Abstract
Epstein-Barr virus (EBV)-associated nasopharyngeal carcinoma (NPC) is highly metastatic, and this malignant feature may be promoted by an EBV oncoprotein, latent membrane protein 2A (LMP2A). Acting as a signal regulator, LMP2A can enhance invasiveness and motility of epithelial cells. Downstream from the LMP2A-triggered signaling events, it is largely unknown what key effector proteins are induced and essentially promote cell invasion. In the present study, we found that in NPC cells, LMP2A upregulated matrix metalloproteinase 9 (MMP9), a metastasis-associated protease. LMP2A increased MMP9 expression at both the mRNA and protein levels. It also activated the MMP9 promoter, in which two AP-1 elements were required for the promoter activation. Among AP-1 transcription factors, Fra-1 was induced by LMP2A and is essential for LMP2A-triggered MMP9 expression. Induction of Fra-1 was dependent on the LMP2A-activated ERK1/2 pathway, and induction of the ERK1/2–Fra-1–MMP9 axis required PY motifs in the amino-terminal domain of LMP2A. Notably, LMP2A-promoted invasion of NPC cells was blocked when MMP9 expression, Fra-1 induction, or ERK1/2 activation was inhibited. In addition, we found an association of LMP2A with MMP9 expression in NPC tumor biopsy specimens, where Fra-1 was a major mediation factor. This study reveals an underlying mechanism of LMP2A-induced cell invasion, from signal transduction to upregulation of a critical protease. Considering that MMP9 can also be upregulated by another EBV oncoprotein, LMP1, this protease may be a pivotal effector at which the EBV-induced, invasion-promoting mechanisms converge, serving as an attractive therapeutic target for NPC treatment.
INTRODUCTION
Nasopharyngeal carcinoma (NPC) is distinguished from other head-and-neck carcinomas by several features, including its strong association with Epstein-Barr virus (EBV) infection, a poorly differentiated phenotype of the epithelial tumor cells, and high incidences of metastasis (52, 61). Most NPC patients suffer from nodal involvement or distal metastasis at initial diagnosis, and the metastasis, especially that recurring after therapies, predicts very poor prognosis (32, 61). EBV oncoproteins may contribute to the highly metastatic phenotype of NPC (51), so clarifying their underlying mechanisms should shed light on therapeutic strategies to block malignant progression of this cancer.
Latent membrane protein 2A (LMP2A) is an EBV oncoprotein generally expressed in NPC and other EBV-associated cancers, such as Hodgkin lymphoma and gastric carcinoma (52). LMP2A is detected in around half of NPC tumor specimens at the protein level and in more than 95% of the tumors at the mRNA level (7, 9, 22). Acting like a ligand-independent receptor on the plasma membrane, LMP2A affects multiple signaling events, mainly through its amino-terminal intracellular domain, which contains an immunoreceptor tyrosine-based activation motif (ITAM), a YEEA motif, and two PPPPY (PY) motifs (44). The ITAM and the YEEA motif mediate loading of some protein tyrosine kinases to LMP2A, while the PY motifs recruit several ubiquitin ligases to regulate protein stability of LMP2A and its binding proteins (18, 19, 29, 71). Depending on the cellular background, various kinase pathways can be triggered by LMP2A, including spleen tyrosine kinase (Syk), phosphatidylinositol 3-kinase (PI-3K)/Akt, and mitogen-activated protein kinases (MAPKs), such as extracellular signal-regulated kinase (ERK) and c-Jun N-terminal kinase (JNK) (13, 18, 38, 64). Also in a cell-dependent manner, the LMP2A-triggered signaling events can further regulate various transcription factors, including β-catenin-associated lymphoid enhancer factor (LEF), nuclear factor κB (NF-κB), signal transducer and activator of transcription 3 (STAT3), and c-Jun, a member of the activator protein 1 (AP-1) family (13, 24, 41, 63).
Through the signaling pathways and transcription factors, LMP2A has been shown to exert some biologic effects associated with lymphomagenesis or carcinogenesis. For example, LMP2A drives constitutive activation of PI-3K/Akt to promote survival of B lymphocytes in the absence of B-cell receptor or to inhibit transforming growth factor β1-induced apoptosis (20, 48). LMP2A also induces a Hodgkin lymphoma-like gene transcription profile in B cells and accelerates myc-induced lymphomagenesis (8, 47). For gastric carcinoma cells, LMP2A promotes their survival through upregulation of survivin and inhibits expression of a tumor suppressor gene, PTEN, through induction of promoter hypermethylation (23, 24). Ectopic expression of LMP2A in some epithelial cell lines enhances anchorage-independent growth in vitro and tumorigenesis in vivo (21, 31, 55). In addition, differentiation of epithelial cells is inhibited by LMP2A (17, 42, 55), which may be relevant to the poor differentiation of NPC tumor cells in vivo.
Of note, several lines of evidence suggest that LMP2A also contributes to the metastatic potential of NPC. LMP2A is frequently detected at the invasive front of NPC tumors, and ectopic expression of LMP2A in NPC cells promotes epithelial-mesenchymal transition, which is associated with increased cell motility and invasiveness (31). For primary epithelial cells and some epithelial cell lines, LMP2A induces cell spreading, migration, or invasion in extracellular matrix (ECM) (2, 13, 38, 46). Depending on the cellular context, the LMP2A-induced cell invasion or migration involves some kinase pathways, including those of ERK, Syk, PI-3K/Akt, and focal adhesion kinase (2, 13, 38, 55). However, the mechanisms downstream from the LMP2A-triggered signaling events that promote cell invasion are largely unknown. It is uncertain whether LMP2A-regulated transcription factors are required therein. The key effector proteins that are induced by LMP2A and essentially promote cell invasion are also unclear, except for findings of a previous study showing that an adhesion molecule, integrin α6, may play an important role (46).
Matrix metalloproteinases (MMPs) form a family of zinc-dependent proteolytic enzymes and are involved in many physiological and pathological events (62). MMPs contribute to malignant progression of cancers through induction of cell invasion, remodeling of ECM, release of growth factors, promotion of angiogenesis, or modulation of local immune responses (15, 30). MMP9 is a well-known MMP that promotes cell invasion and metastasis for many cancers, including NPC. MMP9 proteins are detected in most NPC biopsy specimens, and high MMP9 expression correlates with lymph node metastasis, advanced clinical stage, and poor prognosis of NPC (25, 35). Expression of MMP9 is upregulated by many signaling pathways and transcription factors that are triggered by cytokines, growth factors, or other stimuli (62). Previous studies indicate that an EBV oncoprotein, LMP1, can induce MMP9 (73). However, considering that LMP1 at the protein level is detectable in only a subset of the NPC tumors that are generally positive for EBV infection and MMP9 expression (16, 35, 74), other EBV proteins may also contribute to MMP9 induction in NPC. LMP2A is a candidate since it is frequently detected in NPC tumors and is potentially associated with cell invasion.
In this study, we tested whether and how LMP2A induces MMP9 in NPC cells and whether this protease is involved in LMP2A-induced cell invasion. Our results indicate that LMP2A upregulates MMP9 expression at a transcriptional level and the MMP9 induction requires ERK1/2 activation and a downstream transcription factor, Fra-1. The ERK1/2–Fra-1–MMP9 axis is essential for LMP2A-induced ECM invasion of NPC cells. In addition, the association among LMP2A, Fra-1, and MMP9 is also found in NPC biopsy specimens. This study reveals an underlying mechanism of LMP2A-induced cell invasion, from signal transduction to upregulation of a critical protease. Meanwhile, MMP9 serves as a common invasion-promoting factor induced by LMP2A and LMP1, suggesting that it may be a potential therapeutic target for NPC treatment.
MATERIALS AND METHODS
Cell culture and drug treatment.
EBV-negative NPC cell lines (HONE-1 and NPC-TW01) and other epithelial cell lines (HeLa, SCC15, HBL-100, and HaCaT) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (HyClone Laboratories) at 37°C with 5% CO2. NPC cells stably expressing LMP2A were generated by using a lentiviral pLKO-AS2 system (Academia Sinica) and were selected by treatment with 2 μg/ml puromycin (Sigma). To block the MEK-ERK signaling pathway, cells were treated with 5 μM U0126 (Calbiochem) or 0.5 μM PD184352 (Enzo Life Sciences) for 36 to 48 h. To inhibit de novo protein synthesis, cells were treated with 5 μg/ml cycloheximide (Merck) for 12 h starting at 24 h posttransfection. To block proteasome-mediated protein degradation, cells were treated with 5 μM MG132 (Merck) for 12 h starting at 24 h posttransfection. To inhibit Ras activity, cells were treated with 1 μM manumycin A (Enzo Life Sciences) for 36 h. To inhibit MMP9 activity, cells were treated with 10 μM MMP9 inhibitor I (Calbiochem) for 48 h.
Plasmids and siRNAs.
The pSG5 vector-based plasmid expressing hemagglutinin (HA)-tagged LMP2A, pSG5-LMP2A, has been used previously (38). To further construct plasmids expressing LMP2A with serial amino-terminal truncations, DNA fragments of LMP2A were PCR amplified from the pSG5-LMP2A plasmid by using a common reverse primer (5′-GGGAGATCTACAAGCTAGCGTAATCTGG-3′) and different forward primers (5′-GGGAATTCATGGACCCATATTGGGGCAATG-3′, 5′-GGGAATTCATGCAACACGACGGGAATGAC-3′, and 5′-GGGAATTCATGAATCCAGTATGCCTGCC-3′) and then cloned into the pSG5 vector through the 5′ EcoRI site and the 3′ BglII site. Mutation of LMP2A at PY motifs (change of PPPPY into AAAAA) was carried out by using a QuikChange II XL site-directed mutagenesis kit (Stratagene). The pGL2-based reporter plasmid driven by the MMP9 promoter (−941 to 137) has been used previously (34). To further construct reporter plasmids with serial deletions of the promoter, three MMP9 promoter fragments (−546 to +137, −166 to +137, and −66 to +137) were PCR amplified by using a common reverse primer (5′-CCCAAGCTTTGAGATTGGTTCTCAGGTCT-3′) and different forward primers (5′-GGGGTACCCACTTGCCTGTCAAGGAG-3′, 5′-GGGGTACCGTGGTGTAAGCCCTTTCTC-3′, and 5′-GGGGTACCAGAGGAAGCTGAGTCAAAG-3′) and then cloned into the pGL2-basic vector through the 5′ KpnI site and the 3′ HindIII site. Specific mutation of AP-1 elements (change of the consensus sequence 5′-TGAGTCA-3′ into 5′-AACATCA-3′) in the MMP9 promoter (−546 to +137) was also carried out by using a QuikChange II XL site-directed mutagenesis kit (Stratagene). The small interfering RNAs (siRNAs) targeted against Fra-1 (5′-AUCUGUUCACAAGGCCUUCGACGUA-3′), c-Jun (5′-CAUAGAAGGUCGUUUCCAUCUUUGC-3′), and MMP9 (5′-AAGGUUUGGAAUCUGCCCAGGUCUG-3′) and a control siRNA with comparable GC content were all purchased from Invitrogen.
