Abstract
Streptococcus pneumoniae is a major causative agent of otitis media, pneumonia, bacteremia, and meningitis. Pneumolysin (Ply), a member of the cholesterol-dependent cytolysins (CDCs), is produced by virtually all clinical isolates of S. pneumoniae, and ply mutant strains are severely attenuated in mouse models of colonization and infection. In contrast to all other known members of the CDC family, Ply lacks a signal peptide for export outside the cell. Instead, Ply has been hypothesized to be released upon autolysis or, alternatively, via a nonautolytic mechanism that remains undefined. We show that an exogenously added signal sequence is not sufficient for Sec-dependent Ply secretion in S. pneumoniae but is sufficient in the surrogate host Bacillus subtilis. Previously, we showed that Ply is localized primarily to the cell wall compartment in the absence of detectable cell lysis. Here we show that Ply released by autolysis cannot reassociate with intact cells, suggesting that there is a Ply export mechanism that is coupled to cell wall localization of the protein. This putative export mechanism is capable of secreting a related CDC without its signal sequence. We show that B. subtilis can export Ply, suggesting that the export pathway is conserved. Finally, through truncation and domain swapping analyses, we show that export is dependent on domain 2 of Ply.
INTRODUCTION
Streptococcus pneumoniae, a Gram-positive bacterial pathogen and colonizer of the human nasopharynx is one of the leading causes of otitis media, sinusitis, meningitis, pneumonia, and bacteremia throughout the world (15). In developed countries, pneumonia caused by S. pneumoniae is a significant contributor to mortality in the elderly. Childhood S. pneumoniae infections are generally not fatal but do place a large burden on the medical community and an economic burden on parents and caregivers of children. For example, in the United States, ear infections represent the most frequent cause for a child under 5 years of age to visit a pediatrician, at an estimated cost of $3 billion to $4 billion per year (29). In developing countries, particularly in sub-Saharan Africa and southeast Asia, consequences of childhood S. pneumoniae infections are dire. Every year, over one million children in the developing world die as a result of pneumonia, bacteremia, and meningitis caused by S. pneumoniae (33).
Many virulence factors contribute to the severity and wide variety of disease that S. pneumoniae causes. Among these virulence factors is pneumolysin (Ply), a multifunctional protein that is found in virtually all clinical isolates and contributes to both colonization and disease. Ply is a 53-kDa member of a family of proteins known as the cholesterol-dependent cytolysins (CDCs). CDCs bind cholesterol on a eukaryotic membrane surface, oligomerize, and form ∼400-Å-diameter pores, causing cell lysis and tissue damage. In addition to its ability to cause direct tissue damage, Ply can modulate the immune system in several ways, including by the stimulation of polymorphonuclear leukocytes and complement activation (6, 21, 25). All of these Ply functions require an extracellular localization, but unlike all other known members of the Gram-positive pathogen CDC family (e.g., perfringolysin O, streptolysin O, and listeriolysin O), Ply lacks an N-terminal signal sequence for secretion through the canonical Sec machinery (32). Moreover, Ply lacks other known signals for any of the previously described export systems (32). Early work on Ply reported that it has a cytoplasmic localization in S. pneumoniae (17). That report, along with the finding that Ply lacks a signal sequence and the observation that Ply is not found in culture supernatants until cells have undergone, autolysis suggested a cell lysis-dependent release of Ply (24, 32). The release of Ply into the host milieu upon autolysis has been a working hypothesis in the field for the past 20 years, and therefore, subsequent studies that have analyzed Ply's activity or presence have considered only a cytoplasmic or lysis-mediated extracellular location.
Several lines of evidence contradicting this model have emerged. Certain strains have been reported to release Ply into the culture supernatant in early stationary phase when autolysis of the culture has not yet occurred (4). Additionally, Ply has been reported to be released in the absence of the major autolysin LytA, indicating an autolysis-independent release of Ply (1). Subsequently, we confirmed the autolysis-independent export and, in addition, showed that the exported Ply localizes primarily to the bacterial cell wall (26). Here we build on these observations and show that Ply which is released by in vitro autolysis of stationary-phase cultures does not reassociate with the cell wall of actively growing S. pneumoniae when added exogenously, further supporting the autolysis-independent export and cell wall localization model. We identify a domain of Ply that is required for export. Additionally, we show that a signal sequence is not sufficient for Sec-dependent Ply secretion in S. pneumoniae but is sufficient in the surrogate host Bacillus subtilis. Finally, using the native ply sequence, we show that a signal sequence-less protein export pathway also exists in B. subtilis.
MATERIALS AND METHODS
Strains and growth conditions.
The strains used in this study are listed in Table 1. S. pneumoniae was grown to an optical density at 600 nm (OD600) of 0.3 in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY) plus 5 μl/ml Oxyrase (Oxyrase, Inc., Mansfield, OH). These cultures were mixed with glycerol to a final concentration of 20% glycerol, aliquoted into 300-μl “starter cultures,” and stored at −80°C. Starter cultures were thawed and diluted into THY broth plus Oxyrase and grown to mid-exponential phase (OD600 = 0.4 to 0.5).
Table 1.
Strains used in this study
| Species and strain | Alternative strain name | Relevant genotype or description | Reference or source |
|---|---|---|---|
| S. pneumoniae | |||
| AC353 | TIGR4 | TIGR4, Smr derivative | 11 |
| AC2394 | acaps TIGR4 | AC353 Δcps4A′-′4L | R. Iyer |
| AC4037 | 2394 Δply::spc | AC2394 Δply::spc, Spcr | This study |
| AC4038 | Ply-HA | AC2934, Ply tagged with HA epitope, Cmr | This study |
| AC4039 | N-1 | AC4038, Ply aa 1-21 removed, Cmr | This study |
| AC4041 | N-4 | AC4038, Ply aa 1-197 removed, Cmr | This study |
| AC4043 | C-2 | AC4038, Ply aa 343-471 removed, Cmr | This study |
| AC4044 | C-3 | AC4038, Ply aa 319-471 removed, Cmr | This study |
| AC4045 | SSRrgB-Ply | AC2394, SS from RrgB fused to ply, Cmr | This study |
| AC4046 | PfoNoSS | AC2394, pfo without SS in ply locus, Cmr | This study |
| AC4047 | AC2394, Ply with Pfo D2 in ply locus, Cmr, HA tag | This study | |
| AC4048 | AC2394, Pfo with Ply D2 in ply locus, Cmr, HA tag | This study | |
| AC4049 | D2 | AC2394, Ply's D2 replacement with GGS linkers, Cmr | This study |
| B. subtilis | |||
| AC4050 | AC4052, Ply under control of Pspac* at amyE locus, Cmr | This study | |
| AC4051 | AC4052, SSRrgB-Ply under control of Pspac* at amyE locus, Cmr | This study | |
| AC4052 | JH642 | trpC2 pheA1 | 13 |
| E. coli | |||
| AC1000 | pAC1000, Cmr | 12 | |
| AC578 | pMmeISpc, Spcr | 19 |
The Bacillus subtilis strains were grown on LB plates overnight and were used to inoculate THY broth to a starting OD600 of 0.01 to 0.02. Cultures were grown with shaking at 37°C to an OD600 of 0.4 to 0.5.
Strain construction.
The S. pneumoniae strains listed in Table 1 were constructed using the splicing by overlap extension (SOE) PCR method with the primers listed in Table 2 (14). A marked deletion of ply (AC4037, ply::spc) was constructed in three parts, i.e., “Up,” “Spc,” and “Down,” and transformed into AC2394, an acapsular derivative of TIGR4 (AC353), where it replaced the ply allele by double-crossover homologous recombination. All subsequent mutants were transformed into this new background, such that the spc marker would be replaced with a mutated version of ply and a cat chloramphenicol resistance cassette. AC4038 was constructed in four parts, i.e., “Up,” “ply-HA,” “cat,” and “Down,” using AC1000 as the template for “cat” and AC2394 genomic DNA (gDNA) as the template for all other parts. The construct was transformed into strain AC4037 and replaced the spc allele. Truncation mutants and domain swaps (AC4039, AC4041, AC4047, AC4048, and AC4049) were constructed in three parts, i.e., “Up,” “ply′-HA,” and “cat-down,” using gDNA from strain AC4038. AC4046 was constructed in three parts, i.e., “Up,” “pfo,” and “Down.” Genomic DNA from AC2394 served as the template for “Up” and “Down”; gDNA from Clostridium perfringens (a gift of A. L. Sonenshein) served as the template for “pfo.” AC4048 was constructed in four parts, i.e., “Up,” “pfo with Ply D2 a,” “pfo with Ply D2 b,” and “Down.” Genomic DNA from AC2394 served as the template for “Up” and “Down”; gDNA from AC4046 served as the template for “pfo with Ply D2.”
