Abstract
Asunaprevir (BMS-650032) is a potent hepatitis C virus (HCV) NS3 protease inhibitor demonstrating efficacy in alfa interferon-sparing, direct-acting antiviral dual-combination regimens (together with the NS5A replication complex inhibitor daclatasvir) in patients chronically infected with HCV genotype 1b. Here, we describe a comprehensive in vitro genotypic and phenotypic analysis of asunaprevir-associated resistance against genotypes 1a and 1b using HCV replicons and patient samples obtained from clinical studies of short-term asunaprevir monotherapy. During genotype 1a resistance selection using HCV replicons, the primary NS3 protease substitutions identified were R155K, D168G, and I170T, which conferred low- to moderate-level asunaprevir resistance (5- to 21-fold) in transient-transfection susceptibility assays. For genotype 1b, a higher level of asunaprevir-associated resistance was observed at the same selection pressures, ranging from 170- to 400-fold relative to the wild-type control. The primary NS3 protease substitutions identified occurred predominantly at amino acid residue D168 (D168A/G/H/V/Y) and were associated with high-level asunaprevir resistance (16- to 280-fold) and impaired replication capacity. In asunaprevir single-ascending-dose and 3-day multiple-ascending-dose studies in HCV genotype 1a- or 1b-infected patients, the predominant pre-existing NS3 baseline polymorphism was NS3-Q80K. This substitution impacted initial virologic response rates in a single-ascending-dose study, but its effects after multiple doses were more ambiguous. Interestingly, for patient NS3 protease sequences containing Q80 and those containing K80, susceptibilities to asunaprevir were comparable when tested in an enzyme assay. No resistance-associated variants emerged in these clinical studies that significantly impacted susceptibility to asunaprevir. Importantly, asunaprevir-resistant replicons remained susceptible to an NS5A replication complex inhibitor, consistent with a role for asunaprevir in combination therapies.
INTRODUCTION
Hepatitis C virus (HCV), a positive-strand RNA virus that belongs to the family Flaviviridae, infects an estimated 170 million individuals worldwide, causing over 350,000 deaths annually (47). Progression of chronic HCV infection can gradually evolve into cirrhosis, liver failure, and/or hepatocellular carcinoma. In the United States, these clinical manifestations are the leading indication for liver transplantation and account for significant liver-related morbidity and mortality each year (14). Although a combination regimen of pegylated alfa interferon and ribavirin remains a vital therapeutic option against chronic HCV infection, various host and viral factors are believed to influence the outcome of treatment, and different genotypes are also associated with variable responses to this regimen. Sustained virologic response (SVR) rates of just 40 to 50% are achieved in treatment-naïve patients with HCV genotype 1, whereas higher rates (78 to 86%) have been reported during the course of therapy against HCV genotype 2 and 3 infections (11, 14, 28). This highlights the unmet medical need for novel and more effective HCV therapeutic agents. Consequently, an increasing number of investigational small-molecule inhibitors selectively targeting HCV proteins are currently under clinical development. The most advanced class of these inhibitors—collectively referred to as direct-acting antiviral (DAA) agents—targets the HCV nonstructural 3/4A (NS3/4A) serine protease complex, an essential component of the HCV replication life cycle (37). Recently, two NS3 protease inhibitors (PIs), telaprevir and boceprevir, were approved for the treatment of chronic HCV infection. The inclusion of either one of these agents as part of a triple combination regimen with alfa interferon and ribavirin has so far resulted in improved SVR rates of 66 to 75% in genotype 1-infected, treatment-naïve patients (15, 35).
The arrival of telaprevir and boceprevir represents a significant advance toward the eradication of HCV, although important limitations remain. During clinical development studies, the incidence of anemia (defined as hemoglobin values of ≤10 g/dl) was more common in patients receiving a triple-combination regimen of alfa interferon-ribavirin with either telaprevir (36%) or boceprevir (49%) compared with regimens devoid of these NS3 PIs (17% and 29% for telaprevir- and boceprevir-free alfa interferon-ribavirin combinations, respectively) (7, 8). In phase III trials, nearly half of enrolled patients receiving a boceprevir-based regimen required dose reductions and/or were given erythropoietin, which led to treatment discontinuation attributable to anemia (2, 35). Many patients receiving alfa interferon and ribavirin also experience other adverse events, including psychiatric disorders, flu-like symptoms, and/or hematologic abnormalities such as hemolytic anemia and neutropenia (14). Furthermore, as a result of suboptimal pharmacokinetic properties, both telaprevir and boceprevir require 3-times-daily dosing, and both are associated with rapid development of HCV genotypic resistance. Consequently, there is a need for new NS3 PIs and other mechanistically distinct targeted DAAs with improved side effect profiles and dosing potential. More importantly, these improved therapeutics could become core components of future alfa interferon-sparing regimens.
In this era of DAA-based therapy, the potential development of viral resistance is a concern with the new strategies (1). As anticipated, virologic breakthrough during monotherapy and during therapy with alfa interferon-ribavirin combinations with telaprevir or boceprevir has been associated with the emergence of common substitutions at the NS3 domain sites V36, T54, R155, and A156, among others (38, 40, 44, 45). To this end, asunaprevir (ASV; BMS-650032), a highly active HCV NS3 PI with potential for twice-daily (BID) dosing, demonstrated rapid and robust antiviral activity during single-ascending-dose (SAD) and 3-day multiple-ascending-dose (MAD) clinical studies and was well tolerated in patients chronically infected with HCV genotype 1 (34). After a single 600-mg dose of asunaprevir, the mean maximum HCV RNA level reduction from baseline was 2.87 log10 IU/ml. In the MAD study, adverse events were mild and easily manageable, with a headache being the most commonly reported adverse event (25% of patients). In a phase IIa dose-ranging study with asunaprevir in combination with alfa interferon-ribavirin in patients chronically infected with HCV genotype 1, interim analyses indicated robust antiviral responses at all doses tested (200 mg BID, 600 mg once daily [QD], and 600 mg BID) (4). Asunaprevir was also well tolerated, with generally mild adverse events and no viral breakthroughs reported. Clinically significant elevations in alanine transaminase were apparent in patients assigned to the 600-mg regimens; asunaprevir at 200 mg BID was therefore selected for subsequent studies, as no discontinuations and similar antiviral activity were observed at this dose.
Two other classes of DAAs are also being developed by Bristol-Myers Squibb (BMS), the first-in-class NS5A replication complex inhibitor daclatasvir (DCV; BMS-790052) (12), with a pharmacokinetic profile consistent with QD dosing (33), and the NS5B RNA-dependent RNA polymerase non-nucleoside inhibitor BMS-791325 (16). Recent phase IIa clinical trial results in the United States and Japan (5, 27) demonstrated the potential of combination therapy in the hardest-to-treat HCV patient population, genotype 1 null responders. In the U.S. study, SVR was achieved in 100% of genotype 1 null responders receiving asunaprevir and daclatasvir with alfa interferon and ribavirin for 24 weeks (27). Furthermore, 36% of patients receiving the alfa interferon-ribavirin-free dual-DAA regimen of asunaprevir and daclatasvir achieved SVR, demonstrating for the first time that two DAAs used in an alfa interferon-sparing regimen can cure HCV in some patients. In the Japanese study, 100% of patients receiving asunaprevir and daclatasvir for 24 weeks achieved SVR (5), suggesting that genotype 1 subtype strongly influences virologic outcome, as all patients in the Japanese study were infected with genotype 1b, whereas genotype 1a was predominant in the U.S. study.
In preclinical in vitro studies, asunaprevir demonstrated significant antiviral activity in HCV replicon cell systems representing genotype 1a and genotype 1b, with 50% effective concentration (EC50) values of 4 and 1 nM, respectively (32). In this report, we describe the results from a comprehensive in vitro analysis of resistance associated with asunaprevir in genotype 1a and genotype 1b replicon cells. We also compare genotypic and susceptibility analyses of asunaprevir resistance observed during short-term monotherapy in SAD and MAD clinical studies.
MATERIALS AND METHODS
Cell culture and HCV inhibitors.
Human hepatoma cells of the Huh-7 (Ralf Bartenschlager, University of Heidelberg, Heidelberg, Germany) and Huh-7.5 (APATH, Brooklyn, NY) lines were propagated using Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and l-glutamine (2 mM). Huh-7 cells that stably maintain a HCV subgenomic replicon representing genotype 1a (H77c strain: NCBI reference NC_004102.1) or genotype 1b (Con1 strain: GenBank accession no. AJ238799.1) were propagated as subconfluent monolayers in DMEM supplemented with FBS (10%), l-glutamine (2 mM), and 0.5 mg/ml Geneticin (G418; Invitrogen Corp., Carlsbad, CA) (26, 48). Genotype 1a and 1b replicons used in this study were generated at BMS as previously described (10). Asunaprevir was synthesized by BMS as previously described (41). The NS3 PI VX-950 (telaprevir), and an HCV NS5A replication complex inhibitor (C. Bachand et al., PCT international patent application WO20080219272, 2008) synthesized at BMS served as reference compounds.
