Abstract
The culture supernatant fraction of an Enterococcus faecalis gelE mutant of strain OG1RF contained elevated levels of the secreted antigen SalB. Using differential fluorescence gel electrophoresis (DIGE) the salB mutant was shown to possess a unique complement of exoproteins. Differentially abundant exoproteins were identified using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry. Stress-related proteins including DnaK, Dps family protein, SOD, and NADH peroxidase were present in greater quantity in the OG1RF salB mutant culture supernatant. Moreover, several proteins involved in cell wall synthesis and cell division, including d-Ala-d-Lac ligase and EzrA, were present in reduced quantity in OG1RF salB relative to the parent strain. The salB mutant displayed reduced viability and anomalous cell division, and these phenotypes were exacerbated in a gelE salB double mutant. An epistatic relationship between gelE and salB was not identified with respect to increased autolysis and cell morphological changes observed in the salB mutant. SalB was purified as a six-histidine-tagged protein to investigate peptidoglycan hydrolytic activity; however, activity was not evident. High-pressure liquid chromatography (HPLC) analysis of reduced muropeptides from peptidoglycan digested with mutanolysin revealed that the salB mutant and OG1RF were indistinguishable.
INTRODUCTION
Enterococci are commensals of the animal gut, and several species cause disease in humans, including wound and urinary tract infections, endocarditis, and bacteremia. Enterococcus faecalis and Enterococcus faecium have become increasingly problematic due to rising rates of infection, combined with accumulated resistance to antibiotics (4, 27). E. faecalis is the most virulent species in the genus and the cause of approximately 80% of all enterococcal infections, with virulence factors that include adhesins, proteases, and cytolysin (10, 11, 33). The accumulation of resistance genes is important medically, since intergeneric horizontal transfer of antibiotic resistance genes involving Staphylococcus aureus and Listeria species has been reported (3, 16, 20, 30, 38).
E. faecalis strains with an intact Fsr quorum sensing system temporally express metalloprotease (GelE) and serine protease (SprE) (19), and these are the two most abundant exoproteins in post-exponential-growth-phase cultures (21, 29). GelE, a member of the peptidase M4 family, proteolytically activates the major cell wall N-acetylglucosaminidase, AtlA (35), which contributes to cell separation after cell division (1, 15). The E. faecalis exoprotein SalB is a paralogue of the essential E. faecium cell wall-associated SagA (34) and streptococcal PcsB proteins (23, 24). The enzymatic activity of these proteins has not been unambiguously determined, but bioinformatic analysis supports peptidoglycan hydrolase activity and PcsB was reported to interact with the essential cell division protein, FtsX (28). The genes encoding SalA/SagA/PcsB have a conserved location in each species downstream of genes encoding the cell shape-determining proteins MreC and MreD (13, 23). E. faecalis SalB shares ∼40% primary amino acid sequence identity (∼70% similarity) with streptococcal PcsB.
E. faecalis SalB is not essential for growth. Inactivation of salB increased the capability of the bacterium to bind the extracellular matrix (ECM) molecules fibronectin and collagen type I and enhanced biofilm production when grown with serum, relative to the wild type (17). A lack of SalB activity results in reduced resistance to a range of stressors, including bile salts, detergent, ethanol, peroxide, heat, and low pH (25). Morphological changes including septation anomalies and increased numbers of spherical cells were identified in a salB mutant of E. faecalis strain JH2-2, and the mutant had a growth defect in complex medium relative to its isogenic parent (5). SalB expression is induced by extracellular stress via the two-component signal transduction system CroRS in strain JH2-2 (18).
In this study, SalB was found at elevated levels in the culture supernatant of strain TX5264 (OG1RF gelE). To investigate a potential epistatic relationship between GelE and SalB, double mutant strains Liv1016 (OG1RF gelE salB) and Liv1013 (OG1RF fsrB salB) were constructed and compared with strain Liv729 (OG1RF salB) and the wild-type parent OG1RF strain. Strain Liv729 (OG1RF salB) expresses a distinct set of exoproteins compared with its isogenic parent, potentially indicating a cellular role for SalB.
MATERIALS AND METHODS
Strains and culture conditions.
E. faecalis strains were routinely cultured in brain heart infusion (BHI) broth (Lab M) or BHI solidified with 1.5% (wt/vol) agar (Table 1). Escherichia coli strains were cultured using Luria-Bertani (LB) medium. Antibiotics were added at selective concentrations for the maintenance of plasmids, prior to comparative analyses (Table 1). Culture conditions for E. faecalis were 50 ml BHI in a 250-ml conical flask incubated with shaking at 37°C and with growth monitored at 600 nm.
Table 1.
Strains and plasmids used in this study
| Bacterium strain or plasmid | Strain ID | Characteristics | Marker | Source |
|---|---|---|---|---|
| E. faecalis strains | ||||
| OG1RF | TX4002 | Wild type, Rifr/Fusr | 7 | |
| JH2-2 | Wild type, Rifr/Fusr | 12 | ||
| OG1RF gelE | TX5264 | In-frame deletion of gelE | 27 | |
| OG1RF fsrB | TX5266 | fsrB mutant | 18 | |
| OG1RF fsrB pTEX5249 | Liv305 | fsrB mutant complemented with fsrABDC | Erm | 27 |
| OG1RF salB | Liv729 | salB insertional inactivation using pTEX4577 | Kan | This study |
| OG1RF salB pAT18::salB | Liv883 | Complemented salB mutant | Kan, Erm | This study |
| JH2-2 salB | Liv1012 | salB insertional inactivation using pTEX4577 | Kan | This study |
| OG1RF fsrB salB | Liv1013 | salB inactivation in OG1RF fsrB background | Kan | This study |
| OG1RF gelE salB | Liv1016 | salB inactivation in OG1RF gelE background | Kan | This study |
| E. coli strains | ||||
| BL21(DE3) pLysS | F− ompT hsdSB(rB− mB−) gal dcm (DE3) | Cm | 30 | |
| TOP10 | Cloning host | Invitrogen | ||
| BL21(DE3) pLysS pJAY1 | SalB expression strain | Kan, Cm | This study | |
| Plasmids | ||||
| pTEX4577 | Insertional inactivation plasmid | Kan | 33 | |
| pAT18 | Shuttle vector | Erm | 28 | |
| pET24d (+) | T7 RNA polymerase vector His-tag | Kan | Novagen | |
| pRGW1 | pTEX4577 with 587-bp salB fragment | Kan | This study | |
| pJAY1 | pET24d+ with 1,265-bp fragment of salB | Kan | This study | |
| pJAY2 | pAT18 with 1,645-bp salB operon | Erm | This study |
Molecular cloning.
For insertional inactivation of the salB gene, a 587-bp internal gene fragment was amplified by PCR from the chromosome of E. faecalis OG1RF using primers SalBKO_BamHI (5′-AAGGATCCTCTAAGCCAGCTTTAGAAC) and SalBKO_EcoRI (5′-CCGAATTCCGCTATCAGCAATTGCTAC) (where underlining indicates the restriction enzyme sequence), digested with BamHI/EcoRI, and ligated (11) to digested suicide plasmid pTEX4577 (36) to create pRGW1. The latter plasmid was used to transform OG1RF, TX5264 (OG1RF gelE), and TX5266 (OG1RF fsrB) to create the isogenic mutants Liv729 (OG1RF salB), Liv1016 (OG1RF gelE salB), and Liv1013 (OG1RF fsrB salB). A salB complementation plasmid was made by ligating the BamHI/EcoRI-digested 1,645-bp salB operon of E. faecalis OG1RF, which was amplified by PCR using primers SalBcBamHI (5′-CCACGGATCCGTGAAAACCAGTCGTGAC) and SalBcEcoRI (5′-ACATGAATTCTTAGAATACCACGTTTAGC) to digested plasmid pAT18 (31), producing pJAY2. Strain Liv729 (OG1RF salB) was transformed to create strain Liv883 (OG1RF salB pAT18::salB).
