Abstract
The crystal structure of herpes simplex virus (HSV) gB identifies it as a class III fusion protein, and comparison with other such proteins suggests this is the postfusion rather than prefusion conformation, although this is not proven. Other class III proteins undergo a pH-dependent switch between pre- and postfusion conformations, and a low pH requirement for HSV entry into some cell types suggests that this may also be true for gB. Both gB and gH undergo structural changes at low pH, but there is debate about the extent and significance of the changes in gB, possibly due to the use of different soluble forms of the protein and different assays for antigenic changes. In this study, a complementary approach was taken, examining the conformations of full-length intracellular gB by quantitative confocal microscopy with a panel of 26 antibodies. Three conformations were distinguished, and low pH was found to be a major influence. Comparison with previous studies indicates that the intracellular conformation in low-pH environments may be the same as that of the soluble form known as s-gB at low pH. Interestingly, the antibodies whose binding was most affected by low pH both have neutralizing activity and consequently must block either the function of a neutral pH conformation or its switch from an inactive form to an activated form. If one of the intracellular conformations is the fusion-active form, another factor required for fusion is presumably absent from wherever that conformation is present in infected cells so that inappropriate fusion is avoided.
INTRODUCTION
Entry of herpes simplex virus (HSV) into cells may occur by fusion with the plasma membrane or by endocytosis and fusion with an endosomal membrane, depending on the cell type (29, 50, 55–57, 76). The process involves interactions between various virus glycoproteins, interactions between these glycoproteins and cell receptors, and, in some cases at least, the effects of low pH on one or more glycoproteins. Following attachment to the cell surface via binding to heparan sulfate proteoglycans (72), entry of virions is initiated by binding of gD to one of several coreceptors (12, 16, 28, 43, 51, 70, 72), which triggers a conformational change in gD (22, 44). There are two schools of thought about what happens next, one being that gD, after receptor binding, is able to bind to a preexisting complex of gB and gH/gL (6, 7) and the other being that gD, after receptor binding, is able to trigger the binding of gB to gH/gL (5). There is agreement that the gB-gH/gL interaction is required for fusion, but the molecular details of the interaction and the reason for its requirement are not yet known.
Prior to the availability of three-dimensional (3D) structures for gB and gH/gL, each had been proposed to be the protein with fusogenic activity, with evidence of various kinds coming from studies of the homologs from several herpesviruses in addition to HSV (20, 21, 23, 24, 30, 31, 40, 45, 46, 48, 59, 74). gB became favored as the fusion protein when three-dimensional structures were determined. The ectodomain of HSV-1 gB (36) has a remarkable similarity to the postfusion structure of the G fusion protein of vesicular stomatitis virus (VSV), a rhabdovirus (66), and gB and G were defined together as class III fusion proteins. Although not yet universally accepted, support for the idea that the gB structure represents a postfusion form was provided by the subsequent determination of a prefusion structure for the VSV G protein, in which the domain organization is very different (68). Furthermore, when a structure for Epstein-Barr virus (EBV) gB was determined and was found to be similar to that for HSV-1 gB, a hypothetical prefusion structure was modeled on that of VSV G (9). Two somewhat hydrophobic regions in HSV-1 gB that were hypothesized to be analogous to the fusion loop of class II fusion proteins (1) were shown to insert into membranes and therefore may be essential for the function of gB as a fusion protein (33, 34). In contrast, the HSV-2 gH/gL structure is different from that of any known fusion protein, and binding of gH/gL to gB was therefore proposed to be a positive regulator of the fusion activity of the latter, although whether binding is maintained throughout the fusion process or is transient is unknown (15). Similar conclusions were drawn from the structure of EBV gH/gL (47), but analysis of the pseudorabies virus gH structure identified a “flap” near the C terminus of the ectodomain that, if it were to move, would expose a hydrophobic patch (conserved in HSV-2 and EBV gH) that was postulated to interact with and destabilize the viral membrane (8).
Based on results determined with other class III fusion proteins (the VSV and rabies virus G proteins and the gp64 protein of the Autographa californica multiple nucleopolyhedrovirus [AcMNPV] baculovirus), it seems likely that a characteristic of class III proteins is that the prefusion-to-postfusion conformational change is reversible (26, 38, 66–68, 78). This is in contrast to the conformational changes of class I and II fusion proteins (such as influenza hemagglutinin [HA] and the E protein of flaviviruses, respectively), which are generally irreversible (39, 71, 77). The conformational changes in the rabies virus, VSV, and AcMNPV fusion proteins are all pH dependent (26, 67, 78), and studies of the rabies virus protein with monoclonal antibodies (MAbs) identified both conformations on virions in a pH-dependent equilibrium, together with an intermediate conformation that is antigenically indistinguishable from the prefusion conformation but more hydrophobic (25, 27). The pre- and postfusion forms were also found on the surface of infected cells, but only the postfusion form was found in intracellular locations, suggesting that the protein initially folds in its postfusion conformation, is transported intracellularly in that form to avoid premature activity, and then refolds into the prefusion conformation when it reaches the plasma membrane (26).
For HSV, exposure of virions to low pH after endocytosis is required for infection of some but not all cell types, as mentioned above. In addition, there was an intriguing observation that infectivity is dependent on exposure to low pH in the cells in which virions are produced (35). Whether only one or more than one glycoprotein must respond to low pH is currently the subject of debate, but there is certainly evidence that the conformation of gB is altered by pH in a reversible manner. According to one series of studies, the oligomeric structures of gB in virions and in lysates of infected cells, and of a soluble form of gB lacking the transmembrane anchor (s-gB), are reversibly altered by pH; some epitopes are lost in a partially reversible fashion at low pH, and at least one epitope is lost during entry of virus into cells by endocytosis (17, 18). Their observations led those authors to propose that low pH is an important factor in the activation of gB for fusion with endosomal membranes during virus entry (17). However, in a series of studies of gB730, the soluble form of gB used for crystallization, which lacks the cytoplasmic tail in addition to the transmembrane anchor, and in studies of gB in virions, a second group identified only minor conformational changes as a result of exposure to low pH (11, 73). The possibility was left open that, under some circumstances, full-length gB may undergo a more dramatic pH-triggered conformational change, since the crystal structure of the truncated protein indicates that it is in the same (possibly postfusion) conformation over a range of pH values from 5.5 to 8.5 and thus may be locked in this conformation (73). This suggestion is supported by the subsequent finding that gB730 does not undergo the pH-mediated changes in antigenicity or tryptophan fluorescence spectrum that are seen with s-gB (18). If low pH can indeed trigger an activating conformational change in gB, several possibilities can be envisaged for how inappropriate membrane fusion might be avoided during transport of the protein in infected cells. One possibility is that the conformational change from a prefusion to a fusion-active form (i.e., in an intermediate form between the pre- and postfusion forms) is somehow prevented in low-pH environments in infected cells, possibly by interaction of gB with other viral proteins. Alternatively, intracellular transport of gB may exclusively involve the postfusion conformation, as proposed for the rabies virus G protein. A third possibility is that the prefusion form exists in neutral pH compartments whereas an activated form exists in acidic organelles and that intracellular membrane fusion is avoided by the absence from these organelles of another required factor.
Thus, the purpose of this study was to examine conformations of intracellular gB under conditions where the full-length protein is present and is not subjected to potential influence by other viral proteins or by steps such as extraction and purification. This approach is complementary to those used by others and is aided by the large number of MAbs available for gB. Conformations of gB in transfected HEp-2 cells were first probed using confocal microscopy and a combination of a polyclonal antibody (PAb) and a panel of MAbs. The scans were made under conditions that allowed quantitative determinations of colocalization coefficients for pairs of antibodies, and the results strongly support the idea of existence of at least two conformations for intracellular gB. Simultaneous labeling with three MAbs then provided evidence for a third conformation. Identification of low-pH structures by the use of LysoTrackerRed (LtrRed) in combination with gB antibodies showed that pH is a major factor influencing the conformation of gB within transfected cells and that the intracellular conformation produced at low pH is likely the same as that produced by exposure of virion gB and the s-gB form of soluble gB to low pH (17). Finally, examination of gB in infected cells with a subset of the panel of MAbs produced similar results, namely, an effect of low pH and the presence of three antigenically distinct conformations.
MATERIALS AND METHODS
Cells, plasmids, and anti-gB antibodies.
HEp-2 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Plasmid pMM245, which expresses HSV-2 gB, was described previously (52). R90, a rabbit PAb made against purified native HSV-2 gB, and MAbs DL16, A22, C226, SS10, SS55, SS56, SS63, SS67, SS68, SS69, and SS144 were provided by Gary Cohen and Roselyn Eisenberg. The SS series was produced by immunizing mice with a mixture of native and reduced gB purified from HSV-1- and HSV-2-infected cells, whereas DL16 was produced from a mouse immunized with extracts of infected cells (10). No information is available on the production of A22 and C226. MAb II-125 was provided by Patricia Spear and was produced by immunization with UV-inactivated HSV-1 (60). MAbs H126, H146, H189, H233, H309, H352, H420, H1373, H1375, H1435, H1695, and H1819 (54) were purchased from the Rumbaugh-Goodwin Institute for Cancer Research. Various immunization procedures were used to produce them, but there is no information on which procedure was used for any given MAb (13, 42, 63, 65). MAbs SB1 and SB3 were produced in my laboratory, from mice immunized with UV-inactivated HSV-2 (58).
Transfections.
Cells were grown in Lab-Tek II 2-well coverglass chambers (Nunc), the bottom surface of which is a no. 1.5 coverglass that has the optimal thickness for z-axis resolution during confocal microscopy. They were transfected for 3 h with plasmid pMM245, which expresses HSV-2 gB, by the use of SuperFect (Qiagen), and the staining procedure was begun 18 h later.