Table 2.
Primers used in this study
| Strain | Primer no. | Primer name | Sequence (5′ → 3′) |
|---|---|---|---|
| AC4037 | KP064 | Up F1 | TTCTGGAGTTTCCCGATTTC |
| KP101 | Up R1 | CCGGGCCCCCCCTCGAGGTCTTCTACCTCCTAATAAGTTCCTGGA | |
| KP102 | Spc F2 | CTTATTAGGAGGTAGAAGACCTCGAGGGGGGGCCCGGTACCGAGG | |
| KP103 | Spc R2 | GCATTCTCCTCTCTTATAATTTTTTTAATCTGTTATTTAAATAGTTTATAG | |
| KP104 | Down F3 | CAGATTAAAAAAATTATAAGAGAGGAGAATGCTTGCGACA | |
| KP032 | Down R3 | GCCCATTTACGTCCCATTAG | |
| AC4038 | KP064 | Up F1 | TTCTGGAGTTTCCCGATTTC |
| KP081 | Up R1 | TTCTCCCTGATGGGTCATCTTCTACCTCCTAATAAGTTCCTGGA | |
| KP082 | PlyHA F2 | ATTAGGAGGTAGAAGATGACCCATCAGGGAGAAAGTATTGAA | |
| KP034 | Cat R2 | TTCTCCTCTCTTATAAAAGCCAGTCATTAGGCCT | |
| KP031 | Down F3 | GCTTTTATAAGAGAGGAGAATGCTTGCGACA | |
| KP032 | Down R3 | GCCCATTTACGTCCCATTAG | |
| AC4039 | KP064 | Up F1 | TTCTGGAGTTTCCCGATTTC |
| KP083 | Up R1 | TACAGAAATATCACTCATCTTCTACCTCCTAATAAGTTCCTGGA | |
| KP084 | PlyCatDown F2 | ATTAGGAGGTAGAAGATGAGTGATATTTCTGTAACAGCTACCAAC | |
| KP032 | PlyCatDown R2 | GCCCATTTACGTCCCATTAG | |
| AC4041 | KP064 | Up F1 | TTCTGGAGTTTCCCGATTTC |
| KP089 | Up R1 | GAGATAGACTTGGCGCATCTTCTACCTCCTAATAAGTTCCTGGA | |
| KP090 | PlyHACatDown F2 | ATTAGGAGGTAGAAGATGCGCCAAGTCTATCTCAAGTTGGA | |
| KP032 | PlyHACatDown R2 | GCCCATTTACGTCCCATTAG | |
| AC4043 | KP038 | PlyHA F1 | CAGTCGCCTCTATCCTGGAG |
| KP039 | PlyHA R1 | AGATCCTAAGCATAGTCTGGTACATCGTAGGGGTATCTGTAAGCTGTAACCTTAGTCTC | |
| KP040 | PlyHACatDown F2 | AGCTTACAGATACCCCTACGATGTACCAGACTATGCTTAGGATCTCGAGGTCGACGGTAT | |
| KP032 | PlyHACatDown R2 | GCCCATTTACGTCCCATTAG | |
| AC4044 | KP041 | PlyHA F1 | CAGTCGCCTCTATCCTGGAG |
| KP042 | PlyHA R1 | AGATCCTAAGCATAGTCTGGTACATCGTAGGGGTACGCAACTACATTGTCACGTA | |
| KP043 | PlyHACatDown F2 | TGTAGTTGCGTACCCCTACGATGTACCAGACTATGCTTAGGATCTCGAGGTCGACGGTAT | |
| KP032 | PlyHACatDown R2 | GCCCATTTACGTCCCATTAG | |
| AC4045 | KP064 | Up F1 | TTCTGGAGTTTCCCGATTTC |
| KP191 | rrgB SS R1 | TGCTTTATTTGCCATCCCAGCCGCAAAAACTGTTGCAGCTGAAAACAGGCTACTCGCTGTCAGTAATAAGGCAGCAAGCATTGTTAAAAATTTGTTGATTGATTTCATCTTCTACCTCCTAATAAGTTCCTGG | |
| KP190 | rrgB SS F2 | ATTAGGAGGTAGAAGATGAAATCAATCAACAAATTTTTAACAATGCTTGCTGCCTTATTACTGACAGCGAGTAGCCTGTTTTCAGCTGCAACAGTTTTTGCGGCTGGGATGGCAAATAAAGCAGTAAATGACTT | |
| KP032 | Down R2 | GCCCATTTACGTCCCATTAG | |
| AC4046 | KP064 | Up F1 | TTCTGGAGTTTCCCGATTTC |
| KP201 | ply::noSSPfo R1 | ACTTTGATTCATCTTCTACCTCCTAATAAGTTCCTGG | |
| KP200 | ply::noSSPfo F2 | AGGAGGTAGAAGATGAATCAAAGTATTGATTCTGGAATATCA | |
| KP199 | ply::pfo R2 | GACCTCGAGATCTTAATTGTAAGTAATACTAGATCCAGGG | |
| KP197 | ply::pfo F3 | ACTTACAATTAAGATCTCGAGGTCGACGGTAT | |
| KP032 | Down R3 | GCCCATTTACGTCCCATTAG | |
| AC4047 | KP064 | Up F1 | TTCTGGAGTTTCCCGATTTC |
| KP212 | Ply with Pfo D2 R1 | AAATTTATTACCAGTCTTTTTACCTTCCTTTGGAACAAAACTTTCAATTTTATCTCCATTACTAGCCAAGAGTTTCTTTTTATCGTAATTCA | |
| KP213 | Ply with Pfo D2 F2 | GAAGGTAAAAAGACTGGTAATAAATTTATAGTTGTAGAACGTCAAAAAAGATCCCTTACAACATCACCAAGTGATATTTCTGTAACAGCTACCA | |
| KP214 | Ply with Pfo D2 R2 | AGTTGTTTCTATATAATCTGTTTTATTGTGAACAGCCGCAACTACATTGTCACGTAAA | |
| KP215 | Ply with Pfo D2 F3 | ACAGATTATATAGAAACAACTTCTACAGAGTATTCTAACGGAGATTTACTGCTGG | |
| KP032 | Down R3 | GCCCATTTACGTCCCATTAG | |
| AC4048 | KP064 | Up F1 | TTCTGGAGTTTCCCGATTTC |
| KP216 | Pfo with Ply D2 R1 | AAACTCATCGGGTAGCTGATTACCCTCTTTGATGAAACGATTTTCAATACTTTCTCCCTGATGGGTTAAAACTTCATTTCTATTGTAACTTAAGC | |
| KP217 | Pfo with Ply D2 a F2 | ATCAAAGAGGGTAATCAGCTACCCGATGAGTTTGTTGTTATCGAAAGAAAGAAGCGGAGCTTGTCGACAAATACAGTAGATATATCAATAATTGATTCTGTAAATGAC | |
| KP218 | Pfo with Ply D2 a R2 | AGCTGTAACCTTAGTCTCAACATAGTCTGTACTATTTTGAAAGGTAGCAACTGAGTTATCTTTTAAGAAAAC | |
| KP219 | Pfo with Ply D2 b F3 | ACAGACTATGTTGAGACTAAGGTTACAGCTTACAGAAAGGGAAAAATAAACTTAGATCATAGTG | |
| KP229 | Pfo with Ply D2 b R3 | GTACATCGTAGGGGTAATTGTAAGTAATACTAGATCCAGGG | |
| KP228 | Pfo with Ply D2 F4 | GTATTACTTACAATTACCCCTACGATGTACCAGA | |
| KP032 | Down R4 | GCCCATTTACGTCCCATTAG | |
| AC4049 | KP064 | Up F1 | TTCTGGAGTTTCCCGATTTC |
| KP208 | Ply domain 2 R1 | AGAAATATCACTAGAACCACCAGAACCACCCAAGAGTTTCTTTTTATCGTAATTCATAG | |
| KP209 | Ply domain 2 F2 | AAGAAACTCTTGGGTGGTTCTGGTGGTTCTAGTGATATTTCTGTAACAGCTACC | |
| KP210 | Ply domain 2 R2 | AGTAAATCTCCGTTAGAACCACCAGAACCACCAGAACCACCAGAACCACCAGAACCACCCGCAACTACATTGTCACGTAA | |
| KP211 | Ply domain 2 F3 | AATGTAGTTGCGGGTGGTTCTGGTGGTTCTGGTGGTTCTGGTGGTTCTGGTGGTTCTAACGGAGATTTACTGCTGG | |
| KP032 | Down R3 | GCCCATTTACGTCCCATTAG | |
| AC4050 | KP245 | Ply in B.s. F | TAAGGAGGTGTTATATATGGCAAATAAAGCAGTAAATGACTTT |
| KP246 | Ply in B.s. R | CTAGTCATTTTCTACCTTATCTTCTACCTG | |
| AC4051 | KP244 | SS Ply B.s. F | TAAGGAGGTGTTATATATGAAATCAATCAACAAATTTTTAACAATGC |
| KP246 | Ply in B.s. R | CTAGTCATTTTCTACCTTATCTTCTACCTG |
The B. subtilis strains expressing the signal sequence (SS) from RrgB fused to ply (SSRrgB-Ply) and Ply were constructed by first amplifying the SSRrgB-Ply and Ply sequences from the gDNAs of strains SSRrgB-Ply and AC2394, respectively. These PCR products were cloned into the blunt-ended cloning vector pCRscript (Stratagene) to introduce EcoRV and SacI restriction enzyme sites. The insert was cut from the vector and ligated into the prepared integrative plasmid pAWS8 (28). Plasmid pAWS8 contains the yeast aconitase gene acoI under the control of the constitutively active Pspac* promoter between two segments of the nonessential amyE gene. The acoI gene was removed from pAWS8 by first digesting with HindIII, blunting the plasmid with Klenow, and then digesting the plasmid with SacI, thus removing acoI and creating a blunt end and a SacI overhang. The ply insert (with a blunt 5′ end and a 3′ SacI overhang) was ligated into the cut vector with T4 DNA ligase (NEB) and transformed into Escherichia coli DH5α (for the ply construct) and E. coli JM107 (for the SSRrgB-Ply construct). Colonies were picked, and plasmids were isolated with the QIAquick miniprep kit (Qiagen). The resulting plasmids were used in a transformation into B. subtilis strain JH642 (9, 13). Colonies were tested for double crossover by amylase activity assays, colony PCR, and sequencing.