To obtain the six-histidine-tagged enzyme, the salB gene was amplified and digested to produce a 1,265-bp NcoI/XhoI product using primers SalBOexF (5′-GTACCCATGGACAATGTTGATAAAAAA) and SalBOexR (5′-GTACCTCGAGGGCTGAGTGTCCTACGATTG). This product was ligated to digested pET24d+ (Novagen) to create plasmid pJAY1. The fidelity of the cloned salB gene was confirmed by DNA sequencing (GATC Biotech). The six-histidine-tagged SalB was expressed in strain E. coli BL21 (λDE3) pLysS following induction of log-phase cells in LB containing 0.5 M glucose with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were lysed using sonication, and the protein was purified under native conditions using Probond chelate resin (Invitrogen) according to the manufacturer's instructions.
Exoprotein analysis.
Separation of proteins using one-dimensional (1D) SDS-PAGE was performed after precipitation of cell culture supernatant proteins with 10% (wt/vol) trichloroacetic acid (TCA). Exoprotein purification for 2D SDS-PAGE used a previously described method (6). Exoproteins (500 μg) were focused using a pH 4 to 7 strip (Bio-Rad) and then separated in the second dimension using a 12% (wt/vol), 20 cm by 20 cm polyacrylamide gel. Gels were stained using colloidal Coomassie, scanned using a Gel Imager, and analyzed by using SameSpots software (Progenesis). For comparative proteomics of strains OG1RF and Liv729 (OG1RF salB), 80 μg exoprotein from each was labeled with the Cy dyes (GE Healthcare) Cy3 and Cy5, respectively, according to the manufacturer's instructions, before focusing and separation as for the 2D SDS-PAGE protocol. Dye swap controls were performed to confirm labeling efficiency. As an internal control, 40 μg exoprotein from each strain was pooled and stained with Cy2 dye and separated with the samples. Gel images were captured using a Typhoon gel imager and compiled using Adobe Photoshop software. The cutoff criteria for differential quantification (1.5-fold) were used based upon the manufacturer's literature.
Protein spots of interest were excised from the gels and digested overnight with digest buffer containing Trypsin Gold (20 ng μl−1), mass spectrometry (MS) grade (Promega). The peptide mass fingerprints were derived by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) analysis of tryptic fragments using a Voyager DE Pro (Applied Biosystems). Mass lists were transferred to the Mascot peptide mass fingerprint database (Matrix Science, Ltd.), where they were used to search for matches within the Firmicutes. A peptide tolerance of ±0.5 Da and up to one missed peptide cleavage was allowed. Fixed carboxymethyl modifications and variable oxidation modifications were allowed.
Peptidoglycan hydrolase zymograms.
Peptidoglycan hydrolase activity was visualized after proteins were separated by SDS-PAGE, supplementing the resolving gel with 10 optical density at 600 nm (OD600) units of purified peptidoglycan isolated from the cell walls of E. faecalis or Micrococcus luteus (26, 32). After separation, gels were washed for 30 min in distilled water and then renaturing buffer (10 mM MgCl2, 25 mM Tris-HCl, 10% [vol/vol] Triton X-100). Gels were incubated overnight at 37°C in fresh renaturing buffer and then washed with distilled water prior to staining (0.05% [wt/vol] methylene blue in 1 mM KOH) for 2 h. The gel was destained and then repeatedly washed with distilled water until bands of clearing appeared.
Autolysis assay.
Exponential-phase cells were harvested and washed with phosphate-buffered saline (PBS) prior to their resuspension in PBS containing either 0.1% (vol/vol) Triton X-100 or 0.1 mg ml−1 penicillin G and incubated at 37°C with shaking. Lysis was monitored by measuring OD600.
Imaging and viability staining.
An equal volume of exponential-growth-phase bacterial culture and freshly prepared Live/Dead BacLight (Invitrogen) stain (6 μM Syto9 and 30 μM propidium iodide [PI]) was incubated at room temperature for 15 min in the dark. Ten microliters stained cells was mounted between two coverslips and visualized using a Carl Zeiss LSM 710 laser scanning microscope equipped with dual-emission filters for visualizing Syto9- and PI-stained cells simultaneously. Mean green and red fluorescence levels of Live/Dead Baclight-stained cells from 100,000 events were measured using a flow cytometer. The total fluorescence was gated, and propidium iodide-stained dead cells were plotted as a percentage of total events in the population for comparison.
For electron microscopy thin sections (40 to 70 μm) of epoxy resin-embedded mid-exponential-phase cells were cut using a Leica microtome and transferred to pioloform-coated grids. The thin sections were then visualized in a 120 kV Technai G2 Spirit Biotwin transmission electron microscope (TEM) with images analyzed by Technai G2 software.
Purification of peptidoglycan and muropeptide analysis.
Peptidoglycan was isolated using the procedure of Rosenthal and Dziarski (26). Briefly, cultures of E. faecalis OG1RF or M. luteus were grown to mid-exponential phase, harvested, washed, and autoclaved. Cells were ruptured five times in a French pressure cell at 14,000 lb/in2 (>95% breakage). The ruptured cells were harvested at 16,100 relative centrifugal force (RCF) for 5 min and boiled/washed five times in SDTE buffer (2% [wt/vol] SDS, 20 mM dithiothreitol [DTT], 50 mM Tris-HCl, 1 mM EDTA) at 100°C for 10 min and pelleted at 20,000 RCF for 5 min. The final pellet was boiled and washed 6 times in water. The peptidoglycan was isolated by gentle shaking with water between washes. All the peptidoglycan was pooled and harvested by centrifugation at 20,000 RCF for 5 min and resuspended in 1 ml sterile water. Its turbidity was measured at 600 nm, and the sample was stored at −20°C.
The method for the reduction and separation of muropeptides was modified from previous work on E. faecalis autolysin AtlA (8). Ten OD units of purified peptidoglycan was incubated at 37°C in 200 mM sodium phosphate buffer at pH 6.0, 0.1 mM MgCl2 with 5 μg mutanolysin (Sigma) or with 45 to 50 μg of SalB to a final volume of 250 μl. Following digestion overnight, the peptidoglycan was centrifuged at 16,000 RCF for 30 min. Supernatant (200 μl) containing soluble muropeptides was recovered and transferred to a fresh Eppendorf tube. An equal volume of 250 mM sodium borate buffer was added with 2 mg of solid sodium borohydride and left at room temperature (RT) for 15 min. Excess sodium borohydride was removed by the addition of 5 μl of o-phosphoric acid. Fifty microliters of muropeptides was acidified with 2.5 μl trifluoroacetic acid (TFA) (1% [vol/vol] final concentration) before separation on a Brownlee PepMap C-18 high-pressure liquid chromatography (HPLC) column run on a Beckman-Coulter System Gold HPLC instrument. Muropeptides were separated using a linear gradient of 0 to 45% (vol/vol) acetonitrile in 0.1% (vol/vol) TFA, and elution was monitored at 214 nm.
RESULTS
Inactivation of gelE and salB alters culture supernatant exoproteins.