Infections.
Cells were grown in Lab-Tek II coverglass chambers as described above and infected with HSV-2 strain 333 at a low multiplicity of infection (MOI) for 12 h to maximize the number of cells that had not yet rounded up or progressed to plaque formation.
Labeling of cells for confocal microscopy.
In experiments where the cell surface was biotinylated, this step was carried out before fixation and incubation with antibodies. Cells were washed sequentially with cold phosphate-buffered saline (PBS) and C-PBS (PBS supplemented with 1 mM MgCl2 and 0.1 mM CaCl2) and then incubated for 15 min at 4°C with sulfo-NHS-SS-biotin (0.1 mg/ml in cold C-PBS; Pierce). They were then washed with cold C-PBS, and excess biotin was quenched with C-PBS supplemented with 10 mM glycine. Following washes with cold C-PBS and PBS, the cells were fixed with 3% paraformaldehyde–PBS, washed with DMEM–5% FBS, and incubated with a fluorescent streptavidin Alexa Fluor 647 conjugate in DMEM–5% FBS for 30 min at room temperature (RT). They were then washed with DMEM–5% FBS and then with PBS prior to incubation for 1 h at RT with primary anti-gB antibodies diluted in PBS supplemented with 0.25% bovine serum albumin and 0.1% saponin detergent for permeabilization (PBS/BSA/SAP). Following washes with PBS/BSA/SAP, the cells were incubated for 30 min at RT with combinations of secondary antibodies (goat anti-rabbit IgG and goat anti-mouse IgG1, IgG2a, or IgG2b, as appropriate) conjugated to Alexa Fluor 488, 555, 594, or 633, after which they were again washed with PBS/BSA/SAP and finally with PBS. They were then mounted in SlowFade Gold-DAPI (Invitrogen) and stored in the dark at 4°C until they were scanned. All fluorescent conjugates were obtained from Molecular Probes.
In those cases where LtrRed was used to label low-pH structures, this step was done before biotinylation and fixation. The incubation time and concentration of LtrRed used were the minimum compatible with subsequent detection, in order to minimize the possibility of it acting as a weak base and raising the pH in acidic organelles. Cells were washed with DMEM–5% FBS at 37°C, incubated with LtrRed at 2 μm in DMEM–5% FBS for 10 min at 37°C, and washed once with DMEM–5% FBS for 3 min at 37°C, once with PBS at RT, and once with either C-PBS at 4°C (if the next step was biotinylation) or PBS at 4°C (if the next step was fixation). LtrRed diffuses freely into organelles but becomes trapped when protonated in an acidic environment. Consequently, it remains in place during subsequent washing steps whereas LtrRed elsewhere in the cell is removed; furthermore, it is fixed by paraformaldehyde and therefore is not removed even when the cells are permeabilized with detergent.
Confocal microscopy.
Multicolor sequential scanning (for DAPI and either three Alexa Fluor dyes or two Alexa Fluor dyes and LtrRed) was performed with a Leica TCS SP5 AOBS system using a 100× oil-immersion Plan Apo objective with a numerical aperture of 1.46, and z-stacks with voxel dimensions of 61 by 61 by 400 nm (corresponding to axes x, y, and z) were obtained. Scans for Alexa Fluors 488, 555, 594, and 633 (i.e., for detection of gB) and for LtrRed were performed six times per optical slice and averaged to maximize the signal-to-noise ratio. Scans for DAPI (nucleus) and for Alexa Fluor 647 (cell surface) were performed only once per slice, as a maximal signal-to-noise ratio was not required. The laser power and the sensitivity of the photomultiplier tube (PMT) detectors were set at levels that maximized the dynamic range of signals obtained without causing saturation. Furthermore, the labeling and scanning conditions were optimized for avoidance of bleedthrough between channels.
Quantitation of colocalization for two anti-gB antibodies.
Scans of cells labeled with the rabbit PAb R90 and a MAb were exported to AutoQuant software version X2.1.3 (Media Cybernetics) for 3D blind iterative deconvolution. Quantitative colocalization of the deconvolved Alexa Fluor 488 and Alexa Fluor 594 images (both identifying gB) was then performed with Zeiss LSM 510 software version 4.2 SP1, and weighted colocalization coefficients for each color were determined for the central slice in each z-stack. The colocalization coefficient for color 1 (M1) has a numerator of the summed intensities of pixels containing color 1 and at least some intensity of color 2 and a denominator of the summed intensities of all pixels containing color 1 (37), i.e., the fraction of the total signal for color 1 that is colocalized with any amount of color 2 above background. The coefficient for color 2 (M2) is calculated in a similar manner, and the range for each coefficient is from 0 to 1, such that a value of 1 for both M1 and M2 indicates complete colocalization. This was done for each of 26 MAbs together with R90, and z-stacks of at least six cells were analyzed for each MAb. For display purposes, the deconvolved images were reassembled with nondeconvolved images of DAPI and Alexa Fluor 647 (identifying the cell surface) using the Zeiss software. In addition, the images were brightened for display purposes (but not for quantitation) using the Zeiss software, as nonsaturated images are not bright enough to be seen clearly except in a darkened room. Panels of images were assembled with Photoshop.
Quantitation of gB staining in acidic structures.
The method used for quantitation of dual antibody binding could not be used for quantitating the labeling of gB in acidic structures. First, LtrRed is found in the lumen of organelles, whereas gB is found in the membrane; consequently, with an organelle larger than the limit of resolution (approximately 150 nm in this case), much of the gB staining would be in a concentric ring around the LtrRed. Second, the dynamic range of the PMT detectors is not sufficient to capture the range of signal intensities of LtrRed, with the intensity being determined by the LtrRed concentration, which in turn is dependent on the pH of the organelle. Therefore, scanning parameters that visualize mildly acidic organelles result in nonquantitative saturation of those which are more acidic. The method used was therefore to produce three-dimensional projections from z-stacks and determine the percentage of LtrRed-positive organelles that were also stained with a gB antibody. This was done with 6 cells per antibody. As in the case of the images visualized with pairs of antibodies, the images for display purposes, but not the images used for quantitation, have been brightened using Zeiss LSM 510 software so as to be clearly visible.
RESULTS
Quantitative colocalization of anti-gB MAbs with PAb R90.
To determine if gB has more than one conformation in transfected cells, a dual-labeling procedure was used such that HEp-2 cells transiently expressing gB were incubated simultaneously with pairs of antibodies and examined by quantitative confocal microscopy (as described in Materials and Methods). If there were only one conformation, then simultaneous incubation with any pair of antibodies should produce complete colocalization such that the weighted colocalization coefficients (M1 and M2) for both antibodies would have a value of 1. The only exception would be if two antibodies showed competitive binding and one were used in great excess over the other; this was avoided by titrating them individually such that they gave similar intensities of staining (not shown). Conversely, if there were two or more conformations, a variety of coefficients would be expected, depending on how particular antibodies were affected by the conformational change. A panel of 26 anti-gB MAbs was used, with each one being tested for colocalization with the rabbit PAb R90. A summary of the properties of the MAbs is given in Table 1. They were produced in several laboratories and represent a cross-section of those available, recognizing most of the gB domains identified by use of mutated proteins (10, 53, 54). Furthermore, about half of the selected MAbs have complement-independent neutralizing activity. gB at the cell surface was not examined in these experiments, as it requires higher concentrations of antibodies for detection (due to the relatively low levels) and may also be affected by the cell surface biotinylation used to identify the periphery of each cell.
Table 1.
Properties of anti-gB MAbs
| Antibody | Isotypea | Epitope | Minimal sequence recognized (residues)b | Domain | Neutralization | Reference |
|---|---|---|---|---|---|---|
| SB1 | 2b | Discontinuous | NDc | ND | − | 58 |
| SB3 | 2b | Discontinuous | ND | ND | − | 58 |
| II-125 | 2a | Discontinuous | ND | ND | − | 60 |
| H126 | 2b | Discontinuous | 1–462 | D2a | + | 54 |
| H146 | 2a | Discontinuous | 1–605 | Dd6 | − | 54 |
| H189 | 2a | Oligomer dependent | 1–695 | Dd5b | − | 54 |
| H233 | 2a | Discontinuous | 1–462 | D2b | + | 54 |
| H309 | 2b | Discontinuous | 1–605 | Dd6 | − | 54 |
| H352 | 2a | Discontinuous | 1–480 | D3a | − | 54 |
| H420 | 1 | Oligomer dependent | 1–695 | Dd5a | − | 54 |
| H1373 | 2a | Oligomer dependent | 1–695 | Dd5a | − | 54 |
| H1375 | 1 | Discontinuous | 1–462 | D2a | + | 54 |
| H1435 | 1 | Discontinuous | 1–462 | D2a | + | 54 |
| H1695 | 2a | Oligomer dependent | 1–695 | Dd5a | + | 54 |
| H1819 | 2a | Discontinuous | 1–462 | D2b | + | 54 |
| DL16 | 2a | Oligomer dependent | 1–700 | ND | − | 10 |
| SS10 | 1 | Pseudocontinuous | 615–645 | IV-III | + | 10 |
| SS55 | 1 | Discontinuous | 71–445 | I | + | 10 |
| SS56 | 1 | Discontinuous | ND | I | + | 10 |
| SS63 | 1 | Continuous | 672–700 | V | − | 10 |
| SS67 | 1 | Pseudocontinuous | 615–645 | IV-III | + | 10 |
| SS68 | 1 | Pseudocontinuous | ND | IV-III | + | 10 |
| SS69 | 1 | Pseudocontinuous | ND | IV-III | + | 10 |
| SS144 | 1 | Pseudocontinuous | 672–700 | V | + | 10 |
| A22 | 2a | Pseudocontinuous | 446–615 | III-IV | − | 10 |
| C226 | 1 | Pseudocontinuous | 207–445 | II | + | 10 |
If not determined in the referenced publication, isotype was determined by exclusive reaction with one of three isotype-specific secondary antibodies.
