Abstract
Natural mutations of Plasmodium falciparum dihydrofolate reductase (PfDHFR) at A16V and S108T specifically confer resistance to cycloguanil (CYC) but not to pyrimethamine (PYR). In order to understand the nature of CYC resistance, the effects of various mutations at A16 on substrate and inhibitor binding were examined. Three series of mutations at A16 with or without the S108T/N mutation were generated. Only three mutants with small side chains at residue 16 (G, C, and S) were viable from bacterial complementation assay in the S108 series, whereas these three and an additional four mutants (T, V, M, and I) with slightly larger side chains were viable with simultaneous S108T mutation. Among these combinations, the A16V+S108T mutant was the most CYC resistant, and all of the S108T series ranged from being highly to moderately sensitive to PYR. In the S108N series, a strict requirement for alanine was observed at position 16. Crystal structure analyses reveal that in PfDHFR-TS variant T9/94 (A16V+S108T) complexed with CYC, the ligand has substantial steric conflicts with the side chains of both A16V and S108T, whereas in the complex with PYR, the ligand only showed mild conflict with S108T. CYC analogs designed to avoid such conflicts improved the binding affinity of the mutant enzymes. These results show that there is greater spatial limitation around the S108T/N residue when combined with the limitation imposed by A16V. The limitation of mutation of this series provides opportunities for drug design and development against antifolate-resistant malaria.
INTRODUCTION
Plasmodium falciparum dihydrofolate reductase (PfDHFR) is one of the most well-defined drug targets in malaria chemotherapy with such antifolates as pyrimethamine (PYR) and cycloguanil (CYC; administered as proguanil) among its inhibitors (26). It catalyzes the NADPH-dependent reduction of 7,8-dihydrofolate (DHF) to 5,6,7,8-tetrahydrofolate (THF) in the biosynthesis of deoxythymidylate (dTMP). After decades of antifolate deployment, the parasite has become increasingly resistant to PYR and CYC due to mutations that reduce the binding affinity of inhibitors. While PfDHFR mutations at residues 108, 59, 51, and 164 confer cross-resistance to both PYR and CYC, the double-mutation variant A16V+S108T specifically confers resistance to CYC. The CYC-resistant mutant was postulated to have arisen by the S108T mutation followed by A16V, giving rise to high CYC resistance without a loss in catalytic efficiency (18). Molecular modeling suggests that the A16V mutation causes steric clash between the side chain and one of the 2,2-dimethyl groups of CYC (15). PYR, on the other hand, avoids such a steric clash, presumably due to lack of a group equivalent to the methyl of CYC (15). However, the lack of crystal structure data for the A16V+S108T PfDHFR and its complex with CYC makes such conclusion a tentative one.
Unlike cumulative mutations in the S108N series of PYR resistance-generating mutant PfDHFRs, no other resistant variants in the S108T series apart from A16V+S108T have been identified from the field. Laboratory selection for PYR resistance in a wild-type parasite strain led to the identification of mutants with a single mutation at either A16S or I164M (22). In the same study, selection for increased PYR resistance in the PfDHFR-TS A16V+S108T mutant strain T9/94 demonstrated DHFR-TS gene amplification but no further mutation of the protein (22). Another study of laboratory-induced PYR resistance reported A16V+S108T variants with additional mutations at F223S and D54N (20). The A16V+S108T+D54E+F223S mutant enzyme shows enhanced resistance to CYC and is also resistant to PYR (19).
We investigated here the effect of mutation at residue 16 on PfDHFR catalytic activity and antifolate resistance. In the presence or the absence of the S108T/N mutation, mutants of A16X were catalytically active only when X was an amino acid with a small side chain, suggesting limitation in space around residue 16. Crystal structures of PfDHFR-TS T9/94 variant (A16V+S108T) in complex with CYC and PYR were determined to reveal the different modes of inhibitor binding. The structural information presented here shows confinement of the DHFR binding pocket at subsites containing residues 16 and 108, with the mutual effects of steric constraint in one subsite exerting further confinement on another subsite. Such mutual effects are present in the binding of CYC, which has a 2-methyl group in conflict with the side chain of V16, and a p-chlorine atom in conflict with the side chain of T108. These constraints are absent in the binding of PYR, which lacks an equivalent group in the vicinity of V16. Therefore, the resistance of the A16V+S108T PfDHFR-bearing parasite only to CYC, but not to PYR, is explained. This information should help in the design of effective inhibitors against mutant PfDHFRs and against antifolate-resistant malaria.
MATERIALS AND METHODS
Construction of mutant libraries.
Mutant libraries of A16-saturated mutations were generated by QuikChange site-directed mutagenesis using pET17b containing synthetic PfDHFR–A16V+S108T gene as a template, 5′-C TTT GAC ATC TAC GCT ATC TGC [NNG/C] TGC TGT AAA GTT GAA AGC AAA-3′ as the sense primer, and 5′-TTT GCT TTC AAC TTT ACA GCA [G/CNN] GCA GAT AGC GTA GAT GTC AAA G-3′ as the antisense primer, where the A16X codon is marked by brackets, and N represents all four bases. The mutagenesis reaction contained 40 ng of template, 125 ng of each primer, 1× Pfu buffer, and 10 U of Pfu DNA polymerase (Promega) and was carried out using the following temperature cycling profile: 95°C for 5 min, followed by 20 cycles of 95°C for 30 s, 45°C for 1 min, and 68°C for 8 min, with a final incubation at 68°C for 10 min. The mutagenesis product was digested with 20 U of DpnI at 37°C for 1 h and then transformed into E. coli DH5α. Recombinant clones grown on Luria-Bertani (LB) agar with 100 μg of ampicillin (Amp) ml−1 were randomly selected for sequencing. Generation of A16X+S108 series was performed using the same protocol with an equimolar mixture of the 20 identified A16X+S108T as templates and a pair of primers that allowed mutation of T108 back to S108—5′-GTT GTT ATG GGC CGC ACC TCG TGG GAA TCG ATT CCG AAA AAA TTC-3′ and 5′-GAA TTT TTT CGG AAT CGA TTC CCA CGA GGT GCG GCC CAT AAC AAC-3′—as the sense and antisense primers, respectively. A16X mutants in the N108 series were limited to G, C, S, T, and V. The mutagenesis reactions were performed separately using specific templates from the T108 series and a pair of primers that allowed the mutation of T108N: 5′-GGG CCG CAC CAA CTG GGA ATC GAT-3′ and 5′-ATC GAT TCC CAG TTG GTG CGG CCC-3′. The clones obtained were verified by automated Sanger sequencing to ensure that there were no adventitious mutations in the course of making the site-specific mutations.