Resistance selection.
Genotype 1a and 1b replicon cells were seeded in 6-well assay plates (Becton, Dickinson, Franklin Lakes, NJ) for resistance selection at a density of 4 × 105 and 7.5 × 103 cells per well, respectively. Replicon cells were maintained in DMEM supplemented with FBS (10%), l-glutamine (2 mM), and 0.5 mg/ml G418 in the presence of asunaprevir at concentrations of 10 and 30 times the EC50 values determined in these systems (genotype 1a, 50 or 150 nM final concentration, respectively; genotype 1b, 30 or 90 nM final concentration, respectively). Cells were passaged every 3 to 4 days to maintain a subconfluent monolayer, and asunaprevir diluted in dimethyl sulfoxide (DMSO) was replenished in fresh medium at the desired concentrations. Control replicon cells were maintained in parallel in medium containing 0.5% DMSO. After 3 to 4 weeks, cultures derived from either populations of resistance replicon cells or individual replicon cell colonies were assessed for asunaprevir susceptibility.
Genotypic analysis.
Total RNA was isolated and purified from cells harboring either asunaprevir-selected replicons or the parental wild-type replicon using a Qiagen RNeasy kit (Qiagen, Valencia, CA). NS3 sequences were amplified by reverse transcription-PCR (RT-PCR) using a SuperScript one-step RT-PCR kit with Platinum Taq (Invitrogen, Carlsbad, CA) and specific primers for genotype 1a (forward, 5′-GTCATGGCTCTCCTCAAGC-3′; reverse, 5′-TCGACACGACGACAACATC-3′) and genotype 1b (forward, 5′-TGAAGGATGCCCAGAAGGTACC-3′; reverse, 5′-GCATCACTGATGGCATTCACA-3′). PCR amplicons, encompassing the NS3 coding region from amino acids 1 to 405, were cloned into the TA-TOPO system (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. NS3 nucleotide sequences for the coding region spanning amino acids 1 to 181 were obtained using dye terminator technology, and alignments were performed using Sequencher software (Gene Codes, Ann Arbor, MI).
Phenotypic analysis. (i) Replicons.
To assess the phenotypic profiles of emerging asunaprevir-resistant replicons, amino acid substitutions identified in the NS3 region were introduced into wild-type replicons using a QuikChange mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The susceptibility of wild-type and engineered NS3 variant replicons to HCV replication inhibitors was measured using a cell-based transient-transfection assay. Huh-7 cells cured of an HCV replicon, as described previously (22), were used in all experiments. Cells were seeded at a density of 5 × 105 per well in 6-well assay plates (Becton, Dickinson, Franklin Lakes, NJ) and incubated overnight in DMEM supplemented with FBS (10%) and l-glutamine (2 mM). T7 RNA polymerase “runoff” replicon RNA transcripts were prepared from ScaI-digested plasmid templates using the T7 RiboMax Express large-scale production system (Promega, Madison, WI). HCV replicon RNA (2 μg per well) was transfected into cured Huh-7 cells by using the cationic lipid DMRIE-C (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Following transfection, cells were trypsinized and diluted to 2 × 104 cells per ml with assay medium (DMEM supplemented with FBS [4%] and l-glutamine [2 mM]). Diluted cells (500 μl per well) were added to 24-well assay plates (Becton, Dickinson, Franklin Lakes, NJ). Compound, serially diluted in DMSO, was added per well with a final DMSO concentration of 0.5% in the assay. After 4 days of incubation at 37°C with 5% CO2, inhibition of replicon replication was measured using the Renilla luciferase assay system (Promega, Madison, WI). Briefly, cells were washed once with Dulbecco's phosphate-buffered saline without magnesium and harvested in 125 μl 1× lysis buffer. Cell lysate (50 μl) was transferred to 96-well solid white plates (Perkin Elmer, Waltham, MA), and Renilla substrate (190 μl) was subsequently added to each well. Luminescence was measured immediately using a TopCount NXT counter (Perkin Elmer, Waltham, MA). EC50 values were calculated with XLfit software (version 2.0; IDBusiness Solutions, Burlington, MA) using a 4-parameter logistic equation: y = A + ([B − A]/[1+([C/x]D̂)]). The relative fitness of replicons carrying NS3 substitutions was determined by comparing the level of Renilla luciferase activity from variant and wild-type replicon transfections. Luciferase data were normalized using “no-replication” control transfections, as previously described (9). Briefly, replicons were treated with an NS5A inhibitor at 10,000 times the EC50 (inhibiting >99% of replication) to account for variations in RNA transfection efficiencies.
(ii) NS3/4A protease enzyme complex.
To assess the susceptibility of patient population NS3 protease sequences to inhibition by asunaprevir, modified constructs encoding pET-H77c and pET-Con1 were employed to overexpress protein, as previously described (43). The HCV NS3 protease assay buffer and the determination of 50% inhibitory concentration (IC50) were as previously described (6).
SAD and MAD clinical study populations.
In the SAD clinical study, a total of 20 HCV-infected patients (treatment naïve or experienced) received a single dose of asunaprevir (10, 50, 200, or 600 mg; 5 patients per dose group), and 4 patients received placebo treatment. Patients enrolled in this study included men and women, aged 18 to 49 years, who were chronically infected with HCV genotype 1 and had HCV RNA levels of >105 IU/ml at screening. Serum samples were collected at baseline and 24, 48, and 144 h postdosing. Viral RNA was isolated from all baseline samples and from samples from all time points from patients receiving 200- and 600-mg doses using a QIAamp MiniElute viral vacuum kit (Qiagen, Valencia, CA). The NS3 protease domain was amplified and analyzed by population sequencing. Available baseline samples from patients receiving 10- and 50-mg doses were similarly analyzed. Since minimal or no viral response to asunaprevir was observed at these lower doses, and since asunaprevir-resistant variants were not detected (29), patient baseline polymorphisms are not described here.
In the MAD clinical study, a total of 12 HCV-infected patients (alfa interferon-ribavirin treatment naïve or experienced) received multiple doses of asunaprevir (200, 400, or 600 mg; 4 patients per dose group) BID for 3 days, and 3 patients received placebo treatment. As with the SAD study, patients enrolled in this study included men and women, aged 18 to 49 years, who were chronically infected with HCV genotype 1 and had HCV RNA levels of >105 IU/ml at screening. Serum samples were collected at baseline and days 1 through 10, 14, 21, and 28. Additional longer-term follow-up outpatient visits were scheduled at days 42, 98, and 182 (±7 days).
These studies were approved by institutional review boards in all study centers and conducted in accordance with Good Clinical Practice and in accordance with the ethical principles that have their origin in the Declaration of Helsinki. Informed written consent was obtained from all subjects.
RESULTS
Asunaprevir is a potent NS3 PI with HCV genotype 1a and 1b replicon activity (32). Structurally, it is an acyclic peptide mimetic inhibitor that is more flexible than the NS3 PI macrocycles currently in clinical development. To gain insight into the mechanisms of asunaprevir resistance, the HCV subgenomic replicon system was employed to identify and characterize signature resistance substitutions that may emerge in the clinical setting. Clinically relevant variations causing resistance to the recently approved NS3 PIs telaprevir and boceprevir were also introduced into genotype 1a and 1b replicons, and their susceptibility to inhibition by asunaprevir was evaluated.
Genotype 1a resistance.
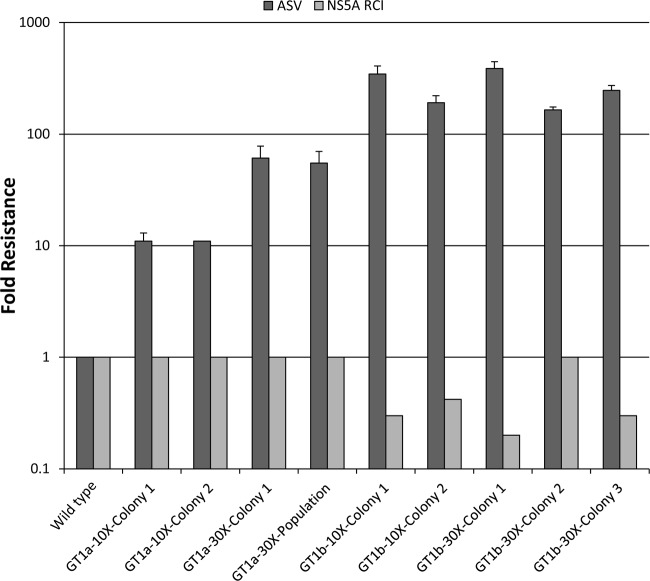
The HCV subgenomic replicon representing genotype 1a carrying an in-frame neomycin phosphotransferase resistance gene (neo) was employed to select for resistance to the NS3 PI asunaprevir. Replicon cells were maintained under selective pressure with asunaprevir at concentrations of 10 and 30 times the EC50 values (50 or 150 nM final concentrations, respectively). Cells were maintained in the presence of G418 in order to eliminate replicons susceptible to the HCV inhibitor enabling drug-resistant variants to emerge. Visual inspection of the cultures under selective pressure showed that widespread cell death had occurred by day 14. By day 23, individual colonies visible in cultures were isolated and expanded for phenotypic characterization. As shown in Fig. 1, the level of asunaprevir resistance varied from 11- to 61-fold relative to wild-type DMSO-treated control cells, depending on drug selection pressure. More importantly, asunaprevir-resistant replicon cell lines retained susceptibility (≤1-fold) to an HCV NS5A replication complex inhibitor relative to the parental replicon cell line, suggesting that the resistance phenotype was specific to asunaprevir.