Transfection with plasmid DNA or siRNA.
Before transfection, cells were seeded into 6-well plates and cultured overnight. HONE-1, NPC-TW01, HeLa, and SCC15 cells were transfected with plasmid DNA or siRNA by using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. For single transfection, 10 μl Lipofectamine reagents mixed with 4 μg DNA or 5 μl Lipofectamine reagents mixed with 500 pmol siRNA were applied to each well. For dual transfection with DNA and siRNA, 10 μl Lipofectamine reagents mixed with 2 μg DNA and 500 pmol siRNA were applied to each well. After incubation for 4 h, the cells were washed and cultured in serum-free medium for 24 to 48 h. HBL-100 and HaCaT cells were transfected with plasmid DNA by using T-Pro nonliposome transfection reagent II (Biopioneer) according to the manufacturer's instructions. For this experiment, 8 μl transfection reagents mixed with 4 μg DNA were applied to each well.
Immunoblotting assay.
Protein extracts were prepared by lysing cells with NP-40 lysis buffer containing 1% NP-40, 1× protease inhibitor cocktail (Roche), and 1 μM Na3VO4 (Calbiochem). Protein concentrations were determined by using a protein assay reagent (Bio-Rad Laboratories). Protein extracts (40 μg of each sample) were heated at 100°C for 10 min, separated by electrophoresis in a 10% polyacrylamide gel, and then transferred onto Hybond-C extra membranes (Amersham Bioscience). The subsequent procedures, including membrane blocking, antibody reaction, and luminescence detection, were performed as described previously (27). Wild-type, deleted, and mutated LMP2A proteins were detected by using an anti-HA antibody (Roche). Antibodies recognizing phosphorylated Fra-1 at Ser265, phosphorylated c-Jun at Ser63, total c-Jun, phosphorylated ERK1/2 at Thr202/Tyr204, total ERK1/2, phosphorylated ERK5 at Thr218/Tyr220, and total ERK5 were purchased from Cell Signaling. An antibody detecting total Fra-1 was from Santa Cruz Biotechnology, and an antibody detecting β-actin was from Chemicon.
Gelatin zymography.
Serum-free culture supernatants were collected from the cells after transfection for 24 to 48 h. The supernatants (16 μl of each sample) were mixed with 4 μl 6× loading buffer (300 mM Tris-HCl [pH 6.8], 60% glycerol, 12% SDS, and 0.6% bromophenol blue) and resolved in an 8% SDS-polyacrylamide gel containing 1 mg/ml gelatin, the substrate of MMP9 and MMP2. After electrophoresis, gels were incubated in 2.5% Triton X-100 for 30 min to remove SDS and then transferred into a developing buffer containing 50 mM Tris-HCl (pH 7.6), 0.05 M NaCl, 5 mM CaCl2, and 0.05% Brij 35 (Sigma). After the incubation overnight at 37°C, the gels were stained with 0.2% Coomassie blue R250 for 1 h and destained in 10% methanol and 10% acetic acid. MMP9 (92 kDa) and MMP2 (72 kDa) were detected as transparent bands with different protein sizes in the blue gels. The medium (16 μl) containing 10% fetal bovine serum was used as a positive control of MMP9 and MMP2.
RNA extraction and quantitative RT-PCR.
Cellular RNAs were extracted by using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Reverse transcription (RT) was carried out by using Superscript III reverse transcriptase (Invitrogen) and oligo(dT)15 primers (Roche). The cDNAs of MMP9, Fra-1, and an internal reference gene, for TATA box-binding protein, were quantified by real-time PCR by using LightCycler reagents and a compatible detection system (Roche). PCR primers for detecting MMP9 were 5′-GAACCAATCTCACCGACAGG-3′ and 5′-GCCACCCGAGTGTAACCATA-3′, and the locked nucleic acid-based probe was 5′-CAGAGGAA-3′. Primers for detecting Fra-1 were 5′-AACCGGAGGAAGGAACTGAC-3′ and 5′CTGCAGCCCAGATTTCTCAT-3, and the probe was 5′-CTTCCTGC-3′. Primers for detecting the internal reference gene were 5′-GCTGGCCCATAGTGATCTTT-3′ and 5′-TCCTTGGGTTATCTTCACACG-3′, and the probe was 5′-CCCAGCAG-3′. Each experiment was done twice independently, and relative mRNA levels were calculated by using the LightCycler software program (Roche).
Reporter gene assay.
Firefly luciferase activity expressed by reporter plasmids was detected as described previously (27). Briefly, in each well of six-well plates, cells were cotransfected with 2 μg reporter plasmid and 2 μg effector plasmid under the serum-free culture condition. At 48 h posttransfection, the cells were harvested and subjected to the luciferase assay by using a Bright-Glo assay kit (Promega) according to the manufacturer's instructions. Each assay was carried out in duplicate, and the whole set of the experiments was performed at least twice independently.
Ras activation assay.
The Ras activation experiment was carried out by using a Ras activation assay kit (Cytoskeleton) according to the manufacturer's instructions. Briefly, 300 μg cellular proteins in 200 μl lysates were incubated with Raf-RBD beads at 4°C for 1 h. The active Ras proteins pulled down by the beads were resolved in a 12% SDS-polyacrylamide gel and detected by an immunoblotting assay using an anti-Ras monoclonal antibody. A sample pretreated with the nonhydrolyzable GTP analog GTPγS was used as a positive control of active Ras.
Matrigel invasion assay.
Invasive activity of NPC cells in ECM was measured by using BD Biocoat Matrigel invasion chambers (Becton, Dickinson). Cells suspended in serum-free medium were plated on the upper chamber, while the medium in the lower chamber contained 10% fetal bovine serum. After incubation at 37°C with 5% CO2 for 24 h, the noninvading cells on the upper chamber were removed using a cotton swab, and the cells invading through the Matrigel layer to the underside of the membrane were fixed and stained with Giemsa dye. Cell invasiveness in each chamber was determined by counting invading cells in five randomly chosen microscopic fields and is expressed as average numbers of invading cells per field.
Immunohistochemical staining.
We obtained formalin-fixed, paraffin-embedded tissue blocks of 71 primary NPC tumor biopsy specimens from Surgical Pathology Laboratory of National Cheng Kung University Hospital. The study protocol and specimen usage were approved by the Institutional Human Experiment and Ethics Committee of National Cheng Kung University Hospital (approval number ER-100-069). All the tumors presented an undifferentiated or poorly differentiated phenotype and were positive for EBV EBER expression (26). Serial tissue sections were deparaffinized, rehydrated, and pretreated with 3% H2O2 in 100% methanol for 15 min and then incubated with a blocking reagent containing normal horse serum (Novocastra Laboratories) at room temperature for 15 min. Heating in a citrate buffer (Invitrogen) at 120°C for 4 or 10 min was used for antigen retrieval, except that LMP1 was retrieved by treatment with 10 μg/ml proteinase K at room temperature for 15 min. Primary antibodies used for immunostaining included a rat monoclonal antibody against LMP2A (clone 15F9; AbD Serotec), a mouse monoclonal antibody against LMP1 (clone S12) (26), and rabbit polyclonal antibodies against MMP9 (Chemicon) or Fra-1 (Santa Cruz Biotechnology). The immunohistochemical staining was carried out by using an LSAB detection kit and a 3,3′-diaminobenzidine (DAB) substrate kit (Dako) and counterstained with hematoxylin. The staining results were examined under a light microscope (Olympus). The percentages of tumor cells that were positively stained for target proteins in each specimen were quantified.
Statistics.
To assess the effect of LMP2A or LMP1 on the expression level of MMP9 and to evaluate how much the effect was mediated through Fra-1, mediation analysis was performed using the Sobel test (49, 59). All the analyses were performed with the SAS macro programs SOBEL and INDIRECT (49, 50) (http://www.afhayes.com/spss-sas-and-mplus-macros-and-code.html#indirect) by using SAS version 9.1. Using the expression percentage of LMP2A or LMP1 in tumor cells as the predictor variable (X), that of MMP9 as the outcome variable (Y), and that of Fra-1 as the mediator (M), three linear regression models were built: (i) Y = i1 + cX; (ii) M = i2 + aX; and (iii) Y = i3+c′X+bM. The coefficient c denotes the overall effect of X on Y, a denotes the effect of X on M, b denotes the effect of M on Y adjusted for X, and c′ denotes the M-independent effect of X on Y. The mediation effect through M is defined as c − c′ = ab. The statistical significance of the mediation effect, ab, was assessed by constructing the 95% confidence interval (CI) using the bootstrap approach (49, 58); the mediation effect was considered statistically significant when its 95% CI did not contain 0. The mediation analysis was initially performed without adjusting for any covariate and was further performed by adjusting for the effect of LMP1 or LMP2A to exclude the possibility that these two viral proteins may confound each other's association with MMP9. Finally, to assess the joint influence between LMP2A and LMP1 on the expression level of MMP9, interaction analysis was performed by including a product term (LMP2A × LMP1) in the regression model and testing for its statistical significance.
RESULTS
LMP2A upregulates MMP9 expression in NPC cells.
