Abstract
Carbapenem-resistant bacteria represent a significant treatment challenge due to the lack of active antimicrobials available. MK-7655 is a novel β-lactamase inhibitor under clinical development. We investigated the combined killing activity of imipenem and MK-7655 against four imipenem-resistant bacterial strains, using a mathematical model previously evaluated in our laboratory. Time-kill studies (TKS) were conducted with imipenem and MK-7655 against a KPC-2-producing Klebsiella pneumoniae isolate (KP6339) as well as 3 Pseudomonas aeruginosa isolates (PA24226, PA24227, and PA24228) with OprD porin deletions and overexpression of AmpC. TKS were performed using 25 clinically achievable concentration combinations in a 5-by-5 array. Bacterial burden at 24 h was determined in triplicate by quantitative culture and mathematically modeled using a three-dimensional response surface. Mathematical model assessments were evaluated experimentally using clinically relevant dosing regimens of imipenem, with or without MK-7655, in a hollow-fiber infection model (HFIM). The combination of imipenem and MK-7655 was synergistic for all strains. Interaction indices were as follows: for KP6339, 0.50 (95% confidence interval [CI], 0.42 to 0.58); for PA24226, 0.60 (95% CI, 0.58 to 0.62); for PA24227, 0.70 (95% CI, 0.66 to 0.74); and for PA24228, 0.55 (95% CI, 0.49 to 0.61). In the HFIM, imipenem plus MK-7655 considerably reduced the bacterial burden at 24 h, while failure with imipenem alone was seen against all isolates. Sustained suppression of bacterial growth at 72 h was achieved with simulated doses of 500 mg imipenem plus 500 mg MK-7655 in 2 (KP6339 and PA24227) strains, and it was achieved in an additional strain (PA24228) when the imipenem dose was increased to 1,000 mg. Additional studies are being conducted to determine the optimal dose and combinations to be used in clinical investigations.
INTRODUCTION
Carbapenem-resistant bacteria represent a significant challenge to clinicians, since carbapenems are often used as the last line of therapy for resistant Gram-negative bacteria. In the last decade, the prevalence of resistance to β-lactam antibiotics mediated by β-lactamases has increased at an alarming rate, especially among the Enterobacteriaceae (1). Specifically, β-lactamases able to hydrolyze the carbapenems, including class A carbapenemases (KPCs) and class B metallo-β-lactamases (VIM-like), have spread rapidly throughout the world (11). As a result of these newer β-lactamases with broad substrate specificity, resistance to virtually all β-lactams can result. The current commercially available β-lactamase inhibitors are not active against these new and emerging enzymes (7).
Aside from carbapenemases seen more commonly in Enterobacteriaceae, carbapenem resistance in Pseudomonas aeruginosa often results from the interplay of several other resistance mechanisms (14). These may include efflux pumps, loss of outer membrane porin proteins (OprD), and overexpression of chromosomal AmpC cephalosporinase. It has been shown that P. aeruginosa isolates with loss of OprD typically require hyperproduction of AmpC in order to achieve complete resistance to imipenem (10, 14). It has also been shown that hyperproduction of AmpC is associated with less favorable clinical outcomes (15). A large survey of carbapenem-resistant P. aeruginosa isolates collected in the United States recently demonstrated that only about 10% (45/452 isolates) of isolates expressed detectable levels of OprD (4). In the same study, 69% (312/452 isolates) of isolates had wild-type levels of AmpC; 15% were characterized as having highly elevated (≥3× increased activity compared to wild-type activity) AmpC production, while 10% had levels characterized as low elevated activity (>1× but <3× increased activity). These data emphasized the need for alternative treatments for infections with carbapenem-resistant isolates.
In view of the multiple and emerging resistance mechanisms mediated by Gram-negative bacteria, carbapenems may no longer be reliable treatment options. Specifically, for isolates resistant to carbapenems, new treatment strategies are needed. One potential option to restore the activity of carbapenems may be combining them with new agents capable of inhibiting a broad spectrum of β-lactamases. MK-7655 is a novel β-lactamase inhibitor under clinical development; in vitro studies have demonstrated its potent inhibition of class A and class C β-lactamases (19). To provide rational guidance on dosing design in clinical studies, we investigated the in vitro combined killing activity of imipenem and MK-7655 against four imipenem-resistant Gram-negative isolates.
