Abstract
Antiviral therapeutics are currently unavailable for treatment of coxsackievirus B3, which can cause life-threatening myocarditis. A modified small interfering RNA (siRNA) containing 5′-triphosphate, 3p-siRNA, was shown to induce RNA interference and interferon activation. We aimed to develop a potent antiviral treatment using CVB3-specific 3p-siRNA and to understand its underlying mechanisms. Virus-specific 3p-siRNA was superior to both conventional virus-specific siRNA with an empty hydroxyl group at the 5′ end (OH-siRNA) and nonspecific 3p-siRNA in decreasing viral replication and subsequent cytotoxicity. A single administration of 3p-siRNA dramatically attenuated virus-associated pathological symptoms in mice with no signs of toxicity, and their body weights eventually reached the normal range. Myocardial inflammation and fibrosis were rare, and virus production was greatly reduced. A nonspecific 3p-siRNA showed relatively less protective effect under identical conditions, and a virus-specific OH-siRNA showed no protective effects. We confirmed that virus-specific 3p-siRNA simultaneously activated target-specific gene silencing and type I interferon signaling. We provide a clear proof of concept that coxsackievirus B3-specific 3p-siRNA has 2 distinct modes of action, which significantly enhance antiviral activities with minimal organ damage. This is the first direct demonstration of improved antiviral effects with an immunostimulatory virus-specific siRNA in coxsackievirus myocarditis, and this method could be applied to many virus-related diseases.
INTRODUCTION
RNA interference (RNAi) is a sequence-specific gene silencing process, which has been extensively investigated as a novel technique for the development of antiviral agents (38, 40). RNAi is mediated by a small interfering RNA (siRNA) comprised of a 19- to 26-nucleotide (nt) RNA duplex. Recognition of target RNA by an siRNA conjugated with an RNA-induced silencing complex (RISC) triggers the catalytic degradation of complementary target RNA (8). Previously, we showed that siRNAs specific to enteroviral RNAs selectively reduce replication of the corresponding enterovirus and cytotoxicity in infected cells (2, 18). Merl et al. also reported that coxsackievirus B3 (CVB3)-specific siRNA attenuates CVB3 replication and cytopathogenicity and increases the animal survival rate (27). However, antiviral efficacies need to be further improved before siRNA-based agents can be applied in clinical settings (5, 21, 33).
Poeck et al. recently demonstrated that a modified siRNA with a triphosphate at the 5′ end (3p-siRNA) complementary to Bcl-2 interfered with Bcl-2 expression by selectively cleaving Bcl-2 mRNA (30). This siRNA differed from conventional siRNAs, which possess an empty hydroxyl group at the 5′ end (OH-siRNA).Unlike the traditional OH-siRNA, 3p-siRNA also activated interferon (IFN) production. Thus, Bcl-2-specific 3p-siRNA simultaneously silenced Bcl-2 and produced IFNs using a single molecule, which induced highly enhanced antitumor effects compared with those of an OH-siRNA, which solely elicited RNAi activity.
CVB3 belongs to an enterovirus genus in the Picornaviridae family and is the most common etiological agent inducing myocarditis, particularly in neonates and young children (28). CVB3 infection can be life threatening due to the risk of heart failure, with a poor prognosis in cases of acute and chronic myocarditis. CVB3 infection results in irreversible cytopathic effects at the cellular level and cardiac injuries at the tissue level, which suggests that direct virus replication-mediated damage causes clinical symptoms (1, 10). No vaccines or antiviral agents are currently available, and treatments are only symptomatic, which means there is an urgent need for an antiviral agent.
Type I IFN, including alpha IFN (IFN-α) and beta IFN (IFN-β), is a key component of the innate immune response and the first line of defense against viral infection (31). Previous reports have shown that disruption of type I IFN activities in IFNAR1−/− and IFN-β−/− mice resulted in much higher virus titers in the heart and mortality following CVB3 infection than was the case with wild-type mice (6, 37). These results strongly suggest that type I IFNs have a key protective role against CVB3-induced myocarditis. In addition to viruses and microbial products, various nucleic acids, including DNA and double- or single-stranded RNA, provoke type I IFN production via germ line-encoded pattern recognition receptors (PRRs) (3, 15). 3p-RNA is sensed by cytosolic PRR retinoic acid-inducible protein I (RIG-I) in a non-sequence-dependent manner (16). Direct binding of 3p-RNA to RIG-I triggers the potent induction of a set of genes, including type I IFNs and antiviral responses (20, 35, 36).
Based on these findings, we aimed to achieve effective antiviral effects by developing 3p-siRNAs specific to CVB3 in a viral myocarditis model. To analyze their potentials as antiviral agents, we generated CVB3-specific siRNAs with (3p-siRNAs) and without (OH-siRNAs) a 5′-triphosphate and compared their antiviral potencies in vitro and in vivo by examining viral replication, cytotoxicity, animal morbidity, and histopathology. We further investigated the underlying mechanism of the selective cleavage of the CVB3 genome and the induction of type I IFNs, and unexpected side effects were also determined.
MATERIALS AND METHODS
Cells, virus, and virus preparation.
HeLa, Vero, MRC5 cells were obtained from the American Type Culture Collection (ATCC) and maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum, GlutaMAX-1 (2 mM), and penicillin (100 IU/ml)-streptomycin (50 μg/ml) at 37°C in a 5% CO2 incubator. CVB3 (Nancy strain) was purchased from ATCC, propagated, and then titrated by conventional methods in Vero cells, as previously described (2).