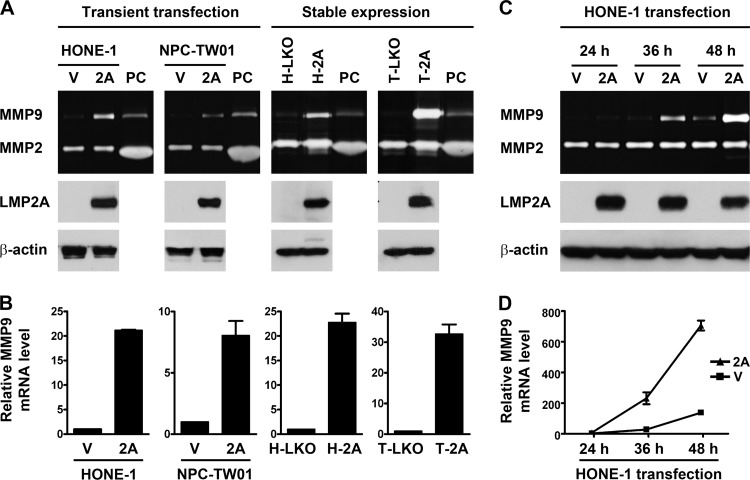
Since the expression of LMP2A proteins in EBV-infected epithelial cell lines is generally undetectable or much lower than that in NPC tumors, these cells may not be suitable for studying LMP2A's functions. Therefore, we chose ectopic expression of LMP2A in EBV-negative NPC cell lines to study the effects of LMP2A on the transformed epithelial cells. Transient LMP2A expression in the NPC cell lines, HONE-1 and NPC-TW01, was carried out by plasmid transfection. We also used a lentiviral expression system to establish H-2A and T-2A, the stable LMP2A-expressing NPC cells derived from HONE-1 and NPC-TW01, respectively. Expression of the LMP2A proteins in these NPC cells was confirmed by using an immunoblotting assay, and production of the MMP9 and MMP2 proteins in the cell culture supernatants was examined by using a gelatin zymography assay. Figure 1A shows that both transient and stable expression of LMP2A increased MMP9 production from NPC cells, while MMP2 production was not affected. The ectopic expression of LMP2A also increased MMP9 mRNA in NPC cells, which was detected by quantitative RT-PCR (Fig. 1B). LMP2A induced MMP9 proteins in a time-dependent manner in that the induction was detectable at 36 h posttransfection and became more prominent at 48 h posttransfection (Fig. 1C). The time-dependent induction of MMP9 proteins coincided with the increase of MMP9 mRNA (Fig. 1D), suggesting that LMP2A upregulates MMP9 expression at a transcriptional level.
Fig 1.
LMP2A upregulates MMP9 expression in NPC cells. (A) HONE-1 and NPC-TW01 cells were transiently transfected with an LMP2A-expressing plasmid (2A) or a vector plasmid (V). H-2A and T-2A were stable LMP2A-expressing cells derived from HONE-1 and NPC-TW01 cells, respectively, and H-LKO and T-LKO were their vector control cells, respectively. Expression of LMP2A and β-actin in cell lysates was examined by using an immunoblotting assay. Production of MMP9 and MMP2 in the cell culture supernatants was detected by using a gelatin zymography assay, where a positive control (PC) of both MMPs was included. (B) The increase of MMP9 mRNA in the NPC cells with transient or stable LMP2A expression was examined by using quantitative RT-PCR analysis. Shown are the relative levels of MMP9 mRNA. (C) HONE-1 cells were transiently transfected with an LMP2A-expressing plasmid or a vector plasmid. At 24, 36, and 48 h posttransfection, expression of indicated proteins was examined as described for panel A. (D) MMP9 mRNA of the cells in panel C was quantified.
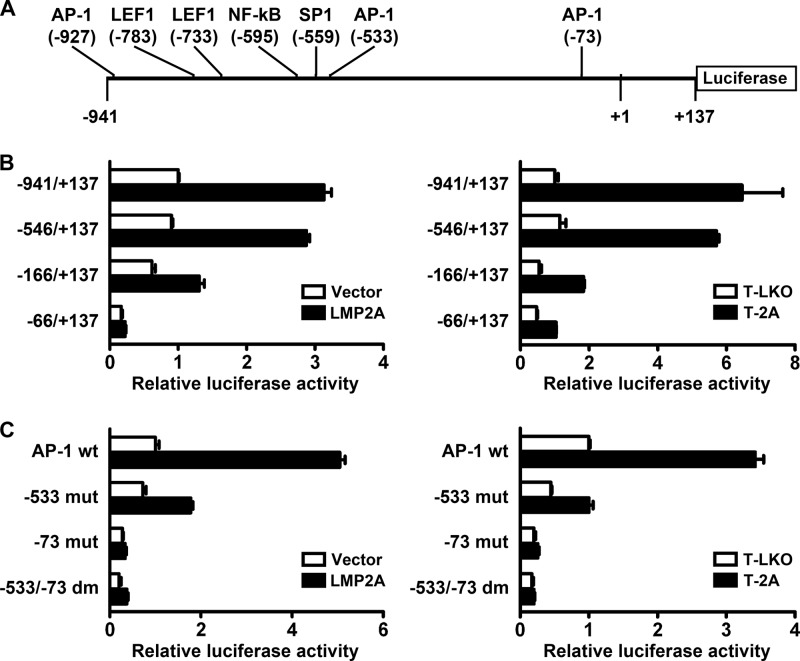
LMP2A-induced activation of the MMP9 promoter requires two AP-1 elements in the promoter.
Within the promoter region (−941 to +137) of the MMP9 gene, we found some potential DNA elements that may be targeted by LMP2A-regulated transcription factors, including the sites responsive to AP-1, β-catenin/LEF1, NF-κB, and SP1 (Fig. 2A). Therefore, we used reporter gene assays to test whether LMP2A can regulate activity of the promoter. Figure 2B shows that the MMP9 promoter (−941 to +137) was considerably activated by transient or stable expression of LMP2A in NPC cells compared with the promoter activity in the vector control cells. Deletion of the promoter region −941 to −547, which removed a distal AP-1 site, two LEF1 sites, an NF-κB site, and an SP1 site, did not significantly affect LMP2A-induced promoter activation (Fig. 2B), indicating that these elements are dispensable herein. Further deletion of the promoter region spanning −546 to −167 remarkably reduced the promoter responsiveness to LMP2A, and additional removal of the downstream region spanning −166 to −67 further impacted the responsiveness (Fig. 2B). Within the LMP2A-responsive region spanning −546 to −67 of the MMP9 promoter, we recognized two AP-1 elements, one at −533 and the other at −73. Disruption of the −533 AP-1 site alone largely impaired promoter responsiveness to LMP2A, and disruption of the −73 AP-1 site alone or of both the AP-1 sites completely abolished responsiveness (Fig. 2C), indicating that these two AP-1 elements are important for LMP2A-induced activation of the MMP9 promoter.
Fig 2.
LMP2A-induced activation of the MMP9 promoter requires two AP-1 elements in the promoter. (A) The MMP9 promoter studied in our reporter gene assays is illustrated. Locations of potential regulatory elements are indicated. (B) The reporter plasmids containing the MMP9 promoters with serial deletion were tested for their responsiveness to LMP2A (black bars) or the vector control (white bars). (C) The reporter plasmids containing the MMP9 promoter (−546 to +137) with wild-type AP-1 elements (AP-1 wt) or with AP-1 mutation at −533 (−533 mut), −73 (−73 mut), or both sites (−533/−73 dm) were tested for their responsiveness to LMP2A (black bars) or to the vector control (white bars). For panels B and C, reporter gene assays were performed by using NPC cells in which LMP2A expression was driven by transient transfection (left) or by the stable expression system (right).
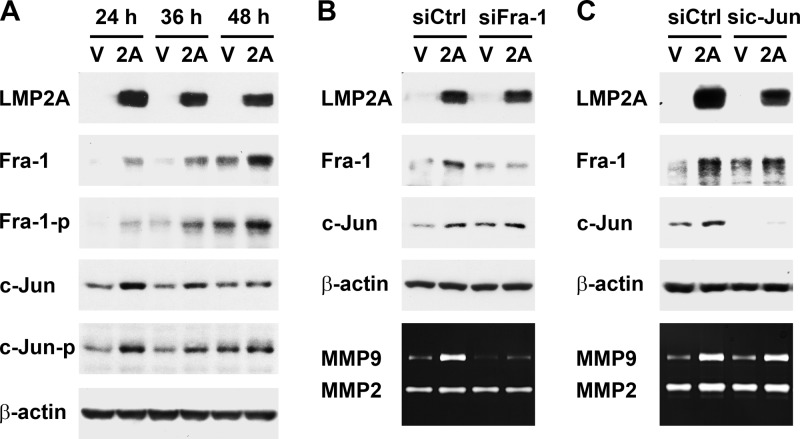
LMP2A upregulates Fra-1, which is required for LMP2A-induced MMP9 expression.
The result in Fig. 2 raises a possibility that LMP2A may upregulate AP-1 transcription factors to induce MMP9 expression. AP-1 proteins act as dimers consisting of Jun-Jun homodimers or Fos-Jun heterodimers (69, 75). Examining protein expression of the Fos family (c-Fos, FosB, Fra-1, and Fra-2) and Jun family (c-Jun, JunB, and JunD) in NPC cells, we found that LMP2A increased Fra-1 and c-Jun at both total and phosphorylated protein levels (Fig. 3A). However, the time courses of the induction of Fra-1 and c-Jun were different. In the transient LMP2A expression study, Fra-1 induction was merely detectable at 24 h posttransfection and became more prominent at 36 h and 48 h posttransfection, when MMP9 production was also induced by LMP2A significantly (Fig. 1C and 3A). In contrast, c-Jun was induced by LMP2A only at 24 h posttransfection, before MMP9 induction was detectable. To test whether the LMP2A-induced AP-1 proteins are required for MMP9 induction, we used siRNA to specifically inhibit expression of Fra-1 or c-Jun. Knockdown of Fra-1, which did not affect c-Jun expression, remarkably blocked LMP2A-induced MMP9 production (Fig. 3B), while knockdown of c-Jun had no effect on the upregulation of Fra-1 and MMP9 (Fig. 3C). Inhibition of Fra-1 induction also blocked the LMP2A-induced increase of MMP9 mRNA (data not shown). Therefore, Fra-1, not c-Jun, is essential for LMP2A-induced MMP9 expression.
