Abstract
ComN (YrzD) is a small, 98-amino-acid protein recently shown to be involved in the posttranscriptional control of the late competence comE operon in Bacillus subtilis. We show here that ComN localizes to the division site and cell poles in a DivIVA-dependent fashion. Yeast two-hybrid and glutathione S-transferase pulldown experiments showed that ComN interacts directly with DivIVA. ComN is not essential for the polar assembly of the core competence DNA uptake machinery. Nevertheless, polar localization of ComN should play some role in competence acquisition because delocalization of ComN leads to a small reduction in competence efficiency. We found that ComN promotes the accumulation of its target comE mRNA to septal and polar sites. Thus, we speculate that localized translation of ComE proteins may be required for efficient competence development. Our results underscore the versatility of DivIVA as a promoter of the differentiation of bacterial poles and demonstrate that the repertoire of polarly localized molecules in B. subtilis is broad, including a regulator of gene expression and its target mRNA. Moreover, our findings suggest that mRNA localization may play a role in the subcellular organization of bacteria.
INTRODUCTION
The surge in the application of cell biology tools and concepts to microbiology in the last decade has revealed that bacterial cells have an elaborate subcellular organization (32, 34). The cell poles, in particular, are highly differentiated portions of the bacterial cell (6). Poles differ from the rest of the cell in terms of geometry, cell wall turnover, and protein composition. In highly asymmetric cells such as Caulobacter crescentus, >5% of the proteins localize to the cell poles (38). Polarly localized proteins are responsible for the assembly of well known polar structures such as flagella, pili, stalks, and chemoreceptor arrays. In addition, polar proteins help establish positional landmarks (e.g., the cell middle), orient the axis of cell growth, and drive cellular differentiation (6).
DivIVA is a key protein involved in the differentiation of the cell poles of Bacillus subtilis. DivIVA localizes to the poles by first going to the division site (7). Because the division site will become the new poles, going to the division site and remaining there after division is finished results in polar localization. Recruitment of DivIVA to the division site requires the assembly of the divisome, the FtsZ-based division machine of bacteria (25). Initially, it was thought that DivIVA localization to the division site depended on an interaction with a component of the divisome. However, several lines of evidence indicate that the subcellular localization of DivIVA is dictated by a physical aspect of the cell rather than by protein-protein interactions (8, 14, 15, 22, 31). In particular, DivIVA is a membrane-associated protein that has affinity for negatively curved (concave) membranes which are found at the division site and cell poles (29).
Once localized, DivIVA serves as a landmark to attract other proteins to the cell poles. One well-studied example is the MinCD regulator of cell division. MinCD is a site-selective inhibitor of cell division: by accumulating at the cell poles it prevents unproductive polar divisions that would result in DNA-less minicells (25). Polar localization of MinCD may also create a gradient of inhibitor that favors division to happen at midcell (24). MinCD localization is strictly dependent on DivIVA; in the absence of DivIVA, the cellular distribution of MinCD becomes unrestricted, leading to inhibition of division at the regular medial sites and filamentation (25). Polar recruitment of MinCD by DivIVA is mediated by MinJ, a membrane protein that serves as a bridge between DivIVA and MinCD (3, 30). Another B. subtilis protein that requires DivIVA for its localization and function is RacA, a centromere-like protein necessary for the correct segregation of chromosomes during sporulation (1, 40). Successful segregation during sporulation requires that chromosomes become elongated and closely associated with the cell poles. RacA is a DNA-binding protein that associates preferentially to sequences around the replication origin of the chromosome and that is also capable of interacting with DivIVA. The simultaneous binding of RacA to the replication origin and to DivIVA thus promotes the association of chromosomes with the cell poles (1, 40).
B. subtilis poles are also the preferred site of localization of the protein complex involved in DNA uptake and transformation of competent cells (12, 19, 20). This protein complex consists of a DNA transport pore associated with a pseudopilus related to components of type II and type IV secretion systems. Proteins involved in the intracellular processing of the incoming DNA, such as RecA and single-stranded binding proteins (SSBs), are also associated with the DNA uptake complex (19, 20).
We show here that ComN, a recently described posttranscriptional regulator of competence gene expression (28), is polarly localized in B. subtilis. ComN localization is mediated by a direct interaction with DivIVA. Interestingly, we found that the ComN-DivIVA interaction led to the accumulation of ComN′s target mRNA (comE) to septal and polar sites. Although ComN is not essential for the polar assembly of the core competence DNA uptake machinery, delocalization of ComN leads to a small but significant reduction in competence efficiency. Our findings underscore the versatility of DivIVA as a promoter of the differentiation of bacterial poles and suggest that localized regulators of gene expression and their target mRNAs may play a role in the targeting of proteins to specific locations in the bacterial cell.
MATERIALS AND METHODS
General methods.
All B. subtilis strains are listed in Table 1 and were derived from the wild-type strain PY79. B. subtilis was grown in Luria-Bertani (LB) medium at 37°C or 30°C. Preparation of competent cells and transformation of B. subtilis was performed according to the two-step procedure (16). When present, the antibiotic concentrations were as follows: chloramphenicol, 5 μg/ml; erythromycin plus lincomycin, 1 and 25 μg/ml; spectinomycin, 100 μg/ml; kanamycin, 10 μg/ml; and tetracycline, 10 μg/ml. When necessary, various concentrations of isopropyl-β-d-thiogalactopyranoside (IPTG) or xylose were used. The exact concentrations in each experiment are given in the figure legends.
Table 1.