The contribution made by GelE activity to temporal expression of the exoprotein fraction of E. faecalis OG1RF was determined by extracting the culture supernatant proteins of TX5264 (OG1RF gelE) after 8 h of growth (determined to be in stationary phase, data not shown) and comparing them to exoproteins from the isogenic parent strain OG1RF (Fig. 1; data not shown). Several novel protein bands were present in the supernatant protein fraction of strain TX5264 (OG1RF gelE), including a 55-kDa protein present at all growth stages, which was identified as SalB following trypsin digest and MALDI-TOF mass spectrometry (seven matching peptides; Mowse score = 52). In the culture supernatant of strain OG1RF, there was an absence of SalB from 5 h (mid-exponential growth phase) concomitant with increased protease (GelE and SprE) expression (data not shown). A potential epistatic relationship between salB and gelE was investigated via insertional inactivation of salB in OG1RF and TX5264 (OG1RF gelE) to produce strains Liv729 (OG1RF salB) and Liv1016 (OG1RF gelE salB). Subsequently, since the enzymatic function of SalB remains undetermined in E. faecalis, the stationary-phase culture supernatant exoproteins produced by strains Liv729 (OG1RF salB) and Liv1016 (OG1RF gelE salB) were compared with those of their isogenic parent strains (Fig. 1). A distinct complement of exoproteins was present in the culture supernatant of strain Liv729 (OG1RF salB), and this was absent in the complementation strain Liv883 (OG1RF salB pAT18::salB) (Fig. 1 and data not shown), indicating that the absence of SalB caused changes to the exoproteome. These changes were investigated to determine whether they might reveal a function for SalB.
Fig 1.
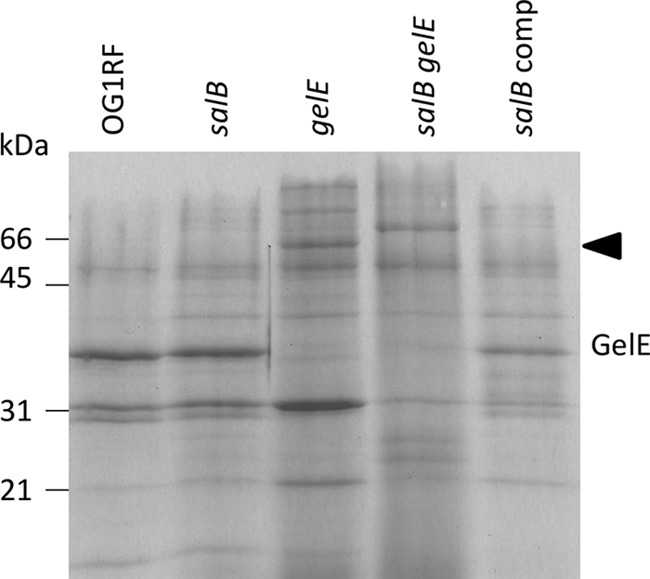
Culture supernatant exoproteins from E. faecalis strains. Exoproteins of OG1RF and its isogenic mutants with mutations in salB (Liv729 [OG1RF salB]), gelE (OG1RF gelE), salB gelE (Liv1016 [OG1RF salB gelE]), and salB comp (Liv883 [OG1RF salB pAT18::salB]), harvested at early stationary (8 h) growth phase. Molecular mass markers (kDa) are shown on the left. The arrowhead indicates SalB.
Identification of the salB mutant exoproteome.
To gain insight into the cellular role of SalB, the exoproteins produced by strain Liv729 (OG1RF salB) were quantitatively compared with those of its isogenic parent strain using 2D differential fluorescence gel electrophoresis (DIGE). Based upon a previous exoproteome analysis (29) stationary-phase culture supernatants (stationary phase was determined to be after 8 h of growth; data not shown) were extracted, 80 μg of protein from each strain was separated, and the proteins were compared. The exoprotein fraction from strain Liv729 (OG1RF salB) could be distinguished from that of OG1RF largely by the presence of a unique protein set with low pI and high molecular mass (Fig. 2A). Differentially abundant or novel proteins from triplicate gels of strain Liv729 (OG1RF salB) were compared using SameSpots software (Progenesis) and quantified relative to OG1RF (Tables 2 and 3). Subsequently, proteins with differential abundance were isolated from a 500-μg exoprotein sample, separated by 2D SDS-PAGE, and oriented relative to the gels from DIGE (Fig. 2A and B). MALDI-TOF mass spectrometry of tryptic digests assigned 24 identities to 31 protein spots that were quantified as being in increased amount in strain Liv729 (OG1RF salB) compared with OG1RF, with a fold difference of at least 1.5 (Tables 2 and 3). Thirteen proteins were quantified from OG1RF exoproteome gels (Fig. 2A and data not shown) (29) as being decreased or absent in the salB mutant strain, with eight spots assigned identities from MALDI-TOF mass spectrometry of tryptic digests (Table 3). Bioinformatic analysis of these identified proteins revealed that many of these have known or proposed roles in the general stress response, cell wall synthesis, cell division, and metabolism.
Fig 2.
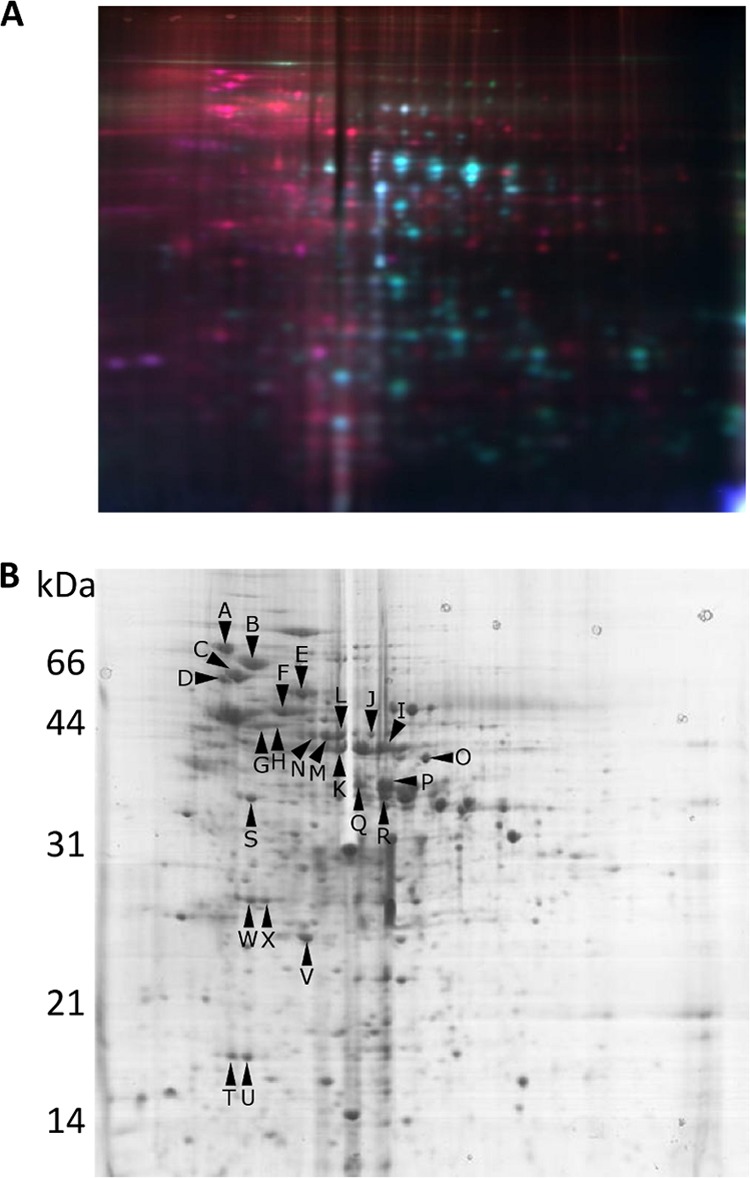
(A) DIGE analysis of culture supernatant exoproteins produced after 8 h of growth by strain Liv729 (OG1RF salB), labeled with Cy3 (yellow-green), and those produced by its parent strain OG1RF, labeled with Cy5 (red). (B) Exoproteins produced by strain Liv729 (OG1RF salB) and separated by 2D SDS-PAGE, with arrowed letters indicating differentially regulated proteins.