Fig 3.
LMP2A upregulates Fra-1, which is required for LMP2A-induced MMP9 expression. (A) HONE-1 cells were transfected with an LMP2A-expressing plasmid (2A) or a vector plasmid (V). At 24, 36, and 48 h posttransfection, protein expression of LMP2A, total Fra-1, phosphorylated Fra-1 at Ser265 (Fra-1-p), total c-Jun, phosphorylated c-Jun at Ser63 (c-Jun-p), and β-actin was examined by using an immunoblotting assay. (B) HONE-1 cells were transfected with an LMP2A-expressing plasmid or a vector plasmid, in combination with a Fra-1-targeted siRNA (siFra-1) or a control siRNA (siCtrl). Expression of indicated proteins was detected by using an immunoblotting assay or a gelatin zymography assay. (C) HONE-1 cells were transfected with an LMP2A-expressing plasmid or a vector plasmid, in combination with a c-Jun-targeted siRNA (sic-Jun) or a control siRNA (siCtrl). Expression of indicated proteins was detected as described for panel B.
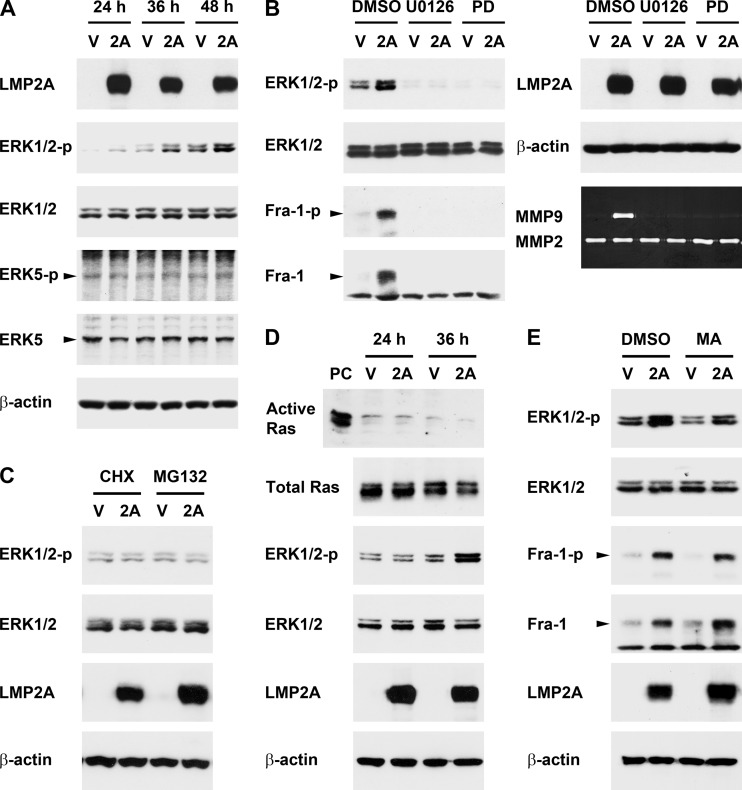
LMP2A activates an ERK1/2 signaling pathway, which is required for LMP2A-triggered induction of Fra-1 and MMP9.
LMP2A did not significantly increase Fra-1 mRNA in NPC cells (data not shown), suggesting that LMP2A may not induce Fra-1 at a transcriptional level. Phosphorylation of the Fra-1 protein at Ser265 has been shown to antagonize proteasomal degradation and thus increase protein stability (3, 11). Since the Ser265-phosphorylated form of Fra-1, as well as total Fra-1, was induced by LMP2A (Fig. 3A), Fra-1 induction may be attributed to protein stabilization. Previous studies indicate that the stabilization-inducing phosphorylation of Fra-1 is caused by the MEK1/2-ERK1/2 or MEK5-ERK5 signaling pathway (11, 65), so we wondered whether LMP2A can induce Fra-1 through the signaling events. We found that LMP2A activated the ERK1/2 but not ERK5 pathway in NPC cells (Fig. 4A). While the total protein level of ERK1/2 was not changed, the phosphorylation (i.e., activation) of ERK1/2 was induced by LMP2A, and the time course of the ERK1/2 activation was consistent with the induction of Fra-1 and MMP9 (Fig. 1C, 3A, and 4A). Treatment with U0126 (an inhibitor of MEK1/2) or PD184352 (an inhibitor of ERK1/2) not only blocked activation of the ERK1/2 pathway but also inhibited LMP2A-triggered induction of phosphorylated and total Fra-1 (Fig. 4B, left panel). In addition, treatment with the inhibitors blocked LMP2A-induced MMP9 production (Fig. 4B, right panel). In contrast, treatment with inhibitors of PI-3K, JNK, or p38 MAPK did not significantly affect MMP9 induction (data not shown). These results indicate that the LMP2A-activated ERK1/2 pathway is an upstream signaling event essential for induction of Fra-1 and MMP9.
Fig 4.
LMP2A activates the ERK1/2 signaling pathway, which is required for LMP2A-triggered induction of Fra-1 and MMP9. (A) HONE-1 cells were transfected with an LMP2A-expressing plasmid (2A) or a vector plasmid (V). At 24, 36, and 48 h posttransfection, protein expression of LMP2A, phosphorylated ERK1/2 at Thr202/Tyr204 (ERK1/2-p), total ERK1/2, phosphorylated ERK5 at Thr218/Tyr220 (ERK5-p), total ERK5, and β-actin was examined by using an immunoblotting assay. (B) A MEK inhibitor, U0126, an ERK inhibitor, PD184352 (PD), and their solvent control, dimethyl sulfoxide (DMSO), were used to treat the cells transfected with an LMP2A-expressing plasmid or a vector plasmid. Protein expression of LMP2A, phosphorylated ERK1/2, total ERK1/2, phosphorylated Fra-1 (Fra-1-p), total Fra-1, and β-actin was detected by using an immunoblotting assay. Production of MMP9 and MMP2 from the cells was detected by using a gelatin zymography assay. (C) A translation inhibitor cycloheximide (CHX) and a proteasome inhibitor (MG132) were used to treat LMP2A- or vector-transfected cells at 24 h posttransfection for 12 h. At 36 h posttransfection, cells were harvested for detection of indicated proteins. (D) Cells were transfected with an LMP2A-expressing plasmid or a vector plasmid for 24 or 36 h. Active Ras was detected by using a Ras activation assay, where a positive control (PC) was included. Expression of total Ras and other indicated proteins was detected by using an immunoblotting assay. (E) A Ras inhibitor, manumycin A (MA), and its solvent control, DMSO, were used to treat LMP2A- or vector-transfected cells. Expression of indicated proteins was detected as described for panel B.
LMP2A-induced ERK1/2 activation requires de novo protein synthesis and proteasome-mediated protein degradation but is independent of Ras.
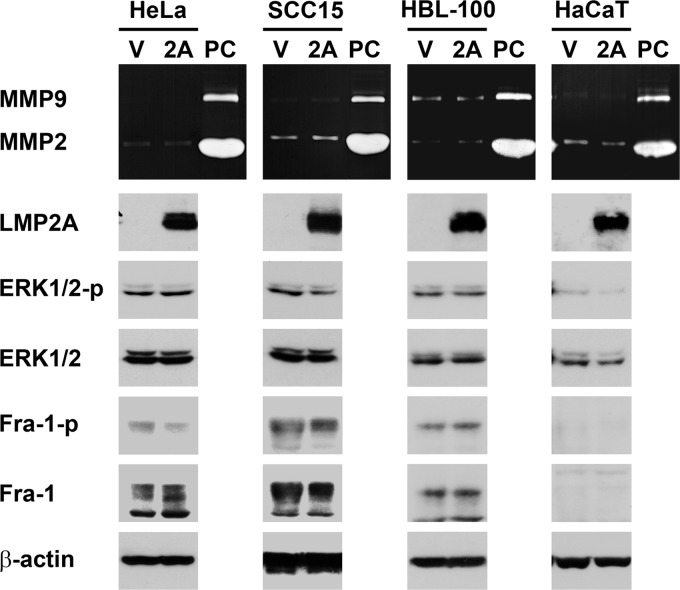
We wondered how LMP2A activates the ERK1/2 pathway. At 24 h posttransfection, when LMP2A expression had reached a plateau, ERK1/2 activation was barely detectable; the activation became prominent 12 h later, at 36 h posttransfection (Fig. 4A). The delay of LMP2A-triggered ERK1/2 activation implies that it may take time to accumulate some positive regulators or to remove some negative regulators. To test this possibility, we treated cells with a translation inhibitor, cycloheximide, or a proteasome inhibitor, MG132, at 24 h posttransfection, when LMP2A was highly expressed but ERK1/2 had not been activated yet. After a 12-h treatment with either inhibitor, LMP2A was unable to activate ERK1/2 (Fig. 4C), indicating that the activation requires both de novo protein synthesis (presumably of certain positive regulators) and proteasome-mediated degradation (presumably of certain negative regulators). On the other hand, previous studies show that LMP2A can activate Ras (21, 48), which is one of the potential activators of ERK1/2. In our study, however, Ras activation was not detectable before or when ERK1/2 was activated by LMP2A (Fig. 4D), and treatment with a Ras inhibitor, manumycin A, did not affect LMP2A-induced ERK1/2 activation and Fra-1 upregulation (Fig. 4E). Therefore, Ras is unlikely to mediate ERK1/2 activation in this context. Meanwhile, we found that LMP2A-triggered induction of the ERK1/2–Fra-1–MMP9 axis is cell dependent, since the effect of LMP2A was not detected in some non-NPC epithelial cell lines, including HeLa (derived from cervical carcinoma), SCC15 (from oral squamous cell carcinoma), and two immortalized, nonmalignant cell lines, HBL-100 (from breast tissue) and HaCaT (from keratinocytes) (Fig. 5).
Fig 5.
