and 2 canine) and one-third of the non-O75 isolates (6 human and 2 canine), underwent broth microdilution fluoroquinolone MIC determination with enrofloxacin, ciprofloxacin, moxifloxacin, and pradofloxacin (40), a new veterinary fluoroquinolone. Methods and interpretive criteria were as specified by the Clinical and Laboratory Standards Institute (CLSI) (5, 6) for all antibiotics except pradofloxacin, for which the epidemiological breakpoint of 2 μg/ml was used (9).
Detection of fluoroquinolone resistance genes.
For the same 23 isolates, fluoroquinolone resistance-associated chromosomal gene mutations were determined by PCR amplification and pyrosequencing of the quinolone resistance-determining regions (QRDRs) within gyrA and parC (31). Screening for the plasmid-mediated quinolone resistance (PMQR) genes qnrA, qnrB, qnrS, and qepA was performed by multiplex PCR (31). A second PCR was done to identify isolates containing the aac(6′)-Ib-cr variant (31). The degree of homogeneity (or lack thereof) in the fluoroquinolone resistance genes may also aid as a marker for clonality and may be used to indicate divergence among FQr isolates.
Efflux pump activity testing.
Two methods were used to indicate the level of efflux pump activity on the 23 representative isolates: MIC testing in the presence of an efflux pump inhibitor and an organic solvent tolerance test. For the former, susceptibility to 4 fluoroquinolones (enrofloxacin, ciprofloxacin, moxifloxacin, and pradofloxacin) was determined in duplicate in the presence of 64 μg/ml of l-phenylalanyl-arginyl-β-naphthylamide (PAβN; P 4157; Sigma) using broth microdilution (5, 6). Organic solvent tolerance was assessed as described previously (31).
Statistical methods.
Comparisons of proportions were tested using Fisher's exact test. Resistance and virulence scores were compared using Mann-Whitney U tests. Analyses were performed using Stata (version 10.0) software (Statacorp, College Station, TX). Diversity of resistance types and virulence types was compared using Simpson's index of diversity (1-D) (38). The criterion for assuming statistical significance was a P value of <0.05.
RESULTS
O typing and susceptibility testing.
Of the 49 non-ST131 FQr group B2 clinical isolates (45 human, 4 canine), approximately half (n = 26; 24 human, 2 canine) represented O-type O75. All 26 O75 isolates were confirmed to be FQr by disc diffusion and broth microdilution MIC determination. Coresistance to non-FQ antimicrobial agents was common among the 26 O75 isolates, with >75% of isolates exhibiting resistance to ampicillin (96%), piperacillin (96%), streptomycin (96%), trimethoprim-sulfamethoxazole (88%), and tetracycline (85%). Resistance to gentamicin (58%), cephalothin (8%), and amoxicillin-clavulanic acid (4%) was less prevalent. In contrast, all 26 O75 isolates were susceptible to amikacin, aztreonam, cefepime, cefoxitin, ceftazidime, chloramphenicol, imipenem, nitrofurantoin, and piperacillin-tazobactam. Resistance scores ranged from 3 to 10 (median, 8). Among the 26 isolates, 8 different resistance profiles were demonstrated (Table 1).
Table 1.
