Abstract
We conducted a prospective multicenter study in Spain to characterize the mechanisms of resistance to amoxicillin-clavulanate (AMC) in Escherichia coli. Up to 44 AMC-resistant E. coli isolates (MIC ≥ 32/16 μg/ml) were collected at each of the seven participant hospitals. Resistance mechanisms were characterized by PCR and sequencing. Molecular epidemiology was studied by pulsed-field gel electrophoresis (PFGE) and by multilocus sequence typing. Overall AMC resistance was 9.3%. The resistance mechanisms detected in the 257 AMC-resistant isolates were OXA-1 production (26.1%), hyperproduction of penicillinase (22.6%), production of plasmidic AmpC (19.5%), hyperproduction of chromosomic AmpC (18.3%), and production of inhibitor-resistant TEM (IRT) (17.5%). The IRTs identified were TEM-40 (33.3%), TEM-30 (28.9%), TEM-33 (11.1%), TEM-32 (4.4%), TEM-34 (4.4%), TEM-35 (2.2%), TEM-54 (2.2%), TEM-76 (2.2%), TEM-79 (2.2%), and the new TEM-185 (8.8%). By PFGE, a high degree of genetic diversity was observed although two well-defined clusters were detected in the OXA-1-producing isolates: the C1 cluster consisting of 19 phylogroup A/sequence type 88 [ST88] isolates and the C2 cluster consisting of 19 phylogroup B2/ST131 isolates (16 of them producing CTX-M-15). Each of the clusters was detected in six different hospitals. In total, 21.8% of the isolates were serotype O25b/phylogroup B2 (O25b/B2). AMC resistance in E. coli is widespread in Spain at the hospital and community levels. A high prevalence of OXA-1 was found. Although resistant isolates were genetically diverse, clonality was linked to OXA-1-producing isolates of the STs 88 and 131. Dissemination of IRTs was frequent, and the epidemic O25b/B2/ST131 clone carried many different mechanisms of AMC resistance.
INTRODUCTION
Escherichia coli is an important etiologic agent for both nosocomial- and community-acquired infections in humans (9, 13, 22). Amoxicillin-clavulanate (AMC) is one of the most widely used antibiotics in many countries (3, 11, 15). In Spain, a 34.7% increase in community use of AMC was recorded from 2000 to 2006 (20). Recently, blood isolates of E. coli nonsusceptible to AMC increased from 9.3% (2003) to 25.9% (2010) in Spain, according to the European Antimicrobial Resistance Surveillance Network (EARS-Net [http://ecdc.europa.eu/en/activities/surveillance/EARS-Net/database/Pages/database.aspx]) (20).
Enzymatic mechanisms of E. coli resistance to AMC include hyperproduction of plasmid-mediated class A β-lactamases such as TEM-1 and SHV-1 (18, 31), plasmid-mediated AmpC-type β-lactamase (p-AmpC) (21), chromosomal AmpC β-lactamase (c-AmpC) (21), production of inhibitor-resistant TEM (IRT) β-lactamases (17, 27), plasmid-mediated β-lactamase OXA-1 (32), and complex mutant TEM (CMT) enzymes than combine IRT- and extended-spectrum β-lactamase (ESBL)-type substitutions (26).
In spite of the significant increase in AMC use in the last several years, there is little recent information available about the prevalence of AMC resistance mechanisms in E. coli; most previous studies analyzed strains isolated more than 10 years ago from single hospitals in the United States (12, 28), France (16), and Spain (19, 24).
Accordingly, the aim of this prospective Spanish national multicenter study was to investigate the epidemiology and mechanisms of AMC resistance in clinical isolates of E. coli causing both community and nosocomial infections.
MATERIALS AND METHODS
Study design and bacterial isolates.
A prospective multicenter study was designed to obtain E. coli isolates resistant to AMC (MIC of ≥ 32/16 μg/ml and/or disk inhibition zone of ≤13 mm according to the Clinical and Laboratory Standards Institute [CLSI]) (7) from clinical samples collected between January 2010 and May 2010. Seven university hospitals of six Spanish Autonomous Communities and members of the Spanish Network for Research in Infectious Diseases (REIPI) participated. Investigators at these hospitals were asked to collect up to 22 consecutive community- and 22 nosocomial-acquired, nonduplicated isolates of E. coli resistant to AMC.