The numbers have been converted from those mapped in HSV-1 gB to the corresponding regions in HSV-2 gB, which has an additional eight residues split between three sites. In addition, the signal sequence, which differs in length between HSV-1 gB and HSV-2 gB, has been omitted from the numbers.
ND, not determined.
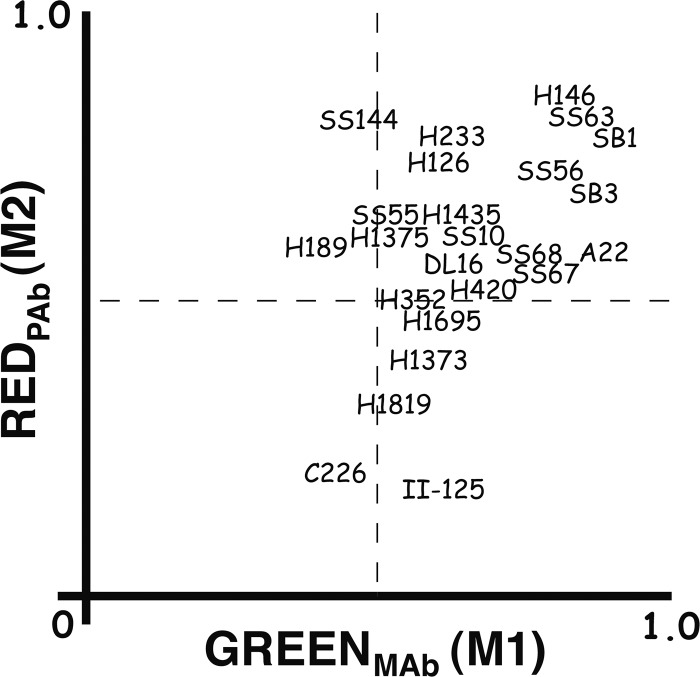
Examples of the results, selected to illustrate their diversity, are shown in Fig. 1. The weighted colocalization coefficients (M1 for the MAb and M2 for the R90 PAb) are shown in Table 2 and graphed for easier comparison in Fig. 2. MAbs H146, SS63, and SB1 all showed nearly complete colocalization with R90 (i.e., M1 and M2 were both close to 1.0) and therefore recognized all conformations that R90 recognizes and vice versa. Sixteen additional MAbs are within the upper right quadrant (M1 and M2 both above 0.5), indicating that more than half the gB that they recognized was also recognized by R90 and vice versa. However, seven MAbs were outliers. H189 and SS144 recognized more than half of the gB seen by R90, but R90 recognized less than half of the gB seen by each MAb. Four MAbs (H1373, H1695, H1819, and II-125) recognized less than half of the gB seen by R90, but R90 recognized more than half of the gB seen by each MAb. Finally, MAb C226 recognized less than half of the gB seen by R90, and R90 recognized less than half of the gB seen by C226. These results are incompatible with the existence of only one conformation of gB in cells and cannot be explained by competition between antibodies (see Discussion). Furthermore, they indicate that the PAb did not recognize all of the gB present in the cell. For the most part, MAbs previously identified as binding to the same gB domain, for example, SS10, SS67, SS68, and SS69 (domain IV-III; see reference 10) and H126, H1375, and H1435 (domain D2a; see references 41 and 54, had similar M1 and M2 values.
Fig 1.
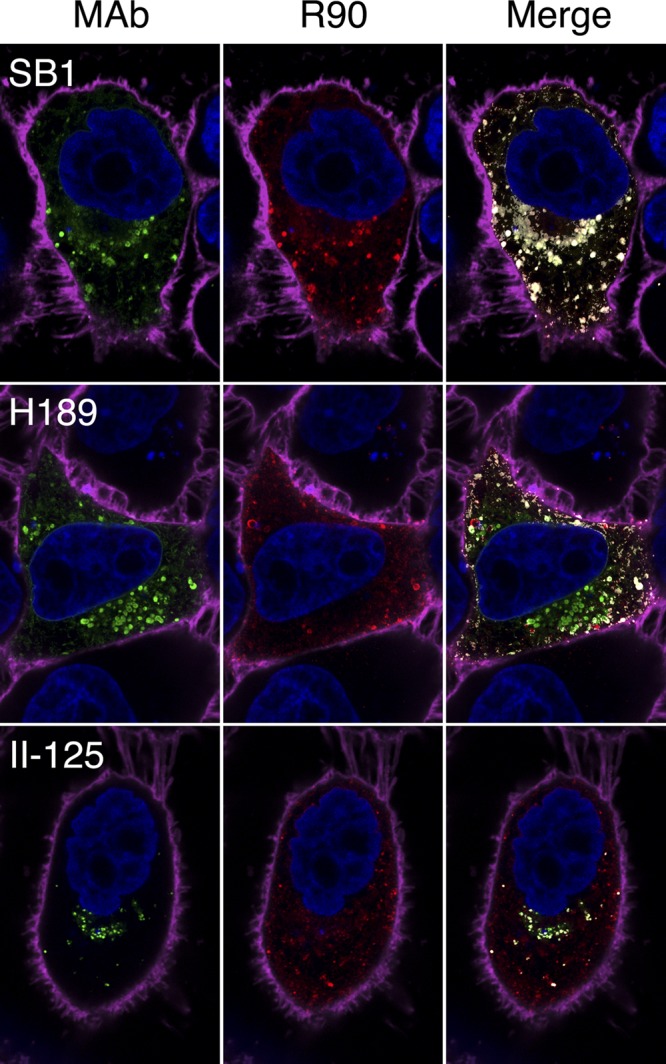
Confocal slices of gB-expressing cells labeled simultaneously with an anti-gB MAb (SB1, H189, or II-125 plus Alexa Fluor 488; green) and an anti-gB PAb (R90 plus Alexa Fluor 594; red). Colocalization in the merged images is indicated by white and does not imply that labeling with the anti-gB MAb and labeling with the anti-gB PAb were equal at these points (see Materials and Methods for details). Cell nuclei are blue, and the cell periphery is magenta. As described in Materials and Methods, these images and those in other figures have been brightened for visibility in a published format, but quantitation was done using the original images.
Table 2.
Colocalization of anti-gB MAbs with PAb R90 and with LtrRed
| Antibody | Colocalization coefficient (± SD) |
% LtrRed colocalization (± SD) | |
|---|---|---|---|
| M1a | M2b | ||
| SB1 | 0.93 ± 0.02 | 0.78 ± 0.06 | 58.3 ± 10.0 |
| SB3 | 0.90 ± 0.06 | 0.69 ± 0.16 | 67.3 ± 11.0 |
| II-125 | 0.60 ± 0.21 | 0.15 ± 0.08 | 70.6 ± 4.9 |
| H126 | 0.62 ± 0.10 | 0.75 ± 0.07 | 15.5 ± 3.8 |
| H146 | 0.85 ± 0.08 | 0.84 ± 0.05 | 70.1 ± 4.5 |
| H189 | 0.38 ± 0.10 | 0.61 ± 0.12 | 22.7 ± 4.7 |
| H233 | 0.64 ± 0.19 | 0.78 ± 0.08 | 58.5 ± 10.0 |
| H309 | 0.89 ± 0.08 | 0.58 ± 0.15 | 68.4 ± 4.9 |
| H352 | 0.54 ± 0.11 | 0.51 ± 0.16 | 28.2 ± 5.5 |
| H420 | 0.71 ± 0.17 | 0.53 ± 0.15 | 64.6 ± 8.0 |
| H1373 | 0.57 ± 0.20 | 0.41 ± 0.09 | 71.5 ± 8.6 |
| H1375 | 0.52 ± 0.13 | 0.64 ± 0.15 | 78.2 ± 6.5 |
| H1435 | 0.63 ± 0.10 | 0.67 ± 0.15 | 59.5 ± 16.1 |
| H1695 | 0.61 ± 0.15 | 0.48 ± 0.07 | 77.9 ± 4.7 |
| H1819 | 0.52 ± 0.19 | 0.34 ± 0.21 | 14.3 ± 7.6 |
| DL16 | 0.64 ± 0.09 | 0.57 ± 0.13 | 40.6 ± 11.9 |
| SS10 | 0.66 ± 0.18 | 0.65 ± 0.11 | 80.9 ± 7.4 |
| SS55 | 0.51 ± 0.16 | 0.67 ± 0.14 | 47.0 ± 11.5 |
| SS56 | 0.81 ± 0.11 | 0.72 ± 0.13 | 61.0 ± 8.4 |
| SS63 | 0.88 ± 0.06 | 0.83 ± 0.05 | 57.6 ± 7.8 |
| SS67 | 0.74 ± 0.09 | 0.57 ± 0.14 | 60.3 ± 6.8 |
| SS68 | 0.80 ± 0.08 | 0.58 ± 0.08 | 66.2 ± 6.4 |
| SS69 | 0.81 ± 0.08 | 0.58 ± 0.08 | 64.2 ± 10.8 |
| SS144 | 0.46 ± 0.09 | 0.83 ± 0.08 | 49.5 ± 7.8 |
| A22 | 0.89 ± 0.05 | 0.58 ± 0.11 | 55.3 ± 7.4 |
| C226 | 0.44 ± 0.17 | 0.19 ± 0.08 | 54.9 ± 6.9 |
M1 has a numerator of the summed intensities of pixels containing color 1 (MAb) and at least some intensity of color 2 (PAb) and a denominator of the summed intensities of all pixels containing color 1.