Resistance profiles of 26 O75 isolates and 7 non-O75 isolates from humans and dogs
| Resistance profilea |
No. of isolates with profile |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Penicillins |
Other beta-lactams |
FQs |
Misc. |
Antifolate |
|||||||||||
| AMP | PIP | AMC | AZ | KF | FOX | CAZ | CIP | ENR | GM | C | T | S | SXT | O75 (n = 26) | Non-O75 (n = 7) |
| + | + | − | − | − | − | − | + | + | + | − | + | + | + | 12 | 0 |
| + | + | − | − | − | − | − | + | + | − | − | + | + | + | 8 | 1 |
| + | + | − | + | − | − | + | + | + | + | − | + | + | + | 1 | 0 |
| + | + | − | − | + | − | − | + | + | + | − | + | + | + | 1 | 0 |
| + | + | + | − | + | − | − | + | + | + | − | − | + | + | 1 | 0 |
| + | + | − | − | − | − | − | + | + | − | − | − | + | − | 1 | 0 |
| + | + | − | − | − | − | − | + | + | − | − | − | − | − | 1 | 0 |
| − | − | − | − | − | − | − | + | + | − | − | − | + | − | 1 | 0 |
| + | + | + | − | + | − | − | + | + | + | − | + | − | − | 0 | 1 |
| + | + | − | − | − | − | − | + | + | + | + | + | − | + | 0 | 1 |
| + | + | + | − | + | − | − | + | + | − | − | − | + | + | 0 | 1 |
| + | − | − | − | + | + | − | + | + | − | − | − | − | + | 0 | 1 |
| + | − | + | − | + | − | − | + | + | − | − | + | − | − | 0 | 1 |
| + | − | − | − | + | + | − | + | + | − | − | − | − | − | 0 | 1 |
| 33 | 29 | 4 | 1 | 7 | 2 | 1 | 33 | 33 | 17 | 1 | 26 | 26 | 26 | 26 | 7 |
Misc., miscellaneous; AMP, ampicillin; AMC, amoxicillin-clavulanic acid; AZ, aztreonam; KF, cephalothin; CAZ, ceftazidime; FOX, cefoxitin; C, chloramphenicol; CIP, ciprofloxacin; ENR, enrofloxacin; GM, gentamicin; PIP, piperacillin; T, tetracycline; SXT, trimethoprim-sulfamethoxazole; S, streptomycin; +, resistant; −, sensitive.
Among the non-O75 isolates examined (n = 8; 2 canine, 6 human), 1 canine isolate showed intermediate susceptibility to both enrofloxacin and ciprofloxacin by disk diffusion, whereas MIC testing indicated susceptibility at the breakpoint for both fluoroquinolones; therefore, this isolate was excluded from further study. Data were therefore collated for six randomly selected non-O75 human isolates and one non-O75 canine isolate. All seven isolates were resistant to ampicillin, the only non-FQ agent to which a >75% resistance prevalence was detected among these isolates. Other less prevalent resistances included cephalothin (71%). piperacillin (57%), tetracycline (57%), trimethoprim-sulfamethoxazole (57%), amoxicillin-clavulanic acid (43%), cefoxitin (29%), gentamicin (29%), streptomycin (29%), and chloramphenicol (14%). All seven non-O75 isolates were susceptible to amikacin, aztreonam, cefepime, ceftazidime, imipenem, nitrofurantoin, and piperacillin-tazobactam. Resistance scores ranged from 5 to 8 (median, 7; versus O75 isolates, P = 0.25). Compared with the O75 isolates (1-D = 0.71), resistance profiles were more diverse among the seven non-O75 isolates (1-D = 1), each of which had a unique profile (Table 1).
Virulence genotypes and lactose fermentation.
Among the 26 O75 isolates, 17 different virulence markers were detected overall, with individual isolates having a median virulence score of 12 (range, 11 to 13). Virulence genotypes were highly similar, with pairwise similarity values ranging from 96.5% to 100%. The O75 isolates' consensus virulence genotype included the F10 papA allele (P fimbria structural subunit variant), iha (adhesin-siderophore receptor), fimH (type 1 fimbriae), sat (secreted autotransporter toxin), vat (vacuolating autotransporter toxin), fyuA (yersiniabactin receptor), iutA (aerobactin receptor), kpsMII (group 2 capsule), usp (uropathogenic specific protein), ompT (outer membrane protease), and malX (pathogenicity island marker), with variation limited to presence/absence of the K1 and K5 group 2 capsule variants (Table 2). None of the detected virulence markers differed in prevalence between human and companion animal O75 isolates.
Table 2.