Screening of active DHFR mutants by bacterial complementation assay.
Plasmids from the pET-PfDHFR mutants were retransformed into E. coli BL21(DE3)/pLysS. The transformants were grown on LB agar with 100 μg of Amp ml−1 and 34 μg of chloramphenicol (Cam) ml−1. The mutant clones were screened for PfDHFR activity by streaking them on minimal medium containing 100 μg of Amp ml−1 and 34 μg of Cam ml−1 with or without 4 μM trimethoprim. After overnight incubation at 37°C, transformed bacteria are only able to grow in the presence of trimethoprim if the PfDHFR has adequate activity; mutants with low activity do not support growth (4).
Expression and purification of mutant PfDHFRs.
All of the mutants were expressed in E. coli BL21(DE3)/pLysS by cultivation in LB supplemented with 100 μg of Amp ml−1 and 34 μg of Cam ml−1 and 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) induction at 20°C for 24 h. The expression levels of all mutants were confirmed by SDS-PAGE. The relative PfDHFR activity of each mutant compared to wild-type enzyme was determined from crude protein extracts. Those with relative crude activity higher than 3% were further purified by methotrexate-Sepharose affinity chromatography (17) and anion exchange as previously described (2, 9).
Enzyme kinetic analysis and inhibition by antifolates.
DHFR activities were determined spectrophotometrically using a Hewlett Packard UV-VIS spectrophotometer (HP8453) in a 1-ml reaction containing 100 μM concentrations each of dihydrofolate and NADPH in 50 mM TES (pH 7.0), 75 mM β-mercaptoethanol, 1 mg of bovine serum albumin ml−1, and the appropriate amount of purified enzyme (1 to 5 mU) to initiate the reaction. PYR, CYC, WR99210, and a series of CYC analogues (15, 27) were 2-fold serial diluted in dimethyl sulfoxide in 96-well plates (200 μl, final well volume), and binding constants were determined using the protocol described previously (21). Kinetic parameters were calculated by a nonlinear least-square fit of the data to the Michaelis-Menten equation using the software Kaleidagraph 3.51 as previously described (9, 21).
Crystallization, data collection, and structure determination.
The bifunctional DHFR-TS enzymes of both wild-type strain TM4 and the A16V+S108T mutant strain T9/94 of P. falciparum were cloned, expressed, and purified as previously described (2, 28). Protein was cocrystallized with either 2 mM CYC or 2 mM PYR in the presence of NADPH and dUMP. X-ray diffraction data were collected at the CuKα wavelength (1.5418 Å) on a Bruker-Nonius FR591 X-ray generator equipped with a κCCD detector. The data were indexed, integrated and scaled using the Denzo/Scalepack program in the HKL-2000 package (14). The data were directly refined at 3.0-Å resolution using PfDHFR-TS TM4 (PDB 1J3I) as a template in the Crystallography and NMR System (1, 28). The models were iteratively rebuilt and adjusted in program O (8) and gradually refined to the highest resolution in CNS (1). Ligands and water molecules were added to a positive Fourier difference electron density map (Fo − Fc). The final models were verified by PROCHECK (11) with Ramachandran parameters listed in Table 1. Structural figures were prepared with PyMOL (16).
Table 1.
Data collection and refinement statistics of crystal structures
| Parametera | Enzyme and ligand |
||
|---|---|---|---|
|
PfDHFR-TS T9/94 |
PfDHFR-TS TM4 (CYC) | ||
| PYR | CYC | ||
| Data collection | |||
| Space group | P212121 | P212121 | P212121 |
| Unit cell parameters (Å) | |||
| a | 56.529 | 56.696 | 58.761 |
| b | 155.119 | 155.549 | 157.538 |
| c | 165.257 | 164.962 | 164.509 |
| Resolution* (Å) | 30–2.40 (2.49–2.40) | 30–2.65 (2.74–2.65) | 50–2.60 (2.69–2.60) |
| Rmerge* (%) | 6.5 (35.1) | 7.9 (35.5) | 4.1 (25.8) |
| I/σI* | 15.5 (3.0) | 19.2 (4.2) | 20.3 (4.3) |
| Completeness* (%) | 98.7 (95.0) | 99.9 (99.7) | 98.4 (93.2) |
| Redundancy | 4.6 | 8.1 | 4.4 |
| Refinement | |||
| Resolution (Å) | 30–2.40 | 30–2.65 | 50–2.60 |
| No. reflections (unique) | 56,048 | 43,280 | 47,165 |
| Rwork/Rfreeb | 20.2/25.4 | 20.3/25.7 | 21.2/25.8 |
| No. of atoms | |||
| Protein | 9,058 | 9,048 | 9,046 |
| PYR or CYC | 34 | 34 | 34 |
| NADPH | 96 | 96 | 96 |
| dUMP | 40 | 40 | |
| Water | 781 | 371 | 456 |
| Phosphate | 10 | ||
| Avg B factor (Å2) | 46.9 | 48.5 | 52.5 |
| Ramachandran plot (%) | |||
| Favored region | 85.6 | 84.6 | 87.3 |
| Additional allowed region | 14.2 | 15.2 | 12.3 |
| Generously allowed region | 0.2 | 0.2 | 0.2 |
| Disallowed region | 0 | 0 | 0.2c |
| RMS deviations | |||
| Bond length (Å) | 0.006 | 0.007 | 0.008 |
| Bond angle (°) | 1.4 | 1.4 | 1.5 |
*, Values in parentheses are for the highest-resolution shell. RMS, root mean square.
