Abstract
Because of their rapid evolution, genetic diversity, broad host range, ongoing circulation in birds, and potential human-to-human transmission, H5N1 influenza viruses remain a major global health concern. Their high degree of genetic diversity also poses enormous burdens and uncertainties in developing effective vaccines. To overcome this, we took a new approach, i.e., the development of immunogens based on a comprehensive serologic study. We constructed DNA plasmids encoding codon-optimized hemagglutinin (HA) from 17 representative strains covering all reported clades and subclades of highly pathogenic avian influenza H5N1 viruses. Using DNA plasmids, we generated the corresponding H5N1 pseudotypes and immune sera. We performed an across-the-board pseudotype-based neutralization assay and determined antigenic clusters by cartography. We then designed a triclade DNA vaccine and evaluated its immunogenicity and protection in mice. We report here that (sub)clades 0, 1, 3, 4, 5, 6, 7.1, and 9 were grouped into antigenic cluster 1, (sub)clades 2.1.3.2, 2.3.4, 2.4, 2.5, and 8 were grouped into another antigenic cluster, with subclade 2.2.1 loosely connected to it, and each of subclades 2.3.2.1 and 7.2 was by itself. Importantly, the triclade DNA vaccine encoding HAs of (sub)clades 0, 2.3.2.1, and 7.2 elicited broadly neutralizing antibody responses against all H5 clades and subclades and protected mice against high-lethal-dose heterologous H5N1 challenge. Thus, we conclude that broadly neutralizing antibodies against all H5 clades and subclades can indeed be elicited with immunogens on the basis of a comprehensive serologic study. Further evaluation and optimization of such an approach in ferrets and in humans is warranted.
INTRODUCTION
Influenza vaccines are a cost-effective way to prevent and control influenza virus infection. Influenza vaccines elicit potent neutralizing antibody responses to the vaccine strains and closely related isolates but rarely extend to more divergent strains within a subtype or to other subtypes. Because of this, current influenza vaccines are prepared annually on the basis of the World Health Organization (WHO) forecasts on the most probable influenza virus strains thought to be circulating in the next seasonal outbreak (1). However, selecting appropriate vaccine strains presents many challenges and sometimes results in suboptimal protection (6). Moreover, predicting the next pandemic virus, including when and where it will arise, is currently impossible. Thus, developing “universal” vaccines that elicit antibody response capable of neutralizing diverse influenza A virus strains would eliminate much of the uncertainty associated with strain selection and impede emerging pandemic viruses.
Since the emergence of highly pathogenic avian influenza (HPAI) H5N1 viruses in 1996, outbreaks have continued in a variety of domestic and wild birds, as well as sporadic human transmission in southeast Asia, Eurasia, and Africa (17). As of 22 September 2011, the World Organization for Animal Health highlighted thousands of HPAI H5N1 virus infection outbreaks in poultry and wild birds in 63 countries (17, 41). As of 12 March 2012, 596 human H5N1 virus infections have been confirmed, resulting in 350 deaths (40).
On the basis of hemagglutinin (HA) genealogy, H5N1 viruses have evolved into 10 clades in various host species (29, 38, 39). Among them, clade 2 is divided into the five subclades 2.1, 2.2, 2.3, 2.4, and 2.5, and clade 7 is divided into the two subclades 7.1 and 7.2 (22, 23, 38, 39). Subclade 2.1 is further divided into subclades 2.1.1, 2.1.2, 2.1.3, 2.1.3.1, 2.1.3.2, and 2.1.3.3. Subclade 2.2 is further divided into subclades 2.2.1 and 2.2.1.1. Finally, subclade 2.3 is further divided into subclades 2.3.1, 2.3.2.1, 2.3.3, 2.3.4, 2.3.4.1, 2.3.4.2, and 2.3.4.3 (38, 39). Thus far, the circulating HPAI H5N1 viruses of human isolates fall into clades 0, 1, 2, and 7 (40, 42), and the other clades that are circulating in avian species may be potentially transmitted to humans either directly from avian species or indirectly through so-called “mixing-vessel” species, such as pigs. Therefore, it is important that a vaccine developed against H5N1 virus not only protect from H5 clades and subclades that have already infected humans but also from potential new emerging H5 clades and subclades to humans.