Virulence gene profiles of Australian O75 FQr isolates from humans and dogs and historical FQ-susceptible human O75 isolatesa
| PFGE cluster | Source | No. of strains | F type | Gene profileb |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| papACEFG | papG allele | afa, dra | iha | fimH |
kpsM |
hlyA,hlyD | sat | vat | fyuA | iutA | ireA | usp | traT | ompT | malX | clbB, clbN | ||||||
| II | K1 | K5 | ||||||||||||||||||||
| I | Australian | 10 | F10 | − | − | − | + | + | + | + | − | − | + | + | + | + | − | + | − | + | + | − |
| II | Australian | 14 | F10 | − | − | − | + | + | + | − | + | − | + | + | + | + | − | + | − | + | + | − |
| II | Australian | 1 | F10 | − | − | − | − | + | + | − | + | − | + | + | + | + | − | + | + | + | + | − |
| II | Australian | 1 | F10 | − | − | − | + | + | + | − | + | + | + | + | + | + | − | + | − | + | + | − |
| II | Historical | 1 | F8/F10 | + | II | − | + | + | + | − | + | − | + | + | + | + | + | + | + | + | + | − |
| Historical | 1 | F10 | − | − | − | + | + | + | − | + | − | + | + | + | + | − | + | − | + | + | + | |
| Historical | 1 | F10 | − | − | − | + | + | + | − | + | − | + | + | + | + | − | + | − | + | + | + | |
| Historical | 1 | F10 | − | − | + | + | + | + | − | + | − | + | + | + | + | − | + | + | + | + | + | |
Isolates tested included 24 human and 2 dog O75 FQr isolates and 4 historical FQ-susceptible human O75 isolates.
papACEFG, pyelonephritis-associated fimbriae; papG allele, papG allele variants; afa, dra, afimbrial Dr-binding adhesions; iha, adhesin-siderophore receptor; fimH, type I fimbriae; kpsMII K1 or K5, group 2 capsule variants; hlyA, hlyD, hemolysins; sat, secreted autotransporter toxin; vat, vacuolating autotransporter toxin; fyuA, yersiniabactin receptor; iutA, aerobactin receptor; ireA, siderophore receptor; usp, uropathogenic specific protein; traT, serum resistance associated; ompT, outer membrane protease; malX, pathogenicity island marker; clbB, clbN, colibactin synthesis system.
In contrast, among the non-O75 isolates, 41 (79%) of the 52 virulence genes were detected, with isolates having a median virulence score of 13 (range, 4 to 20). Virulence profiles were much more diverse among the non-O75 isolates than among the O75 isolates, with pairwise virulence genotype similarity values ranging from as low as 60% to no higher than 94%. Compared with the O75 isolates (1-D = 0.61), resistance profiles were more diverse among the seven non-O75 isolates (1-D = 1), each of which had a unique profile.
All 26 Australian O75 isolates were lactose nonfermenters. In contrast, all 7 non-O75 comparison isolates fermented lactose (P < 0.001).
Pulsed-field gel electrophoresis.
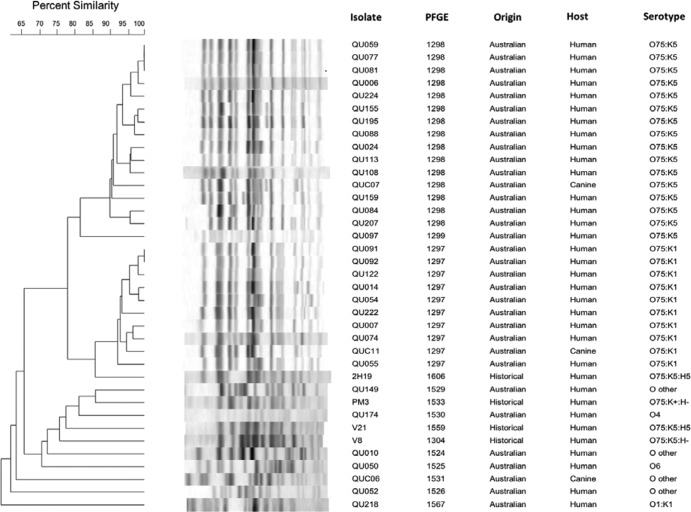
In a dendrogram based on XbaI PFGE profiles for the 26 analyzed Australian O75 isolates (24 human, 2 canine), all but 2 of the isolates fell within two distinct clusters which exhibited >90% within-cluster profile similarity and >82% between-cluster profile similarity (Fig. 1) with no temporal clustering. Supporting the clusters' biological validity was their internal homogeneity for the presence of either the K1 (cluster I, pulsotype 1297) or K5 (cluster II, pulsotype 1298) capsule variants for the Australian isolates. The outlier O75 isolates exhibited <81% similarity to the two main clusters. Notably, both canine O75 isolates (one K1, one K5) fell into the cluster corresponding with the isolate's K type; each was >91% similar to human isolates within that cluster.