To quantify progeny virus production in vitro, HeLa cells were infected with CVB3 at a multiplicity of infection (MOI) of 1 for 1 h in the presence or absence of siRNAs. After the virus inoculum was washed, the cells were cultured with medium for 8 h. Medium and cells were harvested, and the resultant cell lysates were subjected to virus titration by 50% tissue culture infective dose (TCID50) (32). To quantify the levels of infectious viruses in hearts, tissue lysates were prepared using an HT10 homogenizer (IKA-Korea) and subjected to titration.
Preparation of OH-siRNAs/3p-siRNAs and transient transfection.
Three siRNAs targeting the CVB3 region were designed using the CAPSID software program (26). OH-siRNAs with a hydroxyl group at the 5′ end were manufactured by Bioneer using the “ready-to-use” option. 3p-siRNAs with a triphosphate group at the 5′ end were purchased from Genolution using the “ready-to-use” option. The control siRNA had no sequence match with any genes, as previously described (2). None of the siRNAs induced cytotoxicity under experimental conditions in vitro (data not shown). For transient transfection, cells were incubated for 4 h with 100, 10, or 1 nM siRNA using Oligofectamine reagent (Invitrogen) under no-serum conditions, before fresh medium containing 10% serum was added, with further incubation for the specified period.
5′ RACE analysis.
5′ rapid amplification of cDNA ends (RACE) was performed according to the GeneRacer manual (Invitrogen) with some modifications. When sequence-specific viral RNA cleavage by virus-specific siRNA (OH-#5 and 3p-#5) occurred, a PCR product of 150 bp in size was obtained. Five micrograms of total RNA was extracted using TRIzol reagent and ligated to 250 ng of the GeneRacer RNA adaptor using T4 ligase (NEB). Reverse transcription (RT) was conducted as described in the real-time RT-PCR method given below, using a CVB3-specific antisense primer (#5-AS1; 5′-ATG AGC CCA CCA CAC TGG-3′) and a SuperScript III RT kit (Invitrogen). The RT products were amplified using a GeneRacer-specific sense primer (5′RACE-S1; 5′-CGA CTG GAG CAC GAG GAC ACT G-3′) and a CVB3-specific antisense primer (#5-AS1; 5′-ATG AGC CCA CCA CAC TGG-3′). A second round of nested PCR was conducted using a GeneRacer-specific nested sense primer (#5-NST-S1; 5′-GGA CTG AAG GAG TAG AAA GGA GG-3′) and a CVB3-specific nested antisense primer (#5-AS2; 5′-GCA TTC TCT TGG TGG GTG TGC-3′). The initial PCR with high-fidelity Taq polymerase (Invitrogen) was carried out using the following conditions: 94°C for 2 min (1 cycle), 94°C for 30 s and 72°C for 1 min (5 cycles), 94°C for 30 s, 66°C for 30 s, and 68°C for 30 s (25 cycles), and 68°C for 10 min. The second PCR was performed using the following conditions: 94°C for 2 min (1 cycle), 94°C for 30 s, 65°C for 30 s, and 68°C for 1 min (25 cycles), and 68°C for 10 min (1 cycle). The PCR products were cloned into the pCR2.1 vector using a TA cloning kit (Invitrogen) and sequenced.
Animal care and virus and siRNA administration.
We obtained BALB/c mice, which were up to 4 weeks old, from Central Laboratory Animal Inc. All animal experiments were ethically conducted in accordance with the guidelines of the Animal Care Center of the Asan Institute. The animal use protocols were reviewed and approved by the committee in this center. Viral doses of 8 × 105 PFU/200 μl were injected intraperitoneally. After 8 h, siRNA (2.5 mg/kg of mouse body weight) was intravenously injected using Jet polyethyleneimine (PEI) (Polyplus-transfection Inc.). Mouse body weight was measured at 2-day intervals.
Histopathology.
Animals were sacrificed on days 7 and 14 after siRNA administration, and the heart, pancreas, liver, and spleen were harvested. Tissues were fixed in 10% formalin, embedded in paraffin, and stained with hematoxylin and eosin (H&E) and Sirius red. Viral protein expression was determined by immunohistochemistry using VP1 antibody (Leica Biosystems Newcastle Ltd.). The stained sections were subjected to image analysis with a Leica microscope and analyzed by an investigator who was blinded to the treatment. Degrees of inflammation after H&E staining were graded using a scale of 0 to 4, where a score of 0 represented no myocarditis and 4 represented widespread and confluent inflammation. The degree of fibrosis was determined after Sirius red staining as the percentage of positively stained area.
RT-PCR using SYBR green I.