Fig 1.
Resistance of genotype 1a and 1b replicon cells selected with asunaprevir (ASV). Selected cell colonies or populations were assayed for sensitivity to asunaprevir and an HCV NS5A replication complex inhibitor (as described in Materials and Methods; NS5A replication complex inhibitor EC50s of 3.3 pM and 1.6 pM against genotype 1a and 1b, respectively). The change in EC50s was plotted as the EC50 of inhibitor for the resistant cells divided by the EC50 for DMSO-treated control cells. The data are mean average fold reduction values obtained from ≥3 independent experiments with asunaprevir (except for genotype 1a 10× colony 2 and all NS5A replication complex inhibitor dose-response values, which were tested only in duplicate). Error bars show standard deviations. GT, genotype; RCI, replication complex inhibitor.
As summarized in Table 1, genotypic sequence analyses of asunaprevir-selected colonies revealed the presence of amino acid substitutions in the NS3 domain previously reported to confer resistance to various NS3 PIs currently in clinical development (39). Sequencing of individual NS3 cDNA clones indicated that mutations coding for R155K (present in 18 of 37 clones), D168G (3/37), and I170T (10/37) emerged during the selection. Substitution R155K is of particular interest, as it has been associated with reduced responses to telaprevir and boceprevir during clinical development studies (44, 45; T. L. Kieffer, S. De Meyer, D. J. Bartels, J. C. Sullivan, I. Dierynck, N. Adda, A. Kwong, I. M. Jacobson, K. E. Sherman, S. Zeuzem, and G. Picchio, presented at the International Workshop on HIV & Hepatitis Virus Drug Resistance and Curative Strategies, Los Cabos, Mexico, 7 to 10 June 2011). Other amino acid substitutions were identified less frequently and, in general, occurred at positions previously described as either cell culture-adaptive variants (G176E) or growth-compensatory substitutions (18, 25, 43). Clinically relevant substitutions at residues R155 and D168 were prioritized for characterization based on findings with a prior BMS NS3 PI (BMS-605339) (31). The observed I170T substitution, together with other resistance-associated substitutions (including those at positions V36, F43, and Q80) (39) reported to emerge in the presence of diverse peptidomimetic NS3 PIs, were also investigated. To further assess the relevance of these variants in the resistance phenotype, individual substitutions were introduced into the wild-type genotype 1a replicon background and analyzed in transient-replicon-transfection assays for susceptibility to asunaprevir (Table 2). The individual amino acid substitutions F43L, Q80K/L, R155K, and I170T conferred minimal to modest levels of resistance to asunaprevir, with reduced susceptibility ranging from 1 to 21 times the wild-type control level (Table 2). The clinically relevant R155K substitution yielded 21-fold resistance to asunaprevir while demonstrating low-level cross-resistance to telaprevir (5-fold). As previously reported (23, 40), the R155K substitution had a significant impact on replication capacity of the genotype 1a replicon. Combination of the nonconservative substitutions Q80K and Q80L with the R155K variant restored the replication capacity of the replicons, suggestive of compensatory roles. In contrast, substitutions introduced at position D168 resulted in much greater variability in the variants' resistance phenotype profile. The levels of resistance were substantially greater with D168A/E/G/V/Y substitutions, with increases in EC50 values ranging from 14 to 622 times the wild-type control level. The D168Y variant conferred the highest level of resistance to asunaprevir (622-fold). Furthermore, these D168 variants showed no cross-resistance to telaprevir, in accordance with previously reported in vitro studies (23). The mutation of residue V36 to A or M, when combined with the R155K mutation, enhanced resistance to asunaprevir (58- or 55-fold over the wild-type control level, respectively). The combination of the Q80K substitution with R155K (60-fold) or D168V/E (713/242-fold) also resulted in a significant increase in the resistance phenotype of the replicons compared with their individual variant counterparts. Emergence of substitutions at residue Q80 appears to play a compensatory role in the replicon, as these changes significantly improved the replication capacity of the R155K and D168V/E variants.
Table 1.
Clonal sequence analysis of NS3 protease domain from genotype 1a replicon cells selected in the presence of asunaprevir
| Clone or population | Amino acid replacing wild-type residue |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V36 | T40 | Q41 | V51 | R62 | L72 | D79 | T95 | V107 | I114 | R123 | P131 | I132 | G152 | R155 | R161 | D168 | I170 | V172 | N174 | L175 | G176 | |
| ASVc 10× clones | ||||||||||||||||||||||
| 1-1 | T | |||||||||||||||||||||
| 1-2 | K | |||||||||||||||||||||
| 1-3 | K | |||||||||||||||||||||
| 1-4 | K | |||||||||||||||||||||
| 1-5 | E | K | Y | |||||||||||||||||||
| 1-6 | K | |||||||||||||||||||||
| 1-7 | A | K | ||||||||||||||||||||
| 1-8 | K | |||||||||||||||||||||
| 1-9 | A | K | ||||||||||||||||||||
| ASV 10× clones | ||||||||||||||||||||||
| 2-1 | G | |||||||||||||||||||||
| 2-2 | V | G | ||||||||||||||||||||
| 2-3 | G | |||||||||||||||||||||
| 2-4 | T | |||||||||||||||||||||
| 2-5 | T | |||||||||||||||||||||
| 2-6 | E | |||||||||||||||||||||
| 2-7 | K | |||||||||||||||||||||
| 2-8 | T | |||||||||||||||||||||
| 2-9 | T | |||||||||||||||||||||
| ASV 30× clones | ||||||||||||||||||||||
| 1-1 | K | |||||||||||||||||||||
| 1-2 | T | |||||||||||||||||||||
| 1-3 | T | P | ||||||||||||||||||||
| 1-4 | ||||||||||||||||||||||
| 1-5 | T | |||||||||||||||||||||
| 1-6 | T | Q | ||||||||||||||||||||
| 1-7 | G | T | ||||||||||||||||||||
| 1-8 | A | K | ||||||||||||||||||||
| 1-9 | K | |||||||||||||||||||||
| 1-10 | K | |||||||||||||||||||||
| ASV 30× populations | ||||||||||||||||||||||
| 1 | Rb | |||||||||||||||||||||
| 2 | K | |||||||||||||||||||||
| 3 | S | V | K | S | E | Rb | ||||||||||||||||
| 4 | K | |||||||||||||||||||||
| 5 | Ia,b | Ia | K | |||||||||||||||||||
| 6 | A | V | ||||||||||||||||||||
| 7 | Ia,b | Ia | K | |||||||||||||||||||
| 8 | Rb | |||||||||||||||||||||
| 9 | Ia,b | Ia | K | |||||||||||||||||||
Present in the wild-type cell line.
Published cell culture-adaptive mutation.
ASV, asunaprevir.
Table 2.