LMP2A is unable to induce MMP9, ERK, and Fra-1 in some non-NPC epithelial cell lines. HeLa, SCC15, HBL-100, and HaCaT cells were transfected with an LMP2A-expressing plasmid (2A) or a vector plasmid (V). Production of MMP9 and MMP2 was detected by using a gelatin zymography assay, where a positive control (PC) of both MMPs was included. Protein expression of LMP2A, phosphorylated ERK1/2 (ERK1/2-p), total ERK1/2, phosphorylated Fra-1 (Fra-1-p), total Fra-1, and β-actin was examined by using an immunoblotting assay. In HaCaT cells, phosphorylated Fra-1 and total Fra-1 were undetectable.
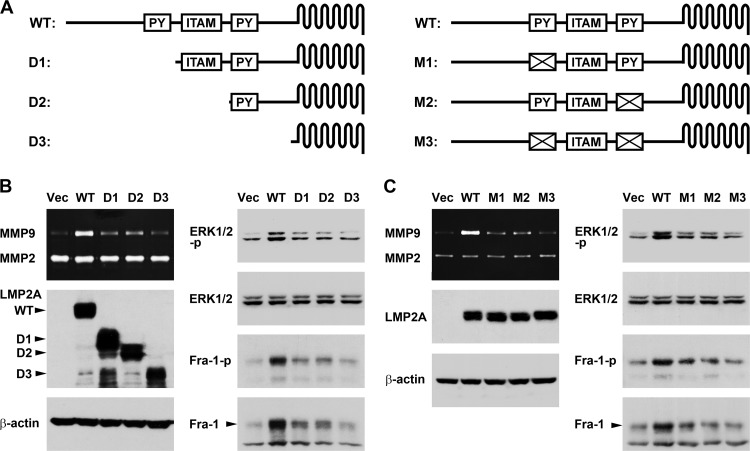
PY motifs of LMP2A are required for ERK1/2 activation, Fra-1 upregulation, and MMP9 induction.
To identify the motifs of LMP2A that are responsible for induction of the ERK1/2–Fra-1–MMP9 axis, several plasmids expressing truncated or mutated LMP2A were constructed and tested (Fig. 6A). Figure 6B shows the effects of serially truncated LMP2A proteins. For LMP2A D1, in which the amino-terminal region containing the first PY motif was deleted, the ability to induce ERK1/2 activation, Fra-1 upregulation, and MMP9 production was considerably reduced. For LMP2A D2, in which the ITAM was further removed, the extent of ERK1/2–Fra-1–MMP9 induction was similar to that for D1, suggesting that ITAM is not important herein. For LMP2A D3, in which the whole amino-terminal intracellular domain, including the second PY motif, was deleted, the induction of ERK1/2, Fra-1, and MMP9 was entirely abolished. This result prompted us to further test the effects of PY motif-mutated LMP2A proteins. Disruption of either of the PY motifs reduced induction of the ERK1/2–Fra-1–MMP9 axis, and the LMP2A mutant losing both PY motifs had no induction effect (Fig. 6C). Therefore, the PY motifs of LMP2A are essential for triggering ERK1/2 activation and the downstream induction of Fra-1 and MMP9.
Fig 6.
PY motifs of LMP2A are required for ERK1/2 activation, Fra-1 upregulation, and MMP9 induction. (A) Protein constructs of wild-type (WT) LMP2A and its derivatives with various truncations (D1 to D3) or mutations (M1 to M3) are illustrated schematically. PY motifs and ITAM in the amino-terminal domain of LMP2A are shown. (B) Plasmids expressing wild-type or serially truncated LMP2A proteins were transfected into HONE-1 cells. Protein expression of LMP2A, phosphorylated ERK1/2 (ERK1/2-p), total ERK1/2, phosphorylated Fra-1 (Fra-1-p), total Fra-1, and β-actin was detected by using an immunoblotting assay, and production of MMP9 and MMP2 in the cell culture supernatants was detected by using a gelatin zymography assay. (C) Plasmids expressing wild-type or PY motif-mutated LMP2A proteins were transfected into HONE-1 cells. Detection of the indicated intracellular proteins and secreted MMPs was performed as described for panel B.
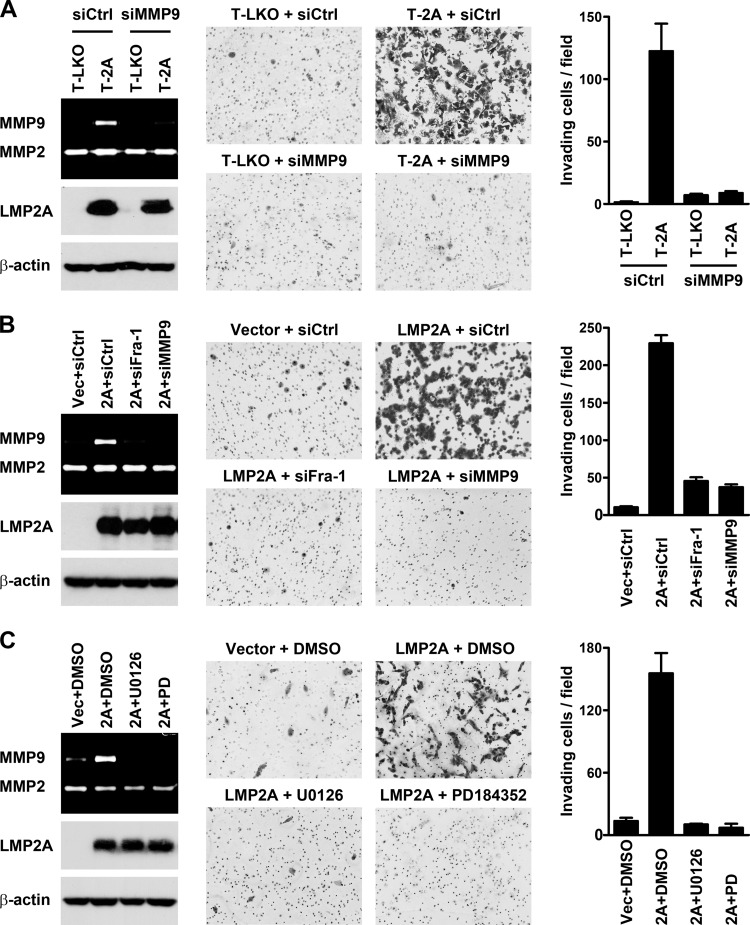
MMP9, Fra-1, and ERK1/2 are required for LMP2A-induced invasion of NPC cells.
Next, we examined whether MMP9, Fra-1, and ERK1/2 are important for LMP2A-induced cell invasion. The invasiveness of NPC cells in ECM was tested by using a Matrigel invasion assay. The LMP2A-expressing T-2A cells were more invasive than the vector control T-LKO cells, and the LMP2A-induced invasion was blocked by an MMP9-targeted siRNA (Fig. 7A). The invasiveness of T-2A was also suppressed by treatment with an MMP9 inhibitor (data not shown). Knockdown of MMP9 also inhibited cell invasion induced by transient LMP2A expression (Fig. 7B). Knockdown of Fra-1, which blocked MMP9 induction, exerted the same inhibitory effect on cell invasion (Fig. 7B). In addition, LMP2A-induced invasion was abolished by treatment with the inhibitors that blocked ERK1/2 activation and MMP9 production (Fig. 7C). According to these results, MMP9 may be a critical effector protein that is upregulated by LMP2A-induced ERK1/2 and Fra-1 and essentially contributes to LMP2A-promoted invasion of NPC cells.
Fig 7.
MMP9, Fra-1, and ERK1/2 are required for LMP2A-induced invasion of NPC cells. (A) LMP2A-expressing T-2A cells and the vector control T-LKO cells were transfected with an MMP9-targeted siRNA (siMMP9) or a control siRNA (siCtrl). (B) HONE-1 cells were transfected with an LMP2A-expressing plasmid (2A) or a vector plasmid (Vec), in combination with a Fra-1-targeted siRNA (siFra-1), an MMP9-targeted siRNA (siMMP9), or a control siRNA (siCtrl). (C) U0126, PD184352, and the solvent control, DMSO, were used to treat HONE-1 cells transfected with an LMP2A-expressing plasmid or a vector plasmid. Expression of LMP2A and β-actin was examined by using an immunoblotting assay, and production of MMP9 and MMP2 was detected by using a gelatin zymography assay. ECM invasiveness of the NPC cells was examined by using a Matrigel invasion assay; shown are the microscopic observations of invading cells and the average numbers of invading cells per field.
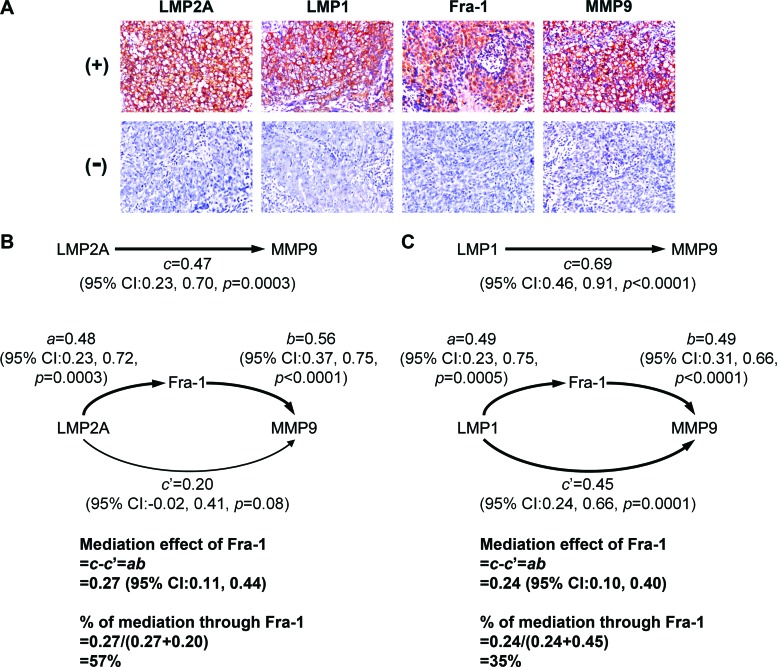
LMP2A, Fra-1, and MMP9 are associated in NPC tumor biopsy specimens.