To deal with this genetic diversification, the WHO is creating additional vaccine seed strains when new viruses emerge. As a result, the current tally of such seed strains in stock is 20, with 3 more in development. These strains cover (sub)clades 1, 2.1, 2.2, 2.3.2, 2.3.4, 4, and 7.2 (38, 39, 47). Not only does this create tremendous economic burdens to produce vaccines from these seed strains, but it is also becoming less clear which seed strains are the most relevant to a given geographic region. To overcome the burden and uncertainties, several new approaches have been evaluated (1, 3, 7, 10, 13, 14, 18, 20, 26, 27, 32). For example, Chen et al. (7) demonstrated that a consensus H5 HA-based DNA vaccine protects mice against divergent H5N1 from (sub)clades 1, 2.1, 2.2, 2.3.2, and 2.3.4. Ducatez et al. (13) demonstrated that reconstructed ancestral H5N1 influenza vaccine based on a resolved phylogenetic topology elicited cross-clade protective immunity against (sub)clades 1, 2.1, 2.2, 2.3.4, and 4 in ferrets. Forrest et al. (18) compared single- versus multiple-clade H5N1 vaccines to induce cross-protection in ferrets and found that the multiple-clade vaccine was broadly immunogenic against clade 1 and 2 viruses. Rao et al. (27) demonstrated that a DNA vaccine encoding multiple H5 HA elicited antibody responses in mice and chickens that could neutralize multiple strains of HPAI H5N1 when given in combination. Prabakaran et al. (26) showed that the expression of HA proteins of selected vaccine strains on the baculovirus surface could elicit neutralizing antibody responses against viruses from clades 1, 2.1, 2.2, 4, 7, and 8 of H5N1 viruses and conferred protection against challenge with (sub)clades 1, 2.1, and 7 of H5N1 strains. Thus, although these approaches have been shown to elicit more cross-reactive antibody responses than an antigen derived from a single strain, most of these studies were based on phylogenetic data or limited antigenic data and only evaluated a limited number of H5 clades and subclades.
We hypothesized that by systematically analyzing the cross-reactivity of neutralizing antibody responses among all H5 clades and subclades, one may determine antigenic clusters among H5 clades and subclades. One can then design immunogens based on antigenic clusters that may elicit broadly neutralizing antibody responses and immune protection against strains of all H5 clades and subclades.
To test this hypothesis, we constructed DNA plasmids encoding codon-optimized HA from 17 representative strains covering all reported H5 clades and subclades. Using the DNA plasmids, we generated corresponding H5N1 pseudotypes and elicited corresponding immune sera. We then performed an across-the-board pseudotype-based neutralization assay and determined antigenic clusters by cartography. Based on the cross-neutralization data and antigenic clusters determined by cartography, a triclade DNA vaccine was designed, and its immunogenicity and immune protection were evaluated in mice.
MATERIALS AND METHODS
Cell lines.
The packaging cell line 293T was maintained in complete Dulbecco modified Eagle medium (complete DMEM; high-glucose DMEM supplemented with 10% fetal bovine serum [FBS], 2 mM l-glutamine, 1 mM sodium pyruvate, penicillin [100 U/ml], and streptomycin [100 μg/ml]; Invitrogen Life Technologies). The Madin-Darby canine kidney (MDCK) cell line was maintained in complete DMEM.
Animals.
All animal experiments were carried out at biosafety level 3 (BSL3) containment facilities complying with the Ethics Committee regulations of the Institut Pasteur, Paris, France, in accordance with EC directive 86/609/CEE and were approved by the Animal Ethics Committee of the Institut Pasteur in Cambodia (permit VD100820). Female BALB/c mice (Mus musculus) at the ages of 6 to 8 weeks were purchased from Charles River Laboratories (L'Arbresle, France) and housed in microisolator cages ventilated under negative pressure with HEPA-filtered air. Virus challenge studies were conducted in a BSL3 facility at the Institut Pasteur in Cambodia. Before each inoculation or euthanasia procedure, the mice were anesthetized by intraperitoneal (i.p.) injection of pentobarbital sodium (65 mg/kg; Sigma).
H5N1 viruses.
HPAI H5N1 viruses A/Shenzhen/406H/06 and A/Cambodia/P0322095/05 were originally isolated from human patients at the Donghu Hospital in Shenzhen, China, and at the Institut Pasteur in Cambodia, respectively (4, 47). Viruses were propagated in MDCK cells, and virus-containing supernatants were pooled, clarified by centrifugation, and stored in aliquots at −80°C. The 50% tissue culture infection doses (TCID50) and the 50% mouse lethal doses (MLD50) of the viruses were determined in MDCK cells and in BALB/c mice, respectively, and were calculated by the method of Reed and Muench (28) as described previously (12).