Rfree is calculated from 5% of reflections chosen randomly in each of the 10 resolution bins.
Lys49 is located on a flexible loop of each molecule of PfDHFR-TS.
PDB accession codes.
Coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession codes 3UM5, 3UM6, and 3UM8.
RESULTS
Screening of active A16X+S108T/N PfDHFR mutants.
Constructs of wild-type and 45 A16X mutants with S108 or S108N/T mutations were successfully generated and verified by DNA sequencing. These mutants included 19 from the A16X+S108 series, 20 from the A16X+S108T series, and 6 from the S108N series. For the S108N series, only the A16 mutants that are capable of complementing bacterial growth in the S108T series were created, i.e., G, A, C, S, T, and V.
The activities of the wild-type and mutant PfDHFRs in E. coli BL21(DE3)/pLysS were screened by complementation assay. Only six mutants from S108T series showed adequate activity for growth, namely, S108T, A16C+S108T, A16G+S108T, A16S+S108T, A16T+S108T, and A16V+S108T. Hence, only the amino acids with small side chains at position 16 when combined with S108T were functionally active. Moreover, two mutants with slightly larger side chains at position 16, namely, A16I+S108T and A16M+S108T, could also complement host DHFR under the same conditions, albeit poorly (Table 2). Further investigation of the S108 series showed that only the wild type and three mutants (A16G, A16S, and A16C) were found to be catalytically active by bacterial complementation assay, while the others were not. In the S108N series, only alanine was tolerated at position 16 among the six mutants tested.
Table 2.
Summary of bacterial complementation assay of PfDHFR activities in E. coli BL21(DE3)/pLysS on a minimal medium plate in the presence of trimethoprima
| Amino acid at residue 16 | Type | Residue volb (Å3) | Surface areac (Å2) | Bacterial complementation |
||
|---|---|---|---|---|---|---|
| S108 | T108 | N108 | ||||
| G | Aliphatic | 60.1 | 75 | + | + | – |
| A (wild type) | Aliphatic | 88.6 | 115 | + | + | + |
| P | Aliphatic | 112.7 | 145 | – | – | NAd |
| V | Aliphatic | 140.0 | 155 | – | + | – |
| M | Aliphatic | 162.9 | 185 | – | +/– | NA |
| I | Aliphatic | 166.7 | 175 | – | +/– | NA |
| L | Aliphatic | 166.7 | 170 | – | – | NA |
| S | Polar, uncharged | 89.0 | 115 | + | + | – |
| C | Polar, uncharged | 108.5 | 135 | + | + | – |
| N | Polar, uncharged | 114.1 | 160 | – | – | NA |
| T | Polar, uncharged | 116.1 | 140 | – | + | – |
| Q | Polar, uncharged | 143.8 | 180 | – | – | NA |
| H | Positively charged | 153.2 | 195 | – | – | NA |
| K | Positively charged | 168.6 | 200 | – | – | NA |
| R | Positively charged | 173.4 | 225 | – | – | NA |
| D | Negatively charged | 111.1 | 150 | – | – | NA |
| E | Negatively charged | 138.4 | 190 | – | – | NA |
| F | Aromatic | 189.9 | 210 | – | – | NA |
| Y | Aromatic | 193.6 | 230 | – | – | NA |
| W | Aromatic | 227.8 | 255 | – | – | NA |
Crude extracts of wild-type PfDHFR and 45 mutants with A16X and S108 or S108N/T were prepared in 50 ml of LB broth with IPTG induction in order to examine their expression levels and DHFR activities. SDS-PAGE analysis of the crude extracts revealed that all of the constructs expressed PfDHFR similarly to the wild-type construct (data not shown). However, these crude extracts exhibited a wide range of enzyme specific activities. The relative DHFR activity of crude protein extracts of the active mutants, defined from the ability to complement bacterial DHFR in the presence of trimethoprim in bacterial complementation assay, ranged from 1.4 to 62.4% of the wild-type enzyme (Table 3), while the activities of the inactive mutants were negligible. The results are in line with our previous reports, wherein only constructs expressing relative DHFR activity higher than 1.3% of the wild-type enzymes could support bacterial growth on trimethoprim plates (4, 9).
Table 3.