Nosocomial-acquired isolates were defined as those acquired at least 48 h after hospital admission. Putative community-acquired isolates were those isolated in the community or within 48 h of hospital admission.
Susceptibility testing.
The disk diffusion and/or microdilution susceptibility tests were performed using different automated systems in each participating laboratory. All isolates were submitted to the antibiotic laboratory of the Centro Nacional de Microbiología (Majadahonda, Madrid, Spain), where additional confirmatory antibiotic susceptibility testing was performed with the agar dilution method according to the CLSI guidelines (6). Control strains used were E. coli ATCC 25922 and E. coli ATCC 35218. The production of extended-spectrum β-lactamases (ESBLs) was studied by the double-disc synergy test and/or Etest ESBL using cefotaxime and ceftazidime as substrates (AB Biodisk, Solna, Sweden).
Molecular characterization of mechanisms of resistance to AMC.
The blaTEM gene and its promoter region were assessed by PCR and sequencing (10). Sequences were compared to those of the blaTEM-1 gene and its promoter region in public databases (GenBank accession number AB194682) (29). For isolates in which the promoter region could not be amplified with these primers, the possibility of linkage of blaTEM alleles to IS26-like elements, as suggested previously (1), was assessed with primers IS26-F (5′-GCG GTA AAT CGT GGA GTG AT-3′) and TEMi-R (5′-TCT TTT ACT TTC ACC AGC GTT-3′).
Genes coding for p-AmpC β-lactamases (CIT, DHA, ACC, EBC, MOX, and FOX) and blaOXA-1 were characterized by PCR amplification with specific primers and sequencing (2, 23, 25) in all isolates. E. coli isolates that had negative results for p-AmpC β-lactamases but that displayed a resistance phenotype consistent with AmpC production on the basis of their resistance to AMC and cefoxitin and inhibition with phenyl boronic acid and cloxacillin (cefotetan/cefotetan-cloxacillin Etest; AB bioMérieux, Solna, Sweden) were categorized as c-AmpC.
Isolates carrying only the blaTEM-1 or blaSHV-1 genes with resistance to ampicillin, AMC, and cefazolin but susceptibility to the remaining β-lactam antibiotic families were considered penicillinase hyperproducers. In the case of SHV-1, an increase in the ceftazidime MIC was also considered compatible with hyperproduction of this enzyme (18).
Genes coding ESBL enzymes (CTX-M, SHV-type, and TEM-type) were studied by PCR and sequencing in all AMC-resistant E. coli isolates with phenotypes consistent with ESBL production on the basis of their resistance to the extended-spectrum cephalosporins whose activity was recovered in the presence of clavulanate (23).
Phylogenetic groups and detection of the O25b type.
The phylogenetic groups of AMC-resistant E. coli isolates were determined by a multiplex PCR assay described by Clermont et al. (4). To search for an ST131/B2/O25b E. coli clone, the detection of the O25b type was performed with an allele-specific PCR (5).
Molecular epidemiology.
The genetic relationship between the AMC-resistant E. coli isolates was determined by pulsed-field gel electrophoresis (PFGE) after total chromosomal DNA digestion with XbaI (23).
A selected sample of 43 AMC-resistant E. coli isolates, representing the two major PFGE clusters (19 isolates each) and 5 IRT-producing isolates belonging to serotype O25b phylogroup B2 isolates, was studied further by multilocus sequence typing (MLST), according to the University College Cork (Éire) scheme for E. coli (http://mlst.ucc.ie/mlst/dbs/Ecoli; data last accessed on 20 July 2011).
Statistical analyses.
Differences in the prevalence of mechanisms of resistance and phylogroups between different groups were assessed by Fisher's exact test. Association was determined by calculation of the odds ratio (OR) with 95% confidence intervals (CIs). The null hypothesis was rejected for values of P of <0.05. Statistical analyses were performed using GraphPad Prism, version 3.02, software (GraphPad Software, Inc., San Diego, CA).