Generation of a panel of DNA plasmids encoding codon-optimized H5 HA genes.
To generate a panel of DNA plasmids encoding codon-optimized H5 HA genes, a total of 17 codon-optimized H5 HA using human-preferred codons (45) were synthesized by recursive PCR and overlap PCR, inserted into a mammalian expression vector pCMV/R as described previously (34). The resulting DNA plasmids were designated as pHK156 (clade 0), pVN and pCA (clade 1), pID (subclade 2.1.3.2), pTK (subclade 2.2.1), pHK5052 (subclade 2.3.2.1), pSZ (subclade 2.3.4), pGX12 (subclade 2.4), pKR (subclade 2.5), pHKSF (clade 3), pGY (clade 4), pGX1378 (clade 5), pHB (clade 6), pBJ (subclade 7.1), pSX (subclade 7.2), pHN (clade 8), and pST (clade 9), respectively (Table 1). The rationale for selecting these strains was as follows. For a given clade or subclade, if the World Health Organization (WHO) recommended candidate vaccine strains, we selected them; for other clades and subclades, we compared amino acid sequences of several strains for a given clade or subclade and then selected relatively conservative strains.
Table 1.
Panel of codon-optimized H5 HAs used for eliciting mouse immune sera and for generating H5N1 pseudotypesa
| Strain | Abbreviation | (Sub)clade | Accession no. |
|---|---|---|---|
| A/Hong Kong/156/97 | pHK156 | 0 | AAC40508 |
| A/Viet Nam/1203/2004 | pVN | 1 | ABP51977 |
| A/Cambodia/p0322095/05 | pCA | 1 | P0322095b |
| A/Indonesia/5/2005 | pID | 2.1.3.2 | ABP51969 |
| A/Turkey/65596/2006 | pTK | 2.2.1 | ABQ58925 |
| A/Common Magpie/Hong Kong/5052/2007 | pHK5052 | 2.3.2.1 | ACJ26242 |
| A/Shenzhen/406H/2006 | pSZ | 2.3.4 | ABO36644 |
| A/Chicken/Guangxi/12/2004 | pGX12 | 2.4 | ABD14809 |
| A/Chicken/Korea/es/2003 | pKR | 2.5 | ABP51986 |
| A/Silky Chicken/Hong Kong/SF189/01 | pHKSF | 3 | AAO52864 |
| A/Goose/Guiyang/337/2006 | pGY | 4 | ABJ96698 |
| A/Duck/Guangxi/1378/2004 | pGX1378 | 5 | ABC66526 |
| A/Duck/Hubei/wg/2002 | pHB | 6 | ABI94747 |
| A/Beijing/01/2003 | pBJ | 7.1 | ABQ58979 |
| A/Chicken/Shanxi/2/2006 | pSX | 7.2 | ABK34764 |
| A/Chicken/Henan/16/2004 | pHN | 8 | AAX53508 |
| A/Goose/Shantou/1621/05 | pST | 9 | ABE68931 |
All H5N1 pseudotypes expressing the same N1 NA were derived from the A/Thailand/1(KAN-1)/04 virus.
The sequence is available from the database of the Los Alamos National Laboratory.
Generation of a panel of corresponding H5N1 pseudotypes.
To generate a panel of H5N1 pseudotypes, 293 T packaging cells were cotransfected with transfer vector pHR′CMV-Luc, packaging vector pCMVRΔ8.2, and pCMVR vector encoding one of the above described codon-optimized H5 HA and N1 neuraminidase (NA) [A/Thailand/1(KAN-1)/04] as described previously (34). Briefly, 4.5 × 106 293T packaging cells were cotransfected with 14 μg of pHR′CMV-Luc, 14 μg of pCMVΔR8.2, 2 μg of CMV/R-HA, and 0.5 μg of CMV/R-NA using a calcium phosphate precipitation method. As a control 293T cells were also cotransfected with 14 μg of pHR′CMV-Luc, 14 μg of pCMVΔR8.2, and 5 μg of DNA plasmid encoding vesicular stomatitis virus protein (VSV-G). After overnight incubation, the cells were washed once with phosphate-buffered saline (PBS) and cultured in 10 ml of complete DMEM supplemented with 100 μM sodium butyrate (Sigma, St. Louis, MO) for 8 h. The cells were then cultured in 10 ml of complete DMEM. The pseudotype-containing supernatants were harvested after 16 to 20 h and stored at −80°C in a freezer in aliquots until used. The relative luciferase activity (RLA) of HA and NA pseudotype stocks were determined in MDCK cells as described previously (34).