(Portions of this study were presented at the Interscience Conference on Antimicrobial Agents and Chemotherapy, Boston, MA, 12 to 15 September 2010, and the European Congress of Clinical Microbiology and Infectious Diseases, Milan, Italy, 7 to 10 May 2011.)
MATERIALS AND METHODS
Antimicrobial agents.
Imipenem and MK-7655 powders were supplied by Merck & Co., Inc. (Rahway, NJ). Prior to MIC determination and time-kill studies, stock solutions of each agent were prepared in sterile water, stored at −70°C, and diluted to the appropriate concentrations prior to use. For in vitro hollow-fiber infection models (HFIM), fresh solutions of each agent were made to the appropriate concentrations needed, frozen at −70°C, and thawed immediately prior to administration.
Microorganisms.
Isolates were supplied by Merck & Co., Inc. (Rahway, NJ). Four geographically diverse, imipenem-resistant clinical isolates were used in the study. A Klebsiella pneumoniae isolate (KP6339; also known as CL6339) obtained from Florida and harboring the KPC-2 carbapenemase and TEM- and SHV-like extended-spectrum β-lactamases was used (13). Outer membrane protein analysis determined that this isolate lacked the OmpK35 porin and had reduced expression of OmpK36. The three Pseudomonas aeruginosa isolates (PA24226 [also known as CLB24226; from Colorado], PA24227 [also known as CLB24227; from Oklahoma], and PA24228 [also known as CLB24228; from New Jersey]) used in the study all overexpressed AmpC and lacked the OprD porin (unpublished data on file; Merck). The bacteria were stored in Protect storage vials (Key Scientific Products, Round Rock, TX) at −70°C and were subcultured twice on 5% blood agar plates (Hardy Diagnostics, Santa Maria, CA) for 24 h at 35°C prior to each experiment.
Susceptibility studies.
Imipenem MICs (alone and in the presence of 4 mg/liter MK-7655) were determined by a modified broth macrodilution method as described by the CLSI (2). The final concentration of bacteria in each macrodilution tube was approximately 5 × 105 CFU/ml of cation-adjusted Mueller-Hinton broth (Ca-MHB) (BBL, Sparks, MD). Serial 2-fold dilutions of the drug were used. The MIC was defined as the lowest concentration of drug that resulted in no visible growth after 24 h of incubation at 35°C in ambient air. The MIC determinations were conducted in triplicate and were repeated at least once on a separate day.
Time-kill studies and mathematical modeling.
Time-kill studies were conducted in triplicate, using imipenem and MK-7655 alone at different and escalating concentrations. The bacterial inoculum was prepared by inoculating a flask containing prewarmed Ca-MHB and placing it in a shaker water bath at 35°C until the culture reached log-phase growth. This bacterial suspension was then diluted with Ca-MHB according to the absorbance at 630 nm. Fifteen milliliters of the resulting suspension was transferred to a 50-ml sterile conical flask containing 1 ml of a drug dilution at 16 times the target concentration. The final concentration of the bacterial suspension in each flask was approximately 1 × 105 to 5 × 105 CFU/ml. Twenty-four-hour time-kill studies were conducted in a shaker water bath set at 35°C. Total bacterial populations after 24 h of drug exposure were quantified by spiral plating 10× serial dilutions of the samples onto Mueller-Hinton agar plates (Spiral Biotech, Bethesda, MD). The medium plates were incubated in a humidified incubator (35°C) for 18 to 24 h, and the bacterial density in each sample was determined by visual inspection. The mean killing effect at 24 h was characterized by an inhibitory sigmoid maximum effect (Emax) model, using the ADAPT II program (3). The time-kill studies were repeated using 25 concentration combinations in a 5-by-5 array as previously described (9). The concentration ranges examined were constrained to those clinically achievable in human serum (up to 40 mg/liter for both imipenem [5, 12] and MK-7655 [unpublished data on file]); specific concentrations used were based on the best-fit parameter estimates and optimal sampling for the most precise characterization of bacterial killing. The total bacterial burden at 24 h (in triplicate) was mathematically modeled using a three-dimensional response surface as described in detail elsewhere (Mathematica 5.2; Wolfram Research, Inc., Champaign, IL) (18). Volumes under the plane (VUP) of the observed (VUPobserved) and expected (VUPexpected) surfaces were computed by interpolation and double integration, respectively. Ninety-five percent confidence intervals (95% CIs) for VUPobserved were computed with mean data points ± 1.96 standard deviations (SD). Synergy and antagonism were defined by interaction indices (VUPobserved/VUPpredicted) of <1 and >1, respectively.