RESULTS AND DISCUSSION
Bacterial isolates and mechanisms of AMC resistance.
A total of 257 clinical AMC-resistant E. coli isolates were collected; 110 of them (43%) produced nosocomial-acquired infections, and 147 (57%) putatively produced community-acquired infections; 170 of the isolates (65.9%) were from urine, 30 (12%) were from wound, 19 (7.4%) were from blood, and 38 (14.7%) were from other clinical samples.
During the study period, the participant hospitals had a prevalence of AMC resistance in E. coli (MIC ≥ 32/16 μg/ml) of 9.3% (range, 3.3% to 13.5%).
The mechanisms of resistance detected were production of OXA-1 (67 isolates, or 26.1%), hyperproduction of penicillinase (58 isolates, or 22.6%; 53 TEM-1 and 5 SHV-1), production of p-AmpC (50 isolates, or 19.5%), hyperproduction of c-AmpC (47 isolates, or 18.3%), and production of IRTs (45 isolates, or 17.5%). In one AMC-resistant isolate with the ESBL phenotype, blaTEM-12 was detected linked to an IS26 element upstream of blaTEM. In two isolates, no enzymatic mechanism of resistance to AMC was identified. Two different mechanisms of AMC resistance were present in 13 isolates: OXA-1 with p-AmpC (7 isolates, or 2.7%) and OXA-1 with c-AmpC (6 isolates, or 2.3%).
The IRTs identified in this study were TEM-40 (15 isolates, or 33.3%), TEM-30 (13 isolates, or 28.9%), TEM-33 (5 isolates, or 11.1%), TEM-32 (2 isolates, or 4.4%), TEM-34 (2 isolates, or 4.4%), TEM-35 (1 isolate, or 2.2%), TEM-54 (1 isolate, or 2.2%), TEM-76 (1 isolate, or 2.2%), TEM-79 (1 isolate, or 2.2%), and the new TEM-185 (4 isolates, or 8.8%), first described in this study.
Of the 50 p-AmpC β-lactamases detected in our study, 37 (74%) were CMY-2, 11 (22%) were DHA-1, 1 (2%) was CMY-30, and 1 (2%) was CMY-42.
There were some relevant geographical differences in the prevalence of the AMC resistance mechanisms. For instance, the p-AmpC mechanism was more prevalent in the isolates from the Sant Pau Hospital, Catalonia, Spain (45.7%; P = 0.0001) than in isolates from the other hospitals; c-AmpC was also more prevalent in isolates from the Vall d'Hebron Hospital, Catalonia, Spain (37.5%; P = 0.022); OXA-1 was more prevalent in isolates from the Complejo Hospitalario A Coruña, Galicia, Spain (46.5%; P = 0.0019); IRT was more prevalent in isolates from the Gregorio Marañón Hospital, Madrid, Spain (30%; P = 0.038).
Previous studies detected the hyperproduction of penicillinases (12, 24, 28), mainly TEM-1, and AmpC production (16, 19) as the most common mechanisms of resistance to AMC in E. coli. A 2011 study showed that of 50 ampicillin-sulbactam-resistant E. coli isolates from the United States, 96% produced blaTEM-1, 8% produced blaCMY-2, and 8% produced blaOXA-1 (31).
Some information about p-AmpC production in AMC-resistant E. coli has been published previously (16, 19, 24, 28); one study from the United States reported that of 69 E. coli isolates resistant to AMC, 5.8% produced CIT (12).
Production of OXA-1 in E. coli has been sporadically described previously (12, 16, 19, 24). The highest-reported OXA-1 prevalence (15.3%) was observed in isolates from the United States collected between 1990 and 1994 (28); by the end of the last century, this figure was 7.6% (24) and 2.6% (19) in two Spanish hospitals.
The prevalence of IRTs found in this study is among the highest reported in AMC-resistant E. coli isolates (12, 19, 24, 28). However, of 255 E. coli isolates resistant to AMC studied in France in 1996 to 1998, 37.7% produced IRTs (16). Recently, in a single Spanish hospital (17) 11.8% of AMC-resistant E. coli isolates produced IRTs.