To normalize the amount of HA/p24 among all pseudotypes, the p24 level was measured from different viral stocks using an HIV-1 p24 antigen assay kit (Beckman Coulter, Fullerton, CA), and the HA value was measured using an HA assay with chicken red blood cells. The HA/p24 ratio varied by no more than 1- to 2-fold.
Generation of a panel of corresponding immune sera against H5 clades and subclades.
To generate immune sera against a panel of H5 HA, BALB/c mice (six mice per group) were immunized intramuscularly (i.m.) three times with 100 μg of the DNA plasmids listed in Table 1. The immunizations were carried out on days 0, 28, and 56. At 14 days after the immunization, serum samples collected from the same group of mice were combined, heat inactivated at 56°C, and stored in aliquots at 4°C until use for the pseudotype-based neutralization assay (see below). To ensure the levels of the expression among these HAs, DNA plasmids were also transiently transfected into NIH 3T3 cells, a murine cell line. At 24 h posttransfection, the cells were stained with human monoclonal antibody F10, followed by fluorescence-activated cell sorting (FACS) analysis. Similar levels of HA expression were observed (P. Zhou et al., data not shown).
HA and NA pseudotype-based neutralization assay.
An HA and NA pseudotype-based neutralization assay used here was described previously (34). Briefly, MDCK cells (3 × 103 cells per well) were seeded onto 96-well culture plate in complete DMEM overnight. Serially 2-fold-diluted serum samples (starting at 1:40 dilution) were incubated with HA and NA pseudotypes equivalent to 6.25 ng of HIV-1 gag p24/ml corresponding to 200,000 to 2,000,000 RLA at the final volume of 100 μl at 37°C for 1 h as described previously (46). Importantly, the different amounts of HA among the input H5N1 pseudotype panel (Zhou et al., data not shown) were within the acceptable range described by Yang et al. (46). The mixture was added onto MDCK cells. RLA was measured after 72 h by a BrightGlo luciferase assay (Promega) according to the manufacturer's instructions. The percentage of inhibition was calculated as follows: (RLA in pseudotypes and medium control − RLA in pseudotypes and immune serum in a given dilution)/RLA in pseudotypes and medium control. The IC50, IC80, and IC90 values were determined as the dilutions of a given immune serum that resulted in 50, 80, and 90% reduction of RLA, respectively.
Antigenic cartography.
Using the serological data (i.e., the IC50 values) from the neutralization assay, antigenic cartography was constructed by AntigenMap, an integrative matrix completion-multidimensional scaling method as recently described (5). AntigenMap has been successfully used in antigenic cartography construction for influenza A viruses (13, 35). The IC50 values were normalized as described previously (5), and an IC50 value was defined as a low reactor if it was <20. An outlier antigen or antiserum was defined as distant from its counterparts by more than two units, i.e., by >4 log2.
Active and passive immunization and challenge.
For active immunization and challenge experiments, female BALB/c mice were randomly divided into two groups (18 mice per group). Mice in group 1 were injected i.m. three times in both hind legs, with a total 200 μg of empty vector plasmid. Mice in group 2 were injected i.m. three times in both hind legs with a total of 200 μg of a mixture of three DNA plasmids (pHK156, pHK5052, and pSX). The DNA injection was done on days 0, 28, and 56. At 2 weeks after the last injection, 12 mice in each group were divided into two subgroups (6 mice per subgroup). One subgroup was challenged intranasally (i.n.) with 100 MLD50 (5,623 TCID50) of heterologous H5N1 virus (A/Shenzhen/406H/06, subclade 2.3.4), and the second subgroup was challenged i.n. with 100 MLD50 (50 TCID50) of heterologous H5N1 virus (A/Cambodia/P0322095/05, clade 1) in a volume of 50 μl. The mice were monitored and recorded daily for signs of illness, such as lethargy, ruffled fur, and weight loss. When mice lost 30% or more of their original weight, they were euthanized and counted as dead. The remaining six mice per group were sacrificed at 10 days after the final injection for intracellular cytokine staining.
