LETTER
Vibrio parahaemolyticus is a major causative agent of gastroenteritis, particularly in areas with high levels of seafood consumption, and it has recently become pandemic due to the emergence of the O3:K6 serotype (12). Tdh and Trh have been implicated as major virulence factors in the strains of V. parahaemolyticus that caused most of the clinical infections (17). An estimated 45,000 (90% confidence interval [CI], 23,000 to 75,000) cases of V. parahaemolyticus infections occur every year in the United States (16). In Hong Kong, V. parahaemolyticus is the leading cause of food-borne illness, although the exact number of cases is not known due to a lack of relevant surveillance systems in Hong Kong (4, 7). V. parahaemolyticus gastroenteritis is self-limiting, but the infections can be fatal to the elderly, immunocompromised patients, or those with medical conditions such as liver disease or diabetes where antibiotic treatment is necessary (9, 17).
In this study, V. parahaemolyticus isolates were collected from raw shrimp samples purchased in markets in four different locations (Hong Kong Island, Hung Hom, Tsuen Wan, and Sai Kung) in Hong Kong from January to April 2010. A total of 128 shrimp samples were collected during the isolation period. Two typical blue-green colonies were acquired from thiosulfate-citrate-bile salts-sucrose (TCBS) agar plates for each sample and subjected to further confirmation by a PCR assay targeting the tl and atp genes (10, 19). Of 128 shrimp samples, 119 (93%) were V. parahaemolyticus positive (with one or two positive isolates), and a total of 208 V. parahaemolyticus isolates were obtained for further characterization (19). The isolation rates for the four locations and 4 months were between 91% and 95% and between 90% and 96%, respectively. Virulence genes thd and trh were detected in 4 and 2 V. parahaemolyticus isolates, respectively, using PCR assays (15), and none of the isolates contained both genes.
The 208 V. parahaemolyticus isolates from shrimp samples were assayed for their antimicrobial susceptibilities by the broth microdilution method as recommended by the Clinical and Laboratory Standards Institute (CLSI) using 13 antibiotics as shown in Table 1 (5). V. parahaemolyticus isolates were resistant to ampicillin (85%), amikacin (62%), tetracycline (53%), and chloramphenicol (35%). Surprisingly, about 6% (12 of 208) of the V. parahaemolyticus isolates also showed resistance to cefotaxime, and 6% of them were resistant to ciprofloxacin. No isolate showed concurrent resistance to both cefotaxime and ciprofloxacin. The resistance to most of the antibiotics observed in this study is consistent with other reports in Hong Kong and around the world except for the emergence of resistance to new front-line antibiotics such as fluoroquinolones and extended-spectrum cephalosporins in V. parahaemolyticus, which is rare and has been reported only once, in Indonesia in 2003 (1, 3, 8, 13, 14, 18). The 12 cefotaxime-resistant V. parahaemolyticus isolates were from 11 independent samples collected from different locations or on difference isolation dates (V137 and V138 were from the same sample), and none of these isolates harbored the virulence gene tdh or trh (Fig. 1). Pulsed-field gel electrophoresis (PFGE) characterization showed that these 12 isolates belonged to two different PFGE types (Fig. 1). V. parahaemolyticus strains of the same PFGE type were isolated from different locations and dates, which suggested that clonal spread of different V. parahaemolyticus strains might play an important role in the emergence of extended-spectrum-β-lactam-resistant V. parahaemolyticus in Hong Kong.
Table 1.