Fig 1.
Dendrogram of XbaI pulsed-field gel electrophoresis profiles of 38 phylogenetic group B2 Escherichia coli isolates, 34 human O75 isolates, 4 canine O75 isolates, 6 human non-O75 isolates, 2 canine non-O75 isolates, and 4 historical O75 isolates.
Multilocus sequence typing.
All 26 Australian O75 study isolates that underwent MLST possessed the same 3-locus haplotype (fumC14, gyrB10, recA10), implying commonality at the ST level. In contrast, the 7 non-O75 isolates (6 human, 1 canine) exhibited 7 distinct allele combinations, evidence of disparate ST backgrounds. All 7 of the Australian O75 isolates (5 human, 2 canine) that underwent 7-locus MLST belonged to ST1193, which is within clonal complex CC14, differing from ST14 by only 1 nucleotide in icd.
Fluoroquinolone resistance characteristics.
Among the 23 FQr isolates that underwent FQ MIC determination (15 O75, 8 non-O75), MICs were lowest for pradofloxacin (median, 4 μg/ml), in the midrange for ciprofloxacin and moxifloxacin (median, 16 μg/ml), and highest for enrofloxacin (median, 32 μg/ml), with no apparent differences between the O75 and non-O75 isolates. All but one of the O75 isolates exhibited identical fluoroquinolone resistance-associated mutations in the QRDR of gyrA and parC, i.e., gyrA S83L (TCG to TTG) and D87N (GAC to AAC) and parC S80I (AGC to AGT), plus a silent gyrA mutation V85V (GTT to GTC), compared to the sequence of wild-type E. coli. The sole exception was human isolate QU108, in which the parC PCR fragment could not be sequenced. In contrast, the non-O75 isolates exhibited diverse mutations in gyrA and parC (not shown).
Additional FQ resistance mechanisms, qnrA, qnrB, qnrS, qepA, and aac(6′)-1b-cr variant, were not detected in any isolate. Likewise, although all 23 isolates exhibited a ≥4-fold decrease in the MICs of enrofloxacin, moxifloxacin, and pradofloxacin in the presence of the efflux pump inhibitor PAβN, only 1 O75 isolate (QU108, human) and 4 of 7 non-O75 isolates (3 human and 1 canine) exhibited various degrees of organic solvent tolerance.
Historical O75 isolates.
According to 7-locus MLST, the 4 historical O75 urosepsis isolates represented ST14 (strain 2H19), ST550 (strains V8 and V21), and ST576 (strain PM3), all of which are within CC14. The virulence genotypes of the historical FQ-susceptible O75/CC14 isolates were highly similar to those of the present FQ-resistant Australian isolates (Table 2). They included all the virulence genes found in the Australian O75 isolates; additionally, V21 contained clbB and clbN, 2H19 contained a complete copy of the pap operon plus traT, and PM3 contained clbB and clbN, traT, and afa-dra. PFGE showed >85% profile similarity between reference isolate 2H19 and the Australian O75:K1 isolates (cluster I, pulsotype 1297) (Fig. 1).