Crude enzyme activity and kinetic properties of purified PfDHFR wild-type and mutant enzymes of A16X with or without the S108 T/N mutation
| Enzyme | Crude enzyme activity |
Kinetic parameters of purified enzymea |
||||
|---|---|---|---|---|---|---|
| Mean sp act (nmol/min/mg) ± SD | Relative activity | Mean Km ± SD (μM) |
kcat (s−1) | kcat/Km | ||
| DHF | NADPH | |||||
| Wild type (A16+S108) | 1,978 ± 284 | 100 | 12.2 ± 2.0 | 5.8 ± 1.7 | 72.9 | 5.9 |
| S108N | 516 ± 147 | 26.1 | 32.5 ± 1.8 | 3.7 ± 0.4 | 36.1 | 1.1 |
| S108T | 1,235 ± 232 | 62.4 | 3.0 ± 0.6 | 2.1 ± 0.6 | 24.7 | 8.2 |
| A16C | 274 ± 33 | 13.8 | 16.4 ± 1.7 | 3.6 ± 0.6 | 37.4 | 2.3 |
| A16G | 77 ± 21 | 3.9 | 15.2 ± 2.3 | 11.7 ± 2.0 | 10.4 | 0.68 |
| A16S | 130 ± 5 | 6.6 | 47.6 ± 1.1 | 4.8 ± 0.3 | 10.8 | 0.23 |
| A16C+S108T | 1,181 ± 198 | 59.7 | 6.6 ± 0.6 | 4.7 ± 1.3 | 62.6 | 9.5 |
| A16G+S108T | 176 ± 33 | 8.9 | 10.2 ± 1.5 | 7.9 ± 1.4 | 16.9 | 1.7 |
| A16S+S108T | 733 ± 195 | 37.1 | 19.4 ± 3.3 | 2.4 ± 0.3 | 16.6 | 0.86 |
| A16T+S108T | 70 ± 7 | 3.5 | 23.8 ± 1.8 | 2.3 ± 0.1 | 1.2 | 0.05 |
| A16V+S108T | 241 ± 49 | 12.2 | 21.3 ± 2.9 | 4.5 ± 0.5 | 18.9 | 0.89 |
| A16M+S108T | 33 ± 7 | 1.7 | ND | ND | ND | ND |
| A16I+S108T | 27 ± 2 | 1.4 | ND | ND | ND | ND |
ND, not determined.
Kinetic parameters of PfDHFR mutants.
Wild-type PfDHFR and 10 active A16X PfDHFR mutants with or without S108T/N mutations were purified to homogeneity and enzyme kinetic parameters determined. As summarized in Table 3, the kcat values of all of the active mutants varied from 1.6 to 86% of that of the wild-type enzyme. Among these mutants, A16C+S108T exhibited the highest kcat value, while A16T+S108T was the lowest. The kcat values of the others were 14 to 51% of the wild-type enzyme.
Mutations of A16 and S108 affected DHF binding to various degrees. The A16S and S108N mutants showed 3- to 4-fold-higher Km values for DHF than those of the wild type, while the Km values for the S108T and A16C+S108T mutants were about 2- to 4-fold less. Unlike DHF, the Km values for NADPH were not much affected by mutations since the values were within a 2-fold range of the wild-type enzyme. Among the 10 active mutants, A16C+S108T and S108T showed the highest catalytic efficiency (kcat/Km), about 1.4- to 1.6-fold that of the wild-type enzyme. The catalytic efficiency of the other mutants, including A16V+S108T, was about 4 to 38% that of the wild-type enzyme and that of A16T+S108T was <1%.
Antifolate sensitivity of PfDHFR mutants.
The inhibition constants (Ki) of PYR, CYC, and WR99210, as well as of three CYC derivatives, namely, C17 (R1, R2, and R3 = H, Me, and p-Cl phenyl, respectively), C88 (R1, R2, and R3 = Me, Me, and m-Cl phenyl, respectively), and C248 (R1, R2, and R3 = H, Me, and m-Cl phenyl, respectively), against the 10 mutants were determined in order to understand the drug resistance pattern of the active mutants. The data are summarized in Table 4.
Table 4.
Inhibition constant (Ki) of PfDHFR wild-type and mutant enzymes of A16X combination with or without the S108T/N mutation
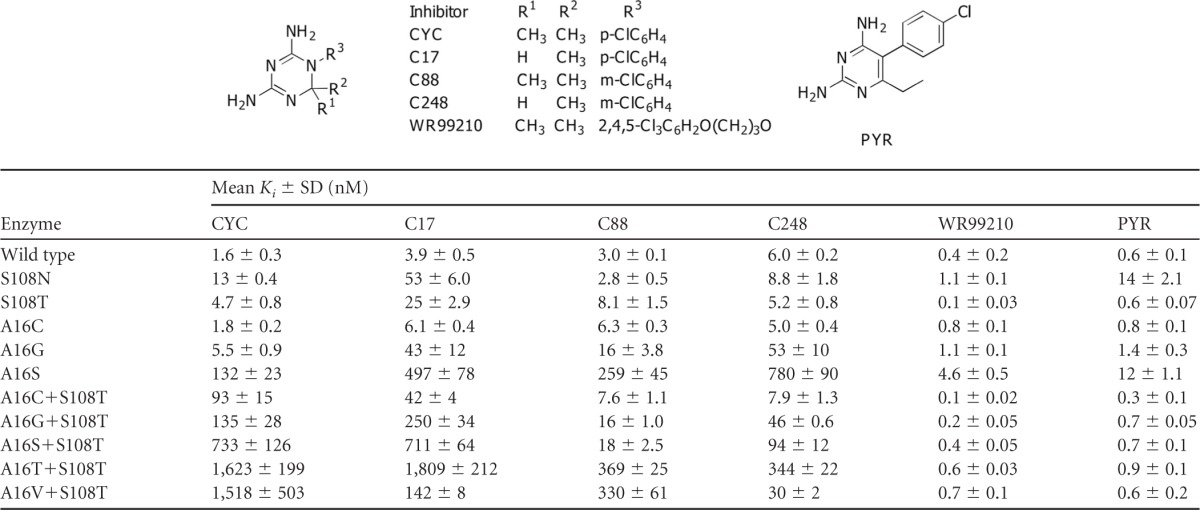
Among the three active A16X single mutants, only A16S was highly resistant to CYC, with Ki values ∼83 times higher than that of the wild type, while the others were about 1- to 3.5 times that of the wild type. In combination with S108T mutation, the Ki values of CYC against all of the active double mutants were markedly increased, i.e., by about 58- to 1,014-fold that of the wild-type and 20- to 345-fold that of the S108T single mutant. In addition, the Ki values of CYC against S108N were 8 times that of the wild-type enzyme and 2.7 times that of S108T. These results suggested that the steric conflicts of the p-Cl group at R3 and a 2-methyl group of CYC with side chains of amino acid residues 108 and 16, respectively, are significant and that the clash with S108N exerted a greater effect on the binding affinity than that of S108T, which is in line with previous observations (18).