M2 has a numerator of the summed intensities of pixels containing color 2 and at least some intensity of color 1 and a denominator of the summed intensities of all pixels containing color 2.
Fig 2.
Weighted colocalization coefficients (M1 and M2) for each MAb used in combination with PAb R90, determined from sets of deconvolved confocal images. M1 indicates how much of the gB recognized by a MAb is also recognized by R90, and M2 indicates how much of the gB recognized by R90 is also recognized by a MAb. SS69 and H309 are not shown, as their coordinates were superimposable with those of SS68 and A22, respectively.
There were obvious differences in the abilities of the MAbs to stain perinuclear gB (i.e., within the inner and/or outer nuclear membranes), although this was not quantitated. Of the 26 MAbs, 23 produced perinuclear staining that was either continuous or nearly so. However, the perinuclear staining obtained with DL16 was punctate and not close to continuous, and C226 and II-125 produced no perinuclear staining at all.
Colocalization of combinations of two anti-gB MAbs.
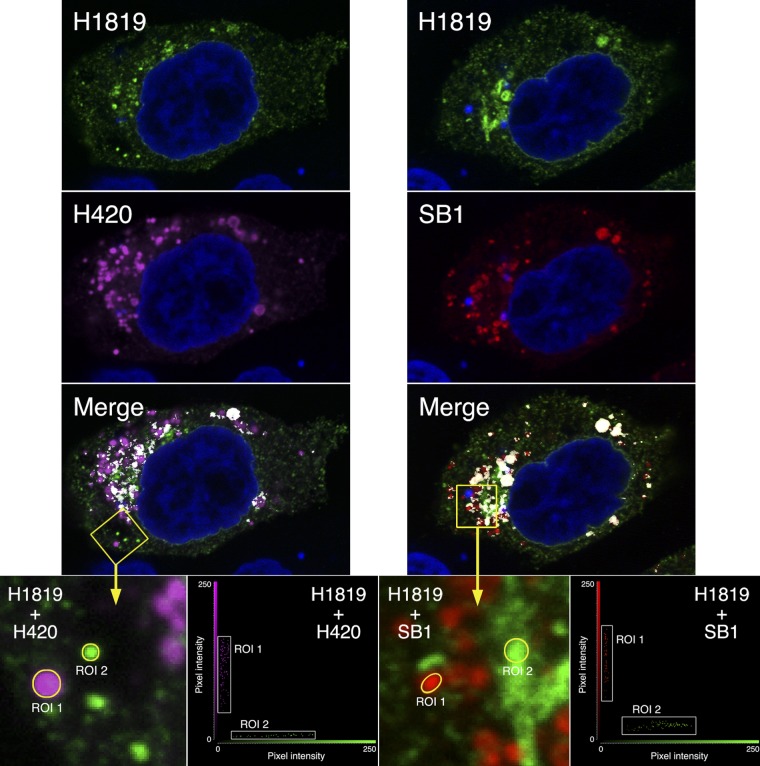
Experiments involving pairs of MAbs (of different isotypes) for dual-labeling and confocal microscopy qualitatively confirmed the existence of at least two conformations of gB in cells. For example, combining MAbs H1819 (IgG2a) and H420 (IgG1) (left set of images in Fig. 3) produced some structures labeled only with the former (green), some only with the latter (magenta), and some with both (white areas in the merged image). This can also be demonstrated by selecting regions of interest (ROIs), as seen in the enlargement, and plotting the corresponding pixel intensities in a fluorogram. ROI 1 contains gB in a conformation that is recognized by H420 but not by H1819 (H1819−/H420+, referred to here as conformation A), whereas ROI 2 contains gB in a conformation that is recognized by H1819 but not by H420 (H1819+/H420−, referred to here as conformation B). Structures labeled with both MAbs might contain a third conformation or a mixture of conformations A and B. To investigate the possible existence of three conformations, an IgG2b MAb was needed for triple-labeling experiments, and H1819 was therefore tested in combination with the four that were available. SB1 was found to be the best candidate, based on the results shown in the right set of images in Fig. 3. ROI 1, as shown in the enlargement from this set of images, contains gB in a conformation that is recognized by SB1 but not by H1819 (H1819−/SB1+), whereas ROI 2 either contains gB in a conformation that is well recognized by H1819 but only very weakly by SB1 (H1819+/SB1low) or alternatively contains an H1819+/SB1− conformation together with a much smaller amount of the H1819−/SB1+ conformation (the low values for red pixel intensity are consistent with both possibilities). These results themselves did not identify a third conformation, as the data are also consistent with the H1819−/SB1+ form being identical to conformation A and the H1819+/SB1− or H1819+/SB1low form being identical to conformation B.
Fig 3.
Confocal slices of gB-expressing cells labeled simultaneously with two anti-gB MAbs (H1819 and H420 in the left set of images and H1819 and SB1 in the right set of images). The conjugates were Alexa Fluor 488 for H1819, Alexa Fluor 555 for SB1, and Alexa Fluor 633 for H420. Colocalization in the merged images is indicated by white, and cell nuclei are blue. Enlargements of the two yellow boxed areas are shown with the merge feature turned off, and intensities of the pixels within the ROIs are plotted in the adjacent fluorograms. Cell peripheries were not labeled in this experiment, as a far-red dye was used for one of the MAbs in anticipation of labeling with three MAbs in the subsequent experiment.
Colocalization of combinations of three anti-gB MAbs.
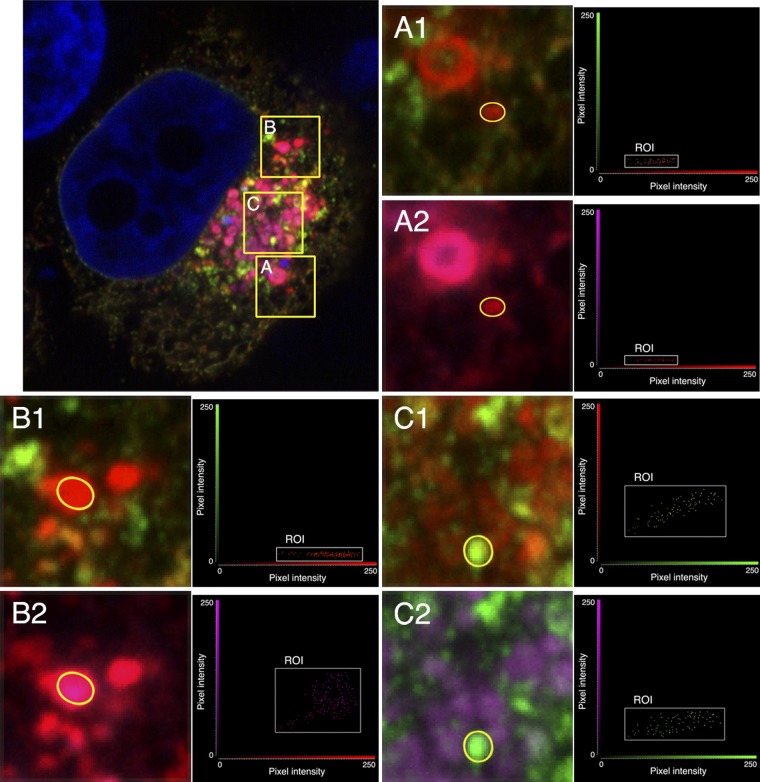
To investigate the hypothesis that three conformations of intracellular gB exist, transfected HEp-2 cells expressing gB were triple-labeled with MAbs H1819 (green), SB1 (red), and H420 (magenta), and a representative example is shown in Fig. 4 (large panel). Each boxed area in the large panel is also shown as two enlargements, with one color channel omitted from each enlargement, such that panel A1, for example, shows the green and red channels for box A whereas A2 shows the red and magenta channels. This allows pixel intensities of the ROIs within the enlarged areas to be represented on two-dimensional fluorograms, and the fluorograms show that the ROI in panels A1 and A2 contains gB that is H1819−/SB1+/H420−. This is different from the properties of conformations A and B and cannot be explained by a combination of the two; therefore, it identifies a third form of the protein, which is referred to here as conformation C. Some other permutations were also seen, two of which are shown because they are informative about conformations A and B. The ROI in panels B1 and B2 is H1819−/SB1+/H420+, for which the most likely explanation (as discussed later) is that conformation A (H1819−/H420+) is also SB1+. The ROI in panels C1 and C2 is H1819+/SB1+/H420+ and therefore must contain either a new conformation or some combination of A, B, and C. The latter explanation is the more likely, and the results also suggest that conformation B is H1819+/SB1low/H420−, as discussed later. Quantitation of the relative amounts of these conformations is not possible, as the ratio of pixel intensity to the number of gB molecules capable of binding a given MAb is different for each MAb.
Fig 4.
Confocal slice of a gB-expressing cell labeled simultaneously with three anti-gB MAbs: H1819 (plus Alexa Fluor 488; green); SB1 (plus Alexa Fluor 555; red); and H420 (plus Alexa Fluor 633; magenta). The main panel shows all three MAbs, whereas the enlargements (A1 and A2 for box A, B1 and B2 for box B, and C1 and C2 for box C), together with the accompanying fluorograms for the ROIs, each show only two MAbs. This is necessary in order to represent the results on two-dimensional fluorograms.
Effect of low pH on gB conformation in transfected cells.