Strains and plasmids
| Strain or plasmid | Revelant genotype | Source or reference |
|---|---|---|
| Strains | ||
| PY79 | Prototroph | 41 |
| FG116 | minD (divIVB1) | Lab stock |
| FG117 | minD (divIVB1) divIVA Δ::spc | Lab stock |
| FG916 | amyE::Pxyl-gfp-comN (cat ble) | This study |
| FG925 | ΔamyE::Pspac-ftsZ-mcherry (spc) amyE::Pxyl-gfp-comN (cat ble) | This study |
| FG931 | minD (divIVB1) divIVA::spc amyE::Pxyl-gfp-comN (cat ble) | This study |
| FG932 | minD::kan amyE::Pxyl-gfp-comN (cat ble) | This study |
| FG985 | ftsZ ΩPspac-ftsZ (spc) amyE::Pxyl-gfp-comN (cat ble) | This study |
| FG994 | ΔminJ amyE::PminJ-minJ (mls) | This study |
| FG1007 | ΔminJ minD::kan amyE::Pxyl-gfp-comN (cat ble) | This study |
| FG1008 | ΔminJ amyE::Pxyl-gfp-comN (cat ble) | This study |
| FG1076 | comN::cat | This study |
| FG1078 | comN::cat amyE::Pxyl-gfp-comN (cat ble) | This study |
| FG1079 | comN::cat minD::kan | This study |
| FG1109 | comGAΩcomGA-cfp (kan) amyE::Pxyl-gfp-comN (cat ble) | This study |
| FG1117 | comGAΩcomGA-cfp (kan) comN::cat | This study |
| FG1253 | divIVA::spc amyE::Pxyl-gfp-comN (cat) | This study |
| FG1254 | comNΩcomN-mcherry (spc) amyE::Pxyl-gfp-divIVA (cat) | This study |
| FG1255 | comN::cat amyE::PcomN-comN (ble) | This study |
| FG1259 | minD (divIVB1) divIVA::spc comGAΩcomGA-cfp (kan) amyE::Pxyl-gfp-comN (cat ble) | This study |
| AB227 | divIVA::spc minD (divIVB1) comGAΩcomGA-cfp (kan) | This study |
| FG1324 | comKΩcomK-lacZ (kan) | This study |
| FG1325 | comKΩcomK-lacZ (kan) minD (divIVB1) divIVA::spc | This study |
| FG1326 | comEAΩcomEA-lacZ (erm) | This study |
| FG1327 | comEAΩcomEA-lacZ (erm) minD (divIVB1) divIVA::spc | This study |
| FG1334 | comK ΩcomK-lacZ (kan) comN::cat | This study |
| FG1335 | comEAΩcomEA-lacZ (erm) comN::cat | This study |
| AB228 | amyE::Pspank-ms2d-gfp (spc) | This study |
| AB229 | amyE::Pspank-ms2d-gfp (spc) comEAΩcomEA-6 × BS (tet) | This study |
| AB230 | amyE::Pspank-ms2d-gfp (spc) comECΩcomEC-6 × BS (tet) | This study |
| AB232 | comN::erm | This study |
| AB234 | amyE::Pspank-ms2d-gfp (spc) comEAΩcomEA-6 × BS (tet) comN::erm | This study |
| AB235 | amyE::Pspank-ms2d-gfp (spc) comECΩcomEC-6 × BS (tet) comN::erm | This study |
| AB238 | amyE::Pspank-ms2d-gfp (spc) comEAΩcomEA-6 × BS (tet) minD (divIVB1) divIVA::cat | This study |
| AB239 | amyE::Pspank-ms2d-gfp (spc) comECΩcomEC-6 × BS (tet) minD (divIVB1) divIVA::cat | This study |
| DH5α | supE44 ΔlacU169 (ϕ80dlacZΔM15) hsdR17 recA1 endA1 gyrA96 thi1 relA1 | |
| Plasmids | ||
| pJR74GFP-comN | amyE::Pxyl-gfp-comN (cat ble) | This study |
| pFG43-comN | 6×His-comN (kan) | This study |
| pGUI6-comN | comN-mcherry (spec) | This study |
| pGBDU-comN | GAL4 binding domain-comN (URA amp) | This study |
| pGADC2-comN | GAL4 activation domain-comN (LEU2 amp) | This study |
| pGADC-divIVA | GAL4 activation domain-divIVA (LEU2 amp) | This study |
| pGBDU-divIVA | GAL4 binding domain-divIVA (URA amp) | This study |
| pGEX4T-1-divIVA | GST-divIVA (amp) | This study |
| pCOM-comN | amyE::PcomN-comN (ble) | This study |
| pAB60 | amyE::Pspank-ms2d-gfp (spc bla) | This study |
| pAB61a | comEA-6 × BS-ter (tet bla) | This study |
| pAB61c | comEC-6 × BS-ter (tet bla) | This study |
DNA methods.
Cloning and DNA manipulations were carried out according to standard methods (33). Taq DNA polymerase and Phusion DNA polymerase (NEB) were used for PCR. Oligonucleotide primers were purchased from IDT (Coralville, IA) and are listed in Table 2. The plasmids and their relevant characteristics are listed in Table 1. The steps involved in plasmid and strain construction are described in detail in the supplemental material. Sequencing of plasmids was carried out by the sequencing service of the Departamento de Bioquímica, IQ-USP.
Table 2.
Oligonucleotides
| Oligonucleotide | Sequence (5′–3′) |
|---|---|
| OFG198 | ACATGCGGCCGCGTGGAGAAGCATCC |
| OFG199 | AAGGATCCCTATCTCAGCAGTTCTTCCAATG |
| OFG274 | GGTCTCGAATTCATCGATAAAAGGAGCGTGGAATTCGTGG |
| OFG275 | GGATCCTCTCAGCAGTTCTTCCAATGCTTCCTG |
| OFG294 | AAGGATCCGCATGCTTATTTGTATAGTTCATCCAT |
| OFG392 | TTCGGGATATCACACCCTAT |
| OFG393 | TCAGCAGATATTTGGTCAAT |
| OFG397 | CATCCTCGAGCGTTTCAGCCATAACAGGG |
| OFG400 | ACTGGATCCTACATATGATGCCATTAACGCCAAATGA |
| OFG401 | TCCCTCGAGAGATCTTAGTTCCTTTTCCTCAAA TACAGC |
| OFG450 | CGAAGCTTGTGCTACGCGCCGCTCTCAT |
| OFG451 | GCCTCGAGGTCATGGCAACGGTTATTCA |
| OFG454 | CGAAGCTTATGAATTGGTTGAATCAGCA |
| OFG463 | TTAAGCTTAAGGAGGTGAACTACTATGGCTTCTAACTTTACT |
| OFG485 | ATATGGATCCTAAGGTACCT |
| OFG486 | GCAGGCGGCCGCCATATGCA |
| OFG487 | AGGTACCTTAGGATCCATATTCTGTTCGCAGCACGCGGAT |
| OFG488 | TGCATATGGCGGCCGCCTGCTCAAAACGGAACGATCCA |
| OFG489 | AGGTACCTTAGGATCCATATCTTTACTGTAATGGAAGACT |
| OFG490 | GCTCCTGCCAAATTAAGCGC |
| OFG491 | CTGAGCGAGGGAGCAGAATTTCAAGAAGACTTGTCATG |
| OFG492 | GTTGACCAGTGCTCCCTGTCGCACAGGTTTAAGGGAGG |
| OFG493 | GCAGTGATAAAGCTTGTCAA |
| oJM28 | TTCTGCTCCCTCGCTCAG |
| oJM29 | CAGGGAGCACTGGTCAAC |
Fluorescence microscopy.
Microscopy was performed on a Nikon TE300 inverted microscope equipped with a ×100 PlanFluor objective lens (oil, NA 1.3) and filters for green fluorescent protein (GFP; Endow GFP set, 41018; Chroma Technology) and mCherry (Texas Red BrightLine set, TXRED4040-B; Semrock). Images were captured with a Roper CoolSnap HQ camera. Exposure times varied from 0.3 to 1 s. Images were processed and analyzed with MetaMorph version 7.1 (Universal Imaging) and ImageJ (http://rsb.info.nih.gov/ij/) software. Microscopy of ComGA and colocalization between ComN-mCherry and GFP-DivIVA was performed on a Nikon Eclipse TiE microscope, with filters for GFP (GFP BrightLine Filter Set) and mCherry (mCherry BrightLine Filter Set), and a Plan APO VC Nikon ×100 objective lens (oil, NA 1.4). Images were captured with an Andor I-Xon EMCCD camera. The images were processed and analyzed with Nikon NIS Elements software (version 3.07) and ImageJ.
To perform microscopy, the cells were grown to exponential phase and mounted on slides covered with a layer of agarose-solidified minimal medium or phosphate-buffered saline (PBS). The membranes were stained with FM5-95 or FM4-64 (Molecular Probes) at a final concentration of 1 μg/ml. Dyes were added directly to cells in medium 5 min before mounting and imaging. For studying the localization of competence proteins, cells were imaged for 1.5 h following resuspension in SpII medium, using the slides and conditions described above.
Yeast two-hybrid assay.