Table 2.
Characteristics of proteins extracted in this study
| Spot ID | Protein description (accession no.) | Fold change | Calculated mass (kDa)/pI value | No. of peptides matched | Sequence coverage (%) |
|---|---|---|---|---|---|
| Group 1, stress-related proteins | |||||
| A | DnaK (EF_1308) | 2.02 | 65.54/4.59 | 12 | 22 |
| C | GroEL (EF_2633) | 4.05 | 55.41/4.66 | 13 | 30 |
| F | NADH peroxidase, Npr (EF_1211) | 2.27 | 49.52/4.83 | 7 | 25 |
| T | Dps family protein (EF_3233) | 1.85 | 17.93/4.56 | 6 | 47 |
| U | Dps family protein (EF_3233) | 3.82 | 17.93/4.56 | 5 | 42 |
| V | Superoxide dismutase (EF_0463) | 2.35 | 15.95/4.96 | 6 | 69 |
| Group 2, proteins of metabolic pathways | |||||
| B | PEP phosphotransferase enzyme I (EF_0710) | 3.03 | 63.13/4.68 | 13 | 21 |
| G | PEP phosphotransferase enzyme I (EF_0710) | 63.13/4.68 | 4 | 16 | |
| E | Dihydrolipoyl dehydrogenase (EF_1356) | 49.11/4.95 | 6 | 28 | |
| I | Glyceraldehyde 3-phosphate dehydrogenase, Gap-2 (EF_1353) | 3.35 | 35.92/5.03 | 6 | 38 |
| K | Acetate kinase, AckA (EF_1983) | 2.72 | 43.49/4.96 | 8 | 37 |
| L | Phosphoglycerate kinase, Pgk (EF_1963) | 2.28 | 42.37/4.9 | 8 | 32 |
| M | Phosphoglycerate kinase (EF_1963) | 5.3 | 42.37/4.9 | 7 | 27 |
| N | Phosphoglycerate kinase (EF_1963) | 2.39 | 42.37/4.9 | 5 | 17 |
| O | Glycosyltransferase (EF_2176) | 3.7 | 29.51/6.36 | 4 | 25 |
| S | Pyruvate dehydrogenase complex, PdhB (EF 1354) | 35.37/4.61 | 9 | 44 | |
| X | Triosephosphate isomerase (EF 1962) | 3.7 | 21.12/4.63 | 6 | 37 |
| Y | Triosephosphate isomerase (EF 1962) | 2.95 | 21.12/4.63 | 6 | 37 |
| Group 3, translation machinery | |||||
| D | tRNA uridine modification enzyme GidA (EF 3311) | 2.49 | 69.64/5.72 | 9 | 21 |
| H | Elongation factor G (EF_0200) | 76.63/4.8 | 7 | 20 | |
| J | Tyrosyl-t-RNA synthetase (EF_0633) | 1.96 | 47.23/5.09 | 5 | 21 |
| Group 4, proteases | |||||
| P | Zn-metalloprotease, GelE (EF_1818) | 1.77 | 55.34/4.99 | 5 | 16 |
| Q | Zn-metalloprotease, GelE (EF_1818) | 1.65 | 55.47/4.99 | 6 | 20 |
| R | Zn-metalloprotease, GelE (EF_1818) | 1.46 | 55.34/4.99 | 15 | 18 |
Table 3.
MALDI-TOF MS protein identity assignations
| Spot no. | Protein description (accession no.) | Fold change | Calculated mass (kDa)/pI value | No. of peptides matched | Sequence coverage (%) |
|---|---|---|---|---|---|
| Group 1, cell division/cell wall synthesis | |||||
| 2 | d-Alanine-d-lactate ligase (EF_2294) | −4.47 | 21.24/4.7 | 6 | 52 |
| 7 | d-Alanyl-d-alanine carboxypeptidase (Q30BF0_ENTFA) | −2.4 | 29.4/5.2 | 9 | 55 |
| 17 | Septation ring formation regulator EzrA (EF_0370) | −1.55 | 68.1/4.8 | 8 | 24 |
| 5 | DnaE protein; DNA Pol III alpha subunit (EF_1044) | 31.3/5 | 5 | 35 | |
| 4 | DNA primase (EF_1521) | −4.47 | 73/5.1 | 6 | 19 |
| Group 2, miscellaneous | |||||
| 1 | GTP-binding protein LepA (EF_2352) | −2.1 | 68.3/5 | 10 | 20 |
| 3 | GTP-binding protein LepA (EF_2352) | −4.47 | 68.3/5 | 5 | 16 |
| 6 | Transcriptional regulator, Cro/CI family (EF_2508) | −8.05 | 20.9/5.8 | 4 | 34 |
Effect of salB inactivation on cell autolysis.
The role of SalB in cell autolysis was explored because of the altered levels of cell wall synthesis and cell division enzymes that were quantified in culture supernatant of the mutant strain; moreover, SalB was hypothesized previously to be a peptidoglycan hydrolase (25). Strain Liv729 (OG1RF salB) displayed an increased rate of autolysis compared to the wild type under challenge from either penicillin G (Fig. 3) or Triton X-100 (data not shown). The autolysis phenotype was complemented in Liv883 (OG1RF salB pAT18::salB) with respect to penicillin G challenge, but only partially for Triton X-100 challenge (data not shown). Inactivation of gelE is reported to increase chain length in strain OG1RF and was therefore proposed to have a role in proteolytic activation of autolysins required for daughter cell separation (37). Limited autolysis was confirmed with strain TX5264 (OG1RF gelE), when challenged with either penicillin G or Triton X-100. The double mutant strain Liv1016 (OG1RF gelE salB) exhibited greater autolysis than did the gelatinase mutant TX5264 (OG1RF gelE). These data indicate that the absence of SalB activity increases autolysis in a gelE mutant, which supports there being no evidence of epistasis with respect to salB and gelE, thereby supporting the proteins having opposing roles in autolysis, at least under the conditions studied here.
Fig 3.
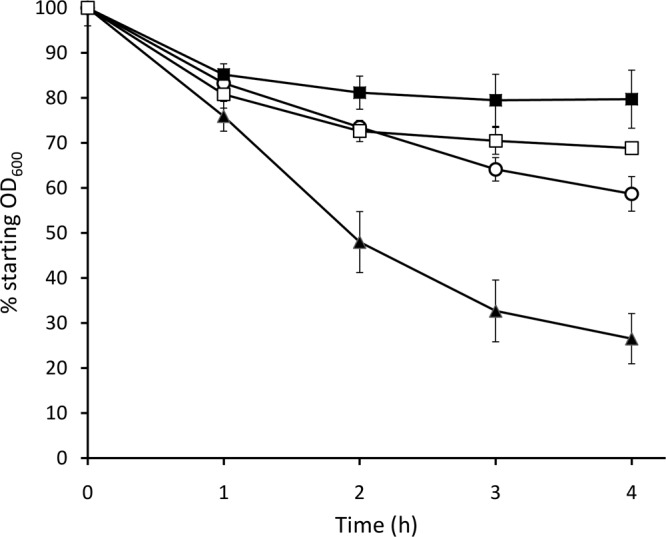
Autolysis of E. faecalis strains after addition of penicillin G. Strains are OG1RF (open circles), TX5264 (OG1RF gelE) (filled squares), Liv729 (OG1RF salB) (filled triangles), and Liv1016 (OG1RF gelE salB) (open squares). Error bars represent standard errors of the means from triplicate experiments. Statistically significant differences (P < 0.05) in autolysis were determined at 2 h, 3 h, and 4 h for Liv729 relative to OG1RF and for Liv1016 relative to TX5264, using Student's t test.