Effects of NS3 protease amino acid substitutions on asunaprevir and telaprevir potency in genotype 1a replicon transient-transfection assaysa
| Amino acid substitutionb | Asunaprevir |
Telaprevir |
Replication level (%) | ||
|---|---|---|---|---|---|
| EC50c (nM) | Fold increase in EC50 | EC50c (nM) | Fold increase in EC50 | ||
| WT | 0.76 ± 0.3 | 1 | 181 ± 87 | 1 | 100 |
| V36A | 2.3 ± 0.7 | 3 | 2,689 ± 241 | 15 | 89 |
| V36L | 1.8 ± 0.6 | 2 | 1,248 ± 427 | 7 | 137 |
| V36M | 1.5 ± 0.5 | 2 | 2,989 ± 1813 | 17 | 171 |
| T40A | 0.52 ± 0.03 | 1 | ND | 41 | |
| Q41E | NR | NR | 0 | ||
| F43L | 2.7 ± 1.1 | 4 | 44 ± 5 | 0.2 | 2 |
| T54A | 0.33 ± 0.1 | 0.4 | 847 ± 209 | 5 | 39 |
| T54S | 0.74 ± 0.2 | 1 | 664 ± 150 | 4 | 36 |
| V55I | 2.2 ± 0.4 | 3 | 269 ± 126 | 2 | 8 |
| R62K | 0.74 ± 0.03 | 1 | ND | 161 | |
| D79E | 0.36 ± 0.08 | 1 | 192 ± 59 | 1 | 2 |
| Q80K | 2.5 ± 1.0 | 3 | 152 ± 75 | 1 | 119 |
| Q80L | 0.90 ± 0.52 | 1 | 146 ± 34 | 1 | 252 |
| S122G | 0.65 ± 0.1 | 1 | 325 ± 34 | 2 | 154 |
| S122N | 0.69 ± 0.1 | 1 | 202 ± 65 | 1 | 112 |
| S122R | 2.6 ± 0.4 | 3 | 36 ± 5 | 0.2 | 112 |
| R123G | NR | NR | 0 | ||
| R155K | 16 ± 3 | 21 | 860 ± 256 | 5 | 7 |
| D168A | 17 ± 4 | 23 | 32d | 0.2 | 1 |
| D168E | 44 ± 17 | 58 | 165 ± 8 | 1 | 66 |
| D168G | 11 ± 4 | 14 | 133 ± 76 | 1 | 2 |
| D168V | 283 ± 4 | 373 | 162 ± 10 | 1 | 33 |
| D168Y | 473 ± 281 | 622 | 58 ± 29 | 0.3 | 23 |
| I170T | 3.6 ± 1.6 | 5 | 647 ± 202 | 4 | 12 |
| I170V | 0.67 ± 0.12 | 1 | 197 ± 13 | 1 | 163 |
| N174Y | 1.0 ± 0.2 | 1 | ND | 134 | |
| L175P | NR | NR | 0 | ||
| V36L+Q80K | 4.0 ± 2 | 5 | 841 ± 252 | 6 | 371 |
| V36A+R155K | 44 ± 12 | 58 | >10,000 | >55 | 6 |
| V36M+R155K | 42 ± 19 | 55 | >10,000 | >55 | 5 |
| Q80K+R155K | 46 ± 18 | 60 | 1,223 ± 320 | 7 | 33 |
| Q80K+D168V | 542 ± 111 | 713 | 130 ± 37 | 1 | 46 |
| Q80K+D168E | 184 ± 27 | 242 | 251 ± 98 | 1 | 127 |
WT, wild type; ND, not determined; NR, no replication.
Substitutions associated with asunaprevir resistance selection in genotype 1a replicons are indicated in bold.
Data are means ± standard deviations from ≥3 independent experiments unless otherwise stated.
Two independent experiments, each carried out in triplicate.
Genotype 1b resistance.
For genotype 1b resistance selection, replicon cells were maintained in the presence of asunaprevir at 10 or 30 times the EC50 values (30 or 90 nM final concentrations, respectively). After 21 days of selective pressure in the presence of G418, phenotypic analysis of the breakthrough replicons showed that the reduced susceptibility to asunaprevir was greater for genotype 1b than for genotype 1a when they are subjected to the same selection pressures. Reduced susceptibility of the asunaprevir-resistant genotype 1b replicon cell lines varied from approximately 165- to 387-fold relative to wild-type control cells, depending on drug selection pressure (Fig. 1). In contrast, the susceptibility to an HCV NS5A replication complex inhibitor in selected replicon cell lines was not affected compared with the wild-type control level. Sequencing of the NS3 protease domain revealed the emergence of ≤3 amino acid substitutions per clonal population. An amino acid change at position D168 was observed in each (selected with 10 and 30 times the asunaprevir EC50) genotype 1b asunaprevir-resistant cell line characterized. Clonal analysis revealed the presence of HCV genomic quasispecies containing NS3 substitutions at amino acid position D168 in both asunaprevir-resistant cell lines (7/10 in the 10× clone and 12/12 in the 30× clone). Amino acid substitutions H, G, V, or Y at residue D168 conferred resistance to asunaprevir but also impaired the replication capacity of the replicon variants. The frequencies of amino acid substitutions observed in the NS3 domain during selection are summarized in Table 3. The D168G and D168V variants were predominant, with the latter being detected only under higher drug-selective pressure. The substitutions Q41R and Q86R were also frequently observed; however, these changes have been described as cell culture-adaptive mutations conferring minimal resistance (3, 13, 46). A substitution at Q80R was detected in only one clone (30× asunaprevir selection); however, substitutions at Q80 have been reported to confer low-level resistance to some NS3 PIs (21, 23). To confirm the phenotypic resistance profiles associated with these emerging variants, D168 and other reported resistance-associated substitutions previously selected in the presence of NS3 PIs currently in clinical development (including at positions V36, T54, Q80, V170, and A156) (39, 42) were introduced into wild-type genotype 1b replicon backgrounds and analyzed in transient-transfection assays for their susceptibility to asunaprevir or telaprevir (Table 4). Individual substitutions introduced at residue D168 resulted in increases in asunaprevir EC50 values ranging from 16- (D168G) to 280-fold (D168V) above the wild-type level without affecting susceptibility to telaprevir. Furthermore, low to moderate levels of resistance (6- to 20-fold more than the wild-type control level) to asunaprevir were observed against replicons containing A156 substitutions (A156 to S, T, and V). In contrast, these A156 variants demonstrated higher levels of resistance to telaprevir (15- to >57-fold over the wild-type control level) in agreement with previous reports (23; Kieffer et al., presented at the International Workshop on HIV & Hepatitis Virus Drug Resistance and Curative Strategies). Minimal or low levels of resistance (1- to 4-fold over wild-type levels) to asunaprevir were observed with replicons containing Q80 substitutions (Q80 to L and R). The reported adaptive substitution at Q41 (Q41 to R) pre-existed in our genotype 1b replicon cell line. Assessment of this substitution when linked with D168V or Q80R on asunaprevir activity revealed minimal- to low-level resistance (1- and 2-fold resistance, respectively) compared with replicons containing only the D168V or Q80R substitutions (Table 4). To explore differences in the genotype 1a and 1b resistance profiles, some of the emerging resistance variants detected in genotype 1a asunaprevir-resistant replicons were introduced into the genotype 1b replicon. Engineered replicon R155K variants demonstrated reduced susceptibility to asunaprevir, which was similar to that seen with genotype 1a; by itself, the R155K variant yielded 27-fold resistance to asunaprevir and low-level resistance to telaprevir (4-fold). However, combination of V36M with R155K enhanced resistance to asunaprevir (72-fold) and telaprevir (26-fold) compared with the single amino acid changes. In contrast, combination of V170A with R155K resulted in a level of resistance comparable to the R155K replicon.
Table 3.
Clonal sequence analysis of the NS3 protease domain from genotype 1b replicon cells selected in the presence of asunaprevir
| Clone | Amino acid at NS3 position |
|||||||
|---|---|---|---|---|---|---|---|---|
| Q41 | Q80 | Q86 | P89 | Y105 | D168 | E173 | E176 | |
| ASVc 10× clones | ||||||||
| 1-1 | Ra | Ra | H | G | ||||
| 1-2 | Ra | Ra | ||||||
| 1-3 | Ra | G | Gb | |||||
| 1-4 | Ra | Ra | ||||||
| 1-5 | Ra | Ra | G | |||||
| 1-6 | Ra | Ra | ||||||
| 1-7 | Ra | Ra | G | |||||
| 1-8 | Ra | Ra | A | |||||
| 1-9 | Ra | G | ||||||
| 1-10 | Ra | G | ||||||
| ASV 30× clone | ||||||||
| 1-1 | Ra | R | Ra | G | ||||
| 1-2 | Ra | V | ||||||
| 1-3 | Ra | V | ||||||
| 1-4 | C | V | ||||||
| 1-5 | V | |||||||
| 1-6 | Ra | G | ||||||
| 1-7 | V | |||||||
| 1-8 | V | |||||||
| 1-9 | G | |||||||
| 1-10 | Ra | Lb | Y | |||||
| 1-11 | Ra | V | ||||||
| 1-12 | Ra | A | ||||||
Present in wild-type cell line.
Published cell culture-adaptive substitution.
ASV, asunaprevir.
Table 4.