Fractionation of S. pneumoniae and B. subtilis.
Cells were grown to an OD600 of 0.4 to 0.5 and centrifuged at 4,000 × g for 7 min. The culture supernatants were saved, and proteins were precipitated with 10% (vol/vol) trichloroacetic acid (TCA) and resuspended in 70 μl 50 mM Tris, pH 7.5. The cell pellets were washed once with THY, resuspended in 70 μl cell wall digestion buffer (1× protease inhibitor cocktail [Roche], 300 U/μl mutanolysin, and 1 mg/ml lysozyme in a 30% sucrose–10 mM Tris [pH 7.5] buffer), and incubated at 37°C for 2 h with rocking. Protoplasts were separated from the cell wall by centrifugation at 13,100 × g for 10 min. Protoplasts were resuspended in 70 μl 10 mM Tris buffer, pH 7.5.
Hemolytic assays.
Hemolytic assays were performed as described previously (1) with some modifications. Samples of culture supernatants (not TCA precipitated), cell walls, and protoplasts were pooled, and 100 μl of each fraction was 2-fold serially diluted in assay buffer (10 mM dithiothreitol [DTT] and 0.1% bovine serum albumin [BSA] in phosphate-buffered saline [PBS]) in a 96-well V-bottom plate. Fifty microliters of triple-washed 2% sheep red blood cells (RBCs) was added to the dilutions and incubated for 1 h at 37°C. Plates were spun at 233 × g for 10 min, and hemolytic units were determined visually. Hemolytic units are equal to the reciprocal of the highest dilution at which there was 100% lysis. Cell wall and protoplast hemolytic units were divided by 10 to normalize them to the same volume as the culture supernatant.
Western blot analysis.
Equal volumes of samples (cell equivalents) were boiled in SDS sample buffer (50 mM Tris-Cl [pH 6.8], 2% SDS, 0.5% bromophenol blue, 10% glycerol, 100 mM β-mercaptoethanol), cooled, loaded onto an SDS-polyacrylamide gel, and run for 25 min at 75 V and for an additional 90 min at 125 V. Proteins were transferred to a nitrocellulose membrane at 30 V for 75 min. Equal loading of samples on each gel was confirmed by staining the membrane with Ponceau S prior to Western blotting (data not shown). Western blotting was performed using the Snap-ID system (Millipore) according to the manufacturer's directions. Membranes were cut and blocked with NAP blocker (GBiosciences) diluted 1:2 in Tris-buffered saline (TBS). Primary antibody to either Ply (Statens Serum Institut) at 1:1,000, CodY (a gift of A. L. Sonenshein) at 1:10,000, RrgB (18) at 1:2,500, Pfo (a gift of R. Tweten) at 1:1,000, or hemagglutinin (HA) (Santa Cruz) at 1:2,500 in NAP blocker diluted 1:4 in TBS was applied to the membrane for 10 min at room temperature. Membranes were washed three times with TBS. Cy5-conjugated antibody (Invitrogen) was applied to the membrane at a 1:2,500 dilution in NAP (diluted 1:4 with TBS) for 10 min. Membranes were washed as described above and scanned with a Fuji FLA-9000 instrument at 635 nm excitation and with the DBR filter.
Reassociation assay.
A TIGR4 culture was allowed to grow overnight and autolyze. The supernatant was separated from the cell debris by centrifugation (4,000 × g for 7 min). The resulting supernatant was filtered through a 0.22-μm filter to remove any remaining cells. This cell-free supernatant was then applied to a ply culture and allowed to incubate with ply cells throughout growth. Wild-type cultures and ply cultures with THY alone served as positive and negative controls for the presence and absence of Ply, respectively. All cultures were grown to an OD600 of 0.6, fractionated, and assayed for the presence of Ply and CodY by Western blotting.
Protein purification.
Ply-HA and D2-HA proteins were purified from strains AC4038 and AC4049, respectively. Strains were grown to mid-exponential phase, harvested by centrifugation, and resuspended in lysis buffer (20 mM Tris [pH 8.0], 150 mM NaCl, 2 mM MgCl2, 1× Roche complete mini-protease inhibitor cocktail). Each sample was treated with lysozyme (1 mg/ml; Sigma) at 37°C for 1 h, after which the cells were broken apart using a Bead Beater and 0.1-mm zirconia beads. Appropriate samples were pooled and treated with Benzonase (1,250 units; Sigma) at 37°C for 30 min. The resulting lysate was cleared by centrifugation at 100,000 × g for 1 h at 4°C using a Beckman Vti 65.2 rotor. The S100 fractions were collected and used as the source of protein for immunoprecipitation with Pierce anti-HA agarose (Thermo Scientific) as described by the manufacturer. Elution was carried out with Pierce HA peptide (Thermo Scientific), and eluted fractions were run on SDS-PAGE and stained with Coomassie brilliant blue to assess purity. Appropriate fractions were pooled, glycerol was added to a final concentration of 20%, and protein was stored at −20°C.
Limited proteolysis.