In vitro HFIM.
In order to evaluate the mathematical model assessments, an HFIM in which the bacteria were exposed to clinically relevant and fluctuating drug concentrations was used. The schematics of this model have been described in detail elsewhere (9, 16). Briefly, the drug(s) was injected directly into the central reservoir to reach clinically achievable peak concentrations. Twenty milliliters of the bacterial suspension, at approximately 1 × 105 to 5 × 105 CFU/ml, prepared as described above, was inoculated into and confined in the extracapillary compartment of the hollow-fiber cartridge (Fibercell Systems, Inc., Frederick, MD). The bacteria were exposed to the fluctuating drug concentrations in the central reservoir by means of an internal circulatory pump in the bioreactor loop. The experiments were conducted for 72 h in a humidified incubator set at 35°C. For imipenem, 30-minute infusions simulating either 500 (low-dose)- or 1,000 (high-dose)-mg doses every 6 h were used. For MK-7655, a dose of 500 mg (given over 30 min) every 6 h was simulated. The steady-state pharmacokinetic profiles of unbound drugs corresponding to these clinical doses were as follows: for imipenem, the maximum concentration of the unbound fraction of the drug (fCmax) was 40 mg/liter (500 mg) or 80 mg/liter (1,000 mg), and the half-life (t1/2) was 1.5 h (5, 12); and for MK-7655, the fCmax was 20 mg/liter and t1/2 was 1.5 h (data on file). For comparison, imipenem alone was used as a control. The effectiveness of the combinations was assessed by the observed bacterial burden over time.
Pharmacokinetic validation and drug assay.
Serial samples were obtained from the infection models to ascertain the simulated pharmacokinetic profiles. Upon collection, the samples were immediately diluted 1:1:1 with a stabilizing solution consisting of 1:1 ethylene glycol–N-morpholineethanesulfonic acid buffer (pH 6.0) and 50% acetonitrile. The stabilized samples were stored at −70°C until analysis. Imipenem and MK-7655 concentrations were assayed simultaneously using a validated high-performance liquid chromatography–tandem mass spectrometry (HPLC-MS/MS) methodology within 2 months. The mass spectrometer (Sciex API 4000) was equipped with a turbo-ion-spray interface operated in the positive ionization mode. MK-7655 and imipenem were detected in multiple reaction monitor mode (MRM), using precursor-to-product-ion transitions of m/z 349.2 to 269.0 and m/z 300.2 to 97.9, respectively. The chromatographic column (Waters Atlantis HILIC column; 2.1 × 50 mm) was operated with a mobile phase composed of 5 mM ammonium acetate, pH 4.5, in acetonitrile-water (80/20 [vol/vol]). Using 50 μl of stabilized broth, the lower limit of quantification (LLOQ) was 0.325 mg/liter, with a calibration range of 0.325 to 150 mg/liter. Intraday precision and accuracy (n = 6) showed coefficients of variation (CV) of 98.0 to 102% and 1.6 to 4.1%, respectively. Interday precision and accuracy (n = 12) showed CV of 94.4 to 99.9% and 3.2 to 4.5%, respectively. Following the drug assays, the concentration-time profiles were modeled by fitting a one-compartment linear model to the observations, using ADAPT II.
Microbiologic response.
Serial samples were also obtained from each infection model at 0 (baseline), 6, 12, 24, 48, and 72 h (predose when applicable). Bacterial burdens were determined in duplicate by quantitative culture to examine the effects of drug exposure(s) on the total bacterial population over time. Colonies from any isolate displaying regrowth in high-dose models were chosen at random. Imipenem susceptibility testing (with and without MK-7655) was repeated to provide insight into the mechanism(s) of regrowth.
RESULTS
Susceptibility.
All isolates were resistant to imipenem, as shown in Table 1. As anticipated, MICs for each isolate were considerably reduced in the presence of MK-7655 (4 mg/liter).
Table 1.