Overexpression of blaTEM-1 has been associated with promoters PaPb, P4, and others (10, 14, 30), but in this study this association was not clearly observed since 50.9% of our TEM-1 producers had the most commonly found P3 promoter (Table 1). However, our findings are consistent with a previous report of a 54.5% prevalence of P3/TEM-1 among ampicillin-sulbactam-resistant E. coli isolates (31). In our study, the most prevalent strong promoter was PaPb, as previously described (31) (Table 1). Among our IRTs, 62.2% had promoters other than P3, most of which were PaPb (Table 1). The implications of the promoter variations in AMC resistance require further elucidation. The occurrence of IS26 located at different positions upstream of the blaTEM gene has been recently described (1), but the influence of this insertion in the expression of the blaTEM is unclear so far.
Table 1.
The frequency of promoter regions and insertions and deletions identified in TEM-producing E. coli isolates
| TEM type (n)a | Promoter class (no. of isolates) |
No. of isolates with: |
|||||
|---|---|---|---|---|---|---|---|
| P3 | PaPb | P4 | Pdel | IS26 insertionb | IS911 insertionc | C31→T39 deletion | |
| TEM-1 (53) | 27 | 18 | 2 | 4 | 1 | 1 | |
| TEM-40 (15) | 6 | 6 | 3 | ||||
| TEM-30 (13) | 5 | 6 | 1 | 1 | |||
| TEM-33 (5) | 2 | 3 | |||||
| TEM-185 (4) | 3 | 1 | |||||
| TEM-32 (2) | 2 | ||||||
| TEM- 34 (2) | 2 | ||||||
| TEM-35 (1) | 1 | ||||||
| TEM-76 (1) | 1 | ||||||
| TEM-79 (1) | 1 | ||||||
n, number of isolates.
The IS26 element was detected upstream of blaTEM, inserted at promoter position 150 (n = 3) or 46 (n = 2) according to the Sutcliffe numbering system (29).
The IS911 element was detected upstream of blaTEM-1, inserted at promoter position 46.
A total of 37 isolates (14.4%) produced ESBLs: 28 (75.7%) produced CTX-M-15, 7 (18.9%) produced CTX-M-14, 1 (2.7%) produced SHV-12, and 1 (2.7%) produced TEM-12. All but one of them had an additional AMC resistance mechanism, mainly OXA-1 (67.6%), p-AmpC (13.5%), c-AmpC (13.51%), and IRTs (5.4%). Complex mutant TEM β-lactamases were not detected.
In relation to the AMC resistance mechanisms, no significant differences were found between community and nosocomial isolates except for blaTEM-1 promoters other than P3, which were more frequent in nosocomial AMC-resistant E. coli (P < 0.05).
Table 2 shows the distribution of MIC50s and MIC90s, MIC ranges, and the percentages of isolates with MICs of >32/16 μg/ml according to the molecular mechanisms of resistance to AMC. OXA-1-producing isolates had AMC MICs lower than isolates with other AMC resistance mechanisms (P < 0.03). Resistance to non-β-lactam antibiotics in relation to the mechanism of AMC resistance is detailed in Table 3. OXA-1-producing isolates were more resistant to ciprofloxacin, cotrimoxazole, and aminoglycosides than IRT, AmpC, or TEM-1 producers (P < 0.001).
Table 2.
MIC values and ranges by resistance mechanism of AMC-resistant E. coli isolates
| Resistance mechanism (n)a | MIC50 (μg/ml) | MIC90 (μg/ml) | MIC range (μg/ml) | % of isolates with a MIC > 32/16 μg/ml |
|---|---|---|---|---|
| OXA-1 (54) | 32/16 | 64/32 | 16/8–128/64 | 40.3 |
| IRT (45) | 64/32 | 128/64 | 16/8–>128/64 | 93.3 |
| p-AmpC (43) | 64/32 | 128/64 | 32/16–>128/64 | 92 |
| c-AmpC (41) | 64/32 | 128/64 | 32/16–128/64 | 83 |
| P3/TEM-1 (27) | 64/32 | 64/32 | 16/8–>128/64 | 66.7 |
| Pdf3/TEM-1 (26)b | 64/32 | >128/64 | 16/8–>128/64 | 84.6 |
| OXA-1+AmpC (13) | 64/32 | 64/32 | 32/16–64/32 | 53.8 |
n, number of isolates.