Approximately equivalent amounts of each purified protein, i.e., Ply-HA, D2-HA, or Ply* (His-tagged Ply toxoid protein, a gift of J. C. Paton), were incubated either alone or in the presence of 0.4 μg/μl μg of trypsin. Trypsin alone was incubated under the same conditions as a control. Equal amounts of each reaction were analyzed by Western blotting with an HA antibody.
Model of Ply.
The molecular model of Ply was made using the ModWeb program and manipulated in MacPyMol software.
RESULTS
Ply released by autolysis cannot reassociate with S. pneumoniae.
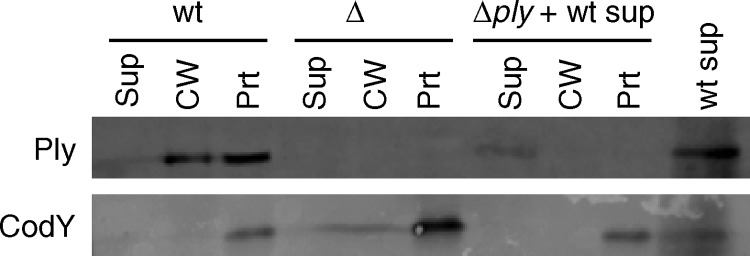
There are several examples of streptococcal proteins that localize to the cell wall despite lacking a signal sequence or cell wall-anchoring motif (16, 22, 23). Among them, enolase has been shown to reassociate with the surface of S. pneumoniae (5). This reassociation leaves open the possibility of an autolysis-dependent release of Ply as opposed to a true mechanism of export from intact, live cells. We have shown previously that Ply localizes to the cell wall in a LytA-independent manner (26), but it is still possible that limited lysis promoted by other, minor lysins releases Ply, which could then reassociate with the cell wall of neighboring, intact cells. To test this possibility, wild-type S. pneumoniae strain TIGR4 was grown overnight in broth to a state of complete autolysis. The supernatant was collected by centrifugation and filtered to ensure that there were no cells remaining. The cell-free supernatant containing Ply released by autolysis was added to a ply mutant culture at the time of inoculation. Cultures of wild-type and ply-deficient strains with no added cell-free supernatant were grown as positive and negative controls for the presence or absence of Ply, respectively. Cultures were grown to mid-exponential phase, fractionated, and assayed by Western blotting for the presence of Ply and CodY, a cytoplasmic control protein. Exogenously added, autolysis-released Ply is detected only in the supernatant and not in the cell wall or protoplast fraction (Fig. 1). This result indicates that Ply released by autolysis cannot reassociate with S. pneumoniae, further supporting the autolysis-independent model for Ply release and suggesting that there is a Ply export pathway that is coupled to cell wall localization.
Fig 1.
Ply released by autolysis cannot reassociate with intact cells. A wild-type (wt) culture was grown overnight and allowed to autolyze. The supernatant was collected, filtered, and added to a Δply culture. Wild-type and Δply cultures without added supernatant from an autolyzed culture were grown as positive and negative controls for the presence or absence of Ply, respectively. All cultures were grown to mid-exponential phase, fractionated into supernatant, cell wall, and protoplast compartments, and assayed for the presence of Ply and CodY by Western blotting. Equal cell equivalents were loaded on the gel. Sup, culture supernatant; CW, cell wall; Prt, protoplast.
In S. pneumoniae, Ply is incompatible with an exogenously added signal sequence.
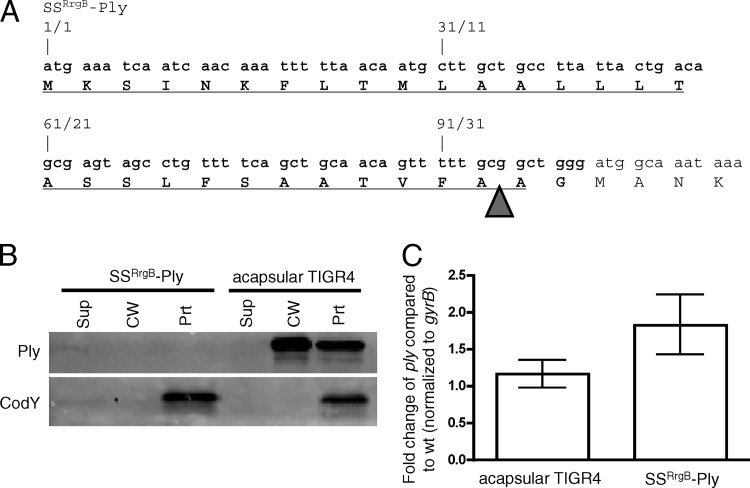
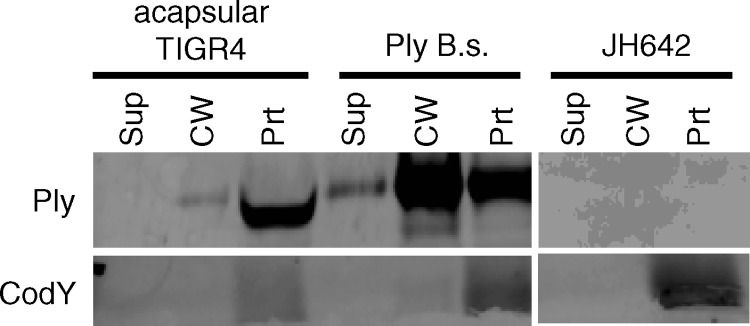
Ply is the only known member of the CDC family that lacks a signal sequence. Since Ply shares extensive sequence identity with the mature portions of other, Sec-secreted CDCs (having, for example, 48% identity with perfringolysin O [Pfo] [31]), we hypothesized that we could create a Sec-secreted derivative if we supplied Ply a signal sequence. To test this hypothesis, the strain SSRrgB-Ply, in which a canonical Sec signal sequence was fused to Ply, was constructed (Fig. 2A). The signal sequence is derived from RrgB, an S. pneumoniae secreted, cell wall-anchored protein. The SSRrgB-Ply strain was grown, fractionated, and assayed for the presence of Ply and CodY by Western blotting. Surprisingly, SSRrgB-Ply was not detected in any fraction (Fig. 2B). The lack of detectable Ply could be indicative of a defect in transcription or a posttranscriptional defect. To test the former possibility, quantitative reverse transcription-PCR (qRT-PCR) was performed on ply in the SSRrgB-Ply fusion strain and in an acapsular derivative of TIGR4, which is known to exhibit wild-type expression and export of Ply (26). The amounts of ply transcript detected in the two strains were comparable (Fig. 2C). Thus, the lack of detectable protein is a posttranscriptional effect and may indicate that the SSRrgB-Ply protein, if it is made, is incompatible with the Sec machinery in such a way that leads to its degradation.
Fig 2.
SSRrgB-Ply fusion mRNA is made but protein is not detected in Streptococcus pneumoniae. (A) N-terminal DNA (top) and amino acid (bottom) sequences of SSRrgB-Ply. Text that is not in bold is Ply sequence. The RrgB signal peptide is in bold and underlined. The arrowhead indicates the predicted site of signal peptidase cleavage. (B) Western blot of cell wall fractionation of SSRrgB-Ply and acapsular TIGR4 cells. Cultures were grown to mid-exponential phase, fractionated into supernatant, cell wall, and protoplast compartments, and assayed for the presence of Ply and CodY by Western blotting. Equal cell equivalents of each fraction were loaded on the gel. A representative Western blot from two independent experiments is shown. Sup, culture supernatant; CW, cell wall; Prt, protoplast. (C) Quantitative RT-PCR on the ply transcript in acapsular TIGR4 and SSRrgB-Ply cells. Shown is the median fold change of the ply transcript of three replicates compared to acapsular TIGR4, normalized to gyrB; error bars indicate the range.
Expression of the SSRrgB-Ply fusion in Bacillus subtilis.