DISCUSSION
The recent worldwide emergence of FQr phylogenetic group B2 ExPEC has occurred primarily through clonal expansion of E. coli ST131 (30, 31, 35). This study sought to determine if non-ST131 FQr phylogenetic group B2 isolates from extraintestinal infections in both humans and dogs also exhibit clonality and whether across-host-species commonality could be detected. We document the apparent emergence of FQr variants of an O75-associated clonal group (ST1193, CC14), accounting for approximately 10% of a large collection of phylogenetic group B2 FQr E. coli isolates from Australia. Although the vast majority (81%) of this collection was previously determined to represent ST131 (31, 32), here we show that approximately half of the remaining FQr isolates represent this newly recognized, FQr O75-associated ST1193 (CC14) clonal group.
Our findings support four main conclusions regarding the O75/ST1193 isolates. First, the O75/ST1193 isolates account for half of the non-ST131 FQr group B2 population in this collection of E. coli isolates from eastern Australia. Second, these isolates are highly clonal, as indicated by both PFGE and MLST and their possession of a relatively homogeneous repertoire of virulence genes, resistance profiles, and FQ resistance mechanisms. Third, although this conclusion is based on a small number of isolates, Australian O75/ST1193 isolates from both humans and dogs are indistinguishable, i.e., are likely to be derived from a recent common ancestor and to have undergone anthroponotic-zoonotic transfer. Lastly, several isolates exhibited close genetic similarity to an historical FQ-susceptible O75 urosepsis isolate from the United States, indicating that rather than being a recently emerged clonal group, the O75 isolates identified in the current study are likely to be descendants of a more long-standing clonal lineage that has more recently acquired FQr mechanisms. Further studies that aim to retrospectively examine FQ-sensitive strain collections for their clonal structure are needed to confirm this conclusion.
Escherichia coli serotype O75 has long been associated with urinary tract infection (UTI) and sepsis in humans (7, 29), virulence-associated phylogenetic group B2 (28), Dr-binding (formerly termed “O75X”) adhesins (27), and ampicillin resistance (27). However, it has not historically been associated with FQ resistance, which could be an important phenotype contributing to recent clonal expansion. Although the extent of clonality within serogroup O75 has not been broadly determined, some studies evaluating the virulence gene repertoire or using methods such as ribotyping point to a strongly clonal population structure (26, 28). However, to the best of our knowledge, previous studies have not simultaneously investigated clonality, virulence, and the occurrence of FQ resistance among O75 isolates.
Our finding that FQr E. coli O75 strains are not host specific to humans is consistent with both the previous evidence that canine UTI isolates commonly belong to human UPEC-associated serogroups, including O75 (22), and the frequent isolation of another successful FQr clonal group, i.e., ST131, from both humans and dogs (16, 31). It appears that fluoroquinolone resistance among phylogenetic group B2 isolates, at least in Australia, is emerging mainly by expansion of two resistant clonal groups—primarily ST131, as previously described (31), and ST1193, as described here—rather than by arising repeatedly by independent mutations in diverse genetic backgrounds.
Historically, FQr extraintestinal infection isolates have tended to represent the less virulent phylogenetic groups D and A (10, 36). In contrast, phylogenetic group B2 E. coli isolates, which tend to possess an enhanced array of virulence genes, typically have been more susceptible to antimicrobials, including FQs (14). In conflict with this classic paradigm, two clonal groups within group B2, i.e., ST131 and the O75-associated ST1193 clonal group described here, possess an extensive virulence gene repertoire (15, 25), together with a multidrug resistance phenotype. Thus, this O75 clonal group and ST131 demonstrate that resistance and enhanced virulence can no longer be regarded as mutually exclusive within E. coli. Concurrence of resistance and virulence within epidemic ExPEC clonal groups was demonstrated previously by the phylogenetic group D-derived clonal groups CC69 (clonal group A; O11/O17/O73/O77:K52:H18) and O15:K52:H1, albeit with resistance to trimethoprim-sulfamethoxazole rather than fluoroquinolones (1, 17, 20). A corollary of this finding is that group B2-derived clonal groups clearly can no longer be presumed to be susceptible to clinically important antimicrobials, as was true until recently.