Susceptibility testing results and assessment of combined killing activity of imipenem (IPM) plus MK-7655 (MK)a
| Isolate | MIC (mg/liter) |
Interactive index (95% CI) | |
|---|---|---|---|
| IPM | IPM with MK (4 mg/liter) | ||
| KP6339 | 128 | 2 | 0.50 (0.42–0.58) |
| PA24226 | 32 | 2 | 0.60 (0.58–0.62) |
| PA24227 | 16 | 2 | 0.70 (0.66–0.74) |
| PA24228 | 32 | 4 | 0.55 (0.49–0.61) |
In all cases, the interaction of IPM with MK was interpreted as synergistic.
Time-kill studies.
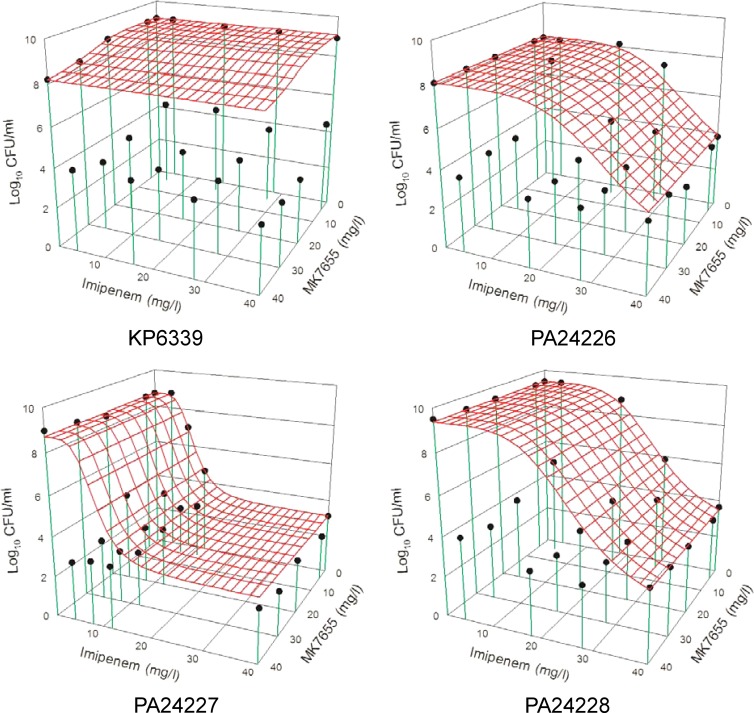
The model fits of all single-drug time-kill studies were satisfactory (r2 > 0.90) (data not shown). In combination time-kill studies, an enhanced or synergistic killing effect was seen against all tested isolates (Fig. 1). The quantitative assessment of the combined killing activity against each isolate is shown in Table 1.
Fig 1.
Comparison of expected and observed killing activities of imipenem plus MK-7655 for each tested isolate. The red mesh surface shows the expected killing activity of the combination, while the black dots show the observed killing. When a black dot is below the mesh, observed killing is more than expected killing (synergism). Conversely, a black dot above the mesh signifies antagonism (observed killing is less than expected killing). Synergism and antagonism are defined by interactive indices (VUPobserved/VUPexpected) of <1 and >1, respectively.
In vitro hollow-fiber infection model studies.