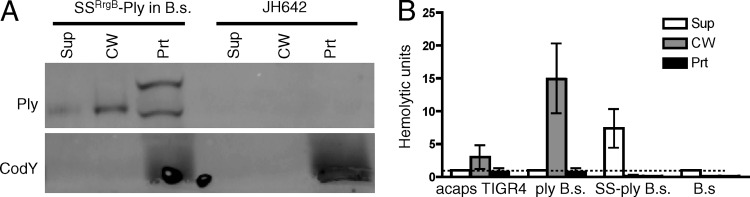
There are two major players in secretion of a protein: the substrate and the export machinery. To examine why the SSRrgB-Ply fusion protein was not detectable, we expressed the fusion in a heterologous host, B. subtilis. We hypothesized that if the fusion protein was intrinsically unstable in S. pneumoniae, then it would be degraded and undetectable in B. subtilis as well. However, if the degradation was instead due to a fault of the Sec machinery of S. pneumoniae, then the SSRrgB-Ply fusion may be detectable and possibly secreted in B. subtilis. To examine these possibilities, the SSRrgB-Ply fusion was placed under the control of the Pspac* constitutively active promoter in the amyE locus on the chromosome of B. subtilis via allelic exchange (9, 28). The strain was grown to mid-exponential phase and fractionated into culture supernatant, cell wall, and protoplast; the presence of Ply was detected by Western blotting, and the activity of Ply was measured by hemolysis assay. SSRrgB-Ply was detected in all three fractions, including the culture supernatant (Fig. 3A), which had the highest level of hemolytic activity (Fig. 3B). In the protoplast fraction, two species of Ply were detected, one at the expected molecular weight of the mature, signal peptidase-cleaved form of the protein and one at the expected molecular weight of the preprotein with its signal sequence still attached (Fig. 3A). In contrast, only the mature, cleaved form was detected in the cell wall and culture supernatant. There was no Ply or cross-reactive species detected in the parent strain B. subtilis JH642. Because the SSRrgB-Ply fusion is cleaved, presumably by signal peptidase, and secreted by B. subtilis into the culture supernatant, this suggests that the SSRrgB-Ply substrate is compatible with the Sec machinery of B. subtilis. Thus, the failure of SSRrgB-Ply to be secreted or even detected in S. pneumoniae is likely due to its Sec machinery being unable to process and secrete the fusion protein.
Fig 3.
SSRrgB-Ply expression in Bacillus subtilis. (A) SSRrgB-Ply can be secreted when expressed in B. subtilis. SSRrgB-Ply was placed under the control of the Pspac* promoter in the amyE locus on the B. subtilis chromosome. All strains were grown to mid-exponential phase, fractionated into supernatant, cell wall, and protoplast compartments, and assayed for the presence of Ply and CodY by Western blotting. SSRrgB-Ply in B.s., SSRrgB-Ply fusion in B. subtilis; JH642, wild-type B. subtilis parent strain. Equal cell equivalents were loaded on the gel. Sup, culture supernatant; CW, cell wall; Prt, protoplast. (B) Hemolytic assay of culture supernatant, cell wall, and protoplast fractions of acapsular S. pneumoniae (acaps TIGR4), B. subtilis with ply (ply B.s.), B. subtilis with SS-ply (SS-ply B.s.), and B. subtilis parent strain JH642 (B.s). The ply in B. subtilis has the same distribution as in S. pneumoniae, with the highest hemolytic activity in the cell wall fraction. The supernatant fraction has the highest hemolytic activity in the SS-ply B. subtilis strain. No hemolytic activity was detected in the B. subtilis parent strain. The dotted line indicates the limit of detection. Bars show the means for three biological replicates; error bars indicate standard errors of the means (SEM).
The Ply homolog Pfo lacking its signal sequence localizes primarily to the protoplast and weakly to the cell wall in S. pneumoniae.
If our hypothesis that the instability of SSRrgB-Ply in S. pneumoniae is due to failure of the Sec machinery to process this substrate is correct, then a closely related homolog of Ply should be similarly incompatible and perhaps even toxic. To test this, we precisely replaced the Ply-coding sequence with that for full-length Pfo. Consistent with our hypothesis, this allele was not tolerated by S. pneumoniae, as the only viable transformants recovered had spontaneous mutations that destroyed the signal sequence. Together with the inability to detect the SSRrgB-Ply fusion in S. pneumoniae, this result suggests that there is a general incompatibility between CDC proteins and Sec in S. pneumoniae.
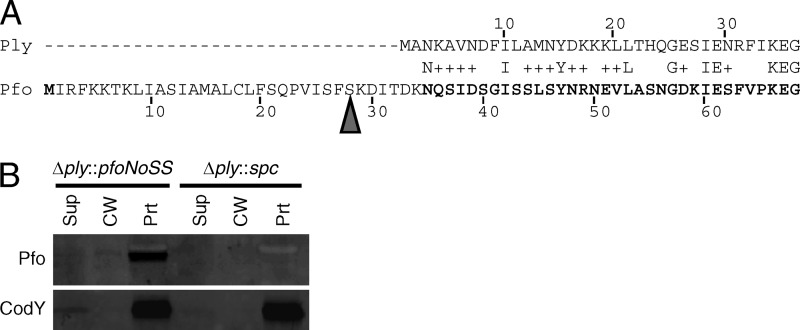
To confirm the incompatibility between Pfo and the S. pneumoniae Sec machinery, a signal sequence-less version of Pfo was expressed from the ply locus (Fig. 4A and the Fig. 4 legend give a description of the mutation). This mutant was not toxic to S. pneumoniae and was detectable with an antibody to Pfo (Fig. 4B). The majority of Pfo localized to the protoplast fraction, and no protein was detected in the culture supernatant. Interestingly, a small portion of Pfo localized to the cell wall fraction (Fig. 4B). This result suggests that the putative Ply export pathway can recognize another mature CDC protein but less efficiently.
Fig 4.
Pfo expression in Streptococcus pneumoniae. (A) Amino acid alignment of Ply (top) and Pfo (bottom). Sequence homology does not begin until the third amino acid of Ply; therefore, fusions were made at this amino acid. The arrowhead indicates the site of signal peptidase cleavage in Pfo. The Pfo sequence not in bold was excluded in the Pfo signal sequence-less construct, thus deleting the Pfo signal peptide and six amino acids after the signal peptidase cleavage that share no homology to Ply. (B) Expression of signal sequence-less Pfo in S. pneumoniae. Signal sequence-less Pfo is expressed in S. pneumoniae, and a small amount localizes to the cell wall. The Pfo signal sequence-less allele (pfoNoSS) was used to replace ply at the native locus (Δply::pfoNoSS). This strain and a negative-control strain in which the ply deletion was replaced with a spectinomycin resistance gene (Δply::spc) were grown to mid-exponential phase, fractionated into supernatant, cell wall, and protoplast compartments, and assayed for the presence of Ply and CodY by Western blotting with anti-Pfo and anti-CodY antibodies. Equal cell equivalents were loaded on the gel.
Domain 2 is necessary for Ply export.
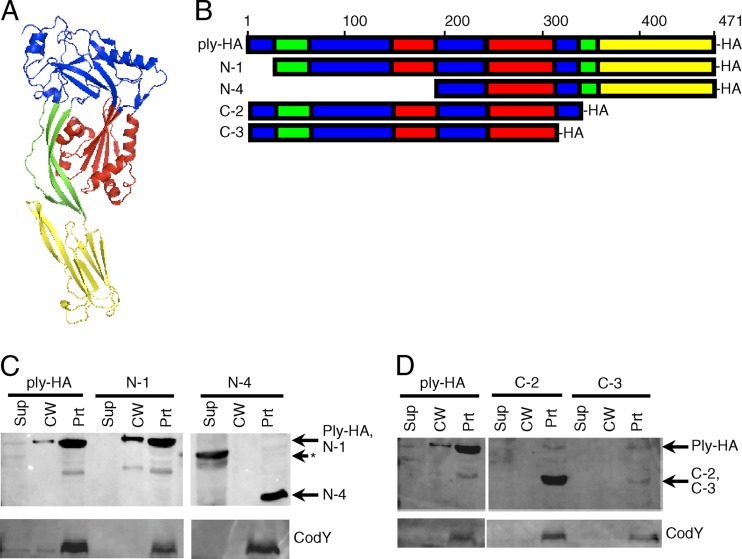
To determine which domain(s) might be important for Ply export and cell wall localization, N- and C-terminal truncation analysis was used. Truncated sequences corresponded to structurally assigned domains of Ply (27). All truncated products have a C-terminal HA epitope tag for detection by Western blotting (a model of Ply structure and a schematic of the truncation mutants are shown in Fig. 5A and B).