Total RNA was extracted from each sample of various cells and heart tissues using TRIzol reagent (Invitrogen). RT was carried out with Superscript III reverse transcriptase using an oligo(dT) primer at 55°C. The synthesized cDNA was amplified by RT-PCR using iQ SYBR green supermix (Bio-Rad). The reaction consisted of 3 min at 95°C for polymerase activation and 39 cycles of 15 s at 95°C (denaturation), 30 s at 60°C (annealing), and 30 s at 72°C (extension). β-Actin was used for normalization in the estimation of relative gene expression. RT-PCR was implemented using the following primers: CVB3-5′NTR-S, ACATGGTGCGAAGAGTCTATTGAG; CVB3-5′NTR-AS, TGCTCCGCAGTTAGGATTAGC; human-IFN-β-S, TGCTTCTCCACTACAGCTCTT; human-IFN-β-AS, TGACACTGAAAATTGCTGCTTCT; human-OAS-S, CTGGATTCTGCTGACCCAGC; human-OAS-AS, GGTCCAGATAACACTGGAGC; mouse IFN-α-S, TCCTGAACCTCTTCACATCAAA; mouse IFN-α-AS, ACAGGCTTGCAGGTGATTGAG; mouse IFN-β-S, GGAGATGACGGAGAAGATGC; mouse IFN-β-AS, CCCAGTGCT GGAGAAATTGT; mouse PKR-S, GGCTCCTGTGTGGGAAGTCA; mouse PKR-AS, TATGCCAAAAGCCAGAGTCCTT; mouse OAS-S, TGAGCGCCCCCCATCTG; and mouse OAS-AS, CATGACCCAGGACATCAAAGG. Relative gene expression was estimated using the GeneXpression Macro chromo4 software program (Bio-Rad).
Statistical analysis.
Data were reported as the means ± standard deviations (SD). For statistical analysis, data were parametrically subjected to analysis of variance (ANOVA) for multiple comparisons. Some data (see Fig. 2b) were further analyzed by paired t test. P values less than 0.05 were considered statistically significant.
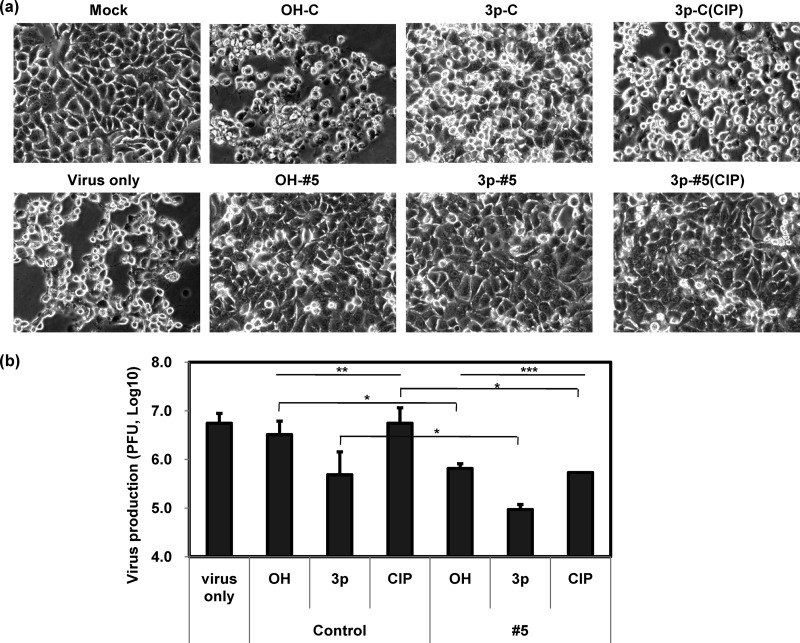
Fig 2.
Influence of CVB3-specific 3p-siRNA on CVB3 infection in HeLa cells. Cells were treated with various siRNAs and CVB3, as shown in Fig. 1b. Cytopathic effects were observed under light microscopy at 10 h p.i. (a) (representatives of 3 independent studies). Cells and culture media were collected at 8 h p.i., and progeny virus production was determined by TCID50 (b) (n = 4). Means ± SD are shown (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
RESULTS
Simultaneous induction of specific gene silencing and type I IFN by CVB3-specific 3p-siRNAs.
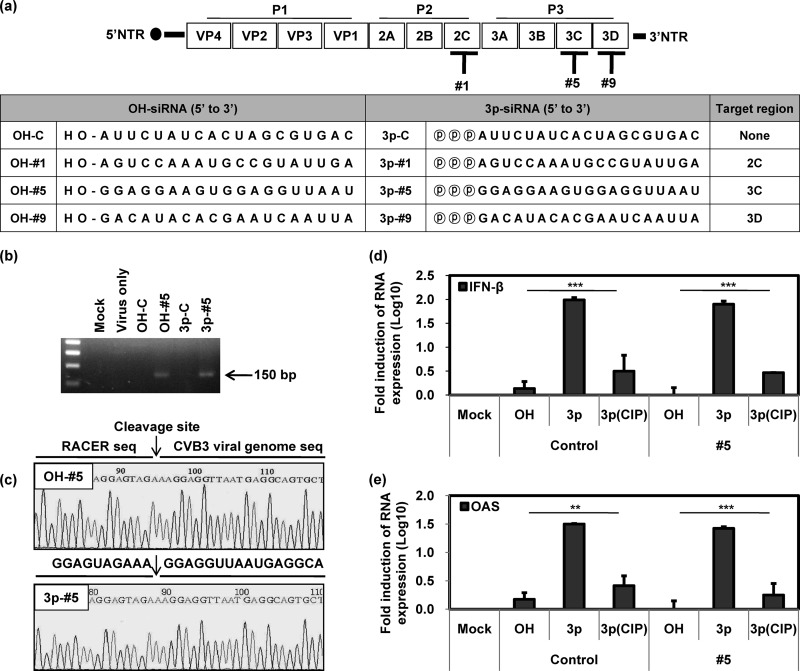
We designed 3 CVB3-specific siRNAs targeting the 2C (#1), 3C (#5), and 3D (#9) regions of the CVB3 genome. Two distinct forms, i.e., a conventional synthetic 5′-free siRNA (OH-siRNAs) and 5′-triphosphate siRNAs (3p-siRNAs) (Fig. 1a), were prepared. Target-specific gene cleavage by siRNAs was verified by RT-PCR using 5′ rapid amplification of cDNA ends (5′ RACE) to exclusively amplify RNA that was specifically cleaved by RNAi. Both of the CVB3-specific siRNAs (OH-#5 and 3p-#5) produced PCR products that corresponded to the virus target genome region (150 bp; Fig. 1b), irrespective of the presence of triphosphate. As expected, no PCR product was detected with nonspecific control siRNAs (OH-C and 3p-C). Sequencing analysis further confirmed that the PCR products were amplified from the cleaved CVB3 genome and that cleavage occurred at the corresponding genome region (5′ … AGU/GGA … 3′) between positions 10 and 11 of the antisense strand of siRNA, which was the expected siRNA-driven cleavage site (Fig. 1a and c).