Cell separation defect and reduced viability of OG1RF salB mutant.
The observed increase in autolysis and the altered exoprotein expression of strain Liv729 (OG1RF salB) support a role for SalB in cell wall homeostasis and/or cell division. Cell separation defects were previously reported to result from salB inactivation in strain E. faecalis JH2-2, which is naturally GelE negative due to an fsr locus deletion (19). A JH2-2 salB mutant was reported to have a reduced growth rate under normal culture conditions (5). In contrast, there was no comparable growth rate reduction observed in this study for strain Liv729 (OG1RF salB) cultured in BHI (data not shown). Strain Liv1012 (JH2-2 salB) was generated in this study by transformation, and the previously described growth rate phenotype was confirmed (data not shown). Thus, salB inactivation results in differing phenotypes between host strains OG1RF and JH2-2, the latter of which contains a large deletion encompassing fsr (19). Based upon optical density measurements, no growth rate differences were observed during culture of strains Liv1016 (OG1RF gelE salB) and Liv1013 (OG1RF fsrB salB) (data not shown); consequently, the basis for the growth rate difference in strain Liv1012 (JH2-2 salB) remains undetermined.
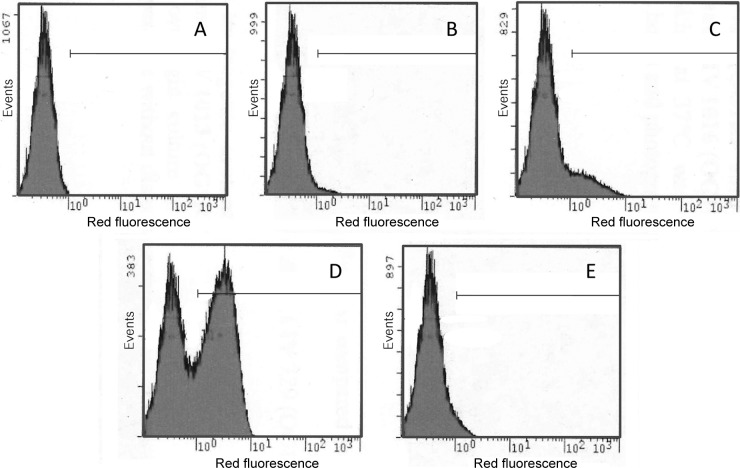
To study the effect of salB inactivation on cell wall synthesis or cell division, the different salB mutant strains were investigated by fluorescently labeling nucleic acids. Assay of the cultures using fluorescence-activated cell sorting (FACS) determined that in mid-exponential phase, strain Liv729 (OG1RF salB) exhibited a 39-fold relative increase in nonviable cells (propidium iodide, red fluorescence) compared with its isogenic parent strain (Fig. 4; Table 4). The mean green fluorescence (Syto-9) was similar for the two strains, with a 1.8-fold increase for Liv729 (OG1RF salB). The effect of gelE inactivation was investigated, and cells of strain TX5264 (OG1RF gelE) displayed an 8-fold increase in nonviable cells (1.4-fold increase in mean green fluorescence). Moreover, strain Liv1016 (OG1RF gelE salB) displayed a 145-fold increase in nonviable cells together with a 5.8-fold increase in mean green fluorescence. A considerable reduction in viability was also observed in strain Liv1013 (OG1RF fsrB salB; data not shown). Partial complementation of the viability defect resulting from salB inactivation was observed in strain Liv1016 (OG1RF salB pAT18::salB) (Fig. 4; Table 4).
Fig 4.
Analysis of viability in cell populations using fluorescence-activated cell sorting (FACS) analysis. Cells were labeled using the Live/Dead viability assay kit (Invitrogen), and 100,000 events were recorded. OG1RF (A), TX5264 (OG1RF gelE) (B), Liv729 (OG1RF salB) (C), Liv1016 (OG1RF salB gelE) (D), and Liv883 (OG1RF salB pAT18::salB) (E) were harvested at exponential (5 h) growth phase. Horizontal lines mark populations of dead cells.
Table 4.
FACS viability results
| E. faecalis strain | Mean green fluorescence (RU)a/fold difference relative to OG1RF | % Dead cells/fold difference relative to OG1RF |
|---|---|---|
| TX4002 (OG1RF) | 1.80/1 | 0.37/1 |
| TX5264 (OG1RF gelE mutant) | 2.60/1.44 | 2.95/7.97 |
| Liv729 (OG1RF salB) | 3.30/1.83 | 14.31/38.67 |
| Liv883 (OG1RF salB pAT18::salB) | 3.06/1.7 | 4.93/13.32 |
| Liv1016 (OG1RF gelE salB) | 10.46/5.81 | 53.91/145.70 |
RU, relative units.
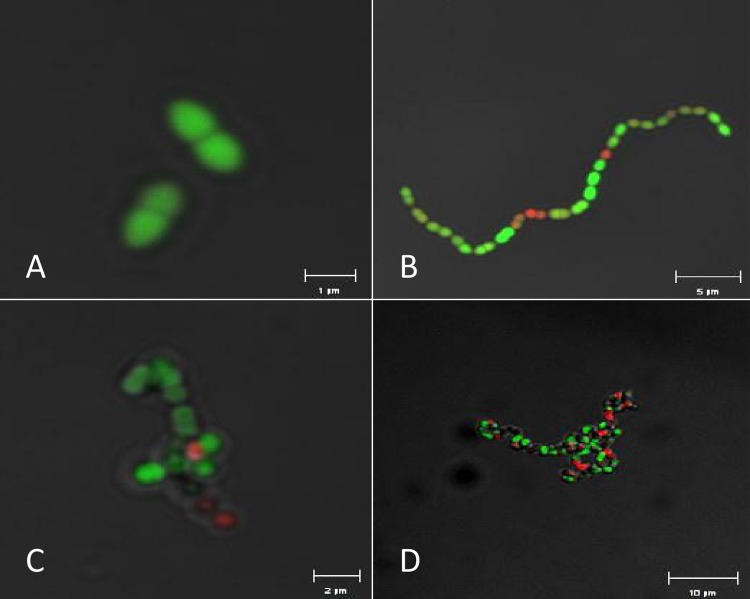
Laser scanning microscopy of fluorescently stained cells was used to examine cell separation and identified that inactivation of either gelE (Fig. 5) or fsrB (data not shown) resulted in chains of cells, as previously described (1, 37). In contrast, inactivation of salB resulted in cell clumps, which were exacerbated in double mutant strains that had salB plus either gelE (Fig. 5) or fsrB (data not shown) or inactivated. The cell separation phenotypes were confirmed by examining relative sedimentation of exponential cultures. After 30 min of incubation, the strains with the greatest mean green fluorescence and reduced cell separation displayed the greatest level of sedimentation (data not shown). Defective septation of mid-exponential-phase cells of strain Liv729 (OG1RF salB) in this study was confirmed by TEM (Fig. 6B) and matched that described for a salB mutant of JH2-2 (4, 24). The septation defect was exacerbated in the double mutant strain Liv1016 (OG1RF gelE salB) relative to the isogenic single mutant parent strains (Fig. 6A and C).