Isolates with blaTEM-1 and with promoters other than P3.
Table 3.
Resistance to non-β-lactam antibiotics in relation to the mechanism of resistance to AMC in E. coli
| Resistance mechanism or source (n)a | No. of isolates (%) with resistance to: |
||||
|---|---|---|---|---|---|
| Ciprofloxacin | Gentamicin | Tobramycin | Amikacin | Cotrimoxazole | |
| TEM-1 (53) | 27 (51.8) | 7 (13.2) | 10 (20.4) | 0 | 30 (56.6) |
| IRTs (45) | 17 (37.8) | 4 (8.9) | 4 (8.9) | 0 | 24 (53.3) |
| OXA-1 (67) | 57 (85.1) | 34 (50.7) | 55 (82.1) | 12 (17.9) | 55 (82.1) |
| AmpC (97) | 51 (52.6) | 17 (17.5) | 23 (23.7) | 1 (1) | 44 (45.4) |
| Total (257) | 149 (58) | 61 (23.7) | 86 (33.5) | 12 (4.7) | 149 (58) |
n, number of isolates.
Phylogenetic groups and detection of the O25b type.
Of the 257 AMC-resistant E. coli isolates, 76 (29.6%) belonged to phylogenetic group A, 32 (12.4%) belonged to group B1, 104 (40.5%) belonged to group B2, and 45 (17.5%) belonged to group D.
Phylogroup A was more frequent in OXA-1 producers (36, or 53.7%; P < 0.0003); phylogroup B2 was more frequent in IRT producers (24, or 53.3%; P = 0.06), p-AmpC producers (21, or 42%; P = 0.87), and TEM-1 producers carrying promoters other than P3 (16, or 61.6%; P < 0.03); and phylogroup D was more frequent in TEM-1 producers with P3 promoters (13, or 48.1%; P < 0.0001).
Of the 104 phylogroup B2 isolates, 56 (21.8% of all isolates) were serotype O25b, which was detected in all seven participant hospitals. Of the O25b/B2 isolates, 22 (39.3%) were OXA-1 producers, 15 (26.8%) were TEM-1 producers with different promoters, 11 (19.6%) were p-AmpC producers, 8 (14.3%) were c-AmpC producers, and 5 (8.9%) were IRT producers. Five of these isolates produced both OXA-1 and AmpC β-lactamases.
Molecular epidemiology.
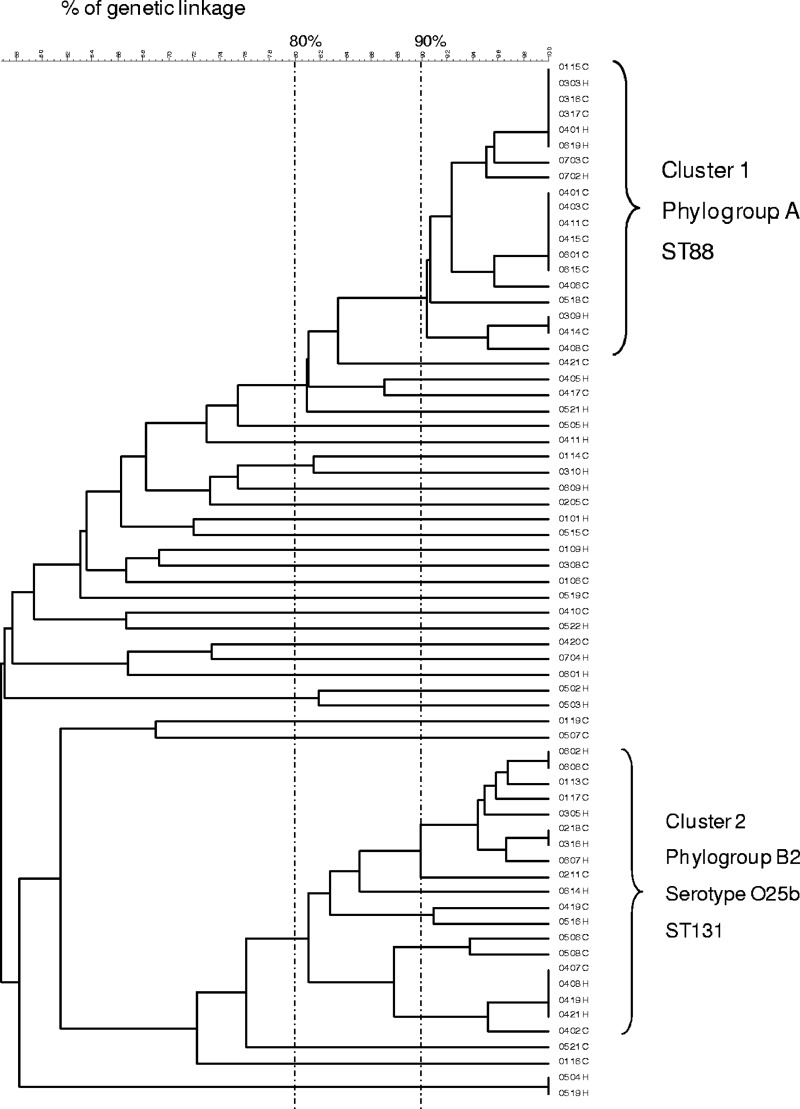
A high degree of genetic diversity was observed according to PFGE as 235 different PFGE patterns were obtained from the 257 AMC-resistant E. coli isolates analyzed. However, two well-defined clusters were detected: cluster C1 (genetic linkage of >90%) consisting of 19 OXA-1-producing isolates of phylogroup A, detected in six of the seven participant