Fusion proteins to the GAL4 DNA-binding domain (BD-protein) or GAL4 transcription activation domain (AD-protein) were expressed in the host strain PJ69-4a, which has two reporter genes for two-hybrid interactions integrated into the genome: HIS3 and ADE2. The method of transformation was that of Gietz et al. (9). PJ69-4a was transformed with combinations of two plasmids (pGBDU-divIVA and pGADC2-divIVA, pGADC2-comN and pGBDU-comN, pGADC2-comN and pGBDU, or pGBDU-comN and pGADC2) and plated in minimal medium (SC) lacking leucine and uracil as a first selection. Colonies that grew in the absence of Leu and Ura were transferred to SC plates lacking Ura, Leu, His, and Ade plus 3-aminotriazole (3AT). The amount of 3AT used varied between 5 and 50 mM. The pair pGBDU-divIVA and pGADC2-divIVA was used as a positive control. To test for self-activation, strains containing pGBDU-comN/pGADC2 and pGADC2-comN/pGBDU combinations were subjected to the same procedures.
Protein pulldown and immunoblot analysis.
All recombinant proteins were expressed in Escherichia coli BL21-CodonPlus (DE3) RIL, transformed with either pGEX-divIVA (GST-DivIVA), pGEX (GST), or pFG43-comN (His-ComN). E. coli cells bearing the different plasmids were grown at 37°C to an optical density at 600 nm (OD600) of 0.6, when IPTG was added to a final concentration of 0.5 mM, and further incubated at 37°C for 4 h. Pulldown of His-ComN was assayed as follows. Whole-cell extracts from E. coli cells expressing GST, GST-DivIVA, and His-ComN were generated in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 [pH 7.4]) and 1 mM phenylmethylsulfonyl fluoride (PMSF) after cellular lysis in a French press (three times) and centrifugation for 40 min at 4°C at 15,000 rpm. Then, 500 μl of glutathione-Sepharose beads (Amersham Biosciences) was incubated with the whole extract of GST and GST-DivIVA in shaking tubes for 2 h at 4°C. Next, the beads were washed three times with 12 ml of PBS and incubated with His-ComN extract for 4 h. Subsequently, the beads were washed three times with 12 ml of PBS, and the bound fraction was eluted with 1 ml of 10 mM reduced glutathione, 50 mM Tris-HCl (pH 8), and 1 mM PMSF. The bound fraction was precipitated with trichloroacetic acid (10%), followed by washing in 100% ethanol and resuspension in protein buffer. The proteins were resolved on SDS-PAGE (15%) and transferred to polyvinylidene difluoride membranes for 3 h at 200 mA. To perform Western blotting, the membranes were incubated with anti-polyhistidine serum (1:5,000; Amersham Biosciences) or with anti-GST serum (1:20,000; Sigma, St. Louis, MO). Immunoblots were developed using the ECL system (Amersham Biosciences).
LacZ assays.
Cells were grown in SpC medium to stationary phase and resuspended in SpII medium, and 1-ml fractions of the culture were collected every half hour for 2 h. For each time point, cell pellets were resuspended in 500 μl of Z buffer (60 mM Na2HPO4·7H2O, 40 mM NaH2PO4·H2O, 10 mM KCl, 1 mM MgSO4 [pH 7.0]), and their OD600s were recorded. Subsequently, 140 μl of each sample was added to the well of a 96 well-plate with 20 μl of Triton X-100 (2%), 3 μl of β-mercaptoethanol, and 40 μl of o-nitrophenyl-β-d-galactoside (4 mg/ml), and the OD420s of the reactions were measured every minute for 40 min in a BioTek Epoch plate reader at room temperature. The measured OD420 slopes (S) were introduced into the formula: Miller units = 1,000 × S/0.14 × OD600.
Localization of comE mRNA.
Constructs for MS2-GFP expression and tagging of the comE mRNA are described in the supplemental material. Fresh colonies were grown in MC competence medium (100 mM potassium phosphate [pH 7.0], 3 mM sodium citrate, 2% glucose, 22 mg of ferric ammonium citrate/ml, 3 mM manganese sulfate, 0.1% casein, 0.2% potassium glutamate) for 2 h, and then IPTG was added to a final concentration of 50 μM to induce MS2d-GFP. Cultures were grown for 2 more hours before being harvested and visualized on the microscope. Membranes were stained with 0.02 mM TMA-DPH (Molecular Probes) and imaged with exposure times of 300 ms. MS2d-GFP fluorescence was captured using 500 ms of exposure time.
RESULTS AND DISCUSSION
ComN localizes to division sites and cell poles.
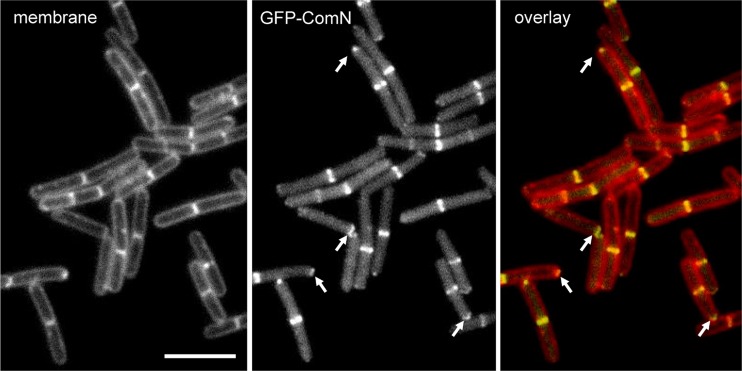
ComN (YrzD) was identified as a potential divisome protein because it displayed a pattern of co-occurrence with the division protein ZapA in the STRING database (37), which identifies functional links between proteins by comparing bacterial genomes. We tested this prediction by constructing a GFP-ComN fusion and determining its subcellular distribution. Fluorescence microscopy of a merodiploid strain expressing GFP-ComN from the Pxyl inducible promoter (FG916) revealed the typical localization pattern of division proteins, with clear accumulation of the protein in midcell bands (Fig. 1). GFP-ComN also seemed to be retained at the newly formed cell poles after septum completion (see arrows in Fig. 1), a pattern commonly seen with late divisome proteins. Localization of the GFP-ComN fusion was independent of the wild-type ComN protein because it also occurred in a strain in which the endogenous comN gene is deleted (FG1078). In addition, tagging of ComN with mCherry at its C terminus via single-crossover integration resulted in a strain that had ComN-mCherry expressed under the endogenous PcomN promoter (FG1254). In this strain, localization of ComN-mCherry was identical to that of the N-terminal GFP fusion expressed from the Pxyl promoter (see Fig. 2B). Thus, the observations that ComN localization is the same irrespective of the fluorescent protein tag (GFP or mCherry), topology (N- or C-terminal) or promoter (Pxyl or PcomN) from which the fusion is expressed suggest that the fluorescent protein fusions are faithfully reporting the localization of the untagged ComN protein.
Fig 1.
ComN localizes to division sites and cell poles. Strain FG916, bearing a GFP-ComN fusion, was grown to mid-log phase (OD600 = 0.3 to 0.5) in LB medium supplemented with 0.5% xylose and imaged as described in Materials and Methods. Note that GFP-ComN localizes both to midcell bands (new septa), as well as to cell poles (old septa). Arrows point to examples of cells in which GFP-ComN is retained at the cell poles. Bar, 5 μm.