Fig 5.
Cell morphology and viability determination of E. faecalis strains. Cells of strains TX4002 OG1RF (A), TX5264 (OG1RF gelE) (B), Liv729 (OG1RF salB) (C), and Liv1016 (OG1RF gelE salB) (D) were labeled with BacLight live/dead stain and visualized using laser scanning microscopy. Bars indicate the scales, which vary among the images due to varied morphology (1, 5, 2, and 10 μm, respectively, for panels A, B, C, and D).
Fig 6.
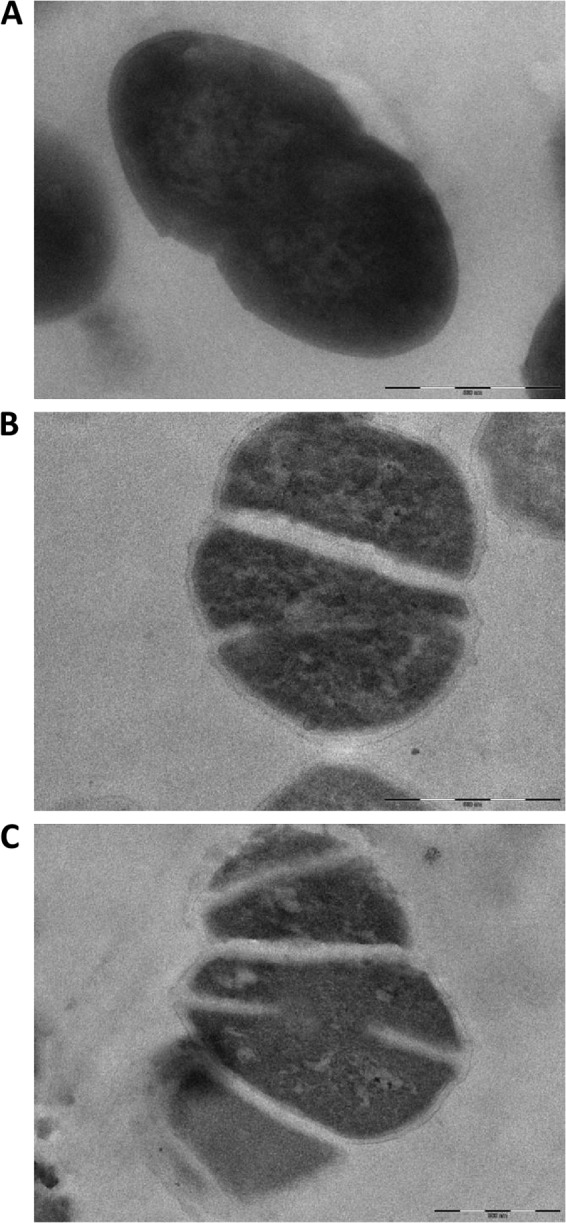
Transmission electron microscopy of thin sections of mid-exponentially growing cells of strains TX5244 (OG1RF) (A), Liv729 (OG1RF salB) (B), and Liv1016 (OG1RF gelE salB) (C). Scale bar, 500 nm.
Absence of SalB peptidoglycan hydrolase activity.
Muralytic activity of SalB was investigated with six-histidine tag-purified enzyme. Initial analyses used SDS-PAGE renaturing zymograms supplemented with 10 OD units of purified peptidoglycan from E. faecalis OG1RF or M. luteus. SalB produced unstained zones of clearing on gels after renaturation and methylene blue staining, indicating either murein hydrolytic or peptidoglycan binding capability (data not shown). An assay of peptidoglycan hydrolase activity with purified enzyme and peptidoglycan from strain OG1RF revealed no change in A540 after prolonged (up to 24-h) incubation (data not shown). In a separate experiment, reverse phase (RP)-HPLC revealed an absence of soluble muropeptides after overnight incubation of SalB with E. faecalis OG1RF peptidoglycan (Fig. 7). Moreover, mutanolysin digestion of peptidoglycan isolated from strain OG1RF and strain Liv729 (OG1RF salB) revealed identical RP-HPLC traces (Fig. 7 and data not shown).
Fig 7.
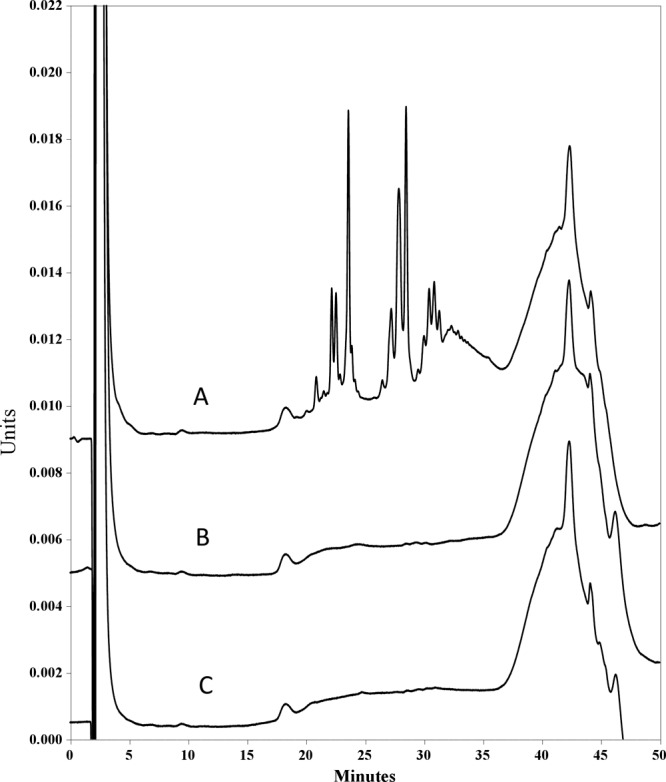
Separation of soluble muropeptides by HPLC after incubation with enzymes. OG1RF peptidoglycan was incubated overnight with 5 μg of mutanolysin (Sigma) (A) or 50 μg of SalB (B) or without enzyme (control) (C).
DISCUSSION
Study of the factors contributing to the culture supernatant protein complement of E. faecalis OG1RF identified that a gelE metalloprotease mutant produced distinct exoproteins. One prominent protein expressed throughout growth in the mutant was identified as SalB. The enzymatic function of SalB from previous studies is not clear, with its inactivation resulting in defective cell morphology and reduced resistance to various environmental stressors in strain JH2-2 (5). In addition, a salB mutant of strain OG1RF is defective for biofilm formation and has increased binding capacity to ECM molecules (17). The link between increased SalB in culture supernatant and gelE inactivation identified here was not clear. To investigate the possibility of epistasis between gelE and salB, a double mutant was generated and the strains were compared by analyzing their exoprotein profile, autolytic activity, cell morphology, and viability.