For passive the immunization and challenge experiment, 24 BALB/c mice were immunized i.m. three times in both hind legs with a total of 200 μg of the same mixture of three DNA plasmids as described above. At 2 weeks after the last injection, postimmune sera were collected and combined. For the controls, the sera from 24 naive BALB/c mice were also collected and combined. Twelve naive female BALB/c mice were i.p. injected with 700 μl of immune sera, and another 12 naive female BALB/c mice were i.p. injected with the same amount of sera collected from naive mice. After 24 h, six mice per group were challenged i.n. with 100 MLD50 (50 TCID50) of A/Cambodia/P0322095/05 strain and remaining 6 mice per group were challenged i.n. with 100 MLD50 (5,623 TCID50) of A/Shenzhen/406H/06 strain in a volume of 50 μl. The mice were monitored and recorded daily for signs of illness, such as lethargy, ruffled fur, and weight loss. When the mice lost 30% or more of their original weight, they were euthanized and counted as dead.
All active and passive immunization and challenge experiments were performed in accordance with the Department of Agriculture guidelines for the Care and Use of Laboratory Animals, the Animal Welfare Act, and Department of Agriculture Biosafety Guidelines in the Microbiological and Biomedical Laboratory.
Intracellular cytokine staining.
CD8+ T-cell responses were evaluated by using intracellular cytokine staining for gamma interferon (IFN-γ), interleukin-2 (IL-2), and tumor necrosis factor alpha (TNF-α) as previously described (21). Briefly, spleens were harvested from triclade DNA- and empty vector-vaccinated mice and gently homogenized into a single-cell suspension. After erythrocyte lysis, the splenocytes (106 cells/well) were stimulated with two HA peptides (IYSTVASSL and LYEKVRLQL [11, 19, 25], 2.5 μg/ml for each peptide) and anti-CD28 and anti-CD49d antibodies (BD Pharmingen, 1 μg/ml) in 24-well tissue culture plates (Costar; Corning) at 37°C. Two hours later, brefeldin A (BD Biosciences, 10 μg/ml) was added. After 6 h of incubation, the cells were refrigerated overnight. The following morning, the cells were washed and incubated with Fc blocker (Becton Dickinson) and then surface stained with anti-CD4 and anti-CD8 antibodies (BD Pharmingen) at 4°C for 1 h. The cells were then washed, fixed, and permeabilized for 20 min with Cytofix/Cytoperm solution (BD Cytofix/Cytoperm fixation/permeabilization kit; BD Biosciences). The cells were washed and stained with the indicated fluorescence-labeled anti-IFN-γ, anti-IL-2, and anti-TNF-α antibodies (BD Pharmingen). After 1 h at 4 °C in the dark, the cells were washed and resuspended in PBS with 1% FBS and 0.1% saponin. All of the samples were run on an 18-color LSR-II cytometer with BD FACS DIVA software and analyzed using FlowJo data analysis software (Tree Star, Inc.).
Statistical analysis.
In animal studies, the response of each mouse was counted as an individual data point. The data obtained from animal studies and pseudotype-based neutralization assay were examined by using one-way analysis of variance from GraphPad; differences were considered significant at P < 0.05.
RESULTS
Cross-reactivity of neutralizing antibody responses and antigenic clusters among all H5 clades and subclades.