Fig 5.
Ply truncation mutants reveal that domain 2 of Ply may be required for export. (A) Model of Ply three-dimensional structure. The model was made with ModWeb software and manipulated with MacPyMol software. Domains are color coded: blue, domain 1; green, domain 2; red, domain 3; yellow, domain 4. (B) Schematic of primary sequences of Ply and truncation mutants. Domains are color coded as in panel A. Truncations of Ply were made from the N and C termini, tagged with an HA epitope at the C terminus of each construct, and crossed into the ply locus on the S. pneumoniae chromosome. Strains were grown to mid-exponential phase, fractionated into cell wall and protoplast compartments, and assayed for the presence of Ply and CodY by Western blotting. (C) N-terminal truncation mutants. The band in the supernatant lane for the N-4 truncation (shown by the asterisk) is of incorrect size and is unexplained. (D) C-terminal truncation mutants. Equal cell equivalents were loaded on the gel. Sup, culture supernatant; CW, cell wall; Prt, protoplast.
Full-length Ply with an HA tag (Ply-HA) was exported and localized to the cell wall like the native Ply (Fig. 5C). The N-1 N-terminal truncation, which lacks a portion of domain 1 (amino acids [aa] 1 to 21), was detected at its predicted molecular mass and was localized to the cell wall and protoplast fractions similarly to the full-length Ply-HA (Fig. 5C). The N-4 mutant, which lacks portions of domains 1, 2, and 3 (amino acids 1 to 197), was detected at its predicted molecular mass but was found only in the protoplast and not in the cell wall or supernatant fraction (Fig. 5C).
For the C-terminal truncations, when domain 4 and a portion of domain 2 (amino acids 343 to 471) were truncated (C-2), the protein was detected at its predicted molecular mass but was found only in the protoplast and not in the cell wall or supernatant fraction (Fig. 5D). Similarly, a further C-terminal truncation of Ply lacking domain 4 and portions of domain 2 and 1 (amino acids 319 to 471) (C-3) was detected at its predicted molecular mass and was found only in the protoplast and not in the cell wall or supernatant fraction (Fig. 5D). The C-3 mutant was weakly expressed, possibly indicating that the protein is unstable.
In both sets of truncations, when a portion of domain 2 was deleted, the mutant no longer localized to the cell wall, was absent from the culture supernatant, and instead was found in the protoplast. This suggests that domain 2, which comprises a beta-sheet between domains 1 and 4 in Ply (Fig. 5A and B), is necessary for export.
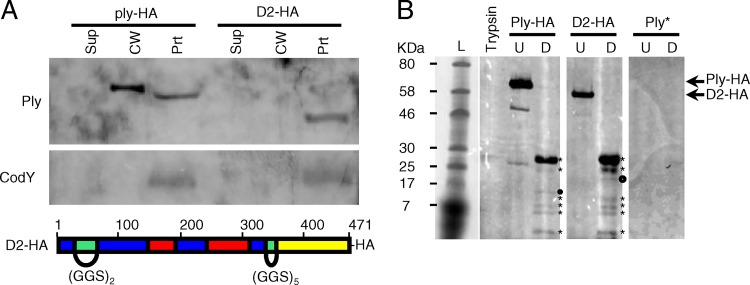
One caveat to the conclusion that domain 2 is necessary for Ply export is that all the truncation mutants lacking a portion of domain 2 are also missing portions of other domains. To more rigorously test this hypothesis, domain 2 was replaced with a flexible linker (Gly-Gly-Ser repeats), leaving the rest of the protein's primary sequence unchanged. The N-terminal portion of domain 2 was replaced with two repeats, while the C-terminal portion of domain 2 was replaced with five repeats. These repeat lengths, which were based on the known length and flexibility of the linker (20), were chosen to maintain the three-dimensional spacing of the remaining domains relative to each other. When analyzed for localization, the domain 2 replacement mutant (D2) was detected at its predicted molecular mass but was found only in the protoplast and not in the cell wall or supernatant fraction (Fig. 6A). Thus, the localization of the D2 mutant recapitulates the truncation analysis, providing further evidence for the role of domain 2 in Ply export.
Fig 6.
Ply with domain 2 replacement does not localize to the cell wall. Domain 2 was replaced with flexible linkers comprised of GGS repeats of appropriate length to maintain the three-dimensional spatial orientation of the remaining domains, tagged with a C-terminal HA epitope, and crossed into the ply locus on the S. pneumoniae chromosome. (A) Top, the ply-HA and D2 strains were grown to mid-exponential phase, fractionated into supernatant, cell wall, and protoplast compartments, and assayed for the presence of Ply and CodY by Western blotting. Equal cell equivalents were loaded on the gel. Sup, culture supernatant; CW, cell wall; Prt, protoplast. Bottom, schematic of the D2 replacement mutant, color coded by domain structure as in Fig. 5: blue, domain 1; green, domain 2; red, domain 3; yellow, domain 4. (B) Limited proteolysis of Ply with the domain 2 linker replacement reveals a wild-type conformation. Ply-HA and D2-HA were purified from strains Ply-HA and D2, respectively, via immunoprecipitation. His-tagged Ply toxoid protein (Ply*) served as a negative control for the anti-HA antibody. Approximately equivalent amounts of each purified protein were incubated either alone (U) or in the presence of identical amounts of trypsin (D). Trypsin alone was incubated under the same conditions. Equal amounts of each reaction product were analyzed for the presence of HA-tagged trypsin-cleaved peptides by Western blotting. The proteolytic degradation products of Ply-HA and D2-HA (marked with asterisks) are nearly identical. The presence of a single unique product in each (marked with circles) suggests a local change in surface accessibility that simultaneously exposes one trypsin cleavage site while deleting or masking another.
To ensure that the domain 2 linker replacement did not result in gross alterations to the three-dimensional structure of the protein, purified full-length Ply-HA and D2-HA were subjected to limited proteolysis (Fig. 6B). Approximately equivalent amounts of each purified protein were incubated either alone or in the presence of identical amounts of trypsin. Trypsin alone and Ply* (polyhistidine-tagged Ply toxoid) plus trypsin served as negative controls. Equal amounts of each reaction product were analyzed by Western blotting with an HA antibody. The proteolytic degradation products of both Ply-HA and D2-HA are nearly identical (Fig. 6B, asterisks), which suggests that the trypsin-accessible sites and thus the three-dimensional structures of the proteins are similar. The presence of a single unique product in each (Fig. 6B, circles) suggests a local change in surface accessibility that simultaneously exposes one trypsin cleavage site while removing another. The latter is consistent with the removal of domain 2, which contains several potential trypsin cleavage sites, and its replacement with a GGS linker that lacks trypsin cleavage sites.
Domain 2 swaps between Ply and Pfo show that Ply domain 2 is necessary for Ply export.
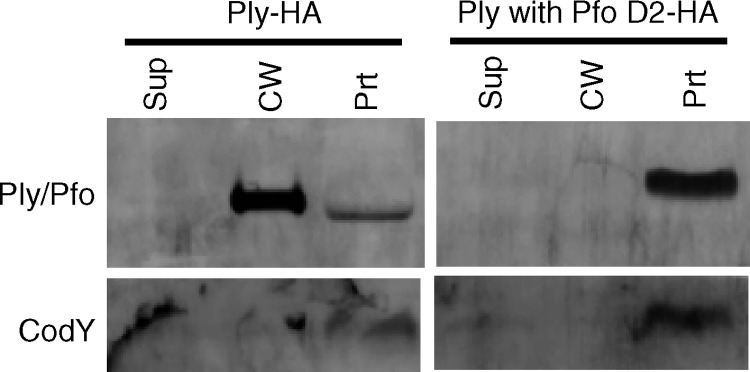
Given that Pfo localizes to the cell wall of S. pneumoniae with low efficiency and that domain 2 is required for Ply export, we hypothesized that domain 2 of Ply is a signal for, or possibly directly involved in, Ply export in S. pneumoniae. To test this hypothesis, domain 2 swaps between Ply and Pfo were constructed and tested. Replacement of Pfo's domain 2 with that of Ply resulted in an unstable protein, as no protein was detected in any fraction using either Pfo or Ply antibodies (data not shown). Additionally, this mutant strain had a low growth rate (data not shown). Taken together these results suggest that the modified Pfo protein is unstable and somewhat toxic to S. pneumoniae. On the other hand, replacement of Ply's domain 2 with that of Pfo resulted in a protein that is detected at its expected molecular mass but is found only in the protoplast and not in the cell wall or supernatant fraction (Fig. 7). This result supports the hypothesis that domain 2 of Ply is necessary for, and possibly directly involved in, Ply export.