Fig 1.
Characteristics of CVB3-specific 3p-siRNAs in vitro. Sequence information for siRNAs and their positions in the viral genome are shown in panel a. HeLa cells were preincubated with various types of siRNAs at 100 nM for 15 h, followed by CVB3 infection at an MOI of 1 for 8 h. RT-PCR products were prepared by target-specific 5′ RACE analysis from total RNA and visualized by agarose gel electrophoresis (n = 2). PCR products are indicated by an arrow (b) and were further analyzed by sequencing (c). After incubating HeLa cells with siRNAs at 100 nM for 24 h, the cytosolic levels of IFN-β (d) and OAS (e) mRNA were determined by RT-PCR (n = 4). NTR, nontranslated region; Mock, mock treated. Means ± SD are shown (**, P < 0.01; ***, P < 0.001).
Type I IFN stimulation by 3p-siRNAs was examined by determining the expression levels of IFN-β and 2′–5′ oligoadenylate synthetase (OAS). HeLa cells were transfected with 3p-siRNAs (3p-C and 3p-#5) regardless of viral specificity, and they exhibited a dramatic increase in the expression of both IFN-β and OAS mRNAs (Fig. 1d and e). IFN-β and OAS expression by 3p-siRNAs sharply decreased following a calf intestine phosphatase (CIP) treatment to eliminate triphosphate. We detected no induction of IFN-β or OAS expression with OH-siRNAs. Taken together, these data imply that CVB3-specific 3p-siRNA induces a target-specific gene silencing activity and it also elicits an innate immune response.
Severe attenuation of CVB3 infection following CVB3-specific 3p-siRNA treatment in vitro.
The antiviral activities of CVB3-specific siRNAs were investigated by testing cytotoxicity and progeny virus production in HeLa cells after siRNA preincubation followed by viral challenge. Infected cells began to exhibit irreversible cytopathic effects as early as 6 h postinfection (p.i.), such as cell rounding and detachment of cells from the culture plate, while progeny virus production reached its maximal level within 8 h p.i. (data not shown). Both of the CVB3-specific siRNAs (OH-#5 and 3p-#5) significantly protected cells from cytopathic effects (Fig. 2a) and progeny virus production (Fig. 2b). The nonspecific control siRNA containing 5′-triphosphate (3p-C) also showed some degree of protective effects. Yet the most significant protection was achieved with the CVB3-specific 3p-siRNA (3p-#5; 63.7 ± 1.8-fold decrease in progeny virus production). The most improved antiviral effects were also observed in the presence of the CVB3-specific 3p-siRNA by increasing IFN-β mRNA expression in human fibroblast MRC5 cells (see Fig. S1 and S2 in the supplemental material). The 3p-siRNAs lost antiviral activity after CIP treatment (Fig. 2). The CVB3-specific 3p-siRNA retained some degree of protective effect even after CIP treatment, whereas the control 3p-siRNA lost most of its antiviral effect. The corresponding 3p-siRNAs also showed enhanced antiviral activity with the other CVB3-specific siRNAs (#1 and #9), along with induction of IFN-β mRNA expression (see Figure S3 in the supplemental material). Antiviral activities following these siRNA treatments lasted up to 48 h and disappeared (data not shown). These results demonstrate that antiviral potency can be significantly enhanced by modifying CVB3-specific siRNA to a form containing 5′-triphosphate, which simultaneously induces CVB3-specific gene silencing and an innate immune response.
Effective attenuation of CVB3 infection following CVB3-specific 3p-siRNA treatment in vivo.
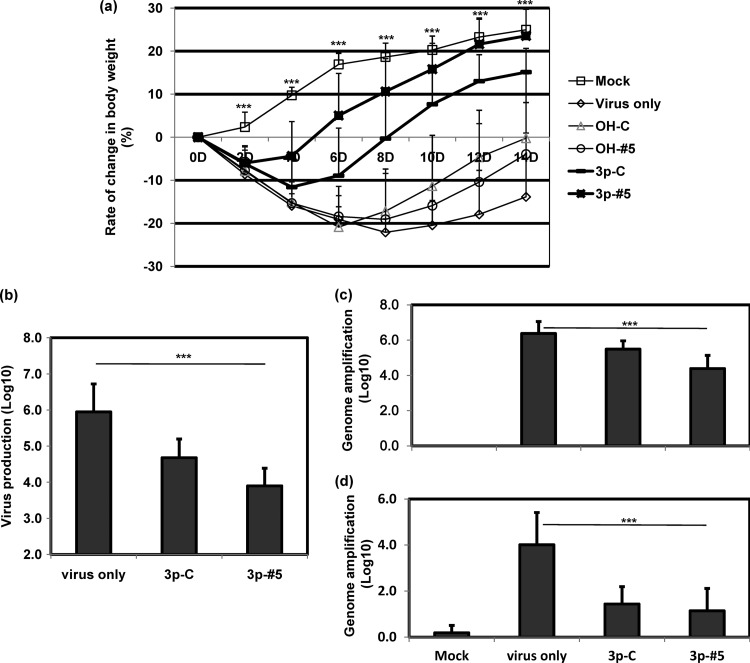
Antiviral potencies of modified siRNAs were examined in vivo using BALB/c mice infected with CVB3 followed by various siRNA injections. In CVB3-infected mice, body weights gradually decreased until day 8 and then slowly recovered to a level similar to their initial body weights (Fig. 3a). In contrast, both of the 3p-siRNA groups gradually gained weight after an initial loss. The initial weight loss was minimal in mice treated with CVB3-specific 3p-siRNA (3p-#5). Body weights began to increase on day 4, and their final body weights were almost the same as those of the uninfected control group. However, OH-siRNA groups lost body weight with kinetics in initial weight loss similar to those for the virus-infected group. Virus replication also did not decrease in OH-siRNA groups (see. Fig. S4 in the supplemental material).