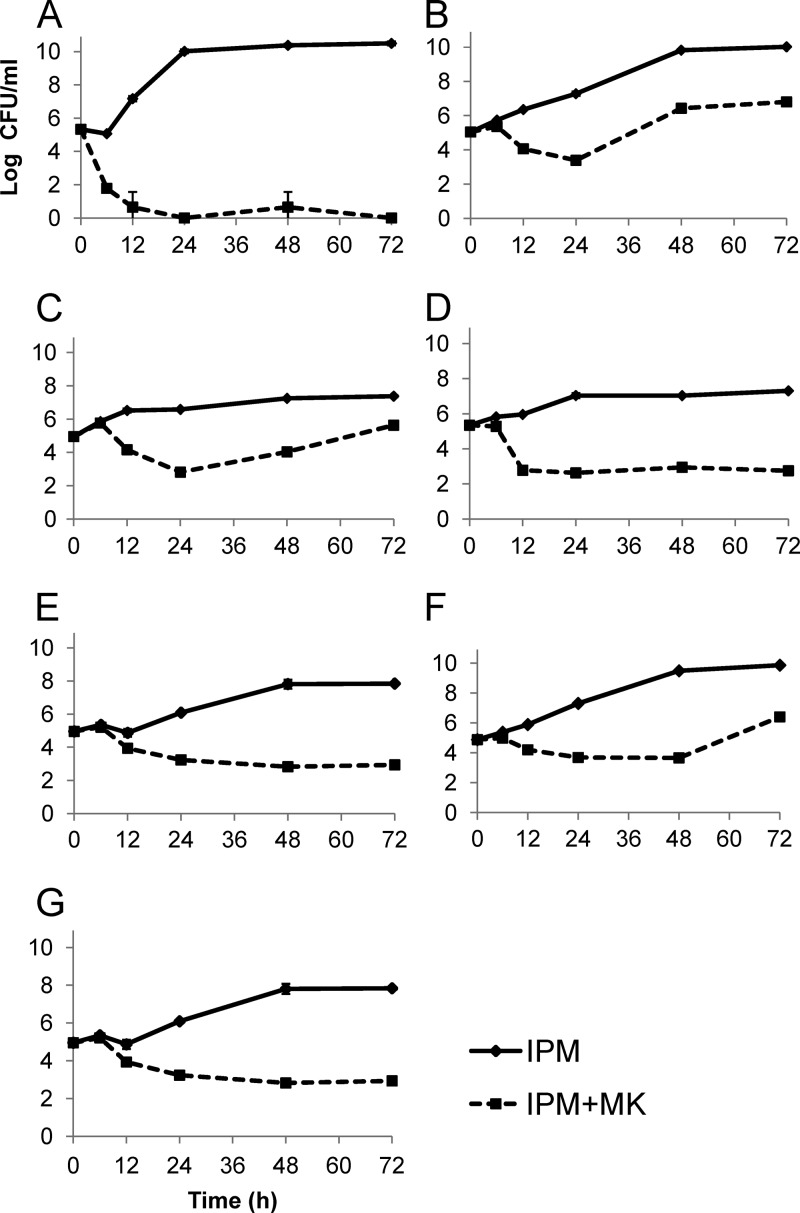
Simulated drug exposures in the HFIM were mostly satisfactory. Against KP6339, imipenem concentrations (in the monotherapy model) could not be detected beyond 6 h, most likely due to hydrolysis of imipenem by KPC. Figure 2 demonstrates the microbiologic responses observed in the infection models. These results are in overall agreement with our assessment of combined killing activity (Table 1). All models demonstrated a significant reduction of bacterial burden at 24 h for the combination of imipenem and MK-7655, while imipenem monotherapy regimens consistently failed to suppress bacterial growth. With the exception of PA24226, effective and sustained suppression of bacterial growth for up to 72 h was also seen with the combination regimen. Against KP6339, bacterial suppression was achieved by 12 h and sustained through 72 h for low-dose combination therapy; high-dose therapy was not investigated further. Against PA24227, sustained suppression of bacterial growth was achieved for 72 h for both low- and high-dose regimens. Against PA24228, bacterial regrowth was seen at 72 h in the low-dose model. In contrast, sustained suppression of bacterial growth was seen over 72 h in the high-dose model.
Fig 2.
Observed microbiologic responses in infection models for the following isolates and drug exposures: KP6339 (A), PA24226 with low-dose exposure(s) (B), PA24226 with high-dose exposure(s) (C), PA24227 with low-dose exposure(s) (D), PA24227 with high-dose exposure(s) (E), PA24228 with low-dose exposure(s) (F), and PA24228 with high-dose exposure(s) (G). Data shown are mean values ± standard deviations. IPM, imipenem; MK, MK-7655.
Results from HFIM models against PA24226 were somewhat unexpected. In both low-dose and high-dose models, bacterial regrowth was seen at 48 h (Fig. 2B and C). The imipenem MIC of the parent strain was unchanged (32 mg/liter), and post-HFIM susceptibility testing of 3 randomly selected colonies from the high-dose model did not reveal a significant elevation in imipenem MIC values (Table 2). Note that the imipenem MICs for the 3 daughter isolates were not reduced as effectively by MK-7655 as those for the parent isolate.
Table 2.