Examination of the exoproteins released into culture supernatant by the OG1RF salB mutant revealed a complement distinct from the wild type. Consequently, as an attempt to gain further insight into the enzymatic role of SalB, a comparative proteomic study of strain Liv729 (OG1RF salB) and the wild-type strain (OG1RF) was undertaken. Relative to its parent, the salB mutant released reduced levels of several proteins with ascribed functions in cell wall metabolism and cell replication into culture supernatant. Higher levels were determined for several general stress response proteins (e.g., GroEL, DnaK, and SOD), glycolytic and translation machinery proteins, and GelE (Table 2). The reduced viability of the salB mutant indicates that cell lysis might contribute to the exoproteome and could explain the differences with OG1RF. Equally, the identified protein abundance differences could be a consequence of a cellular response to the problems encountered with cell division. The protein complement supports known phenotypes of the salB mutant strain; moreover, it potentially defines roles for SalB. Previous studies determined that a salB mutant has reduced sensitivity to environmental stress and salB transcription is directly regulated by the two-component signal transduction system CroRS (14, 18). The stimulus activating CroRS is unknown. The finding in this study that there are elevated levels of general stress proteins, including DnaK, GroEL, Dps, and SOD, expressed by the salB mutant supports the possibility that the salB mutant has an inherent stress defect resulting from gene inactivation. The observed concomitant increased level of glycolytic enzymes could indicate a cellular response to membrane dysfunction, with a resulting requirement for direct energy generation via glycolysis. Definitive evidence for a membrane defect has not been obtained; however, there is a clear association between SalB and cell morphology and division septum formation/placement. Induction of stress genes was similarly observed in a Streptococcus pneumoniae strain with reduced expression of PcsB (2). It was demonstrated by Le Breton et al. (5, 14) that a salB deletion in strain JH2-2 caused morphological defects, which were confirmed in this study for strain OG1RF salB using TEM. The double mutant strain Liv1016 (OG1RF gelE salB) displayed a higher incidence of irregularities in septum formation, including multiple septa, than strain Liv729 (OG1RF salB). Thus, the absence of GelE exacerbates the phenotype described for salB inactivation, although there is not clear evidence that this is due directly to GelE proteolysis of SalB. The high level of GelE in the supernatant of the salB mutant (Table 2) compared to that of strain OG1RF suggests a compensatory aspect to the expression levels of these proteins. A Streptococcus agalactiae mutant of PcsB (SalB homologue) produces cell aggregation similar to that of an E. faecalis salB mutant (23, 24). The role of PcsB has been studied in several streptococci, in which the location of pcsB, immediately downstream of the cell shape-determining operon mreCD, is conserved, similar to the location of salA, the E. faecalis homologue of salB and pcsB. The reduced level of the cell division protein EzrA in culture supernatant of the OG1RF salB mutant lends further support to a theoretical role for SalB in cell division and/or cell shape determination.
The incorrect septum placement in the E. faecalis OG1RF salB mutant is likely to alter the plane of division to produce the cell clumps in the single and multiple mutants studied here. It is tempting to speculate that since GelE regulates the activity of the major cell wall autolysins of E. faecalis OG1RF, the exacerbated phenotype observed in strain Liv1016 (OG1RF gelE salB) supports the linking of SalB to cell division, in part determining the characteristic ovococcal morphology. Certainly, the morphology of a salB mutant and that of a divIVA (trans complementation) mutant of E. faecalis are strikingly similar (22). The modulation of SalB expression by the CroRS two-component signal transduction regulator links cell morphology with intrinsic β-lactam resistance (7, 18). Recent analysis of PcsB in S. pneumoniae has provided the first evidence supporting a role for PcsB/SalB/SagA family proteins in the cell division complex via its interaction with FtsX (28).
An unexpected finding from laser scanning microscopy and flow cytometry of viability stained cells of strain Liv729 (OG1RF salB) was the proportion of red-fluorescent dead cells (14.31%) present in the population compared with OG1RF red-fluorescent dead cells (0.37%) (Table 4). This proportion was greatly increased in strain Liv1016 (OG1RF salB gelE) (53.9%). These measurements do not take account of the fact that changes in cell volume would increase uptake of stain in the mutants, since cell volumes were not measured in this study. The corresponding changes in green fluorescence, however, identify that although there were more cells per event due to defective cell separation in the salB mutants, the relative levels of red (dead) cells were disproportionately high. Complementation of the salB mutant via strain Liv883 (OG1RF salB pAT18::salB) only partially rescued this phenotype.
Previous studies hypothesized that SalB and its homologues might possess peptidoglycan hydrolase activity. Cell autolysis and activity assays of the purified SalB were performed to investigate its function further. In an autolysis assay, a strain under penicillin challenge, Liv729 (OG1RF salB), displayed greater levels of lysis than the wild type. As expected, the absence of GelE in strain TX5264 (OG1RF gelE) resulted in decreased autolysis compared with the wild type (Fig. 3). Similarly, reduced autolysis was observed in the presence of Triton X-100 with strain TX5264 (OG1RF gelE), while strain Liv729 (OG1RF salB) again showed significantly enhanced autolysis. Complementation restored wild-type autolysis rates to strain Liv883 (OG1RF salB pAT18::salB) under penicillin challenge, but not under Triton X-100 challenge (data not shown). Peptidoglycan hydrolysis was tested using both zymograms and HPLC of either muramidase-digested peptidoglycans or wild-type peptidoglycan incubated with purified SalB. Renaturing zymograms performed with an SDS-PAGE gel supplemented with purified peptidoglycan from E. faecalis OG1RF or Micrococcus luteus ATCC 4698 revealed clearing. Since this could indicate either peptidoglycan binding by SalB preventing methylene blue staining or peptidoglycan hydrolysis, the activity of purified peptidoglycan was tested with six-histidine-tagged SalB at a range of pH values in different buffers with several divalent cations. However, no detectable activity was observed by using this assay (data not shown). Moreover, the peptidoglycans from Liv729 (OG1RF salB) and OG1RF were indistinguishable upon mutanolysin digestion and separation by HPLC, indicating that the clearing observed on the zymograms most likely represented peptidoglycan binding by SalB (Fig. 7 and data not shown). A lack of in vitro peptidoglycan hydrolysis was reported for PcsB of S. agalactiae and S. pneumoniae (9, 23, 24). Very recently, evidence was obtained supporting a role for PcsB interacting with the FtsX cell division protein and supporting a potential role for direct or indirect PG hydrolysis during cell division (28).
While the data presented here do not determine the cellular role for SalB or support peptidoglycan hydrolysis activity, the findings are certainly consistent with SalB having a role in cell shape determination and/or cell division. The septation anomalies and the reduced level of EzrA in the salB mutant culture supernatant exoprotein profile relative to the wild type could result from direct interaction of SalB with a cell division protein, in which the coiled-coil domain located in the N-terminal region of SalB has the potential to mediate this interaction.
ACKNOWLEDGMENTS
This research was supported by BBSRC postgraduate award BB/S/K/2004/11198 to R.G.W. and a Dorothy Hodgkin NERC-BP postgraduate award to J.S.
We thank Barbara Murray for kindly providing strains and plasmids that were essential for parts of this study. We are grateful for bioinformatic assistance from Daniel Rigden.