Effects of NS3 protease amino acid substitutions on asunaprevir and telaprevir potency in genotype 1b replicon transient-transfection assays
| Amino acid substitutiona | Asunaprevir |
Telaprevir |
Replication level (%) | ||
|---|---|---|---|---|---|
| EC50b (nM) | Fold increase in EC50 | EC50b (nM) | Fold increase in EC50 | ||
| WT | 0.86 ± 0.3 | 1 | 176 ± 41 | 1 | 100 |
| V36A | 1.6 ± 0.6 | 2 | 933 ± 281 | 5 | 124 |
| V36L | 0.65 ± 0.1 | 1 | 395 ± 19 | 2 | 77 |
| V36M | 1.9 ± 0.9 | 2 | 1,081 ± 224 | 6 | 59 |
| F43S | 3.2 ± 0.2 | 4 | 213 ± 36 | 1 | 79 |
| T54A | 0.35 ± 0.1 | 0.4 | 718 ± 31 | 4 | 101 |
| V55A | 0.79 ± 0.9 | 1 | 205 ± 4 | 1 | 4 |
| Q80K | 5.6 ± 1 | 6.5 | NDc | ND | 139 |
| Q80L | 0.86 ± 0.1 | 1 | 167 ± 14 | 1 | 98 |
| Q80R | 3.5 ± 0.9 | 4 | ND | ND | 102 |
| S122N | 0.7 ± 0.2 | 1 | 134 ± 34 | 1 | 194 |
| S122T | 1.2 ± 0.2 | 1 | 196 ± 11 | 1 | 205 |
| R155K | 23 ± 8 | 27 | 721 ± 75 | 4 | 54 |
| A156S | 5.9 ± 0.9 | 7 | 2,515 ± 624 | 15 | 120 |
| A156T | 5.4 ± 1 | 6 | >10,000 | >57 | 17 |
| A156V | 17 ± 2 | 20 | >10,000 | >57 | 3 |
| D168A | 109 ± 19 | 127 | 21 ± 4 | 0.1 | 37 |
| D168C | 56 ± 4 | 65 | 96 ± 30 | 1 | 25 |
| D168E | 67 ± 11 | 78 | 129 ± 7 | 1 | 25 |
| D168G | 13 ± 4 | 16 | 56 ± 12 | 0.3 | 29 |
| D168H | 85 ± 24 | 98 | 267 ± 105 | 2 | 12 |
| D168V | 241 ± 17 | 280 | 42 ± 7 | 0.2 | 29 |
| D168Y | 205 ± 29 | 238 | 71 ± 43 | 0.4 | 2 |
| V170A | 1.6 ± 0.4 | 2 | 699 ± 237 | 4 | 96 |
| V170I | 0.72 ± 0.01 | 1 | 184 ± 10 | 1 | 121 |
| V36M+R155K | 62 ± 29 | 72 | 4,483 ± 3,402 | 26 | 34 |
| F43S+D168E | 157 ± 30 | 183 | 147 ± 61 | 1 | 90 |
| Q41R+Q80R | 6.9 ± 0.4 | 8 | 229 ± 8 | 1 | 47 |
| Q41R+D168V | 202 ± 7 | 235 | 122 ± 33 | 1 | 20 |
| R155K+V170A | 18 ± 2 | 21 | 1,293 ± 117 | 7 | 84 |
WT, wild type. Substitutions associated with asunaprevir resistance selection in genotype 1b replicons are in bold.
Values are means ± standard deviations from ≥3 independent experiments.
ND, not determined.
Genotypic and phenotypic analysis from SAD and MAD clinical trial studies.
To determine whether there was an in vivo correlation with the in vitro resistance profile of asunaprevir, serum samples from patients receiving asunaprevir in SAD and MAD monotherapy studies were analyzed. Genotypic analysis was performed by population sequencing of the NS3 protease region from baseline and on-treatment patient serum samples. Baseline polymorphisms potentially associated with resistance to NS3 PIs were detected in patients in both the SAD and MAD studies, and the results are summarized in Table 5. The most frequent of these polymorphisms was Q80K, which has been associated with low-level resistance to the HCV NS3 PIs IDX-316 (21) and TMC435 (23). Susceptibility analysis was performed by introducing patient NS3 protease sequences into genotype 1a and 1b NS3/4A protein expression constructs and/or into chimeric replicon backbones as previously described (43). Comparison of changes in susceptibility to inhibition by asunaprevir revealed that baseline patient sequences from the SAD study demonstrated similar potencies to their respective reference strains (Table 6).
Table 5.
Baseline NS3 protease polymorphisms in patients from single- and multiple-ascending-dose studies
| Amino acid substitutiona | No. of patients with NS3 PI resistance-associated substitution |
|
|---|---|---|
| SAD (n = 10) | MAD (n = 15) | |
| V55I | 1 | |
| Q80K | 5 | 5 |
| S122G | 1 | |
| S122T | 1 | |
| I170Vb | 1 | |
| V170Ic | 1 | |
| S122R-V170I | 1 | |
Amino acids monitored included V36, F43, T54, V55, D79, Q80, S122, R155, A156, V158, D168, and V/I170.
Genotype 1a consensus, I170.
Genotype 1b consensus, V170.
Table 6.
In vitro enzyme and replicon susceptibility analysis of HCV NS3 protease sequences derived from patients in the single-ascending-dose study
| Patient | Genotype | Dose (mg) | Isolate time point | Log10 HCV RNA (IU/ml) | Polymorphism(s)a | Fold change (vs. reference) in: |
|
|---|---|---|---|---|---|---|---|
| ASV IC50b | ASV EC50c | ||||||
| 1 | 1a | 200 | Baseline | 6.3 | Q80K | 1 | 3 |
| 24 h | 5.9 | Q80K | |||||
| 2 | 1b | 200 | Baseline | 6.9 | V170V/I | 1 | 1 |
| 24 h | 3.9 | V170V/I | |||||
| 3 | 1a | 200 | Baseline | 7.2 | Q80K | 1 | |
| 24 h | 5.7 | Q80K | |||||
| 4 | 1a | 200 | Baseline | 6.9 | Q80K | 1 | |
| 24 h | 6.1 | Q80K | |||||
| 5 | 1b | 200 | Baseline | 6.0 | S7A, I72T, S122S/T | 1 | 1 |
| 24 hd | 3.2 | S7, L14F, S42T, I72 S122, V132I, V170V/I | 1 | 1 | |||
| 48 h | 3.0 | S7, L14F, I72, S122 V132I, C145Y, V170I | 1 | 0.4 | |||
| 144 h | 4.4 | S7, L14F, S122 | 1 | ||||
| 6 | 1b | 600 | Baseline | 6.3 | 1 | ||
| 24 h | 3.8 | ||||||
| 7 | 1a | 600 | Baseline | 7.2 | Q80K | 1 | |
| 24 h | 5.6 | Q80K | |||||
| 8 | 1a | 600 | Baseline | 6.8 | 1 | 2 | |
| 24 h | 4.1 | ||||||
| 9 | 1a | 600 | Baseline | 6.3 | Q80K | 1 | |
| 24 h | 4.3 | Q80K | |||||
| 10 | 1a | 600 | Baseline | 6.6 | S122S/G | 1 | |
| 24 h | 3.3 | S122, V55A, Q80K | 1 | ||||
| 48 hd | 2.9 | S122, V55A | 1 | ||||
| 48 hd | S122, V55A, Q80K | 1 | |||||
| 144 h | 5.3 | S122 | 1 | ||||
Only polymorphisms at NS3 amino acid positions reported to confer resistance to protease inhibitors are shown for patients where no changes from baseline were detected over 144 h. These positions included 36, 43, 54, 55, 77, 78, 79, 80, 107, 122, 132, 155, 156, 158, 168, 170, and 175.
Recombinant NS3/4A protease assay; the genotype 1a NS3/4A protease reference sequence was H77c; genotype 1b NS3/4A protease reference sequence was Con1. ASV, asunaprevir.
Replicon assay; the genotype 1a replicon reference sequence was H77c; genotype 1b replicon reference sequence was Con1.
To assess assay variability and primer sequence bias, protein was expressed from amplicons generated from different primer sets.