Post-HFIM susceptibility testing results for PA24226 and three random isolates after regrowth was observed
| Isolate | MIC (mg/liter)a |
||||
|---|---|---|---|---|---|
| IPM | IPM with MK (4 mg/liter) | IPM with MK (8 mg/liter) | IPM with MK (16 mg/liter) | IPM with MK (32 mg/liter) | |
| Parent | 32 | 1 | 1 | 0.5 | 0.5 |
| Daughter A | 64 | 4 | 2 | 2 | 2 |
| Daughter B | 64 | 4 | 2 | 2 | 2 |
| Daughter C | 64 | 4 | 2 | 2 | 2 |
IPM, imipenem; MK, MK-7655.
DISCUSSION
Increasing rates of resistance among Gram-negative bacterial species, particularly against carbapenems, continue to limit effective therapeutic options with the current armamentarium of antibiotics (6). Furthermore, currently available clinical data suggest that multidrug-resistant infections may be associated with less favorable outcomes (8, 15, 17). One of the primary mechanisms leading to carbapenem resistance in Gram-negative species is the production of β-lactamases, particularly Klebsiella pneumoniae carbapenemases in Enterobacteriaceae and AmpC in P. aeruginosa (4). The lack of antimicrobials effective against bacteria producing these enzymes is a major challenge to clinicians, due (at least in part) to the use of less effective and/or more toxic second-line antimicrobials (8). The use of a β-lactamase inhibitor able to inhibit emerging β-lactamases in combination with a carbapenem, which could restore the activity of the carbapenem, would be a very promising therapeutic option.
In this study, the imipenem MICs for four imipenem-resistant isolates were considerably reduced in the presence of MK-7655 at 4 mg/liter, suggesting potent efficacy of the combination. This effect was most pronounced for the KPC-producing K. pneumoniae strain (KP6339), resulting in a 64-fold imipenem MIC reduction, from 128 mg/liter to 2 mg/liter. In combination time-kill studies, the combination of imipenem and MK-7655 was consistently found to be synergistic, with interaction indices ranging from 0.5 to 0.7 for each of the 4 isolates used in the study.
HFIM studies simulating fluctuating drug concentrations were used to evaluate the mathematical models over an extended period. Generally speaking, it was more difficult to suppress P. aeruginosa to below the limit of reliable detection, likely due to more complex resistance mechanisms. In 24 h or less, imipenem alone consistently failed to control the bacterial population, while a significant reduction was seen with the combination of imipenem and MK-7655. At least a 2-log reduction in bacterial burden was observed consistently at 24 h, which was highly consistent with the synergistic activity predicted by the mathematical models. This effect was sustained for 72 h against KP6339 and PA24227 with simulated low-dose (500 mg every 6 h) imipenem. Despite the regrowth seen with PA24228 and low-dose imipenem, the bacterial burden remained controlled for 72 h when the imipenem dose was increased to 1,000 mg every 6 h.
Conversely, the results against PA24226 in HFIM experiments were somewhat unanticipated. In both low-dose and high-dose models, regrowth was seen by 48 h. The explanation for this result is not entirely known. Repeat MIC testing of 3 randomly chosen colonies from the post-HFIM (72 h) plates did not demonstrate a significant elevation in imipenem MIC compared to that for the parent isolate. It is unclear whether there is a true difference between the MICs for the parent strain and the daughter isolates if an error of one doubling dilution is considered acceptable. However, MK-7655 concentration-dependent reductions in imipenem MIC appeared to be less dramatic in the isolates with regrowth. The baseline (noninduced) nitrocefin hydrolysis rate of PA24226 was higher than those for both PA24227 (16-fold) and PA24228 (>50-fold) (data not shown). We presume that this may have contributed (at least in part) to the less effective killing against PA24226 than that against PA24227 and PA24228. The effect of supraphysiological exposures of MK-7655 was not investigated due to the lack of perceived clinical relevance. Other resistance mechanisms (e.g., efflux pump overexpression) which could explain the inability of the combination to control the bacterial population are currently being explored.
In this set of HFIM experiments, one of the main objectives was to evaluate the clinical potential of MK-7655 under realistic and fluctuating conditions as a means to guide whether clinical development should proceed. The inoculum (1 × 105 to 5 × 105 CFU/ml) chosen was typical for susceptibility testing and represented the density range likely to be encountered in less severe infections, such as urinary tract infections. In the future, it would be pertinent to study higher-density inocula that are relevant to more severe infections, such as ventilator-associated pneumonia.