Fig 3.
Antiviral effects of CVB3-specific 3p-siRNA in mice. Mice were intraperitoneally injected with 8 × 105 PFU/200 μl of virus and intravenously administered with various siRNAs at 2.5 mg/kg of mouse after 8 h. Body weight was monitored every 2 days (a) (n ≥ 6 for each group). Progeny virus production was tested by TCID50 in heart tissues on day 7 after siRNA treatment (b) (n = 12). The viral genome was quantified on day 7 (c) (n = 12) and day 14 (d) (n = 6) using RT-PCR. Means ± SD are shown (***, P < 0.001).
Progeny virus production was greatly decreased, up to a two-log reduction, in the hearts of the CVB3-specific 3p-siRNA group on day 7 (3.9 ± 0.5 in log10 TCID/g), compared with results for the virus-only group (5.9 ± 0.8) (Fig. 3b). The rate of reduction was about one log (4.7 ± 0.5) in the CVB3-nonspecific group under the same conditions. A similar protective pattern was also determined by virus genome (vg) amplification; an additional one-log reduction in genome amplification (6.4 ± 0.7 in log10 vg in the virus-only group) was achieved by CVB3-specific 3p-siRNA (4.4 ± 0.7) at day 7, compared to results for nonspecific 3p-siRNA (5.5 ± 0.5) (Fig. 3c). In addition, the 3p-siRNA treatments rapidly induced type I IFN-related gene expressions in hearts (data not shown). These results suggest that CVB3-specific 3p-siRNA has dual effects and effectively attenuates pathological effects in vivo by significantly hindering viral replication.
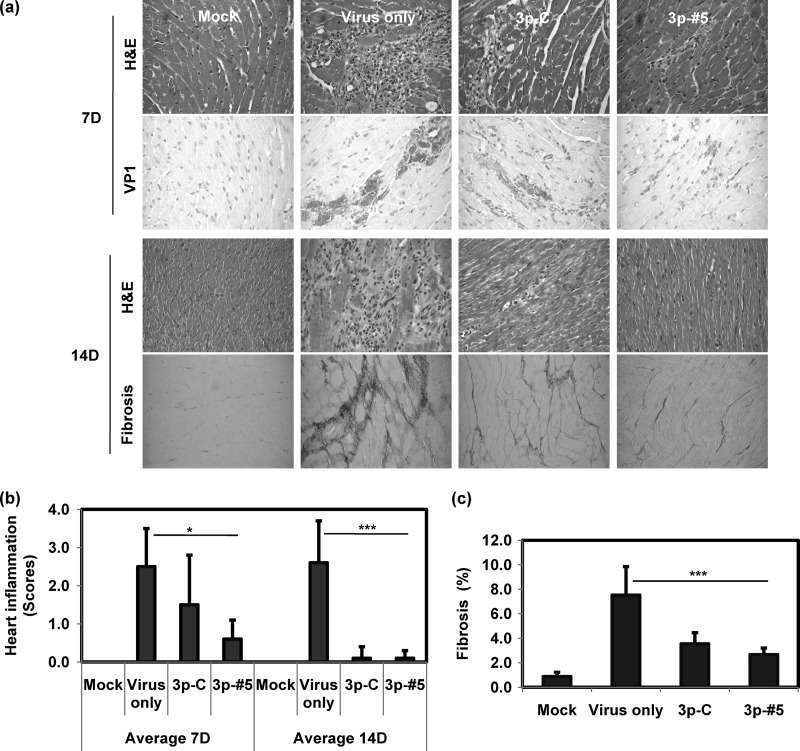
Enhanced protective effects on heart inflammation and fibrosis by CVB3-specific 3p-siRNA treatment.