In the last part of this study, we tested the association among LMP2A, Fra-1, and MMP9 in NPC biopsy specimens. We also examined LMP1 because previous studies indicate that it can induce MMP9 and may also upregulate Fra-1 (12, 73). Expression of these proteins in NPC tissues was detected by using immunohistochemical staining. The positive staining of LMP2A, LMP1, and MMP9 was in the membranes and cytoplasm of tumor cells, and Fra-1 was generally detected in the cytoplasm and nuclei of tumor cells (Fig. 8A). We quantified the expression percentages of target proteins in tumor cells and analyzed their association. Figure 8B presents the mediation analysis results for LMP2A, Fra-1 and MMP9. The overall effect, c, of LMP2A on MMP9 was 0.47 (P = 0.0003), meaning that for every 1% increment in LMP2A, MMP9 was increased by 0.47%. The effect, a, of LMP2A on Fra-1 and the effect, b, of Fra-1 on MMP9 were also positive and significant. The Fra-1-mediated effect, ab, of LMP2A on MMP9 was 0.27 (95% CI, 0.11 to 0.44); the effect was statistically significant since its 95% CI did not contain 0. On the other hand, the Fra-1-independent effect, c′, of LMP2A on MMP9 was relatively weak (95% CI, −0.02 to 0.41). Notably, more than half (57%) of the overall effect of LMP2A on MMP9 was mediated through Fra-1. Figure 8C shows the mediation analysis results for LMP1, Fra-1, and MMP9. The overall effect of LMP1 on MMP9 was significant (c = 0.69; P < 0.0001). Although the Fra-1-mediated effect for the influence of LMP1 on MMP9 was also significant (ab = 0.24; 95% CI, 0.10 to 0.40), it contributed only 35% of the overall effect of LMP1 on MMP9. Meanwhile, the Fra-1-independent effect of LMP1 on MMP9 existed significantly (c′ = 0.45; 95% CI, 0.24 to 0.66), suggesting that other mediation factors are involved therein. To exclude the possibility that LMP2A and LMP1 may confound each other's association with MMP9 or Fra-1, we performed additional mediation analyses by adjusting for each other's effect between LMP2A and LMP1. The adjustment did not change the overall assessment and further strengthened the contrast that most (73%) of the effect of LMP2A on MMP9 was through Fra-1, but Fra-1 mediated only a small portion (26%) of the effect of LMP1 on MMP9 (data not shown). Furthermore, the interaction analysis did not reveal any significant joint influence between LMP2A and LMP1 on MMP9 (coefficient for LMP2A × LMP1 = −0.003; P = 0.56), suggesting that these two viral oncoproteins regulate MMP9 expression independently.
Fig 8.
LMP2A, Fra-1, and MMP9 are associated in NPC tumor biopsy specimens. (A) Shown are the typical patterns for positive (+) and negative (−) immunohistochemical staining of LMP2A, LMP1, Fra-1, and MMP9 in NPC tissue sections. (B) Mediation analysis of LMP2A, Fra-1, and MMP9 was performed. The effective coefficients, 95% CIs, and P values are provided. The overall effect of LMP2A on MMP9 (c), the effect of LMP2A on Fra-1 (a), and the effect of Fra-1 on MMP9 (b) were all positive and statistically significant. The Fra-1-mediated effect between LMP2A and MMP9, ab, was also significant and contributed more than half (57%) of the overall effect of LMP2A on MMP9. On the other hand, the Fra-1-independent effect of LMP2A on MMP9 (c′) was relatively weak. (C) Mediation analysis of LMP1, Fra-1, and MMP9 was performed as described for panel B. The statistical results were similar to those for LMP2A, except that the Fra-1-independent effect of LMP1 on MMP9 (c′) existed significantly.
DISCUSSION
Although previous studies have shown that LMP2A enhances invasiveness of epithelial cells through activation of several signaling pathways (2, 13, 38), it is unclear what effector proteins link the signaling events with enhanced cell invasiveness. LMP2A-induced integrin α6 has been shown to promote invasion of primary epithelial cells, but how this adhesion molecule is upregulated by LMP2A is unknown (46). Here we reveal an underlying mechanism of LMP2A-induced invasion of NPC cells, which requires activation of the ERK1/2 pathway, upregulation of Fra-1, and induction of MMP9. In this context, MMP9 serves as a critical effector protein that bridges the gap between LMP2A-induced signaling transduction and cell invasion. The LMP2A-triggered, Fra-1-mediated induction of MMP9 may also occur in vivo, since the association among LMP2A, Fra-1, and MMP9 was found in NPC biopsy specimens. Considering that we used NPC cell lines and tumor specimens as the study materials, our results reflect a molecular mechanism through which LMP2A may confer a metastasis-prone feature on the established, transformed NPC cells. It remains to be tested whether LMP2A exerts similar effects on epithelial cells at early stages before the carcinoma has developed.
After stimulation by cytokines, growth factors, or other exogenous activators, MMP9 expression is upregulated mostly at the transcriptional level (53, 67, 72). MMP9 transcription is also induced by several oncogenic viruses, such as EBV, Kaposi's sarcoma-associated herpesvirus, human papillomavirus 16, and hepatitis B virus (14, 43, 70, 73). The NF-κB and AP-1 sites in the MMP9 promoter are the major cis-regulatory elements responsive to the upstream stimulation, while other elements, such as an overlapped SP1/Egr-1 site, may also be involved (33, 53, 57). In this study, LMP2A still activated the MMP9 promoter when the NF-κB and SP1/Egr-1 sites were removed, indicating that these elements are not essential. On the other hand, we found that two AP-1 sites at −533 and −73 of the MMP9 promoter are important for LMP2A-induced promoter activation. This finding prompted us to identify the AP-1 transcription factors mediating LMP2A-induced MMP9 expression. Among AP-1 family proteins, Fra-1 and c-Jun are induced by LMP2A at both total and phosphorylated protein levels in NPC cells. Induction of total and phosphorylated c-Jun proteins by LMP2A-triggered ERK has been observed in human embryonic kidney 293 cells (13). Although both Fra-1 and c-Jun have been associated with induction of MMP9 in previous studies (4, 33, 36), our study indicates that only Fra-1 is essential for LMP2A-induced MMP9 production in NPC cells while c-Jun is dispensable. It is likely that Fra-1 is the limiting factor for formation of the AP-1 heterodimers to induce MMP9 expression, but its binding partners can be redundant. This possibility is supported by our finding that other Jun family proteins, i.e., JunB and JunD, were detectable in NPC cells at a constant and substantial level whether LMP2A was expressed or not (data not shown). Meanwhile, Fra-1 may form dimers with other transcription factors containing a basic leucine zipper domain or even interact with other proteins without the domain (75), so we cannot rule out the possibility that LMP2A may induce other non-Jun proteins that serve as essential partners of Fra-1 to regulate MMP9 expression.
Fra-1 is overexpressed in many kinds of epithelial tumors, and its overexpression is associated with lymph node metastasis or poor prognosis of some squamous cell carcinomas (68, 75, 78). Functionally, Fra-1 can promote proliferation, anchorage-independent growth, migration, or invasion of various epithelial cells (1, 10, 54, 75). Expression of Fra-1 can be induced transcriptionally in that the Fra-1 promoter is activated by MAPK–AP-1, Akt-SP1, or β-catenin–LEF pathways (11, 40, 66). Although these pathways are potentially activated by LMP2A, we did not detect an LMP2A-induced increase of Fra-1 mRNA in NPC cells, indicating that LMP2A may not regulate Fra-1 transcription in this context. Instead, LMP2A can induce ERK1/2-dependent phosphorylation of Fra-1 at Ser265, thus increasing the protein level of Fra-1, probably through induction of posttranslational stabilization (3, 11). Notably, ectopic overexpression of Fra-1 alone did not induce MMP9 expression in NPC cells (data not shown), suggesting that in addition to an increase of total Fra-1 proteins, other LMP2A-induced events are required for MMP9 induction. For example, the transactivation activity of Fra-1 depends on ERK-mediated phosphorylation at a carboxyl-terminal threonine residual of Fra-1 (76), so the LMP2A-triggered signaling event may be required for induction of not only the protein level but also the activity of Fra-1. In a breast cancer cell line, Fra-1 alone does not sufficiently transactivate the MMP9 promoter; it requires cooperation with STAT3 to synergistically activate the promoter (60). Although LMP2A activates STAT3 in gastric carcinoma cells (24), it failed to do so in NPC cells (data not shown). It remains to be explored what factors are regulated by LMP2A to cooperate with Fra-1 for MMP9 induction.
LMP2A regulates signaling transduction in a cell-dependent manner, so it is not surprising that different kinase pathways contribute to LMP2A-induced effects in a variety of cells. The PI-3K/Akt pathway plays an important role in LMP2A-induced transformation of some epithelial cell lines (21, 55), but our study indicates that this pathway is unlikely to be important for LMP2A-induced MMP9 expression in NPC cells. Two LEF1 elements in the MMP9 promoter, which are putative target sites of PI-3K/Akt-activated β-catenin, are dispensable for LMP2A-induced promoter activation. The ITAM of LMP2A, which is essential for activation of Syk and PI-3K/Akt (18, 38, 64), is not essential for MMP9 induction. In addition, in our unpublished data, Akt was not activated by LMP2A in NPC cells, and treatment with a PI-3K inhibitor did not significantly reduce MMP9 induction. In contrast, a pivotal role of the ERK1/2 pathway is recognized in this study. The LMP2A-triggered ERK1/2 pathway essentially contributes to Fra-1 induction, MMP9 production, and ECM invasion of NPC cells. Of note, LMP2A can neither activate ERK1/2 nor induce expression of Fra-1 and MMP9 in some non-NPC epithelial cell lines, further supporting that the cellular background may determine LMP2A-induced signaling events and their downstream effects.