In order to reduce the steric clash of inhibitors with A16X mutants, the inhibitor C17 was designed with only one methyl group at the position 2 of the dihydrotriazine ring (either R1 or R2), while maintaining its p-Cl phenyl group at R3. The Ki values of C17 against the wild type and most of mutants were generally similar to those of CYC (0.2- to 2.2-fold) but reduced markedly by 10.7-fold against A16V+S108T compared to CYC. By moving the p-Cl group to the m-position to avoid steric clash with S108T while preserving the 2,2-dimethyl groups of CYC, the inhibitor C88 showed reduction of Ki values against all of the double mutants to an even greater extent than C17, by 4- to 41-fold compared to CYC. With the combination of both features so as to avoid steric conflicts on both sites, C248 (desmethyl and m-Cl phenyl) showed dramatically lower Ki values against A16V+S108T by a factor of 51 compared to that of CYC (from 1,518 nM for CYC to 30 nM for C248). The Ki values against other A16X+S108T mutants were also reduced by factors of 3 to 12 compared to CYC. Unlike CYC and its derivatives, PYR with p-Cl phenyl at R3 remained effective against most of the mutants created. The Ki values of PYR against the active mutants were mostly <1 nM. Only the A16S and S108N mutants showed slightly poorer binding to PYR, with Ki values of 12 and 14 nM, respectively, mainly due to their high DHF Km value. Likewise, WR99210, a flexible antifolate, also remained effective against the entire collection of active PfDHFR mutants. The WR99210 Ki values against these mutants ranged from 0.1 to 1.1 nM, with the highest Ki value of 4.6 nM against the A16S mutant.
Structure analysis of PfDHFR-TS from T9/94 strain.
Four crystal structures of the PfDHFR-TS wild type (TM4, PYR/CYC sensitive) and A16V+S108T (T9/94, PYR sensitive and CYC resistant) in complex with PYR (TM4-PYR [PDB 3QGT] and T9/94-PYR) and CYC (TM4-CYC and T9/94-CYC) were studied in order to understand the basis of A16V+S108T resistance to CYC. Superimposition of TM4-CYC to TM4-PYR (Fig. 1A) showed similar overall structures, with only a slight shift of CYC, as indicated by movement of the p-Cl group of CYC 0.8 Å away from that of PYR in TM4-PYR. The fact that TM4 is sensitive to CYC (Ki value of 1.6 nM, Table 4) suggests that the slight shift does not affect the binding affinity of CYC in the TM4 variant. Similarly, the superposition of T9/94-PYR to TM4-PYR (Fig. 1B) also revealed an unchanged overall structure of the DHFR enzyme active site and only a minor displacement of PYR due to a slight clash of the γ-CH3 group of T108 of T9/94 with the p-Cl group of PYR. This is seen from a shift of the p-Cl group of PYR in T9/94-PYR by 0.8 Å and a slight twist of the phenyl ring of PYR in T9/94-PYR by 6.8° (the torsion angle of the two rings of PYR changed from 111.1° to 104.3° in TM4-PYR and T9/94-PYR, respectively). The displacement of PYR in the T9/94 binding pocket is an adjustment, which does not have any significant effect on the PYR binding affinity, since the T9/94 remains sensitive to PYR (Ki value = 0.6 nM for both, Table 4).
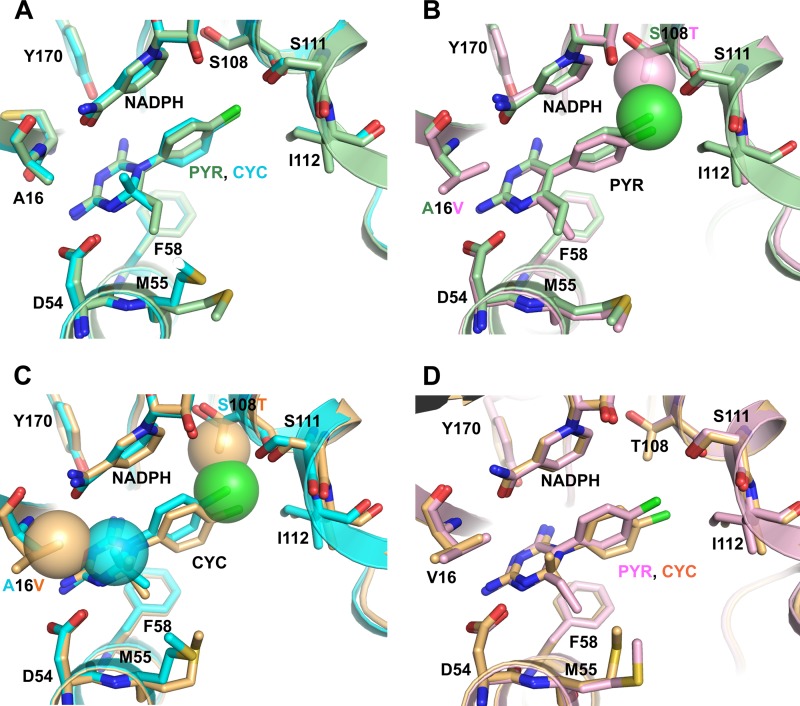
Fig 1.