To determine cross-reactivity of neutralizing antibody responses and antigenic clusters among all H5 clades and subclades, we first constructed DNA plasmids expressing codon-optimized HA from 17 representative strains covering all reported clades and subclades of HPAI H5N1 viruses (Table 1) and then used DNA plasmids to elicit corresponding immune sera in mice and generated corresponding H5N1 pseudotypes (see Materials and Methods for the details). We then performed an across-the-board pseudotype-based neutralization assay and determined the IC50 (Table 2), IC80, and IC90 (Zhou et al., data not shown). We further analyzed the antigenic cartography based on IC50 values (Fig. 1). We observed that, first, all DNA plasmids elicited anti-autologous H5 neutralizing antibody responses with various degrees of potency ranging from 132 in IC50, 68 in IC80, and 48 in IC90 against pBJ, subclade 7.1, to 27,228 in IC50, 4,677 in IC80, and 1,820 in IC90 against pTK, subclade 2.2.1. Second, except for subclades 2.3.2.1 and 7.2, immune sera elicited with clade 0 neutralized H5N1 pseudotypes of all other H5 clades and subclades to different extents, and H5N1 pseudotype derived from clade 0 was neutralized with immune sera elicited with all other H5 clades and subclades. Third, there was reciprocal cross-reactivity of neutralizing antibody responses among clades 0, 1, 3, 4, 5, 6, and 9 and among (sub)clades 0, 2.1.3.2, 2.2.1, 2.4, 2.5, and 8, suggesting that the antigenicity and immunogenicity among H5 clades and subclades within each of these two reciprocal groups correlate well with each other. Fourth, although immune sera elicited with subclades 2.3.4 and 7.1 only neutralized a few H5N1 pseudotypes, pseudotype derived from subclade 2.3.4 was neutralized by most of immune sera except for sera from subclades 2.3.2.1 and 7.2, whereas pseudotype derived from subclade 7.1 was neutralized by most of the immune sera except for sera from (sub)clades 1, 2.2.1, 2.3.2.1, 2.4, and 7.2, suggesting that the antigenicity and immunogenicity of subclade 2.3.4 or 7.1 do not correlate with each other. Finally, subclades 2.3.2.1 and 7.2 each stands on its own, because their immune sera only neutralized very few pseudotypes and their pseudotypes were neutralized by very few immune sera.
Table 2.
Cross-neutralization of a panel of immune sera against corresponding H5N1 pseudotypes
| Serum or virus | Cross-neutralization (IC50)a |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pHK156 (0) | pVN (1) | pCA (1) | pID (2.1.3.2) | pTK (2.2.1) | pHK5052 (2.3.2.1) | pSZ (2.3.4) | pGX12 (2.4) | pKR (2.5) | pHKSF (3) | pGY (4) | pGX1378 (5) | pHB (6) | pBJ (7.1) | pSX (7.2) | pHN (8) | pST (9) | VSVb | |
| pHK156 | 12,521 | 1,432 | 1,466 | 285 | 4,425 | – | 1,008 | 1,380 | 2,536 | 646 | 446 | 6,224 | 1,159 | 867 | – | 616 | 2,088 | – |
| pVN | 4,265 | 3,620 | 3,097 | 322 | 1,253 | – | 50 | 370 | 844 | 851 | 563 | 3,482 | 482 | – | – | 405 | 1,861 | – |
| pCA | 2,228 | 447 | 957 | 1,222 | 351 | – | 757 | – | 373 | 1,429 | 865 | 800 | 353 | 489 | – | 212 | 411 | – |
| pID | 959 | 215 | – | 4,163 | 2,978 | 36 | 503 | 998 | 3,033 | 35 | – | 625 | 55 | 233 | – | 353 | 152 | – |
| pTK | 865 | 164 | 254 | 2,733 | 27,228 | – | 90 | 1,475 | 1,659 | – | – | 99 | – | – | – | 450 | – | – |
| pHK5052 | 292 | – | – | – | 69 | 1,009 | – | – | – | – | – | – | – | 141 | – | – | – | – |
| pSZ | – | – | – | 508 | – | – | 527 | – | – | – | – | 36 | – | 57 | – | – | – | – |
| pGX12 | 1,332 | 195 | – | 1,087 | 3,424 | – | 320 | 4,870 | 3,609 | – | – | – | – | – | – | 564 | 244 | – |
| pKR | 2,429 | 721 | 27 | 2,575 | 6,622 | 511 | 1,356 | 2,642 | 6,084 | 185 | – | 180 | 52 | 58 | – | 1,172 | 202 | – |
| pHKSF | 17,777 | 5,619 | 2,576 | 469 | 435 | – | 147 | 1,050 | 1,124 | 2,758 | 2,501 | 7,279 | 2,255 | 413 | – | 685 | 13,336 | – |
| pGY | 7,775 | 2,591 | 1,710 | 882 | 447 | – | 58 | 258 | 874 | 806 | 2,155 | 2,878 | 994 | 163 | – | 122 | 2,488 | – |
| pGX1378 | 4,490 | 829 | 1,905 | 181 | 551 | – | 306 | – | 269 | 466 | 435 | 2,204 | 539 | 262 | – | 84 | 1,181 | – |
| pHB | 11,098 | 1,026 | 1,119 | 244 | 178 | – | 140 | – | 265 | 1,012 | 642 | 2,808 | 612 | 116 | – | 72 | 1,497 | – |
| pBJ | 169 | – | 387 | – | – | – | 33 | – | – | – | – | 158 | – | 134 | – | – | – | – |
| pSX | – | – | – | 142 | 300 | – | – | – | 65 | – | – | 296 | – | 131 | 1,989 | – | 279 | – |
| pHN | 3,675 | 2,349 | 1,197 | 2,735 | 17,205 | – | 691 | 1,966 | 6,830 | 749 | – | 4,344 | 229 | 95 | – | 1,185 | 2,964 | – |
| pST | 3,202 | 289 | 286 | 68 | 96 | – | – | 162 | 165 | 323 | 386 | 1,840 | 193 | 81 | – | 44 | 1,852 | – |
| VSV | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 14,099 |
The clade or subclade of each HA is indicated in parentheses in the subheadings. Results for antisera against autologous pseudotyped viruses are indicated in boldface. –, Not detected.