hospitals, and cluster C2 (genetic linkage of >80%) consisting of 19 AMC-resistant isolates of phylogroup B2 and serotype O25b, detected in six of the seven participant hospitals. Of these 19 C2 isolates, 12 produced OXA-1 and CTX-M-15, 3 produced OXA-1 only, 3 produced CTX-M-15 and c-AmpC, and 1 produced CTX-M-15 and DHA. Figure 1 shows the PFGE profiles of the 67 OXA-1-producing isolates.
Fig 1.
Dendrogram illustrating the percent linkage of PFGE profiles of 67 OXA-1-producing E. coli isolates resistant to AMC. Isolate numbers are shown to the right of the dendrogram.
By MLST, the PFGE C1 cluster was identified as ST88, and the PFGE C2 cluster was identified as ST131.
Dissemination of OXA-1 β-lactamase in AMC-resistant E. coli isolates in Spain is due in part to the clonal spread of the epidemic ST131 clone producing CTX-M-15 and OXA-1 and to the spread of the ST88 phylogroup A clone producing only OXA-1, an association first described here. ST88 has been previously described in association with c-AmpC production in a French hospital (8). On the website of the University College of Cork (Ireland), 23 ST88 E. coli isolates are registered that were recovered from infections of humans and domestic animals (http://mlst.ucc.ie/mlst/dbs/Ecoli; data last accessed on 20 July 2011).
In 24 O25b/B2 isolates, including 19 C2 isolates and 5 isolates producing IRTs, MLST was performed, and results showed that all of them were ST131. We found that O25b/B2/ST131 E. coli isolates carried not only CTX-M-15 and OXA-1 enzymes as previously reported but also TEM-30, TEM-34, TEM-40, and TEM-54 IRT enzymes. To the best of our knowledge, this is the first description of IRT-producing isolates belonging to the ST131 international clone.
TEM-185 characterization.
The new IRT TEM-185 has a double amino acid substitution at positions 69 (M→L) and 165 (W→L) in comparison with TEM-1 (GenBank accession number 1446016); these positions are frequently modified in IRT enzymes. TEM-185 is similar to TEM-39 except that the latter has an additional substitution at position 276 (N→D) (http://www.lahey.org/studies/temtable.asp; data last accessed on 2 August 2011).