Fig 7.
Ply domain 2 is necessary for export to the cell wall in S. pneumoniae. Domain 2 of Ply was replaced with domain 2 of Pfo and crossed into the ply locus on the S. pneumoniae chromosome. The strains carrying ply-HA and ply with Pfo's D2-HA were grown to mid-exponential phase, fractionated into supernatant, cell wall, and protoplast compartments, and assayed for the presence of Ply and CodY by Western blotting. Equal cell equivalents were loaded on the gel. Sup, culture supernatant; CW, cell wall; Prt, protoplast.
Expression of ply in Bacillus subtilis.
To test whether the pathway that exports Ply in S. pneumoniae is conserved in other Gram-positive bacteria, we generated a B. subtilis strain that expresses native ply under the control of the Pspac* promoter in the same manner as the SSRrgB-Ply fusion. The ply-expressing B. subtilis strain was grown to mid-exponential phase, fractionated, and assayed for the presence of Ply and CodY by Western blotting and for the activity of Ply by hemolysis assays. Ply was detected in all fractions, with the strongest band in the cell wall, which also had the highest level of hemolytic activity (Fig. 8 and 3B). This result suggests that the putative Ply export pathway is conserved in B. subtilis or, alternatively, that Ply is autosecretory.
Fig 8.
Ply localizes to the cell wall when expressed in Bacillus subtilis. The ply gene was placed under the control of the Pspac* promoter in the amyE locus on the B. subtilis chromosome (strain Ply B.s.). The B. subtilis parent strain, JH642, served as a control. Strains were grown to mid-exponential phase, fractionated into supernatant, cell wall, and protoplast compartments, and assayed for the presence of Ply and CodY by Western blotting. Equal equivalents were loaded on the gel. Sup, culture supernatant; CW, cell wall; Prt, protoplast.
DISCUSSION
One commonality to all known functions of Ply is where they take place—outside the bacterial cell (6, 7, 25, 27). Homologs of Ply, which share similar functions, contain signal sequences to direct their secretion through the type II (Sec) secretion system. Ply, in contrast, does not contain a signal sequence (32) and was initially hypothesized to be released by autolysis (24). We have previously shown that Ply is cell wall localized in a lytA-independent manner (26); however, the possibility still remained that limited autolysis caused by other lysins may release Ply, which could then reassociate with cells.
There are other proteins lacking a recognizable signal sequence or cell wall anchoring motif that “moonlight” on the bacterial surface, such as enolase and streptococcal surface GAPDH (SDH) (5, 16, 22, 23). Enolase has been shown to reassociate with the cell surface (5), leaving open the possibility that a limited number of cells lyse and release their cytoplasmic contents, which then stick to the surfaces of nearby intact, unlysed cells. However, a recent study analyzing the localization of SDH in S. pyogenes showed that the protein could be held inside the cell with a hydrophobic tail, lending support to the hypothesis that there is a dedicated export pathway for this protein (16). Here we show that exogenous Ply that has been released by autolysis cannot reassociate with cells. Combined with our previous work showing Ply in the cell wall fraction in a lytA-independent manner (26), we conclude that Ply localized to the cell wall is not the result of autolysis and that there must be an autolysis-independent Ply export pathway.
It is striking that Ply is not secreted through the Sec machinery, in contrast to the case for all other known members of the CDC family. This difference in Ply secretion mechanism could simply be due solely to the lack of a signal sequence in ply or, in addition, to an unknown incompatibility between Ply and the Sec machinery. To address these possibilities, we expressed Ply containing a known functional signal peptide from RrgB, an S. pneumoniae secreted protein (18). When the SSRrgB-Ply fusion was expressed in S. pneumoniae, there was no detectable Ply protein. The transcription of this gene was comparable to that of wild-type ply, indicating that the defect in protein expression is posttranscriptional in nature. Assuming that the RrgB signal peptide directs the fusion protein to the Sec machinery, we hypothesize that SSRrgB-Ply is incompatible with the Sec machinery of S. pneumoniae in such a way that leads to Ply degradation.
This putative signal sequence-dependent degradation of Ply is reminiscent of results with MalE-LacZ fusions from classic studies that identified and defined signal sequences in Escherichia coli (2, 3). According to those experiments, the addition of a signal sequence directed LacZ to the membrane, but the Sec machinery was unable to secrete it. Thus, LacZ is incompatible with the Sec machinery. In our case, two possibilities that could explain the SSRrgB-Ply secretion defect are (i) incompatibility between the SSRrgB-Ply fusion protein and the Sec machinery of Gram-positive bacteria in general (i.e., the defect lies with the substrate) or (ii) incompatibility between SSRrgB-Ply and specifically the S. pneumoniae Sec machinery (i.e., the defect lies with the machinery). To test the first of these possibilities, we expressed SSRrgB-Ply in the heterologous Gram-positive host B. subtilis. Strikingly, the SSRrgB-Ply protein in B. subtilis is expressed and secreted. Thus, SSRrgB-Ply is compatible with the Sec machinery of B. subtilis. This result rules out the hypothesis that the defect of SSRrgB-Ply secretion in S. pneumoniae lies with the substrate and suggests that the defect of SSRrgB-Ply secretion in S. pneumoniae is likely due to components of the S. pneumoniae Sec machinery.
Given that the incompatibility between SSRrgB-Ply and the Sec machinery was likely the Sec machinery of S. pneumoniae, we reasoned that natural signal sequence-containing CDCs might also be incompatible with the Sec machinery in S. pneumoniae. Therefore, we expressed a closely related homolog of Ply, perfringolysin O (Pfo), from the ply locus and determined its localization. We expressed two versions of Pfo, one with and one without its native signal sequence. Similar to the SSRrgB-Ply result, Pfo with its signal sequence not only was undetectable but appeared to be detrimental to cell growth, since all viable transformants had frameshift mutations within the signal sequence or lacked the signal sequence altogether. These results indicate that there is an incompatibility between Pfo and the Sec machinery of S. pneumoniae. In contrast, a construct of Pfo lacking its signal sequence was tolerated by S. pneumoniae and was detected mainly in the protoplast fraction, with a small amount detected in the cell wall fraction. Thus, the putative Ply export pathway appears to be able to recognize and secrete other signal sequence-less CDCs, albeit with lower efficiency. We do not know if this putative Ply export pathway is the same pathway used to export the signal sequence-less proteins enolase and GAPDH. Testing of this possibility awaits identification and inactivation of the putative export pathway.
Though the identity of the hypothesized Ply export pathway remains unknown, we can nonetheless begin to characterize domains of Ply that are required for its export and cell wall localization. To this end, we used a series of terminal truncations and internal domain replacements to deduce that domain 2 is required for proper Ply localization. Domain 2 is a beta-sheet formed by two noncontiguous regions of Ply (amino acids 22 to 57 and 343 to 359). The function of this domain in the biology of Ply is in the prepore-to-pore transition. The alpha-helices of domain 3, which ultimately form the transmembrane hairpins (TMHs) that form the pore in the eukaryotic membrane, are stabilized by domain 2. Once the TMHs have formed, domain 2 is responsible for the vertical collapse of the protein that allows the TMHs to reach the target membrane and pierce it (8, 30). It is therefore possible that in the absence of domain 2, the domain 3 alpha-helices are not stabilized and can form beta-sheets that might be able to insert into the bacterial cell's membrane or result in a conformation not recognized by the putative export pathway, thus preventing its export. However, given that limited proteolyses of Ply-HA and D2-HA yield nearly identical banding patterns, it is unlikely that the D2-HA protein is misfolded, lending more evidence to the hypothesis that domain 2 is directly involved in the export of Ply.