MICs of the ESBL-producing V. parahaemolyticus strains and their transconjugantsa
| Strain | MIC (μg/ml) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CRO | CTX | AZT | MER | CIP | NAL | KAN | GEN | AMI | CHL | TET | STR | |
| J53 | 16 | <1 | <1 | 2 | <0.5 | <0.05 | 8 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V15 | >128 | 64 | >64 | >64 | <0.5 | <0.05 | <4 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V15TC | >128 | >64 | >64 | >64 | <0.5 | 0.125 | 16 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V26 | >128 | 64 | >64 | >64 | <0.5 | <0.05 | <4 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V26TC | >128 | >64 | >64 | >64 | <0.5 | 0.125 | 32 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V36 | >128 | 64 | >64 | >64 | <0.5 | <0.05 | <4 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V36TC | >128 | >64 | >64 | >64 | <0.5 | 0.125 | 32 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V39 | >128 | 64 | >64 | >64 | <0.5 | <0.05 | <4 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V39TC | >128 | >64 | >64 | >64 | <0.5 | 0.125 | 32 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V40 | >128 | 64 | >64 | >64 | <0.5 | <0.05 | <4 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V40TC | >128 | >64 | >64 | >64 | <0.5 | 0.125 | 32 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V41 | >128 | 64 | >64 | >64 | <0.5 | <0.05 | <4 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V85 | >128 | 64 | >64 | >64 | <0.5 | <0.05 | <4 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V85TC | >128 | 64 | >64 | >64 | <0.5 | 0.125 | 32 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V137 | >128 | 32 | 64 | >64 | <0.5 | <0.05 | <4 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V137TC | >128 | 64 | >64 | >64 | <0.5 | 0.5 | >64 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V138 | >128 | 32 | 64 | >64 | <0.5 | <0.05 | <4 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V138TC | >128 | 64 | >64 | >64 | <0.5 | 0.5 | >64 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V189 | >128 | 64 | >64 | >64 | <0.5 | <0.05 | <4 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V189TC | >128 | >64 | >64 | >64 | <0.5 | 0.125 | 32 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V191 | >128 | 64 | >64 | >64 | <0.5 | <0.05 | <4 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V201 | >128 | 64 | >64 | >64 | <0.5 | <0.05 | <4 | <0.5 | <4 | <8 | <4 | <4 | <4 |
| V201TC | >128 | 64 | >64 | >64 | <0.5 | 0.125 | 32 | <0.5 | <4 | <8 | <4 | <4 | <4 |
TC, transconjugant; AMP, ampicillin; CRO, ceftriaxone; CTX, cefotaxime; AZT, aztreonam; MER, meropenem; CIP, ciprofloxacin; NAL, nalidixic acid; KAN, kanamycin; GEN, gentamicin; AMI, amikacin; CHL, chloramphenicol; TET, tetracycline; STR, streptomycin.
Fig 1.
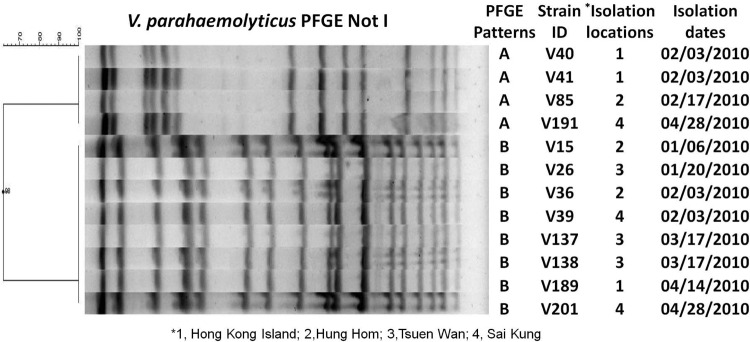
PFGE profile and isolation information for 12 extended-spectrum-β-lactamase (ESBL)-producing V. parahaemolyticus strains. Isolation dates are shown as month/day/year.
A conjugation experiment was performed using the cefotaxime-resistant V. parahaemolyticus isolates with Escherichia coli J53 as the recipient strain as previously described (20) with modifications. Briefly, donor and recipient strains were mixed in a 1:1 ratio, transferred to a filter on an LB agar plate, and incubated for 1 h. The culture on the filter was washed off using saline water, and 100 μl of the washed culture was spread on a selective plate to select for transconjugants. The resistance determinant was found to be encoded on the self-transmissible plasmid and could be transferred to E. coli, except for two isolates with identical strains, V41 and V191. The self-transmissible plasmids were found to be ∼24 kb in size and to belong to the IncN compatibility group, members of which carry a blaPER-1, a class A extended-spectrum β-lactamase gene, by the use of a PCR assay as previously described (2, 6) (Table 1). PER-1 is mostly associated with Gram-negative clinical pathogens such as Pseudomonas aeruginosa, Acinetobacter baumannii, and Salmonella spp. and has never before been reported in Vibrio spp. Surprisingly, transconjugants showed higher (2- to 8-fold) MICs of nalidixic acid than both parental and recipient strains (Table 1). None of the known plasmid-mediated quinolone resistance (PMQR) mechanisms (including qnrA, qnrB, qnrC, qnrD, qnrS, qepA, oqxAB, aac, and qnrVC) was detected on the plasmid (data not shown) (11). Further studies are needed to characterize the novel PMQR mechanisms on the conjugative plasmid.
The increasing prevalence of multidrug-resistant V. parahaemolyticus in seafood may cause public health problems in Hong Kong, since V. parahaemolyticus infections are the leading cause of food-borne illness in Hong Kong (4, 7). More research and surveillance on the multidrug-resistant V. parahaemolyticus should be implemented to improve understanding and control of the progress of multidrug resistance in V. parahaemolyticus.