Perhaps the most obvious factor that might influence the conformation of intracellular gB is pH, which is known to affect the conformation of soluble truncated gB and of virion gB (11, 17, 18, 73) and is required for virus entry into some cell types (56). To investigate this possibility, acidic intracellular organelles were identified by incubating live cells at 37°C with LtrRed fluorescent dye prior to labeling with anti-gB MAbs. Each MAb was tested individually for its ability to label gB in acidic structures in transfected cells; note that the MAbs themselves were not exposed to low pH, as labeling was done at neutral pH after fixing the cells with paraformaldehyde. Examples to indicate the diversity of results are shown in Fig. 5.
Fig 5.
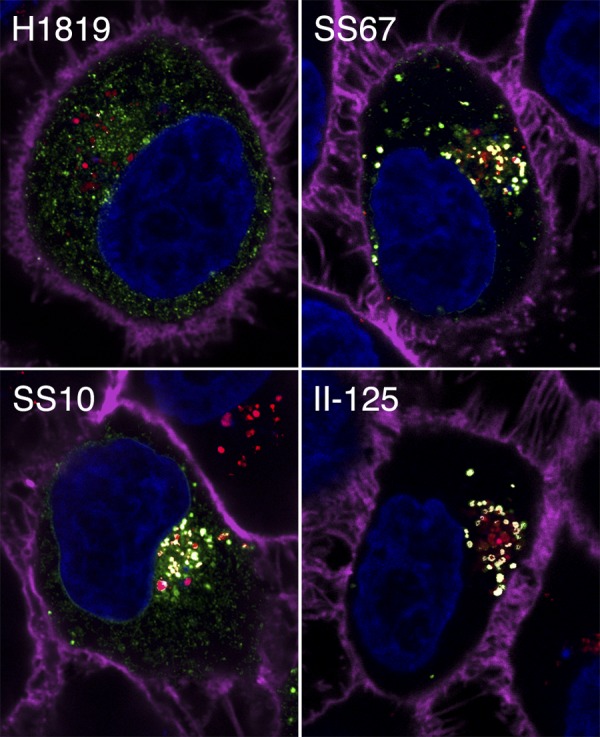
Confocal slices of gB-expressing cells labeled with an anti-gB MAb (plus Alexa Fluor 488; green) and LtrRed (red). Cell nuclei are blue, and the cell periphery is magenta. Each panel presents a merged image, with colocalization of the MAbs and LtrRed indicated by white.
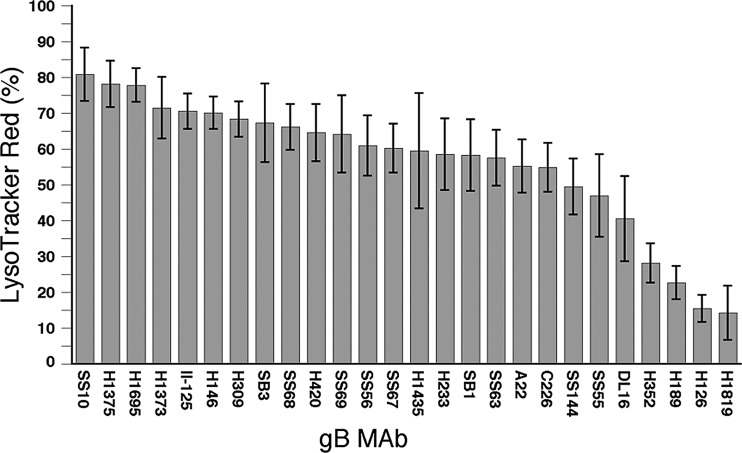
Because colocalization of MAb and LtrRed staining could not be quantitated by the method used in the dual-antibody experiments (see Materials and Methods), an alternative method was used. Three-dimensional projections were made from z-stacks, and the percentage of LtrRed-positive structures that stained with each MAb was determined. The results are shown in Table 2 and in graphed form in Fig. 6. If gB conformation was not changed by exposure to low pH, then all 26 MAbs should have behaved similarly. This was not the case, with values ranging from a high of 80.9% for MAb SS10 to a low of 14.3% for MAb H1819. In interpreting these results, two things should be borne in mind. The first is that a high value indicates a tolerance for low pH and not necessarily a preference for low pH over neutral pH. The second is that all but one MAb (discussed below) stained a much larger number of neutral pH structures, ruling out the possibility that the poor staining of acidic structures by some MAbs was due to a low affinity for a single conformation existing at all pH values. Three MAbs stained about 80% of LtrRed-positive structures, which likely indicates that the epitopes for these MAbs are unaffected by low pH and that the remaining 20% of acidic structures did not contain gB or contained gB that had been degraded. For 16 MAbs, staining of acidic structures ranged from 71.5% down to 54.9%, so a pH-induced conformational change either slightly alters their epitopes or makes them somewhat less accessible. The remaining seven MAbs recognized gB in less than 50% of acidic structures, with four of them giving values below 30%. Their epitopes are more severely affected by low pH, either directly by localized effects on residues within or near the epitope itself or indirectly by a larger conformational change.
Fig 6.
Percentage of LtrRed-positive (low-pH) structures that are labeled with a panel of anti-gB MAbs. The error bars indicate 1 standard deviation.
As alluded to above, MAb II-125 exhibited a unique pattern of staining, in that it appeared to label a conformation of gB found only in acidic structures (as seen in Fig. 5). Quantitation from z-stacks showed that 97% of II-125+ structures were also LtrRed+. As a control, the same quantitation was done for SS10 and H420, and only 10% and 28%, respectively, of labeled structures were also LtrRed+. This indicates that formation or exposure of the II-125 epitope on gB in transfected cells requires a low-pH environment.
gB conformations in infected cells.
To determine if the results obtained with transfected cells would also be true for infected cells, HEp-2 cells were infected with HSV-2 at a low MOI for 12 h, which maximized the number of individually infected cells that had not yet rounded up or progressed to plaque formation. In the first experiment, cells were dually labeled with MAbs H1819 and H420 and examined by confocal microscopy. As shown in panel A of Fig. 7 and the accompanying fluorogram of regions of interest, this produced some structures labeled only with H420 and some labeled only with H1819, as well as some labeled with both. This demonstrates the existence of at least two conformations (H1819−/H420+ and H1819+/H420−), as was found with transfected cells. The possible existence of a third conformation was then examined by triple-labeling with MAbs H1819, H420, and SB1. As shown in Fig. 7 panels B1 and B2 and the accompanying fluorograms, there were some structures containing gB that is H1819−/SB1+/H420−, which cannot be either of the first two conformations or a mixture of them and therefore represents a third conformation. Thus, the three conformations that were identified in transfected cells were also present in infected cells.
Fig 7.
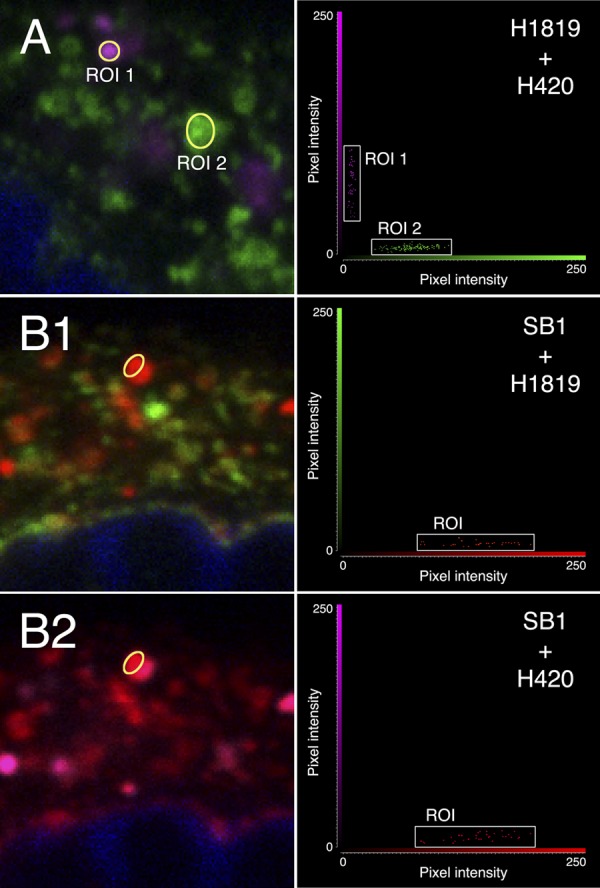
Enlargements of areas within confocal slices of HSV-2-infected cells labeled with combinations of anti-gB MAbs, with accompanying fluorograms for the ROIs. (A) H1819 (plus Alexa Fluor 488; green) and H420 (plus Alexa Fluor 633; magenta). (B1 and B2) (each showing two of the three MAbs from the same triple-labeled slice): SB1 (plus Alexa Fluor 555; red) and either H1819 or H420 with the fluors indicated above.
To investigate the effect of low pH, infected cells were incubated with LtrRed prior to labeling with anti-gB MAbs, and quantitation was performed as described for transfected cells. Four representative MAbs were chosen, and for each one there were fewer MAb-positive low-pH structures than in transfected cells, presumably because there was less gB present. The percentage of positive structures was 30.0% for SS10 (compared to 80.9% in transfected cells), 38.1% for SS67 (compared to 60.3% in transfected cells), 7.8% for H1819 (compared to 14.3% in transfected cells), and 41.0% for II-125 (compared to 70.6% in transfected cells). Although the difference between infected and transfected cells varies somewhat, the important point is that in both cases there was an adverse effect of low pH on binding of MAb H1819 relative to the other MAbs. A further observation in agreement with the results obtained with transfected cells was that binding of II-125 was predominantly to gB in a low pH-environment. A total of 83% of the II-125+ structures also stained with LtrRed, whereas only 12.9% of SS10+ structures did so. Thus, the results are consistent with an effect of low pH on the conformation of gB in infected as well as in transfected cells.