Given that domain 2 is required for Ply export and that a signal sequence-less version of Pfo is somewhat competent for export and localization to the cell wall when expressed in S. pneumoniae, we determined whether domain 2 is necessary and sufficient for cell wall localization of CDCs in S. pneumoniae. Ply with Pfo's domain 2 is not exported, further suggesting that domain 2 is necessary for Ply export. Unfortunately, Pfo with Ply's D2 is undetectable in S. pneumoniae, so we cannot conclude whether Ply's domain 2 is sufficient for export. It may be possible in future studies to identify critical residues in domain 2 by site-specific mutagenesis and thus refine this model.
It appears that there is a domain 2-dependent Ply export pathway in S. pneumoniae. It is possible that this putative export pathway is specific to S. pneumoniae or instead might be conserved among other bacteria. To begin testing how conserved this putative pathway is, a B. subtilis strain that expresses native ply under the control of a constitutively active promoter was constructed. Ply localizes in B. subtilis much like it does in S. pneumoniae, indicating that the putative Ply export pathway is conserved in B. subtilis. However, in the absence of knowledge of the components of this export pathway, we cannot rule out the alternative mechanism of autosecretion of Ply, even though Ply lacks any recognizable autosecretory domain (10).
This work shows that cell wall-localized Ply is exported from the cytoplasm in an autolysis-independent, domain 2-dependent manner. The incompatibility with the Sec machinery exhibited by Ply (and Pfo when expressed in S. pneumoniae) is not observed when Ply is expressed in B. subtilis. These results indicate that the S. pneumoniae Sec machinery cannot accommodate a CDC, which may provide insight as to why Ply, unlike all other CDCs, does not contain a Sec signal sequence.
ACKNOWLEDGMENTS
This work was funded by the Howard Hughes Medical Institute and the Center for Gastroenterology Research on Absorptive and Secretory Processes, NEMC (P30 DK34928).
Footnotes
Published ahead of print 4 May 2012
REFERENCES
- 1. Balachandran P, Hollingshead SK, Paton JC, Briles DE. 2001. The autolytic enzyme LytA of Streptococcus pneumoniae is not responsible for releasing pneumolysin. J. Bacteriol. 183:3108–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bassford P, Beckwith J. 1979. Escherichia coli mutants accumulating the precursor of a secreted protein in the cytoplasm. Nature 277:538–541 [DOI] [PubMed] [Google Scholar]
- 3. Bassford PJ, JR, Silhavy TJ, Beckwith JR. 1979. Use of gene fusion to study secretion of maltose-binding protein into Escherichia coli periplasm. J. Bacteriol. 139:19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benton KA, Paton JC, Briles DE. 1997. Differences in virulence for mice among Streptococcus pneumoniae strains of capsular types 2, 3, 4, 5, and 6 are not attributable to differences in pneumolysin production. Infect. Immun. 65:1237–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bergmann S, Rohde M, Chhatwal GS, Hammerschmidt S. 2001. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40:1273–1287 [DOI] [PubMed] [Google Scholar]
- 6. Braun JS, et al. 2007. Pneumolysin causes neuronal cell death through mitochondrial damage. Infect. Immun. 75:4245–4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Braun JS, Novak R, Gao G, Murray PJ, Shenep JL. 1999. Pneumolysin, a protein toxin of Streptococcus pneumoniae, induces nitric oxide production from macrophages. Infect. Immun. 67:3750–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Czajkowsky DM, Hotze EM, Shao Z, Tweten RK. 2004. Vertical collapse of a cytolysin prepore moves its transmembrane beta-hairpins to the membrane. EMBO J. 23:3206–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dubnau D, Davidoff-Abelson R. 1971. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J. Mol. Biol. 56:209–221 [DOI] [PubMed] [Google Scholar]
- 10. Gardy JL, et al. 2003. PSORT-B: improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res. 31:3613–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hava DL, Camilli A. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389–1406 [PMC free article] [PubMed] [Google Scholar]
- 12. Hava DL, Hemsley CJ, Camilli A. 2003. Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA. J. Bacteriol. 185:413–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoch JA. 1993. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu. Rev. Microbiol. 47:441–465 [DOI] [PubMed] [Google Scholar]
- 14. Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68 [DOI] [PubMed] [Google Scholar]
- 15. Jedrzejas M. 2001. Pneumococcal virulence factors: structure and function. Microbiol. Mol. Biol. Rev. 65:187–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jin H, Agarwal S, Agarwal S, Pancholi V. 2011. Surface export of GAPDH/SDH, a glycolytic enzyme, is essential for Streptococcus pyogenes virulence. mBio 2(3):e00068–11 doi:10.1128/mBio.00068-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson M. 1977. Cellular location of pneumolysin. FEMS Microbiology Lett. 2:243–245 [Google Scholar]
- 18. LeMieux J, Hava DL, Basset A, Camilli A. 2006. RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infect. Immun. 74:2453–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martin B, Prudhomme M, Alloing G, Granadel C, Claverys JP. 2000. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol. Microbiol. 38:867–878 [DOI] [PubMed] [Google Scholar]
- 20. McNew JA, Weber T, Engelman DM, Sollner TH, Rothman JE. 1999. The length of the flexible SNAREpin juxtamembrane region is a critical determinant of SNARE-dependent fusion. Mol. Cell 4:415–421 [DOI] [PubMed] [Google Scholar]
- 21. Mitchell TJ, Andrew PW, Saunders FK, Smith AN, Boulnois GJ. 1991. Complement activation and antibody binding by pneumolysin via a region of the toxin homologous to a human acute-phase protein. Mol. Microbiol. 5:1883–1888 [DOI] [PubMed] [Google Scholar]
- 22. Pancholi V, Fischetti VA. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176:415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pancholi V, Fischetti VA. 1998. α-Enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503–14515 [DOI] [PubMed] [Google Scholar]
- 24. Paton JC, Andrew PW, Boulnois GJ, Mitchell TJ. 1993. Molecular analysis of the pathogenicity of Streptococcus pneumoniae: the role of pneumococcal proteins. Annu. Rev. Microbiol. 47:89–115 [DOI] [PubMed] [Google Scholar]
- 25. Paton JC, Rowan-Kelly B, Ferrante A. 1984. Activation of human complement by the pneumococcal toxin pneumolysin. Infect. Immun. 43:1085–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Price KE, Camilli A. 2009. Pneumolysin localizes to the cell wall of Streptococcus pneumoniae. J. Bacteriol. 191:2163–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rossjohn J, et al. 1998. The molecular mechanism of pneumolysin, a virulence factor from Streptococcus pneumoniae. J. Mol. Biol. 284:449–461 [DOI] [PubMed] [Google Scholar]
- 28. Serio AW, Sonenshein AL. 2006. Expression of yeast mitochondrial aconitase in Bacillus subtilis. J. Bacteriol. 188:6406–6410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stool SE, Field MJ. 1989. The impact of otitis media. Pediatr. Infect. Dis. J. 8:S11–S14 [PubMed] [Google Scholar]
- 30. Tilley SJ, Orlova EV, Gilbert RJ, Andrew PW, Saibil HR. 2005. Structural basis of pore formation by the bacterial toxin pneumolysin. Cell 121:247–256 [DOI] [PubMed] [Google Scholar]
- 31. Tweten RK. 1988. Nucleotide sequence of the gene for perfringolysin O (theta-toxin) from Clostridium perfringens: significant homology with the genes for streptolysin O and pneumolysin. Infect. Immun. 56:3235–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Walker JA, Allen RL, Falmagne P, Johnson MK, Boulnois GJ. 1987. Molecular cloning, characterization, and complete nucleotide sequence of the gene for pneumolysin, the sulfhydryl-activated toxin of Streptococcus pneumoniae. Infect. Immun. 55:1184–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Williams BG, Gouws E, Boschi-Pinto C, Bryce J, Dye C. 2002. Estimates of world-wide distribution of child deaths from acute respiratory infections. Lancet Infect. Dis. 2:25–32 [DOI] [PubMed] [Google Scholar]