In summary, the results of our in vitro studies demonstrate the combination of imipenem and MK-7655 to be synergistic and to display effective killing against a variety of AmpC- and KPC-producing isolates. These in vitro data warrant additional in vivo studies. Currently, additional studies are ongoing to determine the optimal dose and combinations to be used in clinical investigations.
ACKNOWLEDGMENT
This work was supported by an unrestricted grant from Merck & Co., Inc.
Footnotes
Published ahead of print 23 April 2012
REFERENCES
- 1. Castanheira M, Mendes RE, Woosley LN, Jones RN. 2011. Trends in carbapenemase-producing Escherichia coli and Klebsiella spp. from Europe and the Americas: report from the SENTRY antimicrobial surveillance programme (2007–09). J. Antimicrob. Chemother. 66:1409–1411 [DOI] [PubMed] [Google Scholar]
- 2. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial testing; 20th informational supplement. CLSI M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 3. D'Argenio DZ, Schumitzky A. 1997. ADAPT II user's guide: pharmacokinetic/pharmacodynamic systems analysis software. [Google Scholar]
- 4. Davies TA, et al. 2011. Longitudinal survey of carbapenem resistance and resistance mechanisms in Enterobacteriaceae and non-fermenters from the U. S. A. in 2007–09. J. Antimicrob. Chemother. 66:2298–2307 [DOI] [PubMed] [Google Scholar]
- 5. Drusano GL, et al. 1984. Multiple-dose pharmacokinetics of imipenem-cilastatin. Antimicrob. Agents Chemother. 26:715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gaynes R, Edwards JR. 2005. Overview of nosocomial infections caused by gram-negative bacilli. Clin. Infect. Dis. 41:848–854 [DOI] [PubMed] [Google Scholar]
- 7. Hirsch EB, Tam VH. 2010. Detection and treatment options for Klebsiella pneumoniae carbapenemases (KPCs): an emerging cause of multidrug-resistant infection. J. Antimicrob. Chemother. 65:1119–1125 [DOI] [PubMed] [Google Scholar]
- 8. Hirsch EB, Tam VH. 2010. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev. Pharmacoecon. Outcomes Res. 10:441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lim TP, et al. 2008. Quantitative assessment of combination antimicrobial therapy against multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 52:2898–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Livermore DM. 1992. Interplay of impermeability and chromosomal beta-lactamase activity in imipenem-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:2046–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nordmann P, Cuzon G, Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228–236 [DOI] [PubMed] [Google Scholar]
- 12. Norrby SR, et al. 1983. Pharmacokinetics and tolerance of N-formimidoyl thienamycin (MK0787) in humans. Antimicrob. Agents Chemother. 23:293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Painter RE, Young K. 2007. Carbapenem-hydrolyzing beta-lactamases KPC-2 and KPC-3 in Klebsiella pneumoniae isolates from Florida, abstr A-016. Abstr. 107th Gen. Meet. Am. Soc. Microbiol., Toronto, Ontario, Canada [Google Scholar]
- 14. Quale J, Bratu S, Gupta J, Landman D. 2006. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 50:1633–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tam VH, et al. 2009. Impact of AmpC overexpression on outcomes of patients with Pseudomonas aeruginosa bacteremia. Diagn. Microbiol. Infect. Dis. 63:279–285 [DOI] [PubMed] [Google Scholar]
- 16. Tam VH, et al. 2007. Impact of drug-exposure intensity and duration of therapy on the emergence of Staphylococcus aureus resistance to a quinolone antimicrobial. J. Infect. Dis. 195:1818–1827 [DOI] [PubMed] [Google Scholar]
- 17. Tam VH, et al. 2010. Impact of multidrug-resistant Pseudomonas aeruginosa bacteremia on patient outcomes. Antimicrob. Agents Chemother. 54:3717–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tam VH, Schilling AN, Lewis RE, Melnick DA, Boucher AN. 2004. Novel approach to characterization of combined pharmacodynamic effects of antimicrobial agents. Antimicrob. Agents Chemother. 48:4315–4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Young K, et al. 2010. In vitro activity of the class A and C β-lactamase inhibitor MK-7655, abstr F1-2139. Abstr. 50th Intersci. Conf. Antimicrob. Agents Chemother., Boston, MA [Google Scholar]