TEM-185 was detected in four E. coli isolates belonging to phylogroup A; they were isolated in two geographically distant Spanish hospitals with two isolates each. Two of them were closely related (genetic similarity of >85% and same P4 promoter), and the two additional isolates were genetically unrelated and had two different promoters, P4 and Pdel. These findings suggest that several clones can spread TEM-185.
Concluding remarks.
Our findings suggest a complex epidemiological background in which E. coli acquires AMC resistance by several mechanisms, including clonal (ST131 and ST88) and nonclonal spread, dissemination of mobile genetic elements carrying different bla genes, and eventual mutations in individual isolates as a response to selective antimicrobial pressure.
ACKNOWLEDGMENTS
This study was supported by a research grant from Fondo de Investigaciones Sanitarias (FIS PI09/917) and by the Spanish Network for the Research in Infectious Diseases (REIPI C03/14 and RD06/0008).
Footnotes
Published ahead of print 9 April 2012
REFERENCES
- 1. Bailey JK, Pinyon JL, Anantham S, Hall RM. 2011. Distribution of the blaTEM gene and blaTEM-containing transposons in commensal Escherichia coli. J. Antimicrob. Chemother. 66:745–751 [DOI] [PubMed] [Google Scholar]
- 2. Biendo M, et al. 2005. Molecular characterisation and mechanisms of resistance of multidrug-resistant human Salmonella enterica serovar Typhimurium isolated in Amiens (France). Int. J. Antimicrob. Agents 26:219–229 [DOI] [PubMed] [Google Scholar]
- 3. Campos J, et al. 2007. Surveillance of outpatient antibiotic consumption in Spain according to sales data and reimbursement data. J. Antimicrob. Chemother. 60:698–701 [DOI] [PubMed] [Google Scholar]
- 4. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clermont O, et al. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent phylogenetic subgroup. J. Antimicrob. Chemother. 61:1024–1028 [DOI] [PubMed] [Google Scholar]
- 6. Clinical and Laboratory Standards Institute 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 9th ed. Approved standard M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7. Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement. CLSI M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Crémet L, et al. 2010. Occurrence of ST23 complex phylogroup A Escherichia isolates producing extended-spectrum AmpC beta-lactamase in a French hospital. Antimicrob. Agents Chemother. 54:2216–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fluit AC, et al. 2000. Antimicrobial susceptibility and frequency of occurrence of clinical blood isolates in Europe from the SENTRY antimicrobial surveillance program, 1997–1998. Clin. Infect. Dis. 30:454–460 [DOI] [PubMed] [Google Scholar]
- 10. García-Cobos S, et al. 2008. Antibiotic resistance in Haemophilus influenzae decreased, except for beta-lactamase-negative amoxicillin-resistant isolates, in parallel with community antibiotic consumption in Spain from 1997 to 2007. Antimicrob. Agents Chemother. 52:2760–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goossens H, Ferech M, Vander Stichele R, Elseviers M, Project Group ESAC 2005. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet 365:579–587 [DOI] [PubMed] [Google Scholar]
- 12. Kaye KS, et al. 2004. Variety of β-lactamases produced by amoxicillin-clavulanate-resistant Escherichia coli isolate in the northeastern United States. Antimicrob. Agents Chemother. 48:1520–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lark RL, et al. 2001. Four-year prospective evaluation of community-acquired bacteremia: epidemiology, microbiology and patient outcome. Diagn. Microbiol. Infect. Dis. 41:15–22 [DOI] [PubMed] [Google Scholar]
- 14. Lartigue MF, Leflon-Guibout V, Poirel L, Nordmann P, Nicolas-Chanoine MH. 2002. Promoters P3, Pa/Pb, P4 and P5 upstream from blaTEM genes and their relationship to β-lactam resistance. Antimicrob. Agents Chemother. 46:4035–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lázaro BE, Madurga SM, de Abajo FJ. 2002. Evolución del consumo de antibióticos en España, 1985–2000. Med. Clin. (Barc.) 118:561–568 [DOI] [PubMed] [Google Scholar]