Comparison of cocrystal structures of PfDHFR domains with CYC and PYR. (A) Superposition of the wild-type PfDHFR in complex with PYR (TM4-PYR, light green, PDB 3QGT) and CYC (TM4-CYC, cyan, PDB 3UM8) shows no significant difference in overall structure and ligands binding in the active-site pocket. (B) The PYR complex of mutant A16V+S108T PfDHFR (T9/94-PYR, pink, PDB id: 3UM5) was superimposed on that of TM4-PYR (light green). The p-Cl group of PYR (green sphere) in TM4 has only a minor steric conflict with the γ-CH3 group of T108 of T9/94 (pink sphere). Hence, PYR in T9/94 is slightly shifted (0.8 Å), and its ring twisting was observed. (C) T9/94-CYC (light orange, PDB 3UM6) was superimposed on TM4-CYC (cyan). A slight shift of enzyme backbone and CYC displacement/ring twisting were observed due to steric clashes at the p-Cl group of CYC with the γ-CH3 group of T108 of T9/94 and one of the two methyl groups of CYC with the γ-CH3 group of V16 of T9/94, as indicated by spheres. (D) Superposition of T9/94-PYR (pink) and T9/94-CYC (light orange) shows displacement and ring twisting of CYC compared to PYR. The side chains of residues within 3.5 Å around the ligands are shown and labeled. Oxygen, nitrogen, sulfur, and chlorine atoms are colored red, blue, yellow, and green, respectively.
In contrast, in the structure of T9/94-CYC superposed on TM4-CYC (Fig. 1C), a slightly outward shift of the backbone of the T9/94 around V16 and T108 to expand the active site chamber was observed, presumably due to restricted space. Furthermore, the β-branched side chains of S108T and A16V in T9/94-CYC cause steric conflict to the p-Cl group and one of the 2,2-dimethyl groups of CYC, respectively, and hence the displacement of CYC molecule in T9/94-CYC, as evidenced from a 1.3-Å shift of the p-Cl group and a 0.8-Å shift of 2,2-dimethyl groups compared to CYC in the TM4-CYC structure. The p-Cl phenyl ring of CYC is twisted by 10.6° (the torsion angles of the two rings of CYC changed from 108.9° to 98.3° in TM4-CYC and T9/94-CYC, respectively). Superposition of T9/94-CYC to T9/94-PYR (Fig. 1D) clearly confirms that the steric conflict is much more pronounced on CYC than on PYR, as shown by the movement of the side chain of V16 of the T9/94-CYC by 0.4 Å compared to that of T9/94-PYR. The conflict also resulted in a shift of the p-Cl group of CYC by 1.6 Å compared to T9/94-PYR. This would explain the very high Ki value of T9/94-CYC (Ki value = 1,518 nM, Table 4). The higher degree of displacement of CYC in T9/94-CYC, compared to PYR in T9/94-PYR, suggested a less favorable binding interaction with the enzyme, thus causing CYC resistance in the T9/94 strain.
Taken altogether, these results indicated that the steric hindrance caused by A16V and S108T in the active site of T9/94-CYC aggravate the binding of CYC, but not PYR. This is reflected by the poor binding affinity of CYC to A16V+S108T, which results in resistance of T9/94 parasite to CYC, but not to PYR.
DISCUSSION
It has been well established that mutations of PfDHFR are responsible for antifolate resistance in P. falciparum. PfDHFR mutants in the S108N series are resistant to both CYC and PYR, while A16V+S108T, the only member of the S108T series, confers resistance to CYC only. In this report, we confirm that the mutation of residue A16 to V16 in the presence of S108T is indeed the outcome of a compromise between its catalytic activity and CYC resistance.
In combination with or without S108T/N mutation, only 12 mutants of A16X enzymes were catalytically active, as revealed by a bacterial complementation assay in the presence of trimethoprim to inhibit host DHFR (Table 2). These A16X active mutants were confined to those amino acids with small aliphatic side chains, likely due to restriction in space around the substrate binding area for this residue. However, residues with either charged (D and E) or γ/δ-polar side chains (Q and N) also cannot replace A at position 16, indicating that factors other than space around this area are also important.
Among the A16X mutants, A16C and A16C+S108T mutants exhibited the highest catalytic efficiency (kcat/Km) and enzyme turnover rate (kcat). These mutants have not been found in nature, probably due to the fact that the A16C mutation requires changes of all three codon nucleotides (from GCA to UGU/C). Unlike A16C mutants, the A16S and A16G mutants have been found in nature. Despite poor catalytic properties and high PYR sensitivity of the A16G mutant, P. falciparum harboring A16G mutant, Santa Lucia strain, was isolated from El Salvador and adapted in monkeys for vaccine development (5, 6). The A16S mutant, on the contrary, showed resistance to all of the antifolates tested, including PYR. Parasites harboring the A16S mutant could survive under low PYR pressure in vitro (22). In addition, the A16S+C59R mutant parasite has also been identified in Pakistan under Fansidar (PYR–sulfadoxine) pressure without any mutation in the dihydropteroate synthase gene (25). Based on the PfDHFR-TS structure, C59R appears to play a rescuing role on enzyme activity, enhancing substrate binding through ionic attraction with the glutamate moiety (28). The identification of A16S+C59R thus confirms the PYR-resistant phenotype of A16S and supports our finding of antifolate resistance (CYC and PYR) of this mutant. The A16S+S108T double mutant has better kinetic properties and is more CYC resistant than the A16S single mutant. However, this double mutant is more sensitive to PYR than the A16S mutant (Table 4), explaining why it was not found in the nature under PYR or Fansidar pressure. In the background of S108N mutation, none of the five A16X mutants with small amino acid side chains (G, C, S, T, and V) could give rise to active DHFR enzyme. This finding is in line with previous observations, where further mutations of A16V+S108N yielded protein with no DHFR activity (18). These findings suggest that the areas around A16 and S108 in the active- site cleft are crucial for substrate binding. In short, mutation is limited by the ability of the substrate to bind with the enzyme leading to catalytic activity. Although S108N mutation alone confers only a low level of CYC and PYR resistance and no further mutation at A16X is possible, the parasite adopts additional mutations at other residues in the active site such as residues 51, 59, and 164 instead. This narrow evolutionary path of mutation is determined by the fitness of antifolate-resistant variants (7, 12, 18), which explains why these mutations are commonly found in the field (26).