DISCUSSION
Based on the properties of other class III membrane fusion proteins, the purposes of this study were to determine if intracellular HSV gB exists in more than one conformation under conditions where membrane fusion has not yet been triggered and, if it does, to determine if the conformation is changed by exposure to low pH. Studies with two different soluble forms of HSV gB lacking the transmembrane anchor, and performed using different experimental approaches, have produced somewhat conflicting ideas about the extent and importance of conformational changes induced by low pH (11, 17, 18, 73). This suggested the need for an alternative and complementary approach to the issue, by examining full-length gB in transfected cells, in the absence of other viral proteins, and without protein extraction that could conceivably cause conformational changes.
The rationale was that if intracellular gB exists in only one conformation, then dual-labeling of cells with the gB PAb R90 and any MAb that recognizes intracellular gB should produce weighted colocalization coefficients (M1 and M2) of close to 1.0, i.e., the PAb should see all the gB that each MAb does and vice versa. In contrast, if intracellular gB exists in more than one conformation, a variety of colocalization coefficients should be seen, depending on the extent to which the binding characteristics of a given MAb differ between the conformations. The results that were obtained therefore indicate that more than one conformation was present, since only 3 of the 26 MAbs had both coefficients close to 1.0. Competitive binding between MAbs and R90 may occur in some cases and influence the coefficients to some extent, but it does not negate the conclusion about the existence of more than one conformation, because it cannot account for structures labeled only with the PAb and other structures labeled only with a MAb within the same dually labeled cell.
To the extent that comparisons can be made between the colocalization coefficient results and previously published studies on the domains of gB to which MAbs bind (10, 41, 54), there is a good agreement. MAbs binding to the same domain mostly have similar correlation coefficients when compared to the PAb R90, for example SS10, SS67, SS68, and SS69 (domain IV-III), H126, H1375, and H1435 (domain D2a), and H420, H1373, and H1695 (domain Dd5a). Notably, the coefficients for the three Dd5a MAbs, which bind only to gB oligomers (54), are similar to those determined for MAb DL16, which is also oligomer dependent (10). An exception to the agreement between mapping data and correlation coefficients is represented by the pair H233 and H1819, which both map to domain D2b (54); H233 recognizes 78% of the gB recognized by R90 whereas H1819 recognizes only 34%. There are two reasons for believing that it is the behavior of H1819, rather than of H233, that causes this discrepancy. First, the domain D2a MAbs (H126, H1375, and H1435) and domain D2b MAbs (H233 and H1819) bind to the same truncated fragment of gB (amino acids [aa] 1 to 462) and differ only in their abilities to cross-neutralize MAb-resistant virus mutants (62). Second, the M2 correlation coefficient for H233 is similar to those for the domain D2a MAbs, whereas the coefficient for H1819 is quite different, i.e., H233 and the D2a MAbs recognize 64% to 78% of the gB seen by R90, but H1819 recognizes only 34%.
H1819 is one of seven MAbs that are outliers in the panel of 26 in that either they recognize less than 50% of the gB recognized by PAb R90 or R90 recognizes less than 50% of the gB recognized by the MAb or both. The existence of these outliers is interesting for two reasons. First, it indicates that R90, which was made using full-length native HSV-2 gB purified from infected cells, does not recognize all forms of gB found in cells. Second, four of the seven MAbs (C226, SS144, H1695, and H1819) have complement-independent neutralizing activity; two of them, C226 and H1819, have M2 values well below 0.5, indicating that less than half of the intracellular gB recognized by R90 has the conformation that these MAbs bind to in order to inhibit gB activity during virus entry.
Pairs of MAbs of different isotypes were used for dual-labeling of transfected cells, with two objectives. The more straightforward was to confirm that more than one gB conformation can be detected, and this was shown clearly by the absence of complete colocalization. The second objective was to identify MAbs that show mutually exclusive binding to gB. It was impractical to test all possible combinations, so H1819 was chosen as the IgG2a MAb and tested against various IgG1 and IgG2b MAbs in the panel. H1819 was chosen because it was an outlier when compared to R90 and also because it was different from most other MAbs in that it bound poorly to gB in a low-pH environment. Pairing of H1819 with the IgG1 MAb H420 identified some intracellular structures labeled with both but other structures labeled only with H420 (conformation A; H1819−/H420+) or only with H1819 (conformation B; H1819+/H420−). H1819 and H420 were not included in a previous study of competitive MAb binding to soluble truncated gB (10), but their minimum requirements for binding are quite different, consisting of amino acids 1 to 462 for H1819 and the oligomeric form of amino acids 1 to 695 for H420 (54). Pairing of H1819 with the IgG2b MAb SB1 identified some structures labeled strongly with both but other structures labeled only with SB1 (H1819−/SB1+) or strongly with H1819 and only very weakly with SB1 (H1819+/SB1low or a combination of H1819+/SB1− with a much smaller amount of H1819−/SB1+). If the H1819+/H420− and H1819+/SB1− or H1819+/SB1low conformations were identical and the H1819−/H420+ and H1819−/SB1+ conformations were identical, then two conformations would be sufficient to explain the results. If not, then at least one additional conformation must be present.
Combining the three MAbs in a triple-labeling experiment identified a conformation that is H1819−/SB1+/H420−, which is designated conformation C because it is different from conformations A (H1819−/H420+) and B (H1819+/H420−) and cannot be a combination of them. In addition, intracellular structures that were H1819−/SB1+/H420+ and structures that were H1819+/SB1+/H420+ were also identified. Can they be accounted for without invoking a fourth conformation, and do they shed light on the SB1 status of conformations A and B? The simplest explanation for the H1819−/SB1+/H420+ structures is that conformation A and the H1819−/SB1+ conformation in the dual-labeling experiments are identical (i.e., that conformation A is H1819−/SB1+/H420+). This is likely to be correct, because the alternative explanation requires there to be a mixture of conformation C and an H1819−/SB1−/H420+ conformation, which was not detected. The favored explanation for the H1819+/SB1+/H420+ structures, because it is the only one that avoids the need for a fourth conformation, is that they contain a mixture of conformations A and B, where B is now defined as H1819+/SB1low/H420−. One alternative is that they contain a fourth conformation, i.e., H1819+/SB1+/H420+. The other alternative is that they contain A and B, where B is now defined as H1819+/SB1+/H420−. However, this would require the existence of an H1819+/SB1−/H420− conformation (combined with either B or C) to explain the H1819+/SB1low structures observed in the double-labeling experiment and the H1819+/SB1low/H420− structures observed in the triple-labeling experiment. The relevance of the results described above to infected cells was examined by double- and triple-labeling experiments with cells infected with HSV-2 for 12 h. As with the transfected cells, H1819−/H420+, H1819+/H420−, and H1819−/SB1+/H420− conformations were detected, suggesting that the presence of other viral proteins may not influence intracellular conformations of gB to a significant extent.
Before discussing how conformations A, B, and C may relate to forms of gB envisaged or characterized in other studies, there are other possibilities to consider. One is that gB monomers may be one of the three forms detected, by analogy with the VSV G protein, for which there is recent evidence that a monomer is an intermediate between the pre- and postfusion trimers (2, 19). However, dissociation of HSV gB trimers has been seen only as a result of exposure to sodium dodecyl sulfate (SDS) during SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (11, 17), so this possibility is considered unlikely. Another possibility is that the absence of one or more epitopes is caused by proteolytic cleavage of some percentage of gB molecules. Cleavage of a small fraction of the gB in Vero cells has been reported, but there was no cleavage in HEp-2 cells (64), the cell type used for this study, so this possibility is also considered unlikely. A third explanation is that epitopes are gained or lost by interactions of gB with other molecules rather than by changes in gB itself. In this regard, several cell proteins have been reported as receptors for HSV gB, and interactions could conceivably occur if they were to colocalize with gB at some point along the exocytic or endocytic pathways. The first proposed receptor was paired immunoglobulin-like type 2 receptor alpha (69), but this is an unlikely candidate, because it makes cells susceptible to infection by HSV-1 but not by HSV-2 (4). The second proposed receptor was myelin-associated glycoprotein, which was identified in a search for varicella-zoster virus gB receptors and then found to also bind HSV-1 gB (75). However, it was not tested with HSV-2 gB; moreover, it was found to be expressed in glial cells but not in any human cell lines and accordingly can be ruled out with respect to affecting the binding of anti-gB antibodies. The third proposed receptor was nonmuscle myosin heavy chain IIA (3). This protein is expressed in a variety of cell types but was not tested for binding to HSV-2 gB; thus, whether there is any involvement with changes in antibody binding is conjecture. It remains possible that interactions yet to be discovered account for one or more of the observed patterns of MAb binding to gB, but at this point conformational changes within the protein itself appear to be the most credible explanation.
Whether these are large conformational changes such as occur between the pre- and postfusion forms of the VSV G protein, or whether the changes are limited to a few epitopes, cannot be inferred at present, partly because there is no experimental evidence that gB actually needs to undergo such a large change. Furthermore, although differences between two soluble forms of gB (s-gB and gB730) have led to the suggestion that they represent pre- and postfusion forms, respectively (18), a correspondence with the conformations identified herein is not possible, as H1819, SB1, and H420 were not used for characterization of the soluble proteins (10, 17). Identifying an alternative combination of MAbs that would not only distinguish three intracellular conformations but also enable a comparison with s-gB or gB730 was not possible, as no known IgG2b MAbs were used in one previous study (10), and no known IgG2a MAbs were used in the other (17).