The O75 ST1193 clonal group isolates possessed nearly identical virulence genotypes that included the F10 papA allele, iha, fimH, sat, vat, fyuA, iutA, kpsMII, usp, ompT, and malX, differing only for presence of the K1 or K5 group 2 capsule variants. Although according to virulence genotype and PFGE profile our Australian O75 isolates closely resembled an FQ-susceptible O75 urosepsis isolate from Seattle, WA, from the 1980s (isolate 2H19), they lacked traT and most pap operon genes (19), which, in contrast, were present in strain 2H19. Moreover, both 2H19 and the Australian O75 isolates lacked clbB and clbN, which were present in the other historical O75 urosepsis isolates. On a broad scale, most O75 isolates appear to belong to a prevalent clonal complex (CC14) that has been widely geographically distributed for decades, if not longer (23). Certainly, phylogenetic group B2 and serogroup O75 have been associated with urinary tract infection and sepsis for over half a century, and the virulence traits seen in our Australian O75 isolates are known to be broadly disseminated among O75 strains (7, 28, 29). Interestingly, all isolates belonging to the O75/ST1193 clonal group were lactose nonfermenters, which may have implications regarding detection in a clinical microbiology setting.
PFGE analysis resolved two main subgroups (i.e., clusters) among the O75 isolates, one characterized by the K1 capsule (cluster I) and the other (which included historical isolate 2H19) by the K5 capsule (cluster II). Within each cluster, the constituent human and canine isolates were intermingled, without evident species-specific segregation. Although the number of companion animal isolates is low, our identification of highly genetically similar human and veterinary E. coli O75 isolates, which coincides with our previous demonstration of clonality among human and canine ST131 isolates, suggests the potential for transmission between humans and their pets and/or vice versa.
The homogeneity of the Australian FQr O75 ST1193 isolates suggests recent dissemination of successful new FQr clonal variants among both humans and dogs. In contrast, a lesser degree of homogeneity exists among the historical O75 isolates, further evidence that the Australian O75/ST1193 isolates recently emerged and spread from a common ancestor, most likely within the lineage represented by the historical O75 isolate 2H19. This is supported by the identical pattern of a double nonsynonymous mutation (S83L [TCG to TTG] and D87N [GAC to AAC]) plus a single silent mutation (V85V [GTT to GTC]) in gyrA and a single nonsynonymous mutation in parC (S80I [AGC to AGT]) in all Australian FQr O75 isolates compared to the varied gyrA and parC mutations observed in the non-O75 group.
In conclusion, we have documented a high prevalence of clonally related FQr serogroup O75 ExPEC isolates from ST1193 among humans and dogs in Australia. Further studies of current and historical FQr isolate collections are required to determine if this is a local or broader (e.g., global) phenomenon and to pinpoint in time the development of FQ resistance among O75 isolates. Furthermore, there is a global need for strategic surveillance and control schemes for FQr extraintestinal E. coli with high virulence potential, particularly given the emergence and spread of multiresistant group B-derived clonal groups such as ST131 and, now, ST1193 (CC14).
ACKNOWLEDGMENTS
This work was supported by an Australian Research Council Linkage project with Bayer Health Care AG and the University of Queensland (to D.J.T. and R.N.C.) and by the Office of Research and Development, Medical Research Service, U.S. Department of Veterans Affairs (to J.R.J.).
Peter Heisig is a scientific adviser for Bayer Health Care.
The study sponsors imposed no commercial influence in study design, interpretation of data, or submission of the manuscript.
We thank Renu Vohra and Brooke Robinson from QML Pathology, Jenny Robson from S&N Pathology, Susan Moss from UQVDL, Marc Marenda from the University of Melbourne VDL, and Emma Baggs from IDEXX Laboratories for providing the clinical study isolates. Thanks also go to Nadine Emrich from the University of Hamburg for providing reference isolates. Special thanks go to Megan Menard at the VA Medical Center, Minneapolis, MN, and Antje Schnasse of the Institute of Biochemistry and Molecular Biology, University of Hamburg, for their technical advice and assistance.