Hematoxylin-and-eosin (H&E) staining of heart sections indicated that severe inflammatory cell infiltration occurred on days 7 and 14 after CVB3 infection (Fig. 4a). Average heart inflammation scores were estimated as 2.5 ± 1.0 and 2.6 ± 1.1 on days 7 and 14, respectively (Fig. 4b). CVB3-specific 3p-siRNA (3p-#5, 0.6 ± 0.5) treatment attenuated inflammation, and it was statistically superior to nonspecific 3p-siRNA, particularly at day 7 (3p-C, 1.5 ± 1.3). Similarly, Sirius red staining showed that myocardial fibrosis was effectively attenuated on day 14 by CVB3-specific 3p-siRNA to 2.6% ± 0.9%, from 7.3% ± 2.8% (Fig. 4c; 3.3% ± 2.6% with nonspecific 3p-siRNA). Virus protein expression was also determined in heart tissues by virus-specific VP1 immunohistochemistry (Fig. 4a). A dense deposit of VP1 was readily observed in sporadic patterns on day 7 p.i. CVB3-specific 3p-siRNA treatment led to a loss of most of the positive signal, but some of the VP1-positive signal still remained with the nonspecific 3p-siRNA. These data indicate that the CVB3-specific 3p-siRNA treatment was highly effective at reducing myocardial inflammations and that it subsequently attenuated viral illness.
Fig 4.
Histopathological examination of hearts following virus and siRNA treatment. The mice were sacrificed on day 7 or 14, as shown in Fig. 3. Hearts were recovered, and paraffin blocks were prepared for histopathological analysis. Sections were stained with H&E, treated with Sirius red, and immunohistochemically stained using virus-specific VP1 antibody (a). Heart inflammation on days 7 and 14 (b) (n = 6) or fibrosis on day 14 (c) (n = 6) was scored as described in Materials and Methods. Means ± SD are shown (*, P < 0.05; ***, P < 0.001).
No obvious toxicity with 3p-siRNA treatment.
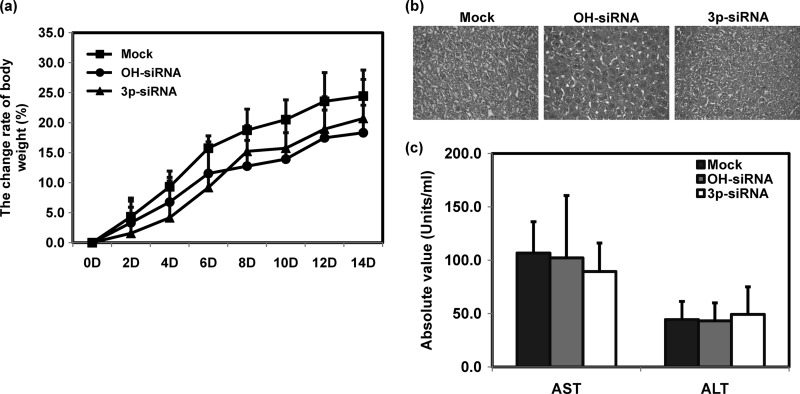
We determined unexpected side effects of 3p-siRNA treatment by examining the influence of siRNA on body weight and liver toxicity. All the mice survived the experimental procedure very well and exhibited no unusual morbidity (data not shown). Body weight increased continuously at similar rates in the siRNA-treated groups (Fig. 5a). The siRNA-treated groups showed normal histological liver features on day 7, with no difference from the control group (Fig. 5b). Serum levels of the liver enzymes alanine aminotransferase (ALT) and aspartate aminotransferase (AST) didn't change on day 14 (Fig. 5c). These data indicate that systemic delivery of 3p-siRNA did not seem to induce significant side effects for the liver.
Fig 5.
Toxicity assessment for siRNA-treated mice. Mice were intravenously administered with siRNAs at 2.5 mg/kg of mouse. Body weight was measured every 2 days (a) (n = 6). Liver tissues were examined on day 7 by H&E staining (b) (n = 3), and liver enzyme activities in sera were determined on day 14 (c) (n = 6). Means ± SD are shown.
DISCUSSION
This study demonstrated that CVB3-specific 3p-siRNA was highly effective in inhibiting viral replication, which resulted in enhanced in vitro and in vivo antiviral effects. Improved protective potency was due to the combined effects of 2 distinct mechanisms: (i) gene silencing by CVB3-specific RNAi and (ii) an innate immune response via type I IFN activation by 5′-triphosphate.
Clinical trials validating the usefulness of siRNA-based therapeutics have been applied to diverse human diseases, including genetic disorders and infectious diseases (5, 24). The first antiviral siRNA clinical trial used an siRNA silencing virus nucleocapsid gene (ALN-RSV01; Alnylam Pharmaceuticals) to treat respiratory syncytial virus, which causes severe bronchiolitis. Randomized, double-blind, placebo-controlled trials were performed either with healthy subjects or with lung transplant recipients (7, 39). These trials consistently demonstrated that ALN-RSV01 is safe and has significant antiviral activity against human RSV infection. Antiviral siRNA protocols for hepatitis B virus, hepatitis C virus, and HIV have been under investigation, also indicating that several siRNA treatments are effective and well tolerated (22, 33). Thus, the initial prospect for these novel therapeutics appears positive and highly attractive.
Yet several limitations must be overcome before the successful introduction of siRNA-based agents into the clinic (21, 24, 29). A major limitation for the development of siRNA-based agents is the strength of gene silencing. The simultaneous application of multiple siRNAs may increase the gene knockdown effect. We made a previous attempt to enhance antiviral effects by cotreatment with several siRNAs targeting CVB3, but the outcome was unsuccessful (2). Similar discouraging results were reported by Gitlin et al. when treating a poliovirus infection (11). Thus, other strategies for improving the efficacy of siRNA-based agents need to be considered. Here we hypothesized that modifying conventional OH-siRNA to 3p-siRNA might provide an additional antiviral property for RNAi. Indeed, CVB3-specific 3p-RNA activated type I IFN signaling with 5′-triphosphate and CVB3-specific gene silencing by RNAi. Consequently, CVB3-specific 3p-siRNA exhibited a dual activity that significantly enhanced antiviral effects with the attenuation of viral replication, cytotoxicity, morbidity, and heart histopathology. Recently, two independent research groups showed that hepatitis B virus (HBV)-specific 3p-siRNAs not only directly inhibit HBV replication but also stimulate innate immunity, implying that HBV-specific 3p-siRNAs are bifunctional and effectively induce antiviral status against HBV infection (9, 13). Together with our data, these findings thus support that immunostimulatory siRNA simultaneously inducing gene silencing with induction of IFN could be readily developed as an effective therapeutic approach to many virus-related diseases.