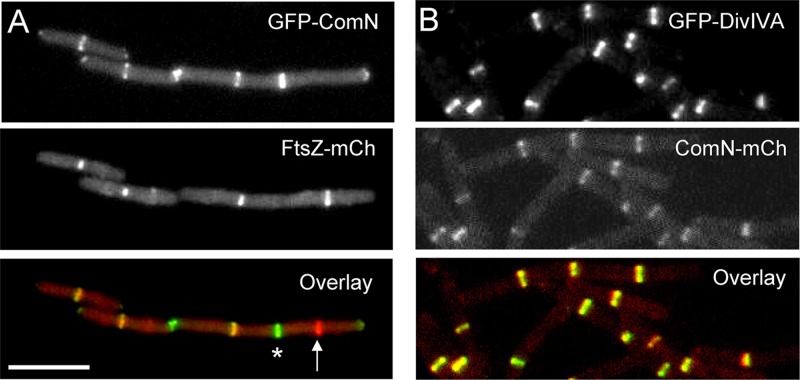
Fig 2.
ComN is a late recruit to the division site and colocalizes with DivIVA. (A) Colocalization of FtsZ and ComN. Strain FG925, expressing FtsZ-mCherry (FtsZ-mCh) and GFP-ComN fusions, was grown to mid-log phase (OD600 = 0.3 to 0.5) in LB medium supplemented with 0.5% xylose and 100 μM IPTG and imaged as described in Materials and Methods. The arrow marks a Z-ring that is not decorated with ComN. These presumably represent young division complexes. An asterisk marks a ComN ring that is not decorated with FtsZ. These should correspond to septa in the final phase of cytokinesis, when FtsZ has already left the division site. Bar, 5 μm. (B) Colocalization of ComN and DivIVA. Strain FG1254, expressing DivIVA-GFP and ComN-mCherry (ComN-mCh), was grown to mid-log phase (OD600 = 0.3 to 0.5) in LB medium supplemented with 0.5% xylose and 100 μM IPTG. The overlay image shows that ComN and DivIVA exhibit essentially perfect colocalization.
To determine when ComN associates with the division site, we colocalized FtsZ and ComN using a strain simultaneously expressing FtsZ-mCherry and GFP-ComN (FG925). Only 40% of the Z rings in an exponentially growing population was decorated with GFP-ComN (Fig. 2A), suggesting that there is a significant delay between the initial assembly of the Z ring and when ComN becomes incorporated into the complex. The frequency of colocalization for ComN and FtsZ is similar to those measured for late divisome proteins such as DivIVA, YpsB, and MinCD (36; G. L. Meira and F. J. Gueiros-Filho, unpublished observations). Thus, ComN is likely a late recruit to the division complex.
ComN is recruited to the division site and cell poles by a direct interaction with DivIVA.
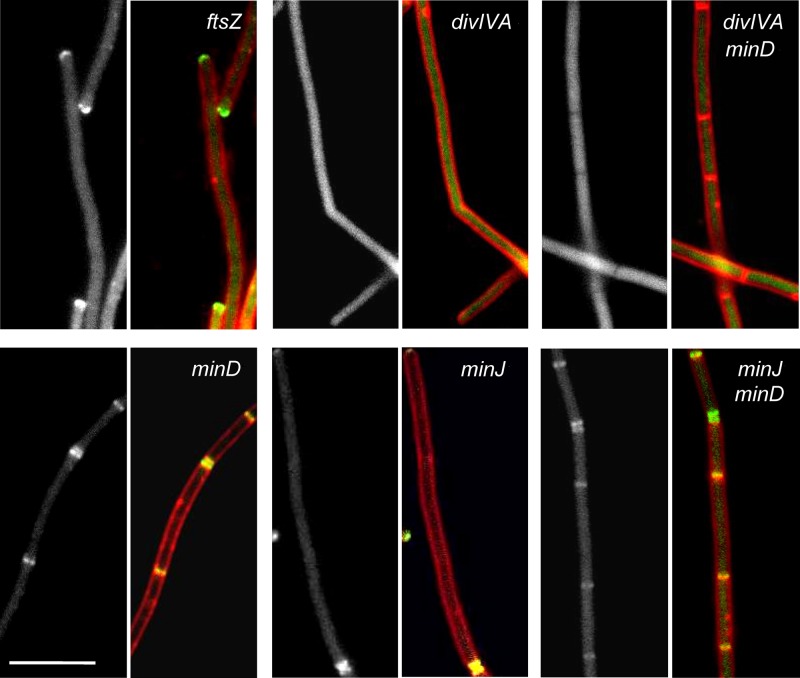
The observations that ComN displays a pattern and kinetics of localization similar to that of DivIVA and the late division proteins that use DivIVA to reach the division site, such as MinCD and MinJ, prompted us to investigate whether ComN localization was dependent on one of these proteins. We initially investigated the pattern of GFP-ComN in a strain depleted of FtsZ and found that the protein was generally delocalized, as expected for a divisomal protein, but intense staining was still associated with the cell poles. This is very much the same pattern seen for DivIVA when FtsZ is depleted (25) and further suggested that ComN could be recruited to the divisome by DivIVA. Indeed, GFP-ComN localization was completely disrupted in a divIVA mutant. In these strains GFP-ComN was evenly distributed in the cytoplasm and, importantly, no sign of polar accumulation remained (Fig. 3). Because elimination of divIVA drastically reduces the frequency of division, we also analyzed the localization of GFP-ComN in a divIVA minD double mutant. Deletion of minD restores division to the divIVA mutant and allows for a clearer assessment of whether ComN requires DivIVA to reach the septum and cell poles. Significantly, GFP-ComN remained completely delocalized in the divIVA minD double mutant, despite the abundant septa made by this mutant. To rule out the possibility that localization of GFP-ComN was not mediated by DivIVA but instead due to the proteins recruited to the division site by DivIVA, we also analyzed the effect of deleting minD or minJ alone or of simultaneously deleting minD and minJ. In all of these cases, GFP-ComN still displayed the septal and polar localization pattern found in the wild-type strain (Fig. 3, panels minD, minJ, and minJ minD). We also tested the localization of GFP-ComN in a strain lacking ypsB (gpsB), a recently characterized paralog of divIVA that associates late with the divisome but independently of DivIVA (4), and in a strain lacking zapA, the division gene that retrieved comN in the STRING database co-occurrence search. However, localization of GFP-ComN was unaffected by either of the mutations (data not shown). Thus, GFP-ComN requires DivIVA, but not other late divisome proteins or ZapA to localize to the division site and cell poles.
Fig 3.
DivIVA is required for ComN localization. The GFP-ComN fusion was introduced into different mutants and localization was assayed in cells grown to mid-log phase. In the case of ftsZ, which is an essential gene, a depletion strain was used and grown for at least five generations in the absence of ftsZ expression before being imaged. For each mutant, a panel corresponding to the GFP-ComN image (gray) and a panel with the overlay between the GFP-ComN channel (green) and membrane channel (red) are shown. ftsZ (FG985), minD (FG932), minJ (FG994), minJ minD (FG1007), divIVA (FG1253), and divIVA minD (FG931) mutant strains are shown as indicated. Scale bar, 5 μm.