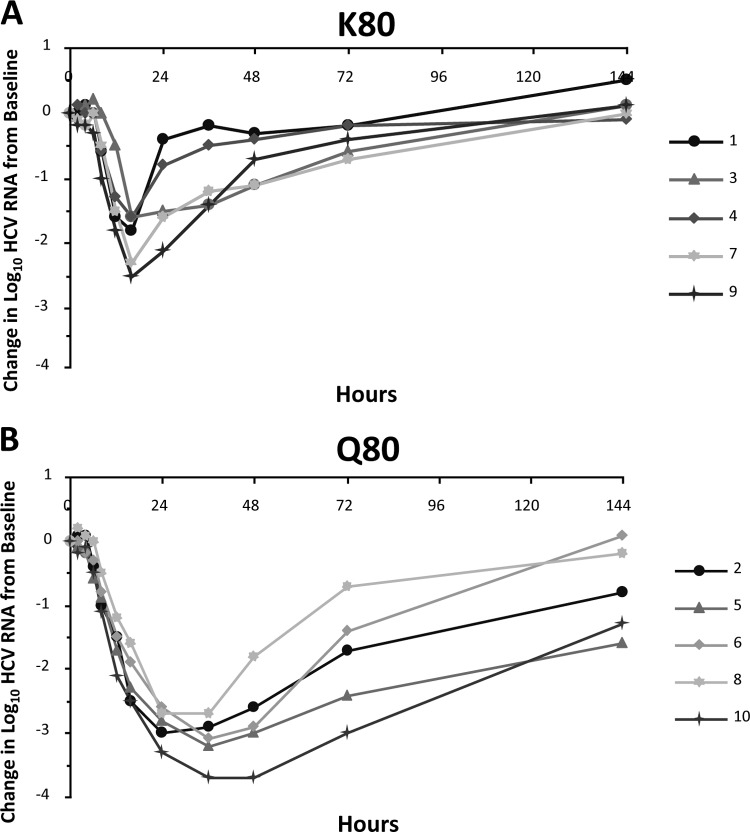
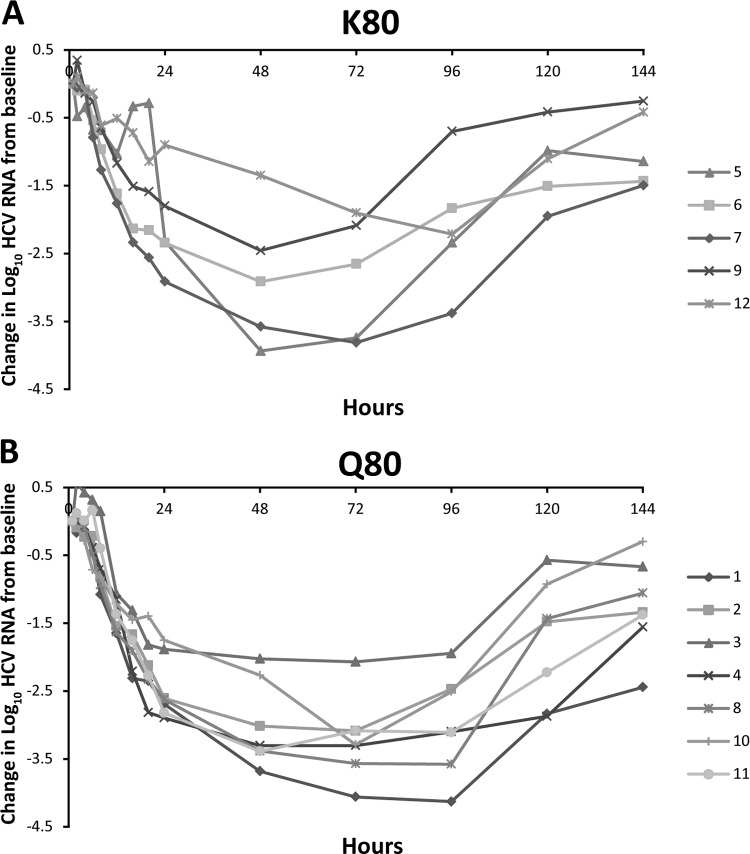
Treatment-emergent transient changes in the NS3 protease sequence were observed in 2 (patients 5 and 10) of 11 patients experiencing robust and rapid HCV RNA declines during the SAD study. Genotypic analysis revealed that patient 5, who was infected with genotype 1b, had a number of NS3 protease polymorphisms (S7A, I172T, and S122S/T) at baseline that changed during treatment with 200 mg asunaprevir. Other emergent substitutions detected were L14F and S42T. Susceptibility analysis revealed that none of these polymorphisms were associated with phenotypic asunaprevir resistance (Table 6). In patient 10 (600-mg cohort), emergent 1a NS3 protease substitutions detected were V55A and Q80K (Table 6). Although genotypic substitutions at these amino acid positions have been associated with resistance to other HCV NS3 PIs (36), in vitro NS3/4A enzyme susceptibility analyses of patient NS3 protease sequences revealed no changes in asunaprevir potency over time (Table 6). Similar findings have also been shown for the MAD study (34). In 2 of 15 patients, one each in the 400- and 600-mg cohorts, the Q80K variant of genotype 1a NS3 emerged. For the patient in the 600-mg cohort, Q80K was shown to persist at the last time point examined (day 42). Susceptibility analyses revealed that these substitutions did not significantly impact the potency of asunaprevir (34). However, a differential effect of baseline NS3-80 polymorphisms on virologic response was observed in the SAD study that was not apparent in the MAD study. Of the 10 patients in the SAD study, 5 (all of whom were infected with genotype 1a) had the K80 polymorphism at baseline and 5 (2 with genotype 1a and 3 with genotype 1b) had Q80. The viral response was found to be more robust in patients with Q80 than those with K80 (Fig. 2). For patients with the K80 polymorphism, mean declines in viral load were 1.29 ± 0.64 log10 at 24 h posttreatment, whereas 2.88 ± 0.31 log10 was observed for patients with the Q80 polymorphism. Interestingly, the susceptibility of patient population sequences containing the genotype 1a K80 NS3 variant to inhibition by asunaprevir was similar to that of sequences with the genotype 1a Q80 NS3 variant when examined in an NS3/4A protease enzyme assay (IC50 [K80] = 1.5 ± 0.2 nM [5 patients]; IC50 [Q80] = 1.1 nM [2 patients]) (29). In the MAD study, 5 of 12 patients had the K80 polymorphism at baseline (all 5 had genotype 1a). The initial log10 change in HCV RNA for these 5 patients appeared to be more variable than that for the 7 patients with Q80 (Fig. 3). Mean viral load declines 24 h after the start of dosing were 2.4 ± 0.45 log10 for patients with the Q80 polymorphism versus 2.06 ± 0.76 log10 for patients with the K80 polymorphism. Although all patients continued to respond during treatment, the mean viral load decline 24 h after the end of the treatment period (96 h after the first dose) was still slightly greater for patients with a baseline Q80 (2.94 ± 0.68 log10) than patients with a pre-existing K80 polymorphism (2.1 ± 0.97 log10). Given the variability in the viral load data in this small MAD study, a potential role for NS3-80 polymorphisms in the antiviral response to asunaprevir was not as apparent as in the SAD study. It should be noted that 2 of the 7 patients with Q80 were infected with genotype 1b in the MAD study, versus 3 of 5 in the SAD study, although the most robust antiviral responses were observed in patients infected with genotype 1a (patient 10 [Fig. 2b] and patient 1 [Fig. 3b]).
Fig 2.
HCV RNA levels in genotype 1a and 1b patients with K80 (A) and Q80 (B) at baseline after the administration of single doses of 200 (patients 1 to 5) or 600 (patients 6 to 10) mg asunaprevir. Patients 2, 5, and 6 were infected with HCV genotype 1b, whereas all other patients were infected with HCV genotype 1a.
Fig 3.
HCV RNA levels in genotype 1a and 1b patients with K80 (A) and Q80 (B) at baseline after the administration of multiple doses of 200, 400, or 600 mg asunaprevir. Patients 1 to 4 received 200 mg, patients 5 to 8 received 400 mg, and patients 9 to 12 received 600 mg. Patients 4 and 10 were infected with genotype 1b, whereas the rest were genotype 1a. Both Q80 and Q80K polymorphisms were detected in patient 12 (∼55% Q80K by clonal analysis).
DISCUSSION
Asunaprevir is a potent inhibitor of the HCV NS3/4A protease, demonstrating EC50 values of 4 and 1 nM against genotype 1a and 1b replicons, respectively (32). Resistance has been reported for all classes of DAAs due to the high rates of viral replication and the error-prone nature of HCV polymerase. In order to understand the resistance profile of asunaprevir and its possible impact in HCV combination therapy, we report the results of a comprehensive analysis of resistance in genotype 1a and 1b replicons and genotypic and phenotypic variants observed during short-term SAD and MAD studies.
For genotype 1a resistance selection, sequencing analysis of the NS3 protease region in asunaprevir-resistant replicons revealed predominant substitutions at residues R155 (R155K), D168 (D168G), and I170 (I170T) (Table 1), which were mutually exclusive. The pattern of substitutions found in asunaprevir-resistant replicons did not appear to differ substantially between drug selection pressures for R155K and I170T, whereas D168G was detected only at a lower drug pressure. When these substitutions were introduced into wild-type genotype 1a replicon background, the R155K substitution conferred moderate resistance to asunaprevir, with a 21-fold reduction in potency compared with 14- and 5-fold reductions for D168G and I170T variants, respectively. The emerging R155K variant has been shown to inhibit all NS3 PIs currently in clinical development, with reported changes in potencies ranging from 30- to 447-fold (BI201335, MK-7009, TMC-435, and ITMN-191) (20, 23, 24). Of the D168 substitutions tested, D168G conferred relatively low resistance to inhibition by asunaprevir compared with other D168 substitutions, with changes in potency of 23, 58, 373, and 622 times the wild-type value demonstrated for D168A, D168E, D168V, and D168Y, respectively. Furthermore, the introduction of an additional substitution at NS3 Q80 (Q80K) reduced susceptibility to inhibition by asunaprevir when linked with either D168E (242-fold increase) or D168V (713-fold increase). Any enhancement in resistance with the linkage of these resistance variants to Q80K was important to characterize, given the natural prevalence of this polymorphism in patients infected with HCV genotype 1a (19). Combination of the R155K substitution with V36A and V36M resulted in 58- and 55-fold decreases in asunaprevir potency, respectively, in transient-transfection assays. In clinical studies with telaprevir, variants with linked V36M and R155K substitutions were observed frequently (40%) in HCV genotype 1a patients who did not achieve sustained virologic responses (Kieffer et al., presented at the International Workshop on HIV & Hepatitis Virus Drug Resistance and Curative Strategies). Comparison of potencies in replicons carrying the V36M and R155K variants revealed asunaprevir EC50 values to be <50 nM compared with >10,000 nM for telaprevir in a transient-transfection replicon assay.