The enhanced antiviral effects of CVB3-specific 3p-RNA also mean that the administered dose of siRNA could be lowered. Previous studies elicited in vivo activity with siRNA by intravenously injecting mice with 10 to 50 mg/kg of siRNA on 2 or 3 occasions (17, 27, 34). In contrast, we achieved antiviral activity with a single administration of 3p-siRNA at a lower dose of 2.5 mg/kg, while conventional OH-siRNA failed to induce obvious antiviral effects at this dosage level. A major concern with most siRNA treatments is the possibility of oversaturation of the RISC by exogenous siRNA, which could lead to interruption of endogenous small-RNA-dependent gene expression (19, 29). Grimm et al. suggested that the high-level expression of short-hairpin RNAs overwhelmed the hepatic RNAi processing machinery and caused hepatic failure and death in mice (12). Such side effects become more severe as the siRNA concentration increases. Thus, the CVB3-specific 3p-RNA strategy provides additional benefits because of the lower effective dosage of siRNA.
CVB3 has detrimental effects on the myocardium, but it has an important role in the pathogenesis of extracardiac diseases, such as hepatitis, pancreatitis, and aseptic meningitis (28). CVB3-specific 3p-RNA efficiently attenuated pancreatitis, and its efficacy was far superior to that of siRNAs that expressed the single activity of RNAi or IFN induction (data not shown; see also Fig. S5 in the supplemental material). Inflammatory cell infiltration and viral protein synthesis was also decreased in the pancreas in the presence of CVB3-specific 3p-RNA. Thus, CVB3-specific 3p-RNA could be developed as a general therapeutic agent for treating CVB-derived diseases.
Synthetic IFNs, such as the PEGylated form of IFN-α, have been clinically applied, and IFN-α is known to induce a sustained and curative virological response in 10 to 90% of hepatitis patients (14, 25). When patients with myocardial enteroviral or adenoviral persistence were treated with IFN-β, ventricular dysfunction was prevented, along with the elimination of viral genomes in most patients (23). However, the usefulness of IFN therapy is limited by substantial side effects, such as flu-like symptoms, fatigue, and depression (4). In contrast to conventional IFN therapies, which directly administer synthetic IFNs, 3p-siRNA acts as a trigger factor for IFN generation in the cell. RNA recognition by RIG-I activates a downstream signaling pathway, and the signal is greatly amplified in multiple steps (35). We observed that a 1/10 concentration of 3p-siRNA activated a much higher level of IFN pathway activity in vitro (data not shown) than was seen with synthetic IFN. Thus, a small amount of 3p-siRNA elicited a large amount of IFN synthesis, thereby producing effective antiviral effects with fewer side effects, as indicated by the absence of organ damage in the 3p-siRNA group.
Excessive activation of innate immunity by an abrupt cytokine surge can cause damaging effects. Kim et al. suggested caution after reporting that more nude mice with an uninhibited innate response died after receiving a sublethal dose of coronavirus than did wild-type mice (22). Nevertheless, our limited toxicity studies indicated no obvious changes in morbidity or liver toxicity following 3p-siRNA treatment. Furthermore, the toxicity of 3p-siRNA was not reported when Hartmann's group intravenously injected nude mice with 2.5 mg/kg of 3p-siRNAs on 3 occasions in a melanoma tumor model (30). Thus, the induction of innate immunity by systematic delivery of 3p-siRNA seems to be well tolerated by both adaptive immune-defective and immunocompetent mice. Yet further detailed toxicity studies would be necessary before moving forward to a clinical program.
As a proof of concept, we have provided clear evidence that CVB3-specific 3p-siRNA simultaneously produced target-specific gene silencing and IFN induction, which enhanced antiviral activity without organ damage. This is the first direct illustration that virus-specific immunostimulatory siRNAs contain 2 distinct functional properties that can enhance antiviral effects in coxsackievirus myocarditis. This strategy for the development of effective antiviral siRNA could be applied generally to other virus-related diseases.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants to H. Lee (NRF-2010-R13-0029521) and Y. K. Kim (2010-0011215) from the National Research Foundation (NRF) and to J. Ahn from the Health & Medical Technology R&D Program (A101470), Republic of Korea.
There is no conflict of interest in this study.