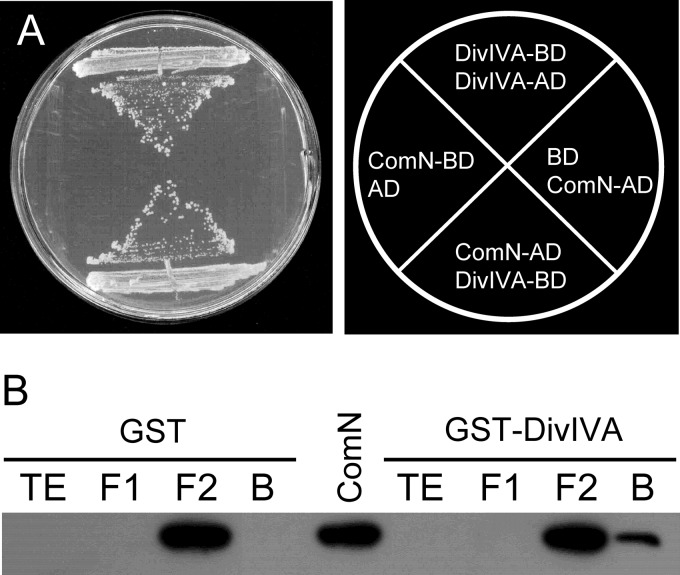
We next used the GAL4 yeast two-hybrid system to investigate whether ComN and DivIVA interact directly. Pairing of ComN and DivIVA fusions in either of the two possible configurations (ComN-AD plus DivIVA-BD or DivIVA-AD plus ComN-BD) resulted in robust growth, indicating the existence of a direct interaction between the two proteins (Fig. 4A). The yeast two-hybrid experiments also showed that ComN is capable of interacting with itself (see Fig. S1 in the supplemental material), suggesting that the physiological state of ComN is an oligomer. To confirm the two-hybrid results, we carried out GST-pulldown experiments. DivIVA was fused to GST and ComN to a 6×His tag and overexpressed in E. coli. The total extract of the strain expressing His-ComN was applied onto a glutathione-agarose column previously loaded with GST-DivIVA. Subsequently, the column was washed and eluted with glutathione, the eluted material was electrophoresed, and Western blotting was performed with anti-His antibodies. As a control, a similar experiment was carried out with GST instead of GST-DivIVA. His-ComN was only detected in the eluate of the GST-DivIVA column, indicating that DivIVA and ComN physically interact in a specific way (Fig. 4B), corroborating the two-hybrid data. Thus, ComN likely interacts directly with DivIVA inside B. subtilis cells.
Fig 4.
ComN and DivIVA interact. (A) Yeast two-hybrid assay. The yeast strain harboring the GAL4 activation domain fused to ComN (AD-ComN) and the GAL4 DNA-binding domain fused to DivIVA (BD-DivIVA) was able to activate transcription of the reporter genes ADE and HIS and grow on SC medium lacking adenine and histidine. Strains containing the AD-ComN fusion and the unfused DNA-binding domain (BD) or the BD-ComN fusion and the unfused activation domain (AD) did not activate transcription and were unable to grow in the absence of adenine and histidine. BD-DivIVA and AD-DivIVA was used as a positive control. (B) Pulldown assay. Total extract from cells expressing either GST or GST-DivIVA (TE) was incubated with glutathione-Sepharose, the flowthrough fraction was collected (F1) and, after washing, the total extract of cells expressing His-ComN (F2) was loaded. The resin was washed again, and the bound fraction (B) was eluted with glutathione. Proteins in each of the fractions were electrophoresed, transferred, and visualized by Western blotting with monoclonal anti-His IgG2a. His-ComN is only pulled down by GST-DivIVA (rightmost lane). ComN in the middle lane represents total extract of cells expressing His-ComN and serves as a size reference.
ComN is not involved in cell division.
A central role of DivIVA in B. subtilis is targeting of the Min proteins to the cell poles (25). Because ComN interacts with DivIVA, ComN could play a role in the polar localization of the Min proteins. We tested this possibility by analyzing the cell division pattern in a comN deletion mutant and found that deletion of the comN gene had no detectable effect on either the frequency or the location of division septa (Fig. 5). Because alterations in MinCD localization lead to minicell formation and filamentation, the regular division pattern of the comN mutant indicates that ComN does not contribute to MinCD polar targeting. Several division proteins appear to have redundant functions and show little effect when deleted (10, 17, 36, 39). However, combining the comN mutation with mutations in other divisome components (zapA, noc, minD, and minJ) did not result in a cell division phenotype (data not shown), suggesting that ComN is not involved in the polar targeting of Min proteins or with any other aspect of cell division.
Fig 5.
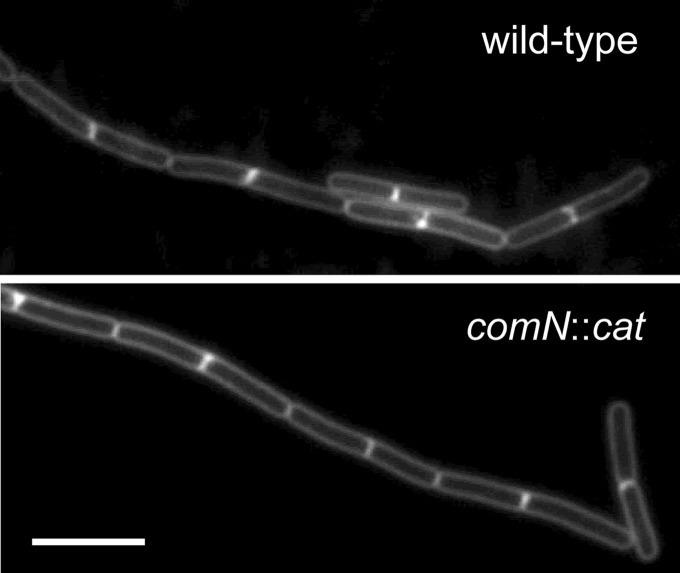
Mutation in comN does not affect the pattern or frequency of division. Fluorescence micrographs of membrane-stained wild-type (PY79) and comN mutant (FG1076) bacteria. Strains were grown to mid-log phase in LB medium at 37°C and imaged as described in Materials and Methods. Bar, 5 μm.
Polar localization of ComN is not essential for competence development.
We next investigated whether the polar localization of ComN was important for its role in competence by measuring the development of competence in a divIVA mutant, where ComN is no longer polarly localized (Fig. 3). Because, as noted above, a divIVA mutant is severely impaired for division, and this mutation is detrimental for homologous recombination and thus to transformability (30), we opted to work with a divIVA minD double mutant, which divides much better than the divIVA single mutant and should not have the transformability problem caused by filamentation. Measuring the transformation efficiency revealed that, as expected, the comN mutant was 100-fold less competent than our wild-type strain [transformation frequencies of (2.6 ± 1.9) × 10−3 for the wild-type strain versus (2.5 ± 2.4) × 10−5 for the comN::cat strain], a result similar to that found by Ogura and Tanaka (28). In contrast, the transformation efficiency of the divIVA minD mutant was just 3-fold lower than that of the wild-type strain [(0.8 ± 1) × 10−3 versus (2.6 ± 1.9) × 10−3]. The low competence of the comN mutant can be explained by a drastic reduction in expression of the comE operon (22). Because comK and comE levels are normal in the divIVA minD mutant (see Fig. S2 in the supplemental material), the mild competence reduction of this mutant cannot be explained by alterations in competence gene expression. To determine whether the reduction in competence of the divIVA minD mutant was due to the delocalization of ComN or whether it was related to other factors such as the absence of MinD or the altered division behavior of this strain, we measured the transformation efficiency of a minD mutant, a strain that has the same division behavior as the divIVA minD mutant, but in which ComN still localizes properly (Fig. 3). The minD mutant exhibited a transformation efficiency similar to that of the wild-type [(4.0 ± 4.5) × 10−3] and higher than that of the divIVA minD mutant. This suggests that the delocalization of ComN does have a negative impact on competence development, although the large variability of the transformation measurements does not give statistical support to this conclusion. We therefore conclude that the polar localization of ComN is not essential but increases the efficiency of competence development. One potential caveat to this conclusion would be if DivIVA had an additional effect on competence besides localizing ComN. In this case, the difference in competence between the divIVA minD double mutant and the minD single mutant could not be ascribed solely to the delocalization of ComN. Even though we currently cannot rule out this possibility, we find it unlikely that DivIVA would play two distinct roles in competence.