For genotype 1b resistance selection, substitutions were detected predominantly at NS3 residue D168, with the majority of clones coding for D168G and D168V substitutions. Selection pressure appeared to influence the emergence of low- and high-level asunaprevir-resistant variants: D168G (predominant at 10-fold concentrations) conferred a modest 16-fold loss in potency, whereas the D168V substitution (detected only at the 30-fold concentration) demonstrated substantially greater resistance (280-fold the EC50 of the wild type [Table 4]). Minor variants detected at very low frequency during resistance selection (not including those present in the wild type or published adaptive mutations) included the Q80R, Y105C, and E173G variants. The Q80R substitution conferred low-level resistance (4-fold the EC50 of the wild type [Table 4]); however, the effects of Y105C and E173G were not determined in transient-transfection assays, and therefore their impact on asunaprevir susceptibility remains unknown. Assessment of genotype 1b replicons containing signature telaprevir-resistant variants identified in clinical studies indicated minimal to moderate cross-resistance to asunaprevir; no resistance was observed with V36A and T54A substitutions, only low resistance to A156S/T and moderate resistance to A156V.
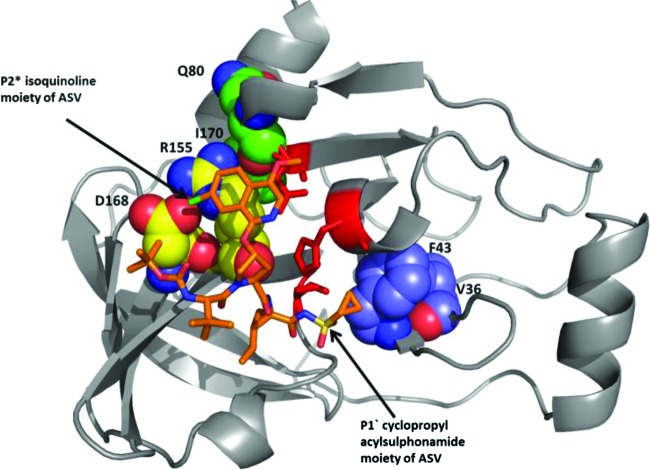
The primary sites of resistance associated with NS3 PIs have been shown to occur in the region of the NS3 substrate binding site, and the extent to which a drug protrudes from this substrate binding site is hypothesized to influence its tendency to resistance (39). X-ray crystal structure analyses have been used extensively to study NS3 PI interactions with the NS3 protease. Residues R155 and A156 have been shown to interact closely with P2 drug moieties, and amino acid substitutions at these residues can confer cross-resistance to most NS3 PIs, including boceprevir, ITMN-191, TMC435, and BILN-2061 (39). Substitutions at D168 demonstrated substantial resistance to asunaprevir in this study. D168 is located between the S2 and S4 pockets of the enzyme active site (17, 37, 39), and the negatively charged D168 forms a salt bridge with the positively charged R155. Substitutions at D168, therefore, can alter this residue to a noncharged state that is unable to form the D168-R155 salt bridge, destabilizing binding to the P2 moiety (in the case of asunaprevir, the isoquinoline moiety [Fig. 4]) and leading to reduced compound sensitivity (32). Similarly, R155 substitutions may also disrupt the D168-R155 salt bridge formation. In addition to the R155 and D168 substitutions, an I170 change to threonine emerged during genotype 1a replicon selection with asunaprevir (Table 1). An amino acid change at residue 170 (V to alanine) was previously reported to confer resistance to the NS3 PIs boceprevir and telaprevir (45). A structural analysis reveals that amino acid I170 interacts with the head group of R155, forming part of the pocket around the R155-D168 salt bridge interaction when asunaprevir is bound. It is thus possible that change of I170 to the more polar threonine may change the environment around R155, leading to a slightly reduced interaction with the P2 isoquinoline moiety of asunaprevir. The V36 residue is in close proximity to F43, which interacts with the P1′ cyclopropyl moiety of asunaprevir (Fig. 4) and other NS3 PIs; inhibitor interaction with F43 may therefore be altered by a V36 substitution. The F43S substitution (in addition to D168) has been associated with resistance to the structurally similar PI BMS-605339 in replicons (unpublished results) and NS3 PIs with P1′ cyclopropyl moieties (23). These substitutions were predicted to increase the hydrophilicity of the S1′ pocket and disrupt interactions with the P1′ moiety of asunaprevir. In this study, replicons containing this F43S substitution demonstrated decreased susceptibility (4-fold) to asunaprevir, whereas V36 substitutions conferred lower-level resistance. However, double substitutions of V36A/M plus R155K resulted in more substantial reductions in susceptibility (Tables 2 and 4). The Q41 residue is also in close proximity to the P1′cyclopropyl moiety, and modification of Q41 to a charged residue could impact asunaprevir binding by changing the S1′ pocket environment. Although no loss in potency was observed when the replicon-adaptive Q41R substitution was linked with D168V, linkage with Q80R resulted in slight reductions in susceptibility (Table 4). Low-level Q41R resistance against other NS3 PIs was reported previously (23). The Q41R substitution has not been detected in NS3 sequences from our baseline clinical samples to date (30, 34).
Fig 4.
Model of asunaprevir (ASV) bound to the active site of truncated tether form of the NS3/4A protease (our unpublished data). Asunaprevir is depicted in orange. The NS3 protease is shown as a ribbon diagram. Residues susceptible to changes that confer minimal to high levels of resistance to asunaprevir are depicted using space-filled atoms and labeled.
Genotypic and phenotypic analyses of HCV clinical isolates from patients at baseline and upon treatment with asunaprevir (30) identified transient NS3 changes (predominantly Q80K substitutions) that were not associated with phenotypic resistance. Genotypic and phenotypic analysis of samples from the SAD and MAD studies found that, despite the emergence and disappearance of Q80K, the effects on asunaprevir susceptibility were minimal over time and similar to EC50 values obtained with the genotype 1a (H77c) reference strains. However, virologic response rates in patients with Q80 were more rapid and robust than those in patients with the K80 polymorphism in the SAD study. This difference in antiviral response was more ambiguous among patients in the MAD studies. However, the viral load responses appeared to be more dependent on the polymorphism at NS3-80 than the administered dose or genotype, with the caveat that these were small studies.
Interestingly, single substitutions with Q80 polymorphisms in engineered genotype 1a and 1b replicons did not significantly influence asunaprevir susceptibility (Tables 2 and 4). Substitutions at residue Q80 have previously been associated with low-level resistance to the NS3 PIs TMC435 and IDX-316 (21, 23) and are believed to result from the disruption of R155-Q80 hydrogen bonding (39). However, Q80 is a natural polymorphism in NS3, with HCV genotype 1a sequences being divided approximately equally by Q80 and K80 residues (19). The clinical relevance of this substitution for asunaprevir activity, therefore, remains to be fully determined. Since the main purpose of the SAD and MAD monotherapy studies was to assess short-term drug tolerability, safety, and pharmacokinetics, treatment duration was too short to confirm a correlation between in vitro and in vivo resistance profiles. To this point, resistance analyses of patients receiving long-term asunaprevir treatment are warranted. Interim analysis of an ongoing phase IIa study in treatment-naïve, chronically infected HCV patients receiving asunaprevir in combination with alfa interferon and ribavirin has so far revealed no virologic breakthrough after 12 weeks of dosing (4). Thus, to understand the impact of pre-existing NS3 polymorphisms on the virologic response rates with asunaprevir, analysis of virologic failures in larger phase IIb clinical studies will be required.
ACKNOWLEDGMENTS
We acknowledge Paul Scola and the chemistry team for synthesizing asunaprevir. We also thank Ramkumar Rajamani and Andrew Good for providing a model of asunaprevir bound to the NS3 protease and Xin Huang and Bernadette Kienzle for sequencing support. Editorial assistance was provided by Stephen Griffiths of ArticulateScience Ltd. and was funded by Bristol-Myers Squibb.