Compounds C17 and C88 (15, 27) were used to investigate the implications of steric clash from both A16X and S108T/N series. C17, a CYC derivative with one methyl group removed, is expected to avoid the hindrance at position 16. However, except for the naturally occurring A16V+S108T mutant, the Ki values of C17 for most of the selected active mutants are higher than those of CYC, which is probably complicated by the nature of the amino acid at position 16. The polar amino acids at position 16 affect not only the space available for DHF or inhibitor binding but also likely interact with the nicotinamide moiety of NADPH, as revealed by lower NADPH Km values (Table 3). Presumably, interaction of A16C/S/T with the amino group of the nicotinamide ring shifts the ring toward the DHF or inhibitor site, resulting in decreased affinity for C17 as the site is in part restricted by NADPH. In comparison to C17, C88 with m-Cl in place of p-Cl has little constraint at position 108, and the Ki values are reduced dramatically in the S108T/N series. C248, which is a desmethyl CYC derivative with an m-Cl group, shows Ki values similar to those of C88 for the A16X and most of A16X+S108T mutants, indicating that the inhibitor is more susceptible to the clash at position 108 than at position 16. However, the A16V+S108T mutant is more sensitive to C248 and C17, suggesting that relaxed binding occurs at position 16 irrespective of whether an m- or p-Cl substituent is present in the vicinity of position 108.
The crystal structure of PfDHFR-TS T9/94 (A16V+S108T) in complex with CYC (T9/94-CYC) confirms that CYC resistance of T9/94 is due to steric conflict of the β-methyl group of A16V and one of 2,2-dimethyl groups of CYC, together with the S108T and p-Cl group of CYC (Fig. 1B). For the S108N mutant, there is space restriction imposed on both PYR and CYC due to the amino group of S108N, which could clash with the p-Cl groups of both compounds. However, the displacement of PYR from the conflict with S108N is less because the side chain of S108N twists, with a push on NADPH, as observed in the structures of PYR complex of PfDHFR-TS mutants (C59R+S108N and N51I+C59R+S108N+I164L) and S58R+S117N PvDHFR (10, 23, 28). In comparison, the side chain of T108 retains its conformation in complexing with PYR or CYC (Fig. 1A), suggesting space restriction. These mutants, e.g., S108N and A16V+S108T, bind reasonably well with DHF in optimal constraint from both positions as revealed from Km and Ki (Tables 3 and 4), whereas A16V+S108N, an inactive mutant, may pose steric clash dramatically with DHF. Hence, it can be concluded that S108T has a more direct effect on the p-Cl group of CYC than that of PYR, whereas S108N affects the binding of both inhibitors. The double mutation A16V+S108T further reduces active-site space available for CYC, with little effect on PYR, which has no steric conflict around the A16V site. On the other hand, WR99210 binds comfortably with the A16 mutant series mainly due to its flexibility, as reflected by the Ki values (Table 4). The steric clash of a 2-methyl group of CYC with A16V side chain in T9/94-CYC supports our previous hypothesis based on molecular modeling and inhibitors testing (15, 27) and gives a more exact explanation of the difference in sensitivity to CYC and PYR.
In a recent modeling study (13), it was predicted that the S108T mutant has increased stability by van der Waals interactions with neighboring residues and greater affinity for NADPH than does the S108N mutant. This prediction is generally supported by our X-ray structural data. Our report further explains why A16V+S108T is the sole member of the S108T series identified from CYC-resistant parasites since 1977 (6, 24). The blind alley for this series is due to its susceptibility to PYR and limitation of further mutation imposed by necessity for residual enzyme activity. The limitation of DHFR mutations described here offers hope for drug development against resistant parasites, highlighting the potential of focusing on the parts of the target where resistant mutation is constrained by its biological activity, which is essential for the survival of the parasite.
ACKNOWLEDGMENTS
This research was supported by grants from the Wellcome Trust and the MMV Programme, WHO/TDR, Synchrotron Light Research Institute (Public Organization), Thailand, Thailand TDR Programme and NSTDA Cluster and Program Management Office to the team. S.K. is an international scholar of Howard Hughes Medical Institute.
We thank Philip Shaw for manuscript editing.