ComN does not colocalize with and is not necessary for polar localization of the competence apparatus.
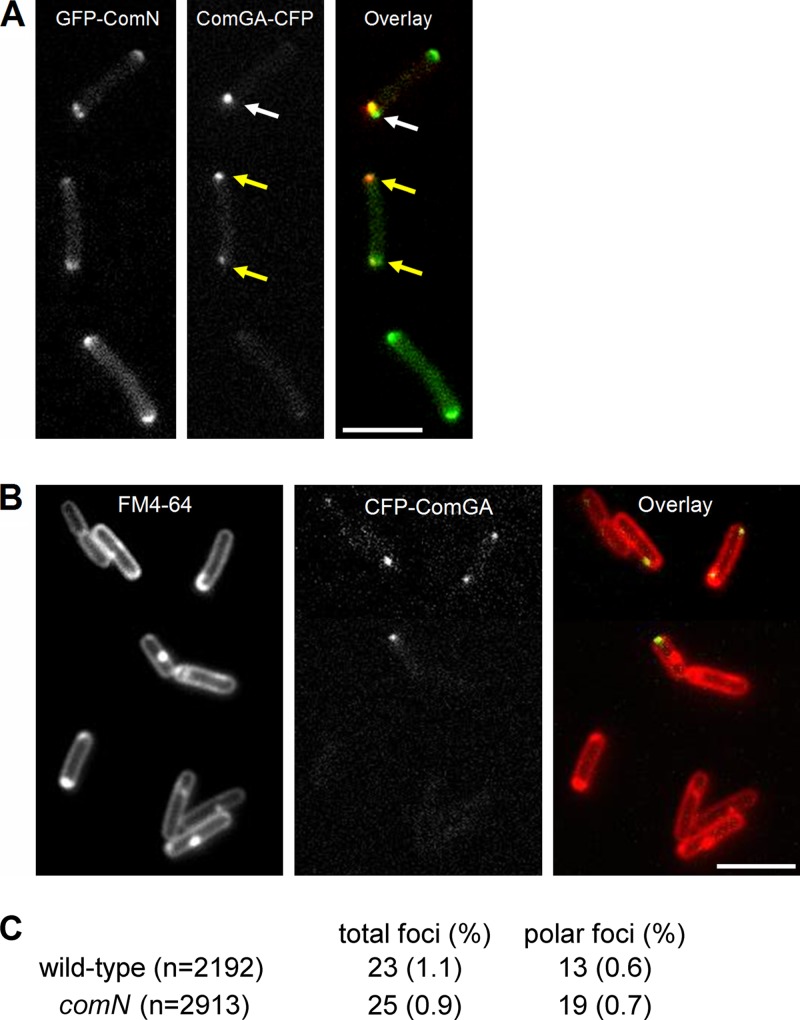
The effect of delocalizing ComN in competence development led us to investigate whether ComN played a role in the polar localization of the competence apparatus. We initially tested whether ComN could be part of the DNA uptake complex by localizing ComN in competent cells and comparing it to the localization of the DNA uptake machinery. We used a ComGA-CFP fusion as a marker for the DNA uptake machinery and combined it with GFP-ComN in strain FG1109. GFP-ComN was imaged with a YFP filter and controls were carried out to show that under our imaging conditions we could separate the signals originating from the ComGA-CFP and GFP-ComN fusions. Fluorescence microscopy of strain FG1109 in competence medium revealed ComGA-CFP foci in about 1% of the cells (Fig. 6A). This is somewhat lower than what has been reported before (12, 19) and may be due to the fact that our wild-type background (PY79) does not develop competence as efficiently as other B. subtilis strains. Among the cells with foci, the majority (∼60%) were polar, but some were localized on the side walls as well. The pattern of localization of GFP-ComN in the same cells was similar to that found for GFP-ComN in growing cells, with clear polar accumulation (Fig. 6A). No septal band of GFP-ComN was found in the competent cells, however, probably because divisome assembly is blocked as a result of competence development (13). Superimposing the ComGA-CFP and GFP-ComN signals (Fig. 6A) revealed that the localization of ComGA and ComN had limited overlap. There were several cases in which ComN staining appeared as bipolar caps, whereas ComGA foci was found in only one pole (Fig. 6A, white arrow). There were also cells in which a ComGA focus was found in a region where little or no ComN staining was present (Fig. 6A, yellow arrow). Furthermore, in the cases where ComGA foci overlapped with ComN polar caps, the staining patterns of the two proteins was dissimilar enough (note the cell marked with the white arrow in Fig. 6A) that they must represent two distinct cellular structures.
Fig 6.
(A) Colocalization of ComGA and ComN. Strain FG1109, bearing ComGA-CFP and GFP-ComN fusions, was grown to competence by the two-step procedure described in Materials and Methods. Cells were harvested 1.5 h after being diluted into SpC medium and imaged. In the overlay, GFP-ComN is pseudocolored green, and ComGA-CFP is pseudocolored red. (B) ComGA-CFP localization in the absence of ComN. Strain FG1117, bearing the ComGA-CFP fusion and the comN::cat deletion, was grown to competence and visualized as described for panel A. Bar, 5 μm. (C) Frequency of comGA foci in wild-type and comN mutant cells.
We have also investigated the effect of the comN mutation on ComGA foci formation. Figure 6B shows that ComGA still forms foci in a comN mutant. The frequency of total ComGA foci was similar in the comN mutant and in the wild type. In addition, deletion of comN did not affect the polar localization of ComGA foci (Fig. 6C). Similar results were obtained when we used a RecA-YFP fusion as a marker of the DNA uptake complex (see Fig. S3 in the supplemental material). We have also evaluated ComGA localization in the divIVA minD mutant, a situation in which ComN is delocalized and found that ComGA was still capable of forming polar foci (see Fig. S4 in the supplemental material). These results indicate that at least part of the components of the DNA uptake and processing apparatus can assemble and localize properly in the absence of ComN or when ComN is delocalized. Polar localization of the DNA uptake apparatus has been proposed to occur by a diffusion and capture mechanism, in which foci form at different locations along the cell membrane and diffuse freely until they reach the cell poles and recognize a still unknown protein that “anchors” the foci there (11). The lack of an effect of the comN deletion indicates that ComN is not this anchor protein and is consistent with a previous report that the polar localization of foci was not dependent on DivIVA (12).
ComN localizes comE mRNA.