All authors are employees of Bristol-Myers Squibb Company.
Footnotes
Published ahead of print 16 April 2012
REFERENCES
- 1. Argentini C, Genovese D, Dettori S, Rapicetta M. 2009. HCV genetic variability: from quasispecies evolution to genotype classification. Future Microbiol. 4:359–373 [DOI] [PubMed] [Google Scholar]
- 2. Bacon BR, et al. 2011. Boceprevir for previously treated chronic HCV genotype 1 infection. N. Engl. J. Med. 364:1207–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blight KJ, Kolykhalov AA, Rice CM. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974 [DOI] [PubMed] [Google Scholar]
- 4. Bronowicki J, et al. 2011. BMS-650032, an NS3 inhibitor (protease), in combination with peginterferon alfa-2a and ribavirin in treatment-naive subjects with genotype 1 chronic hepatitis C infection. J. Hepatol. 54(Suppl 1):S472 [Google Scholar]
- 5. Chayama K, et al. 2012. Dual therapy with the NS5A inhibitor BMS-790052 and the NS3 protease inhibitor BMS-650032 in HCV genotype 1b infected null responders. Hepatology 55:742–748 [DOI] [PubMed] [Google Scholar]
- 6. Drexler D, et al. 2006. Development of an on-line automated sample clean-up method and liquid chromatography-tandem mass spectrometry analysis: application in an in vitro proteolytic assay. Anal. Bioanal. Chem. 384:1145–1154 [DOI] [PubMed] [Google Scholar]
- 7. Food and Drug Administration 2011. Boceprevir prescribing information. http://www.accessdata.fda.gov/drugsatfda_docs/label/2011/202258lbl.pdf
- 8. Food and Drug Administration 2011. Telaprevir prescribing information. Accessed 1 April 2012 http://www.accessdata.fda.gov/drugsatfda_docs/label/2011/201917lbl.pdf
- 9. Fridell RA, et al. 2011. Distinct functions of NS5A in hepatitis C virus RNA replication uncovered by studies with the NS5A inhibitor BMS-790052. J. Virol. 85:7312–7320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fridell RA, Qiu D, Wang C, Valera L, Gao M. 2010. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob. Agents Chemother. 54(9):3641–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fried MW, et al. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975–982 [DOI] [PubMed] [Google Scholar]
- 12. Gao M, et al. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gaudieri S, et al. 2009. Hepatitis C virus drug resistance and immune-driven adaptations: relevance to new antiviral therapy. Hepatology 49:1069–1082 [DOI] [PubMed] [Google Scholar]
- 14. Ghany MG, Strader DB, Thomas DL, Seeff LB. 2009. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology 49:1335–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jacobson IM, et al. 2011. Telaprevir for previously untreated chronic hepatitis C virus infection. N. Engl. J. Med. 364:2405–2416 [DOI] [PubMed] [Google Scholar]
- 16. Kadow JF, et al. 2012. Discovery of BMS-791325, an allosteric NS5B replicase inhibitor for the treatment of hepatitis C, abstr. 23. In Sci. Abstr. 243rd ACS Natl. Meet. Exposition [Google Scholar]
- 17. Kim JL, et al. 1996. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343–355 [DOI] [PubMed] [Google Scholar]
- 18. Krieger N, Lohmann V, Bartenschlager R. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuiken C, Yusim K, Boykin L, Richardson R. 2005. The Los Alamos hepatitis C sequence database. Bioinformatics 21:379–384 [DOI] [PubMed] [Google Scholar]
- 20. Lagace L, et al. 2012. In vitro resistance profile of the HCV NS3 protease inhibitor BI 201335. Antimicrob. Agents Chemother. 56(1):569–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lallos L, et al. 2009. Preclinical profiles of IDX136 and IDX316, two novel macrocyclic HCV protease inhibitors, poster 344. In 44th Annu. Meet. Eur. Assoc. Study Liver (EASL), Copenhagen, Denmark, 22 to 26 April 2009 [Google Scholar]
- 22. Lemm JA, et al. 2005. Replication-competent chimeric hepatitis C virus subgenomic replicons. Intervirology 48:183–191 [DOI] [PubMed] [Google Scholar]
- 23. Lenz O, et al. 2010. In vitro resistance profile of the hepatitis C virus NS3/4A protease inhibitor TMC435. Antimicrob. Agents Chemother. 54:1878–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liverton NJ, et al. 2010. MK-7009, a potent and selective inhibitor of hepatitis C virus NS3/4A protease. Antimicrob. Agents Chemother. 54:305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lohmann V, Korner F, Dobierzewska A, Bartenschlager R. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lohmann V, et al. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 27. Lok AS, et al. 2012. Preliminary study of two antiviral agents for hepatitis C genotype 1. N. Engl. J. Med. 366(3):216–224 [DOI] [PubMed] [Google Scholar]
- 28. McHutchison JG, et al. 2009. Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. N. Engl. J. Med. 361:580–593 [DOI] [PubMed] [Google Scholar]
- 29. McPhee F, et al. 2009. Genotypic and phenotypic analysis of samples from HCV-infected subjects treated with BMS-650032 in a single ascending dose study. Hepatology 50(4):1048A [Google Scholar]
- 30. McPhee F, et al. 2011. No early virologic breakthrough observed with the HCV NS3 protease inhibitor BMS-650032 in multiple dose monotherapy studies and phase 2A combination studies with PEGIFN/RBV. J. Hepatol. 54(Suppl 1):A1223 [Google Scholar]
- 31. McPhee F, et al. 2009. The discovery and early development of the HCV NS3 protease inhibitor BMS-605339. Global Antiviral J. 5(Suppl 1):51 [Google Scholar]
- 32. McPhee F, et al. 2010. Identification and preclinical profile of the novel HCV NS3 protease inhibitor BMS-650032. J. Hepatol. 52(Suppl 1):S296 [Google Scholar]
- 33. Nettles RE, et al. 2011. Multiple ascending dose study of BMS-790052, an NS5A replication complex inhibitor, in patients infected with hepatitis C virus genotype 1. Hepatology 54(6):1956–1965 [DOI] [PubMed] [Google Scholar]
- 34. Pasquinelli C, et al. 2012. Single and multiple ascending dose studies of the NS3 protease inhibitor, asunaprevir, in subjects with or without chronic hepatitis C. Antimicrob. Agents Chemother. 56:1838–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poordad F, et al. 2011. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 364:1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qiu P, et al. 2009. Identification of HCV protease inhibitor resistance mutations by selection pressure-based method. Nucleic Acids Res. 37:e74 doi:10.1093/nar/gkp251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raney KD, Sharma SD, Moustafa IM, Cameron CE. 2010. Hepatitis C virus non-structural protein 3 (HCV NS3): a multifunctional antiviral target. J. Biol. Chem. 285:22725–22731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reesink HW, et al. 2006. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase Ib, placebo-controlled, randomized study. Gastroenterology 131:997–1002 [DOI] [PubMed] [Google Scholar]
- 39. Romano KP, Ali A, Royer WE, Schiffer CA. 2010. Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding. Proc. Natl. Acad. Sci. U. S. A. 107:20986–20991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sarrazin C, et al. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767–1777 [DOI] [PubMed] [Google Scholar]
- 41. Scola P, et al. 2010. Discovery of BMS-650032, an NS3 protease inhibitor for the treatment of hepatitis C, abstr. MEDI-38. In Sci. Abstr. 239th ACS Natl. Meet. Exposition [Google Scholar]
- 42. Sheaffer AK, et al. 2004. Characterisation of HCV replicons resistant to inhibitors of NS3 protease activity, abstr. 290. In Proc. 7th Int. Symp. Positive Strand RNA Viruses [Google Scholar]
- 43. Sheaffer AK, et al. 2011. Development of a chimeric replicon system for phenotypic analysis of NS3 protease sequences from HCV clinical isolates. Antivir. Ther. 16:705–718 [DOI] [PubMed] [Google Scholar]
- 44. Susser S, et al. 2009. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology. 50:1709–1718 [DOI] [PubMed] [Google Scholar]
- 45. Tong X, et al. 2008. Characterization of resistance mutations against HCV ketoamide protease inhibitors. Antiviral Res. 77:177–185 [DOI] [PubMed] [Google Scholar]
- 46. Verbinnen T, et al. 2010. Tracking the evolution of multiple in vitro hepatitis C virus replicon variants under protease inhibitor selection pressure by 454 deep sequencing. J. Virol. 84:11124–11133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. World Health Organization 2011. Hepatitis C key facts. http://www.who.int/mediacentre/factsheets/fs164/en/
- 48. Yanagi M, Purcell RH, Emerson SU, Bukh J. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Nat. Acad. Sci. U. S. A. 94:8738–8743 [DOI] [PMC free article] [PubMed] [Google Scholar]