LETTER
VEB-1-producing Klebsiella pneumoniae strains have recently been identified in Greece (5). We report here on a VEB-1-positive and multidrug-resistant Proteus mirabilis isolate (PM91) recovered from a patient treated in a Greek hospital in 2010. PM91 was resistant to penicillins, penicillin-clavulanate combinations, ceftazidime, cefotaxime, cefoxitin, and imipenem but susceptible to piperacillin-tazobactam, cefepime, meropenem, and aztreonam. The isolate was also resistant to various non-β-lactam antibiotics (Table 1). PCR and sequencing showed that PM91 contained blaVEB-1 along with blaVIM-1, blaOXA-10, and blaTEM-1. Production of the respective enzymes was confirmed by isoelectric focusing (IEF).
Table 1.
Antibiotic susceptibilities of strains harboring VEB-1-encoding plasmids
| Strain | Etest MIC (μg/ml) fora: |
Other resistance markersb | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amp | Amc | Pip | Ptz | Fox | Ctx | Caz | Fep | Atm | Ipm | Mem | ||
| P. mirabilis PM91 | >256 | 32 | 64 | 4 | 16 | 8 | 32 | 2 | 2 | 16 | 1 | Gen, Amk, Tob, Net, Sxt, Tet, Cip |
| E. coli Trc-PM91 | >256 | 64 | 256 | 16 | 64 | 32 | 256 | 2 | 4 | 8 | 0.5 | Gen, Amk, Tob, Net, Sxt, Tet |
| K. pneumoniae KP2741 | NTc | 24 | >256 | 4 | 4 | 1.5 | 256 | 1 | 24 | 0.25 | <0.12 | Gen, Amk, Tob, Net, Sxt, Tet |
| E. coli Trc-KP2741 | >256 | 24 | >256 | 3 | 3 | 3 | 256 | 1.5 | 24 | 0.25 | <0.12 | Gen, Amk, Tob, Net, Tet |
| E. coli (recipient) | 4 | 2 | 2 | 0.25 | 2 | <0.12 | 0.25 | <0.12 | <0.12 | <0.12 | <0.12 | |
Amp, ampicillin; Amc, amoxicillin-clavulanate (2:1); Pip, piperacillin; Ptz, piperacillin-tazobactam (inhibitor fixed at 4 mg/ml); Fox, cefoxitin; Ctx, cefotaxime; Caz, ceftazidime; Fep, cefepime; Atm, aztreonam; Ipm, Imipenem; Mem, meropenem.
Gen, gentamicin; Amk, amikacin; Tob, tobramycin; Net, netlimicin; Sxt, trimethoprim-sulfamethoxazole; Tet, tetracycline; Cip; ciprofloxacin.
NT, not tested.
Most of the resistance traits of PM91 were readily transferred to a rifampin-resistant Escherichia coli strain by conjugation in broth cultures containing rifampin (400 μg/ml) and ampicillin (50 μg/ml) (Table 1). Resistance phenotypes, PCR, IEF, and plasmid content analysis showed a single transconjugant species (Trc-PM91) with a 120-kb plasmid (pPM91) encoding all four β-lactamases of the donor strain. PCR-based replicon typing and sequencing classified pPM91 as an A/C2 plasmid (1). Notably, in the previously described K. pneumoniae 2741 (5), blaVEB-1, as determined here, was also carried by a self-transmissible A/C2 plasmid (pKP2741) conferring to E. coli recipients a multiresistant phenotype (Table 1). We therefore studied the genetic environment of the blaVEB-1 genes in both pPM91 and pKP2741 plasmids to gain information on their evolution, presuming that they were probably related. Additionally, the blaVIM-1-carrying segment in pPM91 was determined. Characterization of the regions flanking bla genes was carried out by PCR mapping and sequencing.
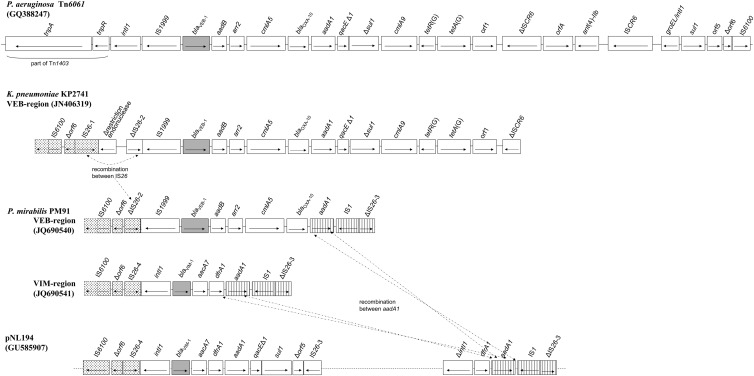
blaVEB-1 in pKP2741 occurred as the first cassette of an integron that also included an array of aadB, arr2, cmlA5, blaOXA-10, and aadA1 gene cassettes. As is common in VEB-1-encoding integrons, an IS1999 was located upstream of blaVEB-1 (2, 3, 6). The 3′ conserved segment (3′CS) of the integron was truncated 528 bp after the start codon of sulI. Moreover, downstream from ΔsulI, there was a cmlA9-tetR(G)-tetA(G) sequence associated with ISCR6. This structure appeared as a part of the blaVEB-1-containing Tn6061 from Pseudomonas aeruginosa (2) (Fig. 1). However, downstream from IS1999, instead of intI1, a ΔIS26 was identified. Upstream of the latter element, there was a region comprising a restriction endonuclease fragment, an intact IS26, and a Δorf6-IS6100 sequence (Fig. 1). Notably, the latter segment has been identified in several IncN plasmids, including the VIM-1-encoding pNL194 (4).