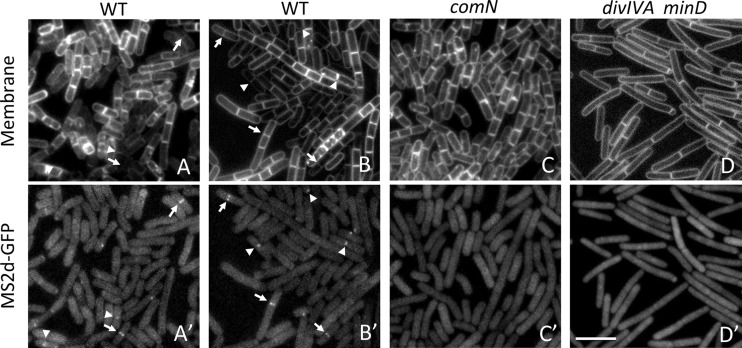
The role of ComN as a posttranscriptional regulator of ComE expression (28) implies that ComN should interact with the comE mRNA and, potentially, localize this mRNA to the nascent septum and cell poles. To investigate this possibility, we tagged the comE mRNA with binding sites for phage MS2 coat protein and visualized the comE mRNA distribution in a strain expressing the MS2 coat protein fused to GFP (MS2-GFP) (2). We created two tagged versions of the predicted comE transcript, one with the tag immediately downstream of the comEA coding region (plasmid pAB61a) and the other with the tag downstream of the comEC coding region (plasmid pAB61c), and used these constructs to replace the wild-type comE gene, such that the expression of the tagged transcripts would be under the control of the native comE promoter. To avoid well-known artifacts of the MS2 system (26), we carried out our experiments at a low level of MS2-GFP induction (50 μM IPTG), which we found does not lead to mRNA-independent aggregation of MS2-GFP (see Fig. S5 in the supplemental material). Localization of comE mRNA in cultures grown in competence medium revealed cells in which the MS2-GFP fluorescence was brighter at nascent septa and new poles (Fig. 7A, A′, B, and B′, arrows). There was also MS2-GFP fluorescence accumulation at older septa/poles (inferred to be old because of being constricted or hemispherical), but it was weaker and less frequent than the septal and new pole localizations (Fig. 7A, A′, B, and B′, arrowheads). This localization pattern was identical whether we used the construct with the MS2 tag downstream of comEA (Fig. 7A and A′) or comEC (Fig. 7B and B′). Because the localization of MS2-GFP was very similar to that detected for the ComN protein (see Fig. 1), this suggested that ComN is indeed capable of localizing comE mRNA. To confirm this hypothesis, we carried out several controls. First, we verified that localization of MS2-GFP was dependent on the presence of the tagged comE transcript, since MS2-GFP was homogenously distributed in the cytoplasm of strains lacking tagged comE constructs (see Fig. S5 in the supplemental material, top panel). Furthermore, we found that cells with localized MS2-GFP never comprised more than ca. 10% of the population. This is expected because only the minority of cells that are developing competence should express the tagged comE transcript. Finally, and most importantly, septal and polar MS2-GFP localization was strictly dependent on ComN and DivIVA: no localization of MS2-GFP could be observed in a comN mutant (Fig. 7C and C′) and in a divIVA minD mutant (Fig. 7D and D′), a situation in which ComN is still present in the cell but delocalized (Fig. 3). Thus, our results strongly support the conclusion that ComN, via its interaction with DivIVA, is capable of localizing the comE mRNA to septa and poles.
Fig 7.
ComN mediates localization of comE mRNA. Strains bearing an IPTG-inducible MS2-GFP fusion (amyE::Pspank-ms2d-gfp) and tagged versions of the comE mRNA (comEA-6×BS and comEC-6×BS) were grown in competence medium and imaged as described in Materials and Methods. The IPTG concentration used for all samples was 50 μM. (A and A′) comEA-6×BS localization in a wild-type background (strain AB229); (B and B′) comEC-6×BS localization in a wild-type background (strain AB230); (C and C′) comEC-6×BS localization in a comN mutant background (strain AB235); (D and D′) comEC-6×BS localization in a divIVA minD mutant background (strain AB239). The accumulation of comE mRNA in septa and new poles are marked with arrows, and accumulation in old poles is marked with arrowheads. Scale bar, 5 μm.
We sought to determine the biological significance of the comE mRNA localization. Targeting of mRNA is often associated with localized translation and asymmetric protein distribution in eukaryotic cells (5, 35). However, we could not demonstrate this to be the case for the proteins encoded by the comE mRNA. ComEA, which is the DNA receptor of the competence apparatus, is distributed roughly evenly in cell membranes (18) and thus would neither require nor benefit from localized translation. On the other hand, ComEC, the integral membrane protein that constitutes the DNA transport pore, is a polarly localized protein (18), and its localization may depend on its mRNA being localized. We attempted to investigate the effect of the comN and divIVA mutations in the polar localization of ComEC, but we found that the ComEC-YFP fusion produced fluorescence that was too weak to generate meaningful data, similarly to what was reported by the Dubnau lab (20). Thus, at this point, we cannot rule out that some of the components of the competence complex, in particular ComEC, will require ComN for proper localization. Other possible roles for septal localization of the comE mRNA would be to enforce equal partitioning of mRNA molecules between daughter cells that are becoming competent or to control the timing of translation by sequestering the mRNA at a subcellular location (in this case, the septum or cell poles), where it would be inaccessible to ribosomes. Developmental programs such as sporulation and competence commence one or more cell cycles before cells commit to differentiation and stop dividing (21, 23). Using the division complex to sequester mRNAs may function as a developmental checkpoint capable of coupling translation of specific messages to the cessation of cell division.
We have demonstrated here that ComN is a protein whose localization is enriched at the septa and poles of B. subtilis cells. Localization of ComN is mediated by a direct interaction with DivIVA, reinforcing the idea that DivIVA is a general pole-marking protein crucial for the spatial organization of a diverse set of processes (division, chromosome segregation, and gene expression and/or protein segregation). One interesting question raised by our observation is how many more proteins find their way to the poles of B. subtilis via DivIVA? Given the variety of processes in which DivIVA is involved in B. subtilis and other Gram-positive bacteria, we suspect that there may be more proteins that exploit DivIVA to reach the cell poles awaiting discovery. Another important question is the physiological significance of ComN localization. ComN localization, and its ability to localize a target mRNA, suggests that production of the proteins controlled by this factor should occur preferentially at the cell poles. Local production of proteins controlled by ComN is not essential for the polar assembly of at least some components of the competence DNA uptake machinery, but it should play a role in competence acquisition because delocalization of ComN leads to a small reduction in competence efficiency. We postulate that this decrease in competence may be related to delocalized production of ComEC, which may prevent its efficient incorporation into polar DNA uptake complexes.