Footnotes
Published ahead of print 4 May 2012
REFERENCES
- 1. Arias CA, Cortes L, Murray BE. 2007. Chaining in enterococci revisited: correlation between chain length and gelatinase phenotype, and gelE and fsrB genes among clinical isolates of Enterococcus faecalis. J. Med. Microbiol. 56:286–288 [DOI] [PubMed] [Google Scholar]
- 2. Barendt SM, et al. 2009. Influences of capsule on cell shape and chain formation of wild-type and pcsB mutants of serotype 2 Streptococcus pneumoniae. J. Bacteriol. 191:3024–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Biavasco F, et al. 1996. In vitro conjugative transfer of VanA vancomycin resistance between enterococci and listeriae of different species. Eur. J. Clin. Microbiol. Infect. Dis. 15:50–59 [DOI] [PubMed] [Google Scholar]
- 4. Bonten MJ, Willems R, Weinstein RA. 2001. Vancomycin-resistant enterococci: why are they here and where do they come from? Lancet Infect. Dis. 1:314–325 [DOI] [PubMed] [Google Scholar]
- 5. Breton YL, et al. 2002. Isolation and characterization of bile salts-sensitive mutants of Enterococcus faecalis. Curr. Microbiol. 45:434–439 [DOI] [PubMed] [Google Scholar]
- 6. Chaussee MA, McDowell EJ, Chaussee MS. 2008. Proteomic analysis of proteins secreted by Streptococcus pyogenes. Methods Mol. Biol. 431:15–24 [DOI] [PubMed] [Google Scholar]
- 7. Comenge Y, et al. 2003. The CroRS two-component regulatory system is required for intrinsic beta-lactam resistance in Enterococcus faecalis. J. Bacteriol. 185:7184–7192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eckert C, Lecerf M, Dubost L, Arthur M, Mesnage S. 2006. Functional analysis of AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis. J. Bacteriol. 188:8513–8519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giefing-Kroll C, Jelencsics KE, Reipert S, Nagy E. 2011. Absence of pneumococcal PcsB is associated with overexpression of LysM domain-containing proteins. Microbiology 157:1897–1909 [DOI] [PubMed] [Google Scholar]
- 10. Gilmore MS, Ferretti JJ. 2003. The thin line between gut commensal and pathogen. Science 299:1999–2002 [DOI] [PubMed] [Google Scholar]
- 11. Hancock LE, Gilmore MS. 2006. Pathogenicity of enterococci, p 301–354 In Fischetti V, Novick R, Ferretti J, Portnoy D, Rood J. (ed), Gram-positive organisms. ASM Press, Washington, DC [Google Scholar]
- 12. Horsburgh MJ, Wiltshire MD, Crossley H, Ingham E, Foster SJ. 2004. PheP, a putative amino acid permease of Staphylococcus aureus, contributes to survival in vivo and during starvation. Infect. Immun. 72:3073–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Land AD, Winkler ME. 2011. The requirement for pneumococcal MreC and MreD is relieved by inactivation of the gene encoding PBP1a. J. Bacteriol. 193:4166–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Le Breton Y, et al. 2003. Molecular characterization of Enterococcus faecalis two-component signal transduction pathways related to environmental stresses. Environ. Microbiol. 5:329–337 [DOI] [PubMed] [Google Scholar]
- 15. Mesnage S, Chau F, Dubost L, Arthur M. 2008. Role of N-acetylglucosaminidase and N-acetylmuramidase activities in Enterococcus faecalis peptidoglycan metabolism. J. Biol. Chem. 283:19845–19853 [DOI] [PubMed] [Google Scholar]
- 16. Miller D, Urdaneta V, Weltman A. 2002. Public health dispatch: vancomycin-resistant Staphylococcus aureus—Pennsylvania. MMWR Morb. Mortal. Wkly. Rep. 51:902. [PubMed] [Google Scholar]
- 17. Mohamed JA, Teng F, Nallapareddy SR, Murray BE. 2006. Pleiotrophic effects of 2 Enterococcus faecalis sagA-like genes, salA and salB, which encode proteins that are antigenic during human infection, on biofilm formation and binding to collagen type I and fibronectin. J. Infect. Dis. 193:231–240 [DOI] [PubMed] [Google Scholar]
- 18. Muller C, et al. 2006. The response regulator CroR modulates expression of the secreted stress-induced SalB protein in Enterococcus faecalis. J. Bacteriol. 188:2636–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakayama J, Kariyama R, Kumon H. 2002. Description of a 23.9-kilobase chromosomal deletion containing a region encoding fsr genes which mainly determines the gelatinase-negative phenotype of clinical isolates of Enterococcus faecalis in urine. Appl. Environ. Microbiol. 68:3152–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paulsen IT, et al. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074 [DOI] [PubMed] [Google Scholar]
- 21. Qin X, Singh KV, Weinstock GM, Murray BE. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183:3372–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ramirez-Arcos S, Liao M, Marthaler S, Rigden M, Dillon JA. 2005. Enterococcus faecalis divIVA: an essential gene involved in cell division, cell growth and chromosome segregation. Microbiology 151:1381–1393 [DOI] [PubMed] [Google Scholar]
- 23. Reinscheid DJ, Ehlert K, Chhatwal GS, Eikmanns BJ. 2003. Functional analysis of a PcsB-deficient mutant of group B Streptococcus. FEMS Microbiol. Lett. 221:73–79 [DOI] [PubMed] [Google Scholar]
- 24. Reinscheid DJ, Gottschalk B, Schubert A, Eikmanns BJ, Chhatwal GS. 2001. Identification and molecular analysis of PcsB, a protein required for cell wall separation of group B Streptococcus. J. Bacteriol. 183:1175–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rince A, et al. 2003. Physiological and molecular aspects of bile salt response in Enterococcus faecalis. Int. J. Food Microbiol. 88:207–213 [DOI] [PubMed] [Google Scholar]
- 26. Rosenthal RS, Dziarski R. 1994. Isolation of peptidoglycan and soluble peptidoglycan fragments. Methods Enzymol. 235:253–285 [DOI] [PubMed] [Google Scholar]
- 27. Seno Y, Kariyama R, Mitsuhata R, Monden K, Kumon H. 2005. Clinical implications of biofilm formation by Enterococcus faecalis in the urinary tract. Acta Med. Okayama 59:79–87 [DOI] [PubMed] [Google Scholar]
- 28. Sham LT, Barendt SM, Kopecky KE, Winkler ME. 2011. Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proc. Natl. Acad. Sci. U. S. A. 108:E1061–E1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shankar J, Walker RG, Ward D, Horsburgh MJ. 2012. The Enterococcus faecalis exoproteome: identification and temporal regulation by Fsr. PLoS One 7:e33450 doi:10.1371/journal.pone.0033450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sievert D, et al. 2002. Staphylococcus aureus resistant to vancomycin—United States. MMWR Morb. Mortal. Wkly. Rep. 51:565–567 [PubMed] [Google Scholar]
- 31. Singh KV, Qin X, Weinstock GM, Murray BE. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416–1420 [DOI] [PubMed] [Google Scholar]
- 32. Smith TJ, Foster SJ. 1995. Characterization of the involvement of two compensatory autolysins in mother cell lysis during sporulation of Bacillus subtilis 168. J. Bacteriol. 177:3855–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tendolkar P, Baghdayan A, Shankar N. 2003. Pathogenic enterococci: new developments in the 21st century. Cell. Mol. Life Sci. 60:2622–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Teng F, Kawalec M, Weinstock GM, Hryniewicz W, Murray BE. 2003. An Enterococcus faecium secreted antigen, SagA, exhibits broad-spectrum binding to extracellular matrix proteins and appears essential for E. faecium growth. Infect. Immun. 71:5033–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thomas VC, et al. 2009. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol. Microbiol. 72:1022–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P. 1991. Shuttle vectors containing a multiple cloning site and a lacZ alpha gene for conjugal transfer of DNA from Escherichia coli to Gram-positive bacteria. Gene 102:99–104 [DOI] [PubMed] [Google Scholar]
- 37. Waters CM, Antiporta MH, Murray BE, Dunny GM. 2003. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J. Bacteriol. 185:3613–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yasmin A, et al. 2010. Comparative genomics and transduction potential of Enterococcus faecalis temperate bacteriophages. J. Bacteriol. 192:1122–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]