Considering the effects of low pH on at least one soluble form of gB and on virion gB in one study (17, 18), and the requirement for exposure to low pH in endosomes for virus entry into some cell types (55–57), low pH was an obvious candidate for changing the conformation of intracellular gB. It would pass through a low-pH environment in the Golgi apparatus during transport to the cell surface and in endosomes when subsequently endocytosed. Examination of MAb binding to gB in organelles with a low pH in transfected cells revealed a wide range of results, with extremes of about 80% acidic organelles positive for SS10, H1375, and H1695 (i.e., binding unaffected by low pH) and only about 15% positive for H126 and H1819. This does not identify the precise pH values at which binding diminishes, as the pH of individual organelles cannot be determined when examining fixed cells, but it does indicate the relative sensitivities of the panel of MAbs. Of the four MAbs with values of less than 30%, only H126 has been tested for reactivity with soluble forms of gB. With gB730, the form used for crystallization, the loss of binding at pH 5.0 in an enzyme-linked immunosorbent assay (ELISA) was only 25%, but H126 was the most severely affected of the 18 MAbs tested (11). More impressively, when s-gB or virions were treated with low pH, H126 binding in a dot blot assay was greatly diminished and SS10 was unaffected (17, 18). Furthermore, MAbs DL16, SS55, and SS144 were also adversely affected by low pH in the latter study, but to a lesser extent than H126, and in the present study they clustered together in the range of 41 to 50% (i.e., were moderately affected). Thus, the results not only build on those already published by demonstrating an effect of low pH on the conformation of intracellular gB but also suggest that the intracellular conformation favored at low pH is the same as that produced by exposure of virion gB and of s-gB to low pH (17). This raises the issue of whether conformation A, B, or C is the dominant form in intracellular low-pH structures, and this is to be the subject of future studies. Examination of a subset of the MAbs for binding to low-pH structures in infected cells showed that the percentage of structures labeled was lower for all of the antibodies, for which the straightforward explanation is that there is less gB present in an infected cell than in a transfected cell. MAbs H1819 and II-125 were again found to have the same unusual behaviors as seen with transfected cells. First, H1819 labeled gB in only a small percentage of low-pH structures while binding well to gB in neutral pH structures. Second, the gB recognized by II-125 was predominantly in low-pH structures. Together, these observations indicate that low pH alters the gB conformation not only in transfected cells but also in infected cells.
Interestingly, the effect of low pH on the ability of MAbs to bind gB does not correlate with their complement-independent neutralizing activity. Some MAbs that are virtually unaffected by low pH (for example, SS10 and H1375) have neutralizing activity, as do some that are dramatically affected (H126 and H1819). This is consistent with the previously published idea that different MAbs neutralize by blocking different functions of gB (10) and may indicate that some neutralize by binding to a prefusion conformation whereas others neutralize by binding to an intermediate conformation triggered by low pH or by another factor such as a receptor interaction. A precedent for triggering of gB conformation changes in a neutral pH environment exists for HCMV gB, which undergoes two changes upon binding of virions to cells, the first one detectable by sensitivity to bis-aryl thiourea drugs and the second by susceptibility to digestion by proteinase K (61). An additional point of note is that if any of the intracellular forms of HSV gB (whether at low or neutral pH) have the fusion-active conformation (likely as an intermediate between pre- and postfusion forms), then some other factor required for fusion is presumably absent from that location in the cell in order for intracellular membrane fusion to be avoided. Candidates to consider would include gH/gL, a gB receptor, or the gK/UL20 complex, which has been shown to interact with gB and which is involved in cell-to-cell spread of the virus (14, 49).
Finally, one or more conformational changes have recently been shown to occur in the gB protein of murine herpesvirus 4 (MuHV-4) during entry into cells by endocytosis (32). The epitope for one MAb is gained (requiring low pH), and the epitope for another MAb is lost (requiring low pH and an additional unidentified trigger). As the authors of that study explain, if these changes occur sequentially, then double-positive structures could represent an intermediate form of gB; alternatively, if they occur simultaneously, then double-positive structures might be the result of a reversible equilibrium between two forms.
ACKNOWLEDGMENTS
This work was supported in part by COBRE grant GM103433 from the National Institute of General Medical Sciences of the NIH (principal investigator [PI], Dennis O'Callaghan) and by the LSUHSC Feist-Weiller Cancer Center.
I thank Gary Cohen and Roz Eisenberg for kindly providing antibodies R90, A22, C226, and DL16 and the SS series of MAbs, Pat Spear for kindly providing MAb II-125, and Lindsey Hutt-Fletcher for numerous invaluable discussions and for critical reading of the manuscript.
Footnotes
Published ahead of print 18 April 2012
REFERENCES
- 1. Ahn A, Gibbons DL, Kielian M. 2002. The fusion peptide of Semliki Forest virus associates with sterol-rich membrane domains. J. Virol. 76:3267–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Albertini AA, et al. 2012. Characterization of monomeric intermediates during VSV glycoprotein structural transition. PLoS Pathog. 8:e1002556 doi:10.1371/journal.ppat.1002556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arii J, et al. 2010. Non-muscle myosin IIA is a functional entry receptor for herpes simplex virus-1. Nature 467:859–862 [DOI] [PubMed] [Google Scholar]
- 4. Arii J, et al. 2009. Entry of herpes simplex virus 1 and other alphaherpesviruses via the paired immunoglobulin-like type 2 receptor α. J. Virol. 83:4520–4527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atanasiu D, et al. 2007. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc. Natl. Acad. Sci. U. S. A. 104:18718–18723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Avitabile E, Forghieri C, Campadelli-Fiume G. 2007. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J. Virol. 81:11532–11537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Avitabile E, Forghieri C, Campadelli-Fiume G. 2009. Cross talk among the glycoproteins involved in herpes simplex virus entry and fusion: the interaction between gB and gH/gL does not necessarily require gD. J. Virol. 83:10752–10760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Backovic M, et al. 2010. Structure of a core fragment of glycoprotein H from pseudorabies virus in complex with antibody. Proc. Natl. Acad. Sci. U. S. A. 107:22635–22640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Backovic M, Longnecker R, Jardetzky TS. 2009. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 106:2880–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bender FC, et al. 2007. Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J. Virol. 81:3827–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cairns TM, et al. 2011. Capturing the herpes simplex virus core fusion complex (gB-gH/gL) in an acidic environment. J. Virol. 85:6175–6184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Campadelli-Fiume G, et al. 2007. The multipartite system that mediates entry of herpes simplex virus into the cell. Rev. Med. Virol. 17:313–326 [DOI] [PubMed] [Google Scholar]
- 13. Chapsal JM, Pereira L. 1988. Characterization of epitopes on native and denatured forms of herpes simplex virus glycoprotein B. Virology 164:427–434 [DOI] [PubMed] [Google Scholar]
- 14. Chouljenko VN, Iyer AV, Chowdhury S, Kim J, Kousoulas KG. 2010. The herpes simplex virus type 1 UL20 protein and the amino terminus of glycoprotein K (gK) physically interact with gB. J. Virol. 84:8596–8606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chowdary TK, et al. 2010. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat. Struct. Mol. Biol. 17:882–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. 1998. The ectodomain of a novel member of the immunoglobulin superfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 72:9992–10002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dollery SJ, Delboy MG, Nicola AV. 2010. Low pH-induced conformational change in herpes simplex virus glycoprotein B. J. Virol. 84:3759–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dollery SJ, Wright CC, Johnson DC, Nicola AV. 2011. Low-pH-dependent changes in the conformation and oligomeric state of the prefusion form of herpes simplex virus glycoprotein B are separable from fusion activity. J. Virol. 85:9964–9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Doms RW, Keller DS, Helenius A, Balch WE. 1987. Role for adenosine triphosphate in regulating the assembly and transport of vesicular stomatitis virus G protein trimers. J. Cell Biol. 105:1957–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duus KM, Hatfield C, Grose C. 1995. Cell surface expression and fusion by the varicella-zoster virus gH:gL glycoprotein complex: analysis by laser scanning confocal microscopy. Virology 210:429–440 [DOI] [PubMed] [Google Scholar]
- 21. Farnsworth A, et al. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. U. S. A. 104:10187–10192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fusco D, Forghieri C, Campadelli-Fiume G. 2005. The pro-fusion domain of herpes simplex virus glycoprotein D (gD) interacts with the gD N terminus and is displaced by soluble forms of viral receptors. Proc. Natl. Acad. Sci. U. S. A. 102:9323–9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Galdiero S, et al. 2005. Fusogenic domains in herpes simplex virus type 1 glycoprotein H. J. Biol. Chem. 280:28632–28643 [DOI] [PubMed] [Google Scholar]
- 24. Galdiero S, et al. 2006. Analysis of synthetic peptides from heptad-repeat domains of herpes simplex virus type 1 glycoproteins H and B. J. Gen. Virol. 87:1085–1097 [DOI] [PubMed] [Google Scholar]
- 25. Gaudin Y, Ruigrok RWH, Knossow M, Flamand A. 1993. Low-pH conformational changes of rabies virus glycoprotein and their role in membrane fusion. J. Virol. 67:1365–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gaudin Y, Tuffereau C, Durrer P, Flamand A, Ruigrok RWH. 1995. Biological function of the low-pH, fusion-inactive conformation of rabies virus glycoprotein (G): G is transported in a fusion-inactive state-like conformation. J. Virol. 69:5528–5534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gaudin Y, Tuffereau C, Segretain D, Knossow M, Flamand A. 1991. Reversible conformational changes and fusion activity of rabies virus glycoprotein. J. Virol. 65:4853–4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618–1620 [DOI] [PubMed] [Google Scholar]