Footnotes
Published ahead of print 23 April 2012
REFERENCES
- 1. Blanco J, et al. 2011. National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain. J. Antimicrob. Chemother. 66:2011–2021 [DOI] [PubMed] [Google Scholar]
- 2. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clermont O, et al. 2009. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J. Antimicrob. Chemother. 64:274–277 [DOI] [PubMed] [Google Scholar]
- 4. Clermont O, Johnson JR, Menard M, Denamur E. 2007. Determination of Escherichia coli O types by allele-specific polymerase chain reaction: application to the O types involved in human septicemia. Diagn. Microbiol. Infect. Dis. 57:129–136 [DOI] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing: twentieth informational supplement M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Clinical and Laboratory Standards Institute 2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals 3rd ed: approved standard M31-A3. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7. Devine DA, Robinson L, Roberts AP. 1989. Occurrence of K1, K5 and O antigens in Escherichia coli isolates from patients with urinary tract infections or bacteraemia. J. Med. Microbiol. 30:295–299 [DOI] [PubMed] [Google Scholar]
- 8. Ewers C, et al. 2010. Emergence of human pandemic O25:H4-ST131 CTX-M-15 extended-spectrum-beta-lactamase-producing Escherichia coli among companion animals. J. Antimicrob. Chemother. 65:651–660 [DOI] [PubMed] [Google Scholar]
- 9. Gibson JS, et al. 2010. Identification of qnr and aac(6′)-1b-cr plasmid-mediated fluoroquinolone resistance determinants in multidrug-resistant Enterobacter spp. isolated from extraintestinal infections in companion animals. Vet. Microbiol. 143:329–336 [DOI] [PubMed] [Google Scholar]
- 10. Johnson JR, Delavari P, Kuskowski M, Stell AL. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183:78–88 [DOI] [PubMed] [Google Scholar]
- 11. Johnson JR, Gajewski A, Lesse AJ, Russo TA. 2003. Extraintestinal pathogenic Escherichia coli as a cause of invasive nonurinary infections. J. Clin. Microbiol. 41:5798–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 51:286–294 [DOI] [PubMed] [Google Scholar]
- 13. Johnson JR, et al. 2008. Virulence genotypes and phylogenetic background of Escherichia coli serogroup O6 isolates from humans, dogs, and cats. J. Clin. Microbiol. 46:417–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson JR, Kuskowski MA, Gajewski A, Sahm DF, Karlowsky JA. 2004. Virulence characteristics and phylogenetic background of multidrug-resistant and antimicrobial-susceptible clinical isolates of Escherichia coli from across the United States, 2000-2001. J. Infect. Dis. 190:1739–1744 [DOI] [PubMed] [Google Scholar]
- 15. Johnson JR, et al. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob. Agents Chemother. 53:2733–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson JR, Miller S, Johnston B, Clabots C, Debroy C. 2009. Sharing of Escherichia coli sequence type ST131 and other multidrug-resistant and urovirulent E. coli strains among dogs and cats within a household. J. Clin. Microbiol. 47:3721–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson JR, et al. 2005. Distribution and characteristics of Escherichia coli clonal group A. Emerg. Infect. Dis. 11:141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnson JR, et al. 2000. Evidence of commonality between canine and human extraintestinal pathogenic Escherichia coli strains that express papG allele III. Infect. Immun. 68:3327–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261–272 [DOI] [PubMed] [Google Scholar]