Finally, the demonstration that ComN mediates targeting of the comE mRNA to septa and poles strengthens the notion that mRNA localization and/or localized translation could play a significant role in the organization of the bacterial cell. Two recent reports showed that bacterial mRNAs are present at specific subcellular locations (26, 27). In one case, C. crescentus mRNAs were shown to stay close to their gene, probably because translation of nascent transcripts by the ribosome prevents their diffusion to other locations (26). In a second report, Nevo-Dinur et al. showed that E. coli mRNAs localized to sites where the proteins encoded by these RNAs should function (the membrane, for integral membrane proteins, and the cell pole in the case of the BglG transcription factor) (27). Such mRNA localization was mediated by cis-acting sequences, but the existence of trans-acting factors capable of binding to the mRNAs and anchoring them in specific cellular locations remained elusive (27). We showed here that ComN functions as one of these trans-acting factors, bolstering the idea that mRNA localization by factors other than the ribosome does indeed occur. Because ComN orthologs are absent from Gram-negative bacteria, other proteins should be responsible for targeting mRNAs to the membrane and poles of E. coli. Identifying these factors and their target mRNAs in E. coli, B. subtilis, and other model bacteria should clarify the breadth and impact of mRNA localization to the cellular organization of prokaryotes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mitsuo Ogura, Dan Kearns, David Rudner, Peter Graumann, and Orna Amster-Choder for plasmids and strains, Carla Columbano de Oliveira for pulldown reagents, and Jonathan Dworkin for critical reading of the manuscript. We are also indebted to Rodrigo T. Ribeiro for technical assistance, José Roberto Tavares for the construction of some of the plasmids used in this study, Márcia Teixeira dos Santos and Marcela Prieto for help with the two-hybrid and pulldown experiments, and Orna Amster-Choder, Keren Nevo-Dinur, and Paula Montero-Llopis for advice on MS2 mRNA localization experiments.
This study was supported by grant 08/58821-1 from the Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP) and grant 478019/2009-2 from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). V.T.D.S. was the recipient of a master's fellowship from the CNPq, and A.W.B.-F. was the recipient of a doctoral fellowship from the FAPESP.
Footnotes
Published ahead of print 11 May 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Ben-Yehuda S, Rudner DZ, Losick R. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299:532–536 [DOI] [PubMed] [Google Scholar]
- 2. Bertrand E, et al. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2:437–445 [DOI] [PubMed] [Google Scholar]
- 3. Bramkamp M, et al. 2008. A novel component of the division site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol. Microbiol. 70:1556–1569 [DOI] [PubMed] [Google Scholar]
- 4. Claessen D, et al. 2008. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol. Microbiol. 68:1029–1046 [DOI] [PubMed] [Google Scholar]
- 5. Du TG, Schmid M, Jansen RP. 2007. Why cells move messages: the biological functions of mRNA localization. Semin. Cell Dev. Biol. 18:171–177 [DOI] [PubMed] [Google Scholar]
- 6. Dworkin J. 2009. Cellular polarity in prokaryotic organisms. Cold Spring Harbor Perspect. Biol. 1:a003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edwards DH, Errington J. 1997. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol. 24:905–915 [DOI] [PubMed] [Google Scholar]
- 8. Edwards DH, Thomaides HB, Errington J. 2000. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J. 19:2719–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gietz RD, Schiestl RH, Willems AR, Woods RA. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355–360 [DOI] [PubMed] [Google Scholar]
- 10. Gueiros-Filho FJ, Losick R. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hahn J, Kramer N, Briley K, Jr, Dubnau D. 2009. McsA and B mediate the delocalization of competence proteins from the cell poles of Bacillus subtilis. Mol. Microbiol. 72:202–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hahn J, Maier B, Haijema BJ, Sheetz M, Dubnau D. 2005. Transformation proteins and DNA uptake localize to the cell poles in Bacillus subtilis. Cell 122:59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haijema BJ, Hahn J, Haynes J, Dubnau D. 2001. A ComGA-dependent checkpoint limits growth during the escape from competence. Mol. Microbiol. 40:52–64 [DOI] [PubMed] [Google Scholar]
- 14. Hamoen LW, Errington J. 2003. Polar targeting of DivIVA in Bacillus subtilis is not directly dependent on FtsZ or PBP 2B. J. Bacteriol. 185:693–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harry EJ, Lewis PJ. 2003. Early targeting of Min proteins to the cell poles in germinated spores of Bacillus subtilis: evidence for division apparatus-independent recruitment of Min proteins to the division site. Mol. Microbiol. 47:37–48 [DOI] [PubMed] [Google Scholar]
- 16. Harwood CR, Cutting S. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 17. Ishikawa S, Kawai Y, Hiramatsu K, Kuwano M, Ogasawara N. 2006. A new FtsZ-interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis. Mol. Microbiol. 60:1364–1380 [DOI] [PubMed] [Google Scholar]
- 18. Kaufenstein M, van der Laan M, Graumann PL. 2011. The three-layered DNA uptake machinery at the cell pole in competent Bacillus subtilis cells is a stable complex. J. Bacteriol. 193:1633–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kidane D, Graumann PL. 2005. Intracellular protein and DNA dynamics in competent Bacillus subtilis cells. Cell 122:73–84 [DOI] [PubMed] [Google Scholar]
- 20. Kramer N, Hahn J, Dubnau D. 2007. Multiple interactions among the competence proteins of Bacillus subtilis. Mol. Microbiol. 65:454–464 [DOI] [PubMed] [Google Scholar]
- 21. Kuchina A, et al. 2011. Temporal competition between differentiation programs determines cell fate choice. Mol. Syst. Biol. 7:557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lenarcic R, et al. 2009. Localization of DivIVA by targeting to negatively curved membranes. EMBO J. 28:2272–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levine JH, Fontes ME, Dworkin J, Elowitz MB. 2012. Pulsed feedback defers cellular differentiation. PLoS Biol. 10:e1001252 doi: 10.1371/journal.pbio.1001252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marston AL, Errington J. 1999. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol. Microbiol. 33:84–96 [DOI] [PubMed] [Google Scholar]
- 25. Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 12:3419–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Montero Llopis P, et al. 2010. Spatial organization of the flow of genetic information in bacteria. Nature 466:77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. 2011. Translation-independent localization of mRNA in E. coli. Science 331:1081–1084 [DOI] [PubMed] [Google Scholar]
- 28. Ogura M, Tanaka T. 2009. The Bacillus subtilis late competence operon comE is transcriptionally regulated by yutB and under posttranscription initiation control by comN (yrzD). J. Bacteriol. 191:949–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oliva MA, et al. 2010. Features critical for membrane binding revealed by DivIVA crystal structure. EMBO J. 29:1988–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patrick JE, Kearns DB. 2008. MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol. Microbiol. 70:1166–1179 [DOI] [PubMed] [Google Scholar]
- 31. Ramamurthi KS, Losick R. 2009. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc. Natl. Acad. Sci. U. S. A. 106:13541–13545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rudner DZ, Losick R. 2010. Protein subcellular localization in bacteria. Cold Spring Harbor Perspect. Biol. 2:a000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed, vol 1 and 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 34. Shapiro L, McAdams HH, Losick R. 2009. Why and how bacteria localize proteins. Science 326:1225–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. St Johnston D. 2005. Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell. Biol. 6:363–375 [DOI] [PubMed] [Google Scholar]
- 36. Tavares JR, de Souza RF, Meira GL, Gueiros-Filho FJ. 2008. Cytological characterization of YpsB, a novel component of the Bacillus subtilis divisome. J. Bacteriol. 190:7096–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. von Mering C, et al. 2005. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 33:D433–D437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Werner JN, et al. 2009. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc. Natl. Acad. Sci. U. S. A. 106:7858–7863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu LJ, Errington J. 2004. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117:915–925 [DOI] [PubMed] [Google Scholar]
- 40. Wu LJ, Errington J. 2003. RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol. Microbiol. 49:1463–1475 [DOI] [PubMed] [Google Scholar]
- 41. Youngman PJ, Perkins JB, Losick R. 1983. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc. Natl. Acad. Sci. U. S. A. 80:2305–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.