- 29. Gianni T, Campadelli-Fiume G, Menotti L. 2004. Entry of herpes simplex virus mediated by chimeric forms of nectin1 retargeted to endosomes or to lipid rafts occurs through acidic endosomes. J. Virol. 78:12268–12276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gianni T, Martelli PL, Casadio R, Campadelli-Fiume G. 2005. The ectodomain of herpes simplex virus glycoprotein H contains a membrane alpha-helix with attributes of an internal fusion peptide, positionally conserved in the herpesviridae family. J. Virol. 79:2931–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gianni T, Piccoli A, Bertucci C, Campadelli-Fiume G. 2006. Heptad repeat 2 in herpes simplex virus 1 gH interacts with heptad repeat 1 and is critical for virus entry and fusion. J. Virol. 80:2216–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Glauser DL, Kratz A-S, Stevenson PG. 2012. Herpesvirus glycoproteins undergo multiple antigenic changes before membrane fusion. PLoS One 2:e30152 doi:10.1371/journal.pone.0030152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hannah BP, et al. 2009. Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops. J. Virol. 83:6825–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hannah BP, Heldwein EE, Bender FC, Cohen GH, Eisenberg RJ. 2007. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J. Virol. 81:4858–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harley CA, Dasgupta A, Wilson DW. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J. Virol. 75:1236–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heldwein EE, et al. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217–220 [DOI] [PubMed] [Google Scholar]
- 37. Hibbs AR, MacDonald G, Garsha K. 2006. Practical confocal microscopy, p 650–671 In Pawley JB. (ed), Handbook of biological confocal microscopy, 3rd ed Springer, New York, NY [Google Scholar]
- 38. Kadlec J, Loureiro S, Abrescia NGA, Stuart DI, Jones IM. 2008. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 10:1024–1030 [DOI] [PubMed] [Google Scholar]
- 39. Kimura T, Ohyama A. 1988. Association between the pH-dependent conformational change of West Nile flavivirus E protein and virus-mediated membrane fusion. J. Gen. Virol. 69:1247–1254 [DOI] [PubMed] [Google Scholar]
- 40. Kinzler ER, Compton T. 2005. Characterization of human cytomegalovirus glycoprotein-induced cell-cell fusion. J. Virol. 79:7827–7837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kousoulas KG, Huo B, Pereira L. 1988. Antibody-resistant mutations in cross-reactive and type-specific epitopes of herpes simplex virus 1 glycoprotein B map in separate domains. Virology 166:423–431 [DOI] [PubMed] [Google Scholar]
- 42. Kousoulas KG, Pellett PE, Pereira L, Roizman B. 1984. Mutations affecting conformation or sequence of neutralizing epitopes identified by reactivity of viable plaques segregate from syn and ts domains of HSV-1(F) gB gene. Virology 135:379–394 [DOI] [PubMed] [Google Scholar]
- 43. Krummenacher C, et al. 2004. Comparative usage of herpesvirus entry mediator A and nectin-1 by laboratory strains and clinical isolates of herpes simplex virus. Virology 322:286–299 [DOI] [PubMed] [Google Scholar]
- 44. Krummenacher C, et al. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 24:4144–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lopper M, Compton T. 2004. Coiled-coil domains in glycoproteins B and H are involved in human cytomegalovirus membrane fusion. J. Virol. 78:8333–8341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maresova L, Pasieka TJ, Grose C. 2001. Varicella-zoster virus gB and gE coexpression, but not gB or gE alone, leads to abundant fusion and syncytium formation equivalent to those from gH and gL coexpression. J. Virol. 75:9483–9492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Matsuura H, Kirschner AN, Longnecker R, Jardetzky TS. 2010. Crystal structure of the Epstein-Barr virus (EBV) glycoprotein H/glycoprotein L (gH/gL) complex. Proc. Natl. Acad. Sci. U. S. A. 107:22641–22646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McShane MP, Longnecker R. 2004. Cell-surface expression of a mutated Epstein-Barr virus glycoprotein B allows fusion independent of other viral proteins. Proc. Natl. Acad. Sci. U. S. A. 101:17474–17479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Melancon JM, Luna RE, Foster TP, Kousoulas KG. 2005. Herpes simplex virus type 1 gK is required for gB-mediated virus-induced cell fusion, while neither gB and gK nor gB and UL20p function redundantly in virion de-envelopment. J. Virol. 79:299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Milne RSB, Nicola AV, Whitbeck JC, Eisenberg RJ, Cohen GH. 2005. Glycoprotein D receptor-dependent, low-pH-independent endocytic entry of herpes simplex virus type 1. J. Virol. 79:6655–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Montgomery RI, Warner MS, Lum BJ, Spear PG. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427–436 [DOI] [PubMed] [Google Scholar]
- 52. Muggeridge MI. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J. Gen. Virol. 81:2017–2027 [DOI] [PubMed] [Google Scholar]
- 53. Muggeridge MI, Roberts SR, Isola VJ, Cohen GH, Eisenberg RJ. 1990. Herpes simplex virus, p 459–481 In Van Regenmortel MHV, Neurath AR. (ed), Immunochemistry of viruses, II. The basis for serodiagnosis and vaccines. Elsevier Biochemical Press, Amsterdam, The Netherlands [Google Scholar]
- 54. Navarro D, Paz P, Pereira L. 1992. Domains of herpes simplex virus 1 glycoprotein B that function in virus penetration, cell-to-cell spread, and cell fusion. Virology 186:99–112 [DOI] [PubMed] [Google Scholar]
- 55. Nicola AV, Hou J, Major EO, Straus SE. 2005. Herpes simplex virus type 1 enters human epidermal keratinocytes, but not neurons, via a pH-dependent endocytic pathway. J. Virol. 79:7609–7616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nicola AV, McEvoy AM, Straus SE. 2003. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J. Virol. 77:5324–5332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nicola AV, Straus SE. 2004. Cellular and viral requirements for rapid endocytic entry of herpes simplex virus. J. Virol. 78:7508–7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Norton DD, Dwyer DS, Muggeridge MI. 1998. Use of a neural network secondary structure prediction to define targets for mutagenesis of herpes simplex virus glycoprotein B. Virus Res. 55:37–48 [DOI] [PubMed] [Google Scholar]
- 59. Omerović J, Lev L, Longnecker R. 2005. The amino terminus of Epstein-Barr virus glycoprotein gH is important for fusion with epithelial and B cells. J. Virol. 79:12408–12415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Para MF, Parish ML, Noble AG, Spear PG. 1985. Potent neutralizing activity associated with anti-glycoprotein D specificity among monoclonal antibodies selected for binding to herpes simplex virions. J. Virol. 55:483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Patrone M, Secchi M, Bonaparte E, Milanese G, Gallina A. 2007. Cytomegalovirus UL131-128 products promote gB conformational transition and gB-gH interaction during entry into endothelial cells. J. Virol. 81:11479–11488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pereira L, Ali M, Kousoulas K, Huo B, Banks T. 1989. Domain structure of herpes simplex virus 1 glycoprotein B: neutralizing epitopes map in regions of continuous and discontinuous residues. Virology 172:11–24 [DOI] [PubMed] [Google Scholar]
- 63. Pereira L, Dondero D, Norrild B, Roizman B. 1981. Differential immunologic reactivity and processing of glycoproteins gA and gB of herpes simplex virus types 1 and 2 made in Vero and HEp-2 cells. Proc. Natl. Acad. Sci. U. S. A. 78:5202–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pereira L, Dondero D, Roizman B. 1982. Herpes simplex virus glycoprotein gA/B: evidence that the infected Vero cell products comap and arise by proteolysis. J. Virol. 44:88–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pereira L, Klassen T, Baringer JR. 1980. Type-common and type-specific monoclonal antibody to herpes simplex virus type 1. Infect. Immun. 29:724–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Roche S, Bressanelli S, Rey FA, Gaudin Y. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313:187–191 [DOI] [PubMed] [Google Scholar]
- 67. Roche S, Gaudin Y. 2002. Characterization of the equilibrium between the native and fusion-inactive conformation of rabies virus glycoprotein indicates that the fusion complex is made of several trimers. Virology 297:128–135 [DOI] [PubMed] [Google Scholar]
- 68. Roche S, Rey FA, Gaudin Y, Bressanelli S. 2007. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 315:843–848 [DOI] [PubMed] [Google Scholar]
- 69. Satoh T, et al. 2008. PILRα is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell 132:935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shukla D, et al. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13–22 [DOI] [PubMed] [Google Scholar]
- 71. Skehel JJ, et al. 1982. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 79:968–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Spear PG. 2004. Herpes simplex virus: receptors and ligands for cell entry. Cell. Microbiol. 6:401–410 [DOI] [PubMed] [Google Scholar]
- 73. Stampfer SD, Lou H, Cohen GH, Eisenberg RJ, Heldwein EE. 2010. Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1. J. Virol. 84:12924–12933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Subramanian RP, Geraghty RJ. 2007. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc. Natl. Acad. Sci. U. S. A. 104:2903–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Suenaga T, et al. 2010. Myelin-associated glycoprotein mediates membrane fusion and entry of neurotropic herpesviruses. Proc. Natl. Acad. Sci. U. S. A. 107:866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wittels M, Spear PG. 1990. Penetration of cells by herpes simplex virus does not require a low pH-dependent endocytic pathway. Virus Res. 18:271–290 [DOI] [PubMed] [Google Scholar]
- 77. Yewdell JW, Gerhard W, Bachi T. 1983. Monoclonal anti-hemagglutinin antibodies detect irreversible antigenic alterations that coincide with the acid activation of influenza virus A/PR/8/34-mediated hemolysis. J. Virol. 48:239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhou J, Blissard GW. 2006. Mapping the conformational epitope of a neutralizing antibody (AcV1) directed against the AcMNPV GP64 protein. Virology 352:427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]