How LMP2A activates the ERK pathway has not been resolved yet. Although a previous study shows that ERK1/2 is associated with the amino-terminal cytoplasmic domain of LMP2A and can phosphorylate LMP2A at Ser15 and Ser102 (45), it is unknown whether the interaction directly results in ERK activation. In this study, LMP2A-induced ERK1/2 activation can be blocked by U0126, which inhibits MEK1/2, the upstream kinase of ERK. Therefore, LMP2A may trigger the ERK1/2 pathway from an upstream event involving MEK1/2 activation. We examined the potential involvement of Ras, but in NPC cells Ras is neither activated by LMP2A nor essential for the LMP2A-induced ERK1/2-Fra-1 pathway. Interestingly, the LMP2A effect on ERK1/2 activation is late and dependent on both de novo protein synthesis and proteasome-mediated protein degradation, suggesting that it needs time to accumulate some stimulatory factors and to remove some inhibitory factors. The requirement for PY motifs of LMP2A in this context provides a supporting clue. The PY motifs interact with class I WW domains rather than SH3 domains, thus recruiting several WW domain-containing ubiquitin ligases that presumably regulate protein stability of certain positive or negative regulators for ERK activation (29, 37, 71). An alternative mechanism is also possible, in which the PY motifs may interact with other WW domain-containing proteins that are not ubiquitin ligases but contribute to ERK activation.
Another EBV oncoprotein, LMP1, induces MMP9 in vitro and is also associated with MMP9 expression in NPC tumor biopsy specimens (25, 73). A potential link between LMP1 and Fra-1 comes from a previous study showing constitutive induction of Fra-1 in LMP1-transgenic mice (12). Therefore, we examined NPC biopsy specimens to assess the effect of LMP2A or LMP1 on MMP9 expression and evaluate how much the effect is mediated through Fra-1. We found that both LMP2A and LMP1 are associated with MMP9 expression and Fra-1 serves as a mediation factor therein. However, Fra-1 may contribute to LMP2A- and LMP1-induced MMP9 expression differentially. The statistical analysis indicates that the effect of LMP2A on MMP9 is mainly through Fra-1, but a large portion of the effect of LMP1 on MMP9 is Fra-1 independent. This result is consistent with the findings in our and other studies that Fra-1 is essential for LMP2A-triggered MMP9 upregulation while LMP1 induces MMP9 expression mainly through NF-κB rather than AP-1 (73). In addition, no synergistic effect of LMP2A and LMP1 on MMP9 was found in the NPC biopsy specimens, suggesting that the two EBV oncoproteins are independent inducers of MMP9. Serving as an oncogenic protease, MMP9 affects several biologic events to promote cancer progression, including cell invasion, angiogenesis, growth factor release, and local immune suppression (5, 39, 56, 77). Selective inhibition of MMP9 or related MMPs has been considered as a potential therapeutic strategy to prevent tumor metastasis (6, 28). Since MMP9 is a common factor induced by two EBV oncoproteins and associated with NPC metastasis, it may be an attractive target for NPC treatment.
ACKNOWLEDGMENTS
We thank Ching-Hwa Tsai for providing the pSG5-LMP2A plasmid and Ju-Ming Wang for providing the reporter plasmid of the MMP9 promoter (−941 to +137).
This study was supported by the National Science Council, Taiwan (NSC99-2628-B-400-001-MY3), the National Health Research Institutes, Taiwan (ID-099-PP-16, ID-100-PP-16, and IV-101-PP-18), and Department of Health, Executive Yuan, Taiwan (DOH101-TD-C-111-003).
This study has no potential conflicts of interest.
Footnotes
Published ahead of print 18 April 2012
REFERENCES
- 1. Adiseshaiah P, Lindner DJ, Kalvakolanu DV, Reddy SP. 2007. FRA-1 proto-oncogene induces lung epithelial cell invasion and anchorage-independent growth in vitro, but is insufficient to promote tumor growth in vivo. Cancer Res. 67:6204–6211 [DOI] [PubMed] [Google Scholar]
- 2. Allen MD, Young LS, Dawson CW. 2005. The Epstein-Barr virus-encoded LMP2A and LMP2B proteins promote epithelial cell spreading and motility. J. Virol. 79:1789–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Basbous J, Chalbos D, Hipskind R, Jariel-Encontre I, Piechaczyk M. 2007. Ubiquitin-independent proteasomal degradation of Fra-1 is antagonized by Erk1/2 pathway-mediated phosphorylation of a unique C-terminal destabilizer. Mol. Cell. Biol. 27:3936–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Belguise K, Kersual N, Galtier F, Chalbos D. 2005. FRA-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene 24:1434–1444 [DOI] [PubMed] [Google Scholar]
- 5. Bergers G, et al. 2000. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat. Cell Biol. 2:737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bonfil RD, et al. 2006. Inhibition of human prostate cancer growth, osteolysis and angiogenesis in a bone metastasis model by a novel mechanism-based selective gelatinase inhibitor. Int. J. Cancer 118:2721–2726 [DOI] [PubMed] [Google Scholar]
- 7. Brooks L, Yao QY, Rickinson AB, Young LS. 1992. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. J. Virol. 66:2689–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bultema R, Longnecker R, Swanson-Mungerson M. 2009. Epstein-Barr virus LMP2A accelerates MYC-induced lymphomagenesis. Oncogene 28:1471–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Busson P, et al. 1992. Consistent transcription of the Epstein-Barr virus LMP2 gene in nasopharyngeal carcinoma. J. Virol. 66:3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Casalino L, et al. 2007. Fra-1 promotes growth and survival in RAS-transformed thyroid cells by controlling cyclin A transcription. EMBO J. 26:1878–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Casalino L, De Cesare D, Verde P. 2003. Accumulation of Fra-1 in ras-transformed cells depends on both transcriptional autoregulation and MEK-dependent posttranslational stabilization. Mol. Cell. Biol. 23:4401–4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Charalambous CT, Hannigan A, Tsimbouri P, McPhee GM, Wilson JB. 2007. Latent membrane protein 1-induced EGFR signalling is negatively regulated by TGFα prior to neoplasia. Carcinogenesis 28:1839–1848 [DOI] [PubMed] [Google Scholar]
- 13. Chen SY, Lu J, Shih YC, Tsai CH. 2002. Epstein-Barr virus latent membrane protein 2A regulates c-Jun protein through extracellular signal-regulated kinase. J. Virol. 76:9556–9561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chung TW, Lee YC, Kim CH. 2004. Hepatitis B viral HBx induces matrix metalloproteinase-9 gene expression through activation of ERK and PI-3K/AKT pathways: involvement of invasive potential. FASEB J. 18:1123–1125 [DOI] [PubMed] [Google Scholar]
- 15. Egeblad M, Werb Z. 2002. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2:161–174 [DOI] [PubMed] [Google Scholar]
- 16. Fahraeus R, et al. 1988. Expression of Epstein-Barr virus-encoded proteins in nasopharyngeal carcinoma. Int. J. Cancer 42:329–338 [DOI] [PubMed] [Google Scholar]
- 17. Fotheringham JA, Mazzucca S, Raab-Traub N. 2010. Epstein-Barr virus latent membrane protein-2A-induced ΔNp63α expression is associated with impaired epithelial-cell differentiation. Oncogene 29:4287–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fruehling S, Longnecker R. 1997. The immunoreceptor tyrosine-based activation motif of Epstein-Barr virus LMP2A is essential for blocking BCR-mediated signal transduction. Virology 235:241–251 [DOI] [PubMed] [Google Scholar]
- 19. Fruehling S, Swart R, Dolwick KM, Kremmer E, Longnecker R. 1998. Tyrosine 112 of latent membrane protein 2A is essential for protein tyrosine kinase loading and regulation of Epstein-Barr virus latency. J. Virol. 72:7796–7806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fukuda M, Longnecker R. 2004. Latent membrane protein 2A inhibits transforming growth factor-β1-induced apoptosis through the phosphatidylinositol 3-kinase/Akt pathway. J. Virol. 78:1697–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fukuda M, Longnecker R. 2007. Epstein-Barr virus latent membrane protein 2A mediates transformation through constitutive activation of the Ras/PI3-K/Akt Pathway. J. Virol. 81:9299–9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heussinger N, et al. 2004. Expression of the Epstein-Barr virus (EBV)-encoded latent membrane protein 2A (LMP2A) in EBV-associated nasopharyngeal carcinoma. J. Pathol. 203:696–699 [DOI] [PubMed] [Google Scholar]
- 23. Hino R, et al. 2008. Survival advantage of EBV-associated gastric carcinoma: survivin up-regulation by viral latent membrane protein 2A. Cancer Res. 68:1427–1435 [DOI] [PubMed] [Google Scholar]
- 24. Hino R, et al. 2009. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 69:2766–2774 [DOI] [PubMed] [Google Scholar]
- 25. Horikawa T, Yoshizaki T, Sheen TS, Lee SY, Furukawa M. 2000. Association of latent membrane protein 1 and matrix metalloproteinase 9 with metastasis in nasopharyngeal carcinoma. Cancer 89:715–723 [DOI] [PubMed] [Google Scholar]
- 26. Hsiao JR, et al. 2009. Endoplasmic reticulum stress triggers XBP-1-mediated up-regulation of an EBV oncoprotein in nasopharyngeal carcinoma. Cancer Res. 69:4461–4467 [DOI] [PubMed] [Google Scholar]
- 27. Hsu M, et al. 2008. Epstein-Barr virus lytic transactivator Zta enhances chemotactic activity through induction of interleukin-8 in nasopharyngeal carcinoma cells. J. Virol. 82:3679–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hua J, Muschel RJ. 1996. Inhibition of matrix metalloproteinase 9 expression by a ribozyme blocks metastasis in a rat sarcoma model system. Cancer Res. 56:5279–5284 [PubMed] [Google Scholar]