- 20. Johnson JR, et al. 2002. Global molecular epidemiology of the O15:K52:H1 extraintestinal pathogenic Escherichia coli clonal group: evidence of distribution beyond Europe. J. Clin. Microbiol. 40:1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson JR, et al. 2000. Analysis of the F antigen-specific papA alleles of extraintestinal pathogenic Escherichia coli using a novel multiplex PCR-based assay. Infect. Immun. 68:1587–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kurazono H, et al. 2003. Distribution of the usp gene in uropathogenic Escherichia coli isolated from companion animals and correlation with serotypes and size-variations of the pathogenicity island. Microbiol. Immunol. 47:797–802 [DOI] [PubMed] [Google Scholar]
- 23. Maslow JN, Mulligan ME, Arbeit RD. 1994. Recurrent Escherichia coli bacteremia. J. Clin. Microbiol. 32:710–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maynard C, et al. 2004. Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. J. Clin. Microbiol. 42:5444–5452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nicolas-Chanoine MH, et al. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273–281 [DOI] [PubMed] [Google Scholar]
- 26. Nimmich W, Voigt W, Seltmann G. 1997. Characterization of urinary Escherichia coli O75 strains. J. Clin. Microbiol. 35:1112–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nowicki B, Barrish JP, Korhonen T, Hull RA, Hull SI. 1987. Molecular cloning of the Escherichia coli O75X adhesin. Infect. Immun. 55:3168–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Obata-Yasuoka M, Ba-Thein W, Tsukamoto T, Yoshikawa H, Hayashi H. 2002. Vaginal Escherichia coli share common virulence factor profiles, serotypes and phylogeny with other extraintestinal E. coli. Microbiology 148:2745–2752 [DOI] [PubMed] [Google Scholar]
- 29. Peddie BA, Little PJ. 1978. Serogroups of Escherichia coli in symptomatic and asymptomatic urinary tract infections in Christchurch. Zentralbl. Bakteriol. Orig. A 240:320–325 [PubMed] [Google Scholar]
- 30. Pitout JD, Gregson DB, Campbell L, Laupland KB. 2009. Molecular characteristics of extended-spectrum-beta-lactamase-producing Escherichia coli isolates causing bacteremia in the Calgary Health Region from 2000 to 2007: emergence of clone ST131 as a cause of community-acquired infections. Antimicrob. Agents Chemother. 53:2846–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Platell JL, et al. 2011. Commonality among fluoroquinolone-resistant sequence type ST131 extraintestinal Escherichia coli isolates from humans and companion animals in Australia. Antimicrob. Agents Chemother. 55:3782–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Platell JL, Cobbold RN, Johnson JR, Trott DJ. 2010. Clonal group distribution of fluoroquinolone-resistant Escherichia coli among humans and companion animals in Australia. J. Antimicrob. Chemother. 65:1936–1938 [DOI] [PubMed] [Google Scholar]
- 33. Pomba C, da Fonseca JD, Baptista BC, Correia JD, Martinez-Martinez L. 2009. Detection of the pandemic O25-ST131 human virulent Escherichia coli CTX-M-15-producing clone harboring the qnrB2 and aac(6′)-Ib-cr genes in a dog. Antimicrob. Agents Chemother. 53:327–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ribot EM, et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 35. Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66:1–14 [DOI] [PubMed] [Google Scholar]
- 36. Sidjabat HE, et al. 2009. Colonisation dynamics and virulence of two clonal groups of multidrug-resistant Escherichia coli isolated from dogs. Microbes Infect. 11:100–107 [DOI] [PubMed] [Google Scholar]
- 37. Sidjabat HE, Derrington P, Nimmo GR, Paterson DL. 2010. Escherichia coli ST131 producing CTX-M-15 in Australia. J. Antimicrob. Chemother. 65:1301–1303 [DOI] [PubMed] [Google Scholar]
- 38. Simpson EH. 1949. Measurement of diversity. Nature 163:688 [Google Scholar]
- 39. Tenover F, et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wetzstein HG. 2005. Comparative mutant prevention concentrations of pradofloxacin and other veterinary fluoroquinolones indicate differing potentials in preventing selection of resistance. Antimicrob. Agents Chemother. 49:4166–4173 [DOI] [PMC free article] [PubMed] [Google Scholar]