- 16. Leflon-Guibout V, Speldooren V, Heym B, Nicolas-Chanoine MH. 2000. Epidemiological survey of amoxicillin-clavulanate resistance and corresponding molecular mechanisms in Escherichia coli isolates in France: new genetic features of blaTEM genes. Antimicrob. Agents Chemother. 44:2709–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martin O, et al. 2010. Population analysis and epidemiological features of inhibitor-resistant-TEM-β-lactamase-producing Escherichia coli isolates from both community and hospital settings in Madrid, Spain. J. Clin. Microbiol. 48:2368–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miró E, et al. 1998. Emergence of clinical Escherichia coli isolates with decreased susceptibility to ceftazidime and synergic effect with co-amoxiclav due to SHV-1 hyperproduction. J. Antimicrob. Chemother. 42:535–538 [DOI] [PubMed] [Google Scholar]
- 19. Miró E, et al. 2002. Prevalence of clinical isolates of Escherichia coli producing inhibitor-resistant β-lactamases at a university hospital in Barcelona, Spain, over a 3-year period. Antimicrob. Agents Chemother. 46:3991–3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oteo J, et al. 2008. Increased amoxicillin-clavulanic acid resistance in Escherichia coli blood isolates, Spain. Emerg. Infect. Dis. 14:1259–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oteo J, et al. 2010. AmpC beta-lactamases in Escherichia coli: emergence of CMY-2-producing virulent phylogroup D isolates belonging mainly to STs 57, 115, 354, 393, and 420, and phylogroup B2 isolates belonging to the international clone O25b-ST131. Diagn. Microbiol. Infect. Dis. 67:270–276 [DOI] [PubMed] [Google Scholar]
- 22. Oteo J, et al. 2005. Antimicrobial-resistant invasive Escherichia coli, Spain. Emerg. Infect. Dis. 11:546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oteo J, et al. 2006. Spread of Escherichia coli strains with high-level cefotaxime and ceftazidime resistance between the community, long-term care facilities, and hospital institutions. J. Clin. Microbiol. 44:2359–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pérez-Moreno MO, et al. 2004. Mechanisms of reduced susceptibility to amoxycillin-clavulanic acid in Escherichia coli strains from the region of Tortosa (Catalonia, Spain). Clin. Microbiol. Infect. 10:234–241 [DOI] [PubMed] [Google Scholar]
- 25. Pérez-Pérez FJ, Hanson ND. 2002. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robin F, et al. 2007. Evolution of TEM-type enzymes: biochemical and genetic characterization of two new complex mutant TEM enzymes, TEM-151 and TEM-152, from a single patient. Antimicrob. Agents Chemother. 51:1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sirot D, et al. 1994. Clinical isolates of Escherichia coli producing multiple TEM mutants resistant to β-lactamase inhibitors. J. Antimicrob. Chemother. 33:1117–1126 [DOI] [PubMed] [Google Scholar]
- 28. Stapleton P, et al. 1995. Incidence and mechanisms of resistance to the combination of amoxicillin and clavulanic acid in Escherichia coli. Antimicrob. Agents Chemother. 39:2478–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sutcliffe JG. 1978. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc. Natl. Acad. Sci. U. S. A. 75:3737–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tristram SG, Nichols S. 2006. A multiplex PCR for β-lactamase genes of Haemophilus influenzae and description of a new blaTEM promoter variant. J. Antimicrob. Chemother. 58:183–185 [DOI] [PubMed] [Google Scholar]
- 31. Waltner-Toews RI, et al. 2011. Clinical characteristics of bloodstream infections due to ampicillin-sulbactam-resistant, non-extended-spectrum-beta-lactamase-producing Escherichia coli and the role of TEM-1 hyperproduction. Antimicrob. Agents Chemother. 55:495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou XY, Bordon F, Sirot D, Kitzis MD, Gutmann L. 1994. Emergence of clinical isolates of Escherichia coli producing TEM-1 derivatives or an OXA-1 β-lactamase conferring resistance to β-lactamase inhibitors. Antimicrob. Agents Chemother. 38:1085–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]