- 29. Ikeda M, Ikeda A, Longnecker R. 2001. PY motifs of Epstein-Barr virus LMP2A regulate protein stability and phosphorylation of LMP2A-associated proteins. J. Virol. 75:5711–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kessenbrock K, Plaks V, Werb Z. 2010. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141:52–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kong QL, et al. 2010. Epstein-Barr virus-encoded LMP2A induces an epithelial-mesenchymal transition and increases the number of side population stem-like cancer cells in nasopharyngeal carcinoma. PLoS Pathog. 6:e1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee AW, et al. 1992. Retrospective analysis of 5037 patients with nasopharyngeal carcinoma treated during 1976–1985: overall survival and patterns of failure. Int. J. Radiat. Oncol. Biol. Phys. 23:261–270 [DOI] [PubMed] [Google Scholar]
- 33. Lin CC, et al. 2009. IL-1β promotes A549 cell migration via MAPKs/AP-1- and NF-κB-dependent matrix metalloproteinase-9 expression. Cell Signal. 21:1652–1662 [DOI] [PubMed] [Google Scholar]
- 34. Lin LF, et al. 2010. ZBRK1 acts as a metastatic suppressor by directly regulating MMP9 in cervical cancer. Cancer Res. 70:192–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Z, et al. 2010. Increased expression of MMP9 is correlated with poor prognosis of nasopharyngeal carcinoma. BMC Cancer 10:270 doi:10.1186/1471-2407-10-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Loesch M, et al. 2010. p38γ MAPK cooperates with c-Jun in trans-activating matrix metalloproteinase 9. J. Biol. Chem. 285:15149–15158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Longnecker R, et al. 2000. WW- and SH3-domain interactions with Epstein-Barr virus LMP2A. Exp. Cell Res. 257:332–340 [DOI] [PubMed] [Google Scholar]
- 38. Lu J, et al. 2006. Syk tyrosine kinase mediates Epstein-Barr virus latent membrane protein 2A-induced cell migration in epithelial cells. J. Biol. Chem. 281:8806–8814 [DOI] [PubMed] [Google Scholar]
- 39. Manes S, et al. 1999. The matrix metalloproteinase-9 regulates the insulin-like growth factor-triggered autocrine response in DU-145 carcinoma cells. J. Biol. Chem. 274:6935–6945 [DOI] [PubMed] [Google Scholar]
- 40. Mann B, et al. 1999. Target genes of β-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc. Natl. Acad. Sci. U. S. A. 96:1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morrison JA, Klingelhutz AJ, Raab-Traub N. 2003. Epstein-Barr virus latent membrane protein 2A activates β-catenin signaling in epithelial cells. J. Virol. 77:12276–12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morrison JA, Raab-Traub N. 2005. Roles of the ITAM and PY motifs of Epstein-Barr virus latent membrane protein 2A in the inhibition of epithelial cell differentiation and activation of β-catenin signaling. J. Virol. 79:2375–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Muhlen S, Behren A, Iftner T, Simon C. 2010. Influence of HPV16 E2 and its localisation on the expression of matrix metalloproteinase-9. Int. J. Oncol. 37:337–345 [DOI] [PubMed] [Google Scholar]
- 44. Pang MF, Lin KW, Peh SC. 2009. The signaling pathways of Epstein-Barr virus-encoded latent membrane protein 2A (LMP2A) in latency and cancer. Cell. Mol. Biol. Lett. 14:222–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Panousis CG, Rowe DT. 1997. Epstein-Barr virus latent membrane protein 2 associates with and is a substrate for mitogen-activated protein kinase. J. Virol. 71:4752–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pegtel DM, et al. 2005. Epstein-Barr-virus-encoded LMP2A induces primary epithelial cell migration and invasion: possible role in nasopharyngeal carcinoma metastasis. J. Virol. 79:15430–15442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Portis T, Dyck P, Longnecker R. 2003. Epstein-Barr virus (EBV) LMP2A induces alterations in gene transcription similar to those observed in Reed-Sternberg cells of Hodgkin lymphoma. Blood 102:4166–4178 [DOI] [PubMed] [Google Scholar]
- 48. Portis T, Longnecker R. 2004. Epstein-Barr virus (EBV) LMP2A mediates B-lymphocyte survival through constitutive activation of the Ras/PI3K/Akt pathway. Oncogene 23:8619–8628 [DOI] [PubMed] [Google Scholar]
- 49. Preacher KJ, Hayes AF. 2004. SPSS and SAS procedures for estimating indirect effects in simple mediation models. Behav. Res. Methods Instrum. Comput. 36:717–731 [DOI] [PubMed] [Google Scholar]
- 50. Preacher KJ, Hayes AF. 2008. Asymptotic and resampling strategies for assessing and comparing indirect effects in multiple mediator models. Behav. Res. Methods 40:879–891 [DOI] [PubMed] [Google Scholar]
- 51. Raab-Traub N. 2002. Epstein-Barr virus in the pathogenesis of NPC. Semin. Cancer Biol. 12:431–441 [DOI] [PubMed] [Google Scholar]
- 52. Rickinson AB, Kieff E. 2007. Epstein-Barr virus, p 2655–2700 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 53. Sato H, Kita M, Seiki M. 1993. v-Src activates the expression of 92-kDa type IV collagenase gene through the AP-1 site and the GT box homologous to retinoblastoma control elements. A mechanism regulating gene expression independent of that by inflammatory cytokines. J. Biol. Chem. 268:23460–23468 [PubMed] [Google Scholar]
- 54. Sayan AE, et al. 2012. Fra-1 controls motility of bladder cancer cells via transcriptional upregulation of the receptor tyrosine kinase AXL. Oncogene 31:1493–1503 [DOI] [PubMed] [Google Scholar]
- 55. Scholle F, Bendt KM, Raab-Traub N. 2000. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J. Virol. 74:10681–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sheu BC, et al. 2001. A novel role of metalloproteinase in cancer-mediated immunosuppression. Cancer Res. 61:237–242 [PubMed] [Google Scholar]
- 57. Shin SY, Kim JH, Baker A, Lim Y, Lee YH. 2010. Transcription factor Egr-1 is essential for maximal matrix metalloproteinase-9 transcription by tumor necrosis factor α. Mol. Cancer Res. 8:507–519 [DOI] [PubMed] [Google Scholar]
- 58. Shrout PE, Bolger N. 2002. Mediation in experimental and nonexperimental studies: new procedures and recommendations. Psychol. Methods 7:422–445 [PubMed] [Google Scholar]
- 59. Sobel ME. 1982. Asymptotic confidence intervals for indirect effects in structural equation models, p 290–312 In Leinhardt S. (ed), Sociological methodology 1982. American Sociological Association, Washington, DC [Google Scholar]
- 60. Song Y, et al. 2008. Fra-1 and Stat3 synergistically regulate activation of human MMP-9 gene. Mol. Immunol. 45:137–143 [DOI] [PubMed] [Google Scholar]
- 61. Spano JP, et al. 2003. Nasopharyngeal carcinomas: an update. Eur. J. Cancer 39:2121–2135 [DOI] [PubMed] [Google Scholar]
- 62. Sternlicht MD, Werb Z. 2001. How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Biol. 17:463–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stewart S, et al. 2004. Epstein-Barr virus-encoded LMP2A regulates viral and cellular gene expression by modulation of the NF-κB transcription factor pathway. Proc. Natl. Acad. Sci. U. S. A. 101:15730–15735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Swart R, Ruf IK, Sample J, Longnecker R. 2000. Latent membrane protein 2A-mediated effects on the phosphatidylinositol 3-kinase/Akt pathway. J. Virol. 74:10838–10845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Terasawa K, Okazaki K, Nishida E. 2003. Regulation of c-Fos and Fra-1 by the MEK5-ERK5 pathway. Genes Cells 8:263–273 [DOI] [PubMed] [Google Scholar]
- 66. Tiwari G, Sakaue H, Pollack JR, Roth RA. 2003. Gene expression profiling in prostate cancer cells with Akt activation reveals Fra-1 as an Akt-inducible gene. Mol. Cancer Res. 1:475–484 [PubMed] [Google Scholar]
- 67. Ueno S, et al. 2011. Asialoglycoprotein receptor promotes cancer metastasis by activating the EGFR-ERK pathway. Cancer Res. 71:6419–6427 [DOI] [PubMed] [Google Scholar]
- 68. Usui A, et al. 25 October 2011. The molecular role of Fra-1 and its prognostic significance in human esophageal squamous cell carcinoma. Cancer [Epub ahead of print.] doi:10.1002/cncr.26652 [DOI] [PubMed] [Google Scholar]
- 69. Verde P, Casalino L, Talotta F, Yaniv M, Weitzman JB. 2007. Deciphering AP-1 function in tumorigenesis: fra-ternizing on target promoters. Cell Cycle 6:2633–2639 [DOI] [PubMed] [Google Scholar]
- 70. Wang L, et al. 2004. The Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) K1 protein induces expression of angiogenic and invasion factors. Cancer Res. 64:2774–2781 [DOI] [PubMed] [Google Scholar]
- 71. Winberg G, et al. 2000. Latent membrane protein 2A of Epstein-Barr virus binds WW domain E3 protein-ubiquitin ligases that ubiquitinate B-cell tyrosine kinases. Mol. Cell. Biol. 20:8526–8535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yen JH, Kocieda VP, Jing H, Ganea D. 2011. Prostaglandin E2 induces matrix metalloproteinase 9 expression in dendritic cells through two independent signaling pathways leading to activator protein 1 (AP-1) activation. J. Biol. Chem. 286:38913–38923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yoshizaki T, Sato H, Furukawa M, Pagano JS. 1998. The expression of matrix metalloproteinase 9 is enhanced by Epstein-Barr virus latent membrane protein 1. Proc. Natl. Acad. Sci. U. S. A. 95:3621–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Young LS, et al. 1988. Epstein-Barr virus gene expression in nasopharyngeal carcinoma. J. Gen. Virol. 69:1051–1065 [DOI] [PubMed] [Google Scholar]
- 75. Young MR, Colburn NH. 2006. Fra-1 a target for cancer prevention or intervention. Gene 379:1–11 [DOI] [PubMed] [Google Scholar]
- 76. Young MR, et al. 2002. Transactivation of Fra-1 and consequent activation of AP-1 occur extracellular signal-regulated kinase dependently. Mol. Cell. Biol. 22:587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yu Q, Stamenkovic I. 2000. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-β and promotes tumor invasion and angiogenesis. Genes Dev. 14:163–176 [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang L, et al. 2010. Fos-related activator-1 is overexpressed in oral squamous cell carcinoma and associated with tumor lymph node metastasis. J. Oral Pathol. Med. 39:470–476 [DOI] [PubMed] [Google Scholar]