Footnotes
Published ahead of print 23 April 2012
REFERENCES
- 1. Brünger AT, et al. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921 [DOI] [PubMed] [Google Scholar]
- 2. Chitnumsub P, et al. 2004. Characterization, crystallization and preliminary X-ray analysis of bifunctional dihydrofolate reductase-thymidylate synthase from Plasmodium falciparum. Acta Crystallogr. D Biol. Crystallogr. 60:780–783 [DOI] [PubMed] [Google Scholar]
- 3. Chothia C. 1976. The nature of the accessible and buried surfaces in proteins. J. Mol. Biol. 105:1–12 [DOI] [PubMed] [Google Scholar]
- 4. Chusacultanachai S, Thiensathit P, Tarnchompoo B, Sirawaraporn W, Yuthavong Y. 2002. Novel antifolate resistant mutations of Plasmodium falciparum dihydrofolate reductase selected in Escherichia coli. Mol. Biochem. Parasitol. 120:61–72 [DOI] [PubMed] [Google Scholar]
- 5. Collins WE, Galland GG, Sullivan JS, Morris CL, Richardson BB. 1996. The Santa Lucia strain of Plasmodium falciparum as a model for vaccine studies. II. Development of Aotus vociferans as a model for testing transmission-blocking vaccines. Am. J. Trop. Med. Hyg. 54:380–385 [DOI] [PubMed] [Google Scholar]
- 6. Collins WE, Warren M, Skinner JC, Chin W, Richardson BB. 1977. Studies on the Santa Lucia (El Salvador) strain of Plasmodium falciparum in Aotus trivirgatus monkeys. J. Parasitol. 63:52–56 [PubMed] [Google Scholar]
- 7. Costanzo MS, Brown KM, Hartl DL. 2011. Fitness trade-offs in the evolution of dihydrofolate reductase and drug resistance in Plasmodium falciparum. PLoS One 6:e19636 doi:10.1371/journal.pone.0019636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jones TA, Zou JY, Cowan SW, Kjeldgaard M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47(Pt 2):110–119 [DOI] [PubMed] [Google Scholar]
- 9. Kamchonwongpaisan S, Vanichtanankul J, Taweechai S, Chitnumsub P, Yuthavong Y. 2007. The role of tryptophan-48 in catalysis and binding of inhibitors of Plasmodium falciparum dihydrofolate reductase. Int. J. Parasitol. 37:787–793 [DOI] [PubMed] [Google Scholar]
- 10. Kongsaeree P, et al. 2005. Crystal structure of dihydrofolate reductase from Plasmodium vivax: pyrimethamine displacement linked with mutation-induced resistance. Proc. Natl. Acad. Sci. U. S. A. 102:13046–13051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 12. Lozovsky ER, et al. 2009. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc. Natl. Acad. Sci. U. S. A. 106:12025–12030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mharakurwa S, et al. 2011. Malaria antifolate resistance with contrasting Plasmodium falciparum dihydrofolate reductase (DHFR) polymorphisms in humans and Anopheles mosquitoes. Proc. Natl. Acad. Sci. U. S. A. 108:18796–18801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 15. Rastelli G, et al. 2000. Interaction of pyrimethamine, cycloguanil, WR99210 and their analogues with Plasmodium falciparum dihydrofolate reductase: structural basis of antifolate resistance. Bioorg. Med. Chem. 8:1117–1128 [DOI] [PubMed] [Google Scholar]
- 16. Schrödinger LLC. 2010. The PyMOL molecular graphics system, version 1.3r1. Schrödinger, Rockville, MD [Google Scholar]
- 17. Sirawaraporn W, Prapunwattana P, Sirawaraporn R, Yuthavong Y, Santi DV. 1993. The dihydrofolate reductase domain of Plasmodium falciparum thymidylate synthase-dihydrofolate reductase. Gene synthesis, expression, and anti-folate-resistant mutants. J. Biol. Chem. 268:21637–21644 [PubMed] [Google Scholar]
- 18. Sirawaraporn W, Sathitkul T, Sirawaraporn R, Yuthavong Y, Santi DV. 1997. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc. Natl. Acad. Sci. U. S. A. 94:1124–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sirawaraporn W, et al. 2002. Mutational analysis of Plasmodium falciparum dihydrofolate reductase: the role of aspartate 54 and phenylalanine 223 on catalytic activity and antifolate binding. Mol. Biochem. Parasitol. 121:185–193 [DOI] [PubMed] [Google Scholar]
- 20. Tanaka M, Gu HM, Bzik DJ, Li WB, Inselburg J. 1990. Mutant dihydrofolate reductase-thymidylate synthase genes in pyrimethamine-resistant Plasmodium falciparum with polymorphic chromosome duplications. Mol. Biochem. Parasitol. 42:83–91 [DOI] [PubMed] [Google Scholar]
- 21. Tarnchompoo B, et al. 2002. Development of 2,4-diaminopyrimidines as antimalarials based on inhibition of the S108N and C59R+S108N mutants of dihydrofolate reductase from pyrimethamine-resistant Plasmodium falciparum. J. Med. Chem. 45:1244–1252 [DOI] [PubMed] [Google Scholar]
- 22. Thaithong S, et al. 2001. Plasmodium falciparum: gene mutations and amplification of dihydrofolate reductase genes in parasites grown in vitro in presence of pyrimethamine. Exp. Parasitol. 98:59–70 [DOI] [PubMed] [Google Scholar]
- 23. Vanichtanankul J, et al. 2011. Trypanosomal dihydrofolate reductase reveals natural antifolate resistance. ACS Chem. Biol. 6:905–911 [DOI] [PubMed] [Google Scholar]
- 24. Volkman SK, et al. 2007. A genome-wide map of diversity in Plasmodium falciparum. Nat. Genet. 39:113–119 [DOI] [PubMed] [Google Scholar]
- 25. Wang P, et al. 1997. Resistance to antifolates in Plasmodium falciparum monitored by sequence analysis of dihydropteroate synthetase and dihydrofolate reductase alleles in a large number of field samples of diverse origins. Mol. Biochem. Parasitol. 89:161–177 [DOI] [PubMed] [Google Scholar]
- 26. Yuthavong Y. 2002. Basis for antifolate action and resistance in malaria. Microbes Infect. 4:175–182 [DOI] [PubMed] [Google Scholar]
- 27. Yuthavong Y, et al. 2000. Development of a lead inhibitor for the A16V+S108T mutant of dihydrofolate reductase from the cycloguanil-resistant strain (T9/94) of Plasmodium falciparum. J. Med. Chem. 43:2738–2744 [DOI] [PubMed] [Google Scholar]
- 28. Yuvaniyama J, et al. 2003. Insights into antifolate resistance from malarial DHFR-TS structures. Nat. Struct. Biol. 10:357–365 [DOI] [PubMed] [Google Scholar]
- 29. Zamyatnin AA. 1972. Protein volume in solution. Prog. Biophys. Mol. Biol. 24:107–123 [DOI] [PubMed] [Google Scholar]