Fig 1.
Schematic representation of the VEB-1- and VIM-1-encoding regions from K. pneumoniae KP2741 and P. mirabilis PM91. The structure of the Tn6061 from P. aeruginosa, as well as parts of pNL194, has been included for comparison. Potential recombination sites are indicated by dashed lines.
In pPM91, blaVIM-1, along with the aacA7, dfrA1, and aadA1 cassettes, comprised the variable region of an integron similar to In-e541 from pNL194 harbored by a K. pneumoniae clinical strain isolated in Greece (4). The 5′CS was disrupted by an IS26-Δorf6-IS6100 sequence as in pNL194. Unlike In-e541, however, the integron did not include a 3′CS due to insertion at the attC site of aadA1 of an IS1 and a ΔIS26 lacking 57 bp of the left inverted repeat (IRL) (Fig. 1). The blaVEB-1 gene in pPM91 was carried by an integron similar to that found in pKP2741. However, the sequences downstream from IS1999 and aadA1 in pPM91 resembled those found in the VIM-1-encoding region described above. The sole difference was the presence of a truncated IS26 downstream from IS1999 in the VEB-1-encoding region instead of an intact IS26 found in the respective blaVIM-1-containing structure. The fact that both IS26 had the same boundary with the 3′CS (Δorf6-IS6100) may indicate that this difference was due to recombination between the two regions (Fig. 1).
The similarities of pPM91 and pKP2741 indicated a common ancestry. The presence of a ΔIS26-IS1999 sequence in both plasmids further supports this notion. The differences in the left-hand extremities in the blaVEB-1-containing regions could be explained by recombination events between IS26 elements that may lead to deletion of the intervening sequence in pPM91. The carriage of at least three pNL194-derived parts (including a VIM-1-encoding region) by pPM91 suggests that the latter plasmid might have evolved through cointegration and resolution event(s) between a VEB A/C2 replicon and the most common VIM IncN plasmid variant in this setting (1, 4). During this process, deletions and recombinations may have resulted in the observed structures.
Nucleotide sequence accession numbers.
Sequences of blaVEB-1-containing regions from P. mirabilis PM91 and K. pneumoniae KP2741 have been assigned GenBank accession numbers JQ690540 and JN406319, respectively. The blaVIM-1-carrying sequence from P. mirabilis PM91 has been assigned GenBank accession number JQ690541.
ACKNOWLEDGMENTS
This study was not funded by external sources.
The authors report no transparency declarations.
Footnotes
Published ahead of print 30 April 2012
Contributor Information
C. C. Papagiannitsis, Department of Microbiology, National School of Public Health, Athens, Greece
E. Tzelepi, Laboratory of Bacteriology, Hellenic Pasteur Institute, Athens, Greece
A. C. Vatopoulos, Department of Microbiology, National School of Public Health, Athens, Greece
E. Petinaki, Department of Microbiology, University Hospital of Larissa, Larissa, Greece
L. S. Tzouvelekis, Laboratory of Bacteriology, Hellenic Pasteur Institute, Athens, Greece
REFERENCES
- 1. Carattoli A, et al. 2006. Replicon typing of plasmids encoding resistance to newer β-lactams. Emerg. Infect. Dis. 12:1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coyne S, Courvalin P, Galimand M. 2010. Acquisition of multidrug resistance transposon Tn6061 and IS6100-mediated large chromosomal inversions in Pseudomonas aeruginosa clinical isolates. Microbiology 156:1448–1458 [DOI] [PubMed] [Google Scholar]
- 3. Girlich D, et al. 2002. Nosocomial spread of the integron-located veb-1-like cassette encoding an extended-spectrum β-lactamase in Pseudomonas aeruginosa in Thailand. Clin. Infect. Dis. 34:603–611 [DOI] [PubMed] [Google Scholar]
- 4. Miriagou V, et al. 2010. Sequence of pNL194, a 79.3-kilobase IncN plasmid carrying the blaVIM-1 metallo-β-lactamase gene in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 54:4497–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Papagiannitsis CC, et al. 2012. Diversity of acquired β-lactamases amongst Klebsiella pneumoniae in Greek hospitals. Int. J. Antimicrob. Agents 39:178–180 [DOI] [PubMed] [Google Scholar]
- 6. Poirel L, Menuteau O, Agoli N, Cattoen C, Nordmann P. 2003. Outbreak of extended-spectrum β-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J. Clin. Microbiol. 41:3542–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]