ACKNOWLEDGMENTS
We acknowledge Chi Chong Chan, Ka Yee Ho, and Hoi Ying So for help with the isolation of Vibrio isolates, the critical reading of the manuscript by Josephine Leung, and the helpful comments from Jianhua Liu.
The research was supported through Hong Kong PolyU internal grant G-U662 to S.C.
Footnotes
Published ahead of print 16 April 2012
REFERENCES
- 1. Baker-Austin C, et al. 2008. Antibiotic resistance in the shellfish pathogen Vibrio parahaemolyticus isolated from the coastal water and sediment of Georgia and South Carolina, USA. J. Food Prot. 71:2552–2558 [DOI] [PubMed] [Google Scholar]
- 2. Carattoli A, et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 3. Chan KY, Woo ML, Lam LY, French GL. 1989. Vibrio parahaemolyticus and other halophilic vibrios associated with seafood in Hong Kong. J. Appl. Bacteriol. 66:57–64 [DOI] [PubMed] [Google Scholar]
- 4. Chan SS, et al. 2003. Acute bacterial gastroenteritis: a study of adult patients with positive stool cultures treated in the emergency department. Emerg. Med. J. 20:335–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2010. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline—2nd ed (M45-A2). Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495 [DOI] [PubMed] [Google Scholar]
- 7. Food and Environmental Hygiene Department 2005. Vibrio species in seafood. Risk assessment studies, report no. 20. Food and Environmental Hygiene Department, The Government of the Hong Kong Special Administrative Region. http://www.cfs.gov.hk/english/programme/programme_rafs/programme_rafs_fm_01_02_vss.html
- 8. Han F, Walker RD, Janes ME, Prinyawiwatkul W, Ge B. 2007. Antimicrobial susceptibilities of Vibrio parahaemolyticus and Vibrio vulnificus isolates from Louisiana Gulf and retail raw oysters. Appl. Environ. Microbiol. 73:7096–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hou CC, et al. 2011. Clinical manifestation and prognostic factors of non-cholerae Vibrio infections. Eur. J. Clin. Microbiol. Infect. Dis. 30:819–824 [DOI] [PubMed] [Google Scholar]
- 10. Izumiya H, et al. 2011. Multiplex PCR assay for identification of three major pathogenic Vibrio spp., Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus. Mol. Cell. Probes 25:174–176 [DOI] [PubMed] [Google Scholar]
- 11. Ma J, et al. 2009. High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac(6′)-Ib-cr, and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrob. Agents Chemother. 53:519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsumoto C, et al. 2000. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38:578–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oh EG, et al. 2011. Antimicrobial resistance of Vibrio parahaemolyticus and Vibrio alginolyticus strains isolated from farmed fish in Korea from 2005 through 2007. J. Food Prot. 74:380–386 [DOI] [PubMed] [Google Scholar]
- 14. Okoh AI, Igbinosa EO. 2010. Antibiotic susceptibility profiles of some Vibrio strains isolated from wastewater final effluents in a rural community of the Eastern Cape Province of South Africa. BMC Microbiol. 10:143 doi:10.1186/1471-2180-10-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pal D, Das N. 2010. Isolation, identification and molecular characterization of Vibrio parahaemolyticus from fish samples in Kolkata. Eur. Rev. Med. Pharmacol. Sci. 14:545–549 [PubMed] [Google Scholar]
- 16. Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. 2011. Foodborne illness acquired in the United States—unspecified agents. Emerg. Infect. Dis. 17:16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shevchuk VB, Gebesh VV, Alekseenko VV, Dobroshtan EV, Padchenko AG. 1986. Clinical aspects of acute intestinal infection caused by Vibrio parahaemolyticus. Vrach. Delo 1986:114–116 (In Russian.) [PubMed] [Google Scholar]
- 18. Tjaniadi P, et al. 2003. Antimicrobial resistance of bacterial pathogens associated with diarrheal patients in Indonesia. Am. J. Trop. Med. Hyg. 68:666–670 [PubMed] [Google Scholar]
- 19. USFDA 2004. Bacteriological analytical manual on line, chapter 9. U.S. Foodand Drug Administration, White Oak, MD: http://www.fda.gov/Food/ScienceResearch/LaboratoryMethods/BacteriologicalAnalyticalManualBAM/ucm070830.htm [Google Scholar]
- 20. Zhao J, et al. 2010. Prevalence and dissemination of oqxAB in Escherichia coli isolates from animals, farmworkers, and the environment. Antimicrob. Agents Chemother. 54:4219–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]