Footnotes
Published ahead of print 16 April 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Ahn J, et al. 2005. All CVB serotypes and clinical isolates induce irreversible cytopathic effects in primary cardiomyocytes. J. Med. Virol. 75:290–294 [DOI] [PubMed] [Google Scholar]
- 2. Ahn J, et al. 2005. A small interfering RNA targeting coxsackievirus B3 protects permissive HeLa cells from viral challenge. J. Virol. 79:8620–8624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801 [DOI] [PubMed] [Google Scholar]
- 4. Borden EC, et al. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6:975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Castanotto D, Rossi JJ. 2009. The promises and pitfalls of RNA-interference-based therapeutics. Nature 457:426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deonarain R, et al. 2004. Protective role for interferon-beta in coxsackievirus B3 infection. Circulation 110:3540–3543 [DOI] [PubMed] [Google Scholar]
- 7. DeVincenzo J, et al. 2010. A randomized, double-blind, placebo-controlled study of an RNAi-based therapy directed against respiratory syncytial virus. Proc. Natl. Acad. Sci. U. S. A. 107:8800–8805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dykxhoorn DM, Novina CD, Sharp PA. 2003. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 4:457–467 [DOI] [PubMed] [Google Scholar]
- 9. Ebert G, et al. 2011. 5′ triphosphorylated small interfering RNAs control replication of hepatitis B virus and induce an interferon response in human liver cells and mice. Gastroenterology 141:696–706.e3 [DOI] [PubMed] [Google Scholar]
- 10. Esfandiarei M, McManus BM. 2008. Molecular biology and pathogenesis of viral myocarditis. Annu. Rev. Pathol. 3:127–155 [DOI] [PubMed] [Google Scholar]
- 11. Gitlin L, Karelsky S, Andino R. 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418:430–434 [DOI] [PubMed] [Google Scholar]
- 12. Grimm D, et al. 2006. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441:537–541 [DOI] [PubMed] [Google Scholar]
- 13. Han Q, et al. 2011. Reversal of hepatitis B virus-induced immune tolerance by an immunostimulatory 3p-HBx-siRNAs in a retinoic acid inducible gene I-dependent manner. Hepatology 54:1179–1189 [DOI] [PubMed] [Google Scholar]
- 14. Heathcote J, Main J. 2005. Treatment of hepatitis C. J. Viral Hepat. 12:223–235 [DOI] [PubMed] [Google Scholar]
- 15. Heil F, et al. 2004. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303:1526–1529 [DOI] [PubMed] [Google Scholar]
- 16. Hornung V, et al. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997 [DOI] [PubMed] [Google Scholar]
- 17. Judge AD, et al. 2006. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol. Ther. 13:494–505 [DOI] [PubMed] [Google Scholar]
- 18. Jun EJ, et al. 2008. Antiviral potency of a siRNA targeting a conserved region of coxsackievirus A24. Biochem. Biophys. Res. Commun. 376:389–394 [DOI] [PubMed] [Google Scholar]
- 19. Khan AA, et al. 2009. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 27:549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim DH, et al. 2004. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 22:321–325 [DOI] [PubMed] [Google Scholar]
- 21. Kim DH, Rossi JJ. 2007. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 8:173–184 [DOI] [PubMed] [Google Scholar]
- 22. Kim KD, et al. 2007. Adaptive immune cells temper initial innate responses. Nat. Med. 13:1248–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuhl U, et al. 2003. Interferon-beta treatment eliminates cardiotropic viruses and improves left ventricular function in patients with myocardial persistence of viral genomes and left ventricular dysfunction. Circulation 107:2793–2798 [DOI] [PubMed] [Google Scholar]
- 24. Lares MR, Rossi JJ, Ouellet DL. 2010. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol. 28:570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lau GK, et al. 2005. Peginterferon Alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N. Engl. J. Med. 352:2682–2695 [DOI] [PubMed] [Google Scholar]
- 26. Lee HS, et al. 2009. A novel program to design siRNAs simultaneously effective to highly variable virus genomes. Biochem. Biophys. Res. Commun. 384:431–435 [DOI] [PubMed] [Google Scholar]
- 27. Merl S, et al. 2005. Targeting 2A protease by RNA interference attenuates coxsackieviral cytopathogenicity and promotes survival in highly susceptible mice. Circulation 111:1583–1592 [DOI] [PubMed] [Google Scholar]
- 28. Pallansch M. 2007. Enteroviruses: poliovirus, coxsackieviruses, echoviruses, and newer enteroviruses. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 29. Pecot CV, et al. 2011. RNA interference in the clinic: challenges and future directions. Nat. Rev. Cancer 11:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Poeck H, et al. 2008. 5′-Triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat. Med. 14:1256–1263 [DOI] [PubMed] [Google Scholar]
- 31. Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seo I, et al. 2007. Mutation variants generated from nonvirulent coxsackievirus B3 acquire virulence phenotypes by active virus replication. Intervirology 50:447–453 [DOI] [PubMed] [Google Scholar]
- 33. Shah PS, Schaffer DV. 2011. Antiviral RNAi: translating science towards therapeutic success. Pharm. Res. 28:2966–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Soutschek J, et al. 2004. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432:173–178 [DOI] [PubMed] [Google Scholar]
- 35. Takahasi K, et al. 2008. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell 29:428–440 [DOI] [PubMed] [Google Scholar]
- 36. Takeuchi O, Akira S. 2009. Innate immunity to virus infection. Immunol. Rev. 227:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wessely R, et al. 2001. Cardioselective infection with coxsackievirus B3 requires intact type I interferon signaling: implications for mortality and early viral replication. Circulation 103:756–761 [DOI] [PubMed] [Google Scholar]
- 38. Whitehead KA, Langer R, Anderson DG. 2009. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 8:129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zamora MR, et al. 2011. RNA interference therapy in lung transplant patients infected with respiratory syncytial virus. Am. J. Respir. Crit. Care Med. 183:531–538 [DOI] [PubMed] [Google Scholar]
- 40. Zhou J, Rossi JJ. 2011. Progress in RNAi-based antiviral therapeutics. Methods Mol. Biol. 721:67–75 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.