Vesicular stomatitis virus (VSV) was used here as the negative control.
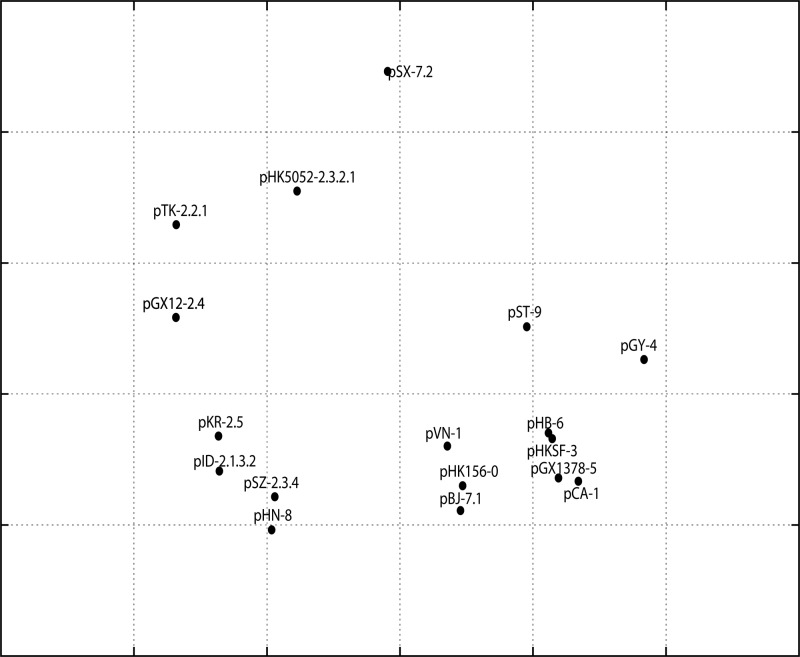
Fig 1.
Antigenic cartography of a panel of immune sera against corresponding influenza virus H5N1 pseudotypes. The antigenic cartography is a geometric representation of IC50 data shown in Table 2 and was constructed using AntigenMap (http://sysbio.cvm.msstate.edu/AntigenMap) (5). The relative distance among each serum sample in the map represents the similarity of antigenicity among various HAs. In the graph, one grid represents a 2-fold change in the pseudotype-based neutralization assay. The virus is represented by strain-(sub)clade, and the abbreviations for the H5N1 strains are listed in Table 1.
To confirm the findings with the HA and NA pseudotype-based neutralization assay (Table 2), we also tested the hemagglutination inhibition (HI) activity of a panel of immune sera. Similar trends were observed when a panel of H5N1 viruses were examined (Chen et al., data not shown).
Figure 1 shows antigenic cartography based on IC50 values. Accordingly, (sub)clades 0, 1, 3, 4, 5, 6, 7.1, and 9 were grouped into antigenic cluster 1, (sub)clades 2.1.3.2, 2.3.4, 2.4, 2.5, and 8 were grouped into another, with subclade 2.2.1 loosely connected to it, and each of subclades 2.3.2.1 and 7.2 was by itself. Interestingly, among (sub)clades in cluster 1, pHK156 clade 0 is closest to cluster 2. This is why immune sera elicited with pHK156 clade 0 cross-neutralize (sub)clades from both clusters 1 and 2.
Broadly neutralizing antibody responses elicited with a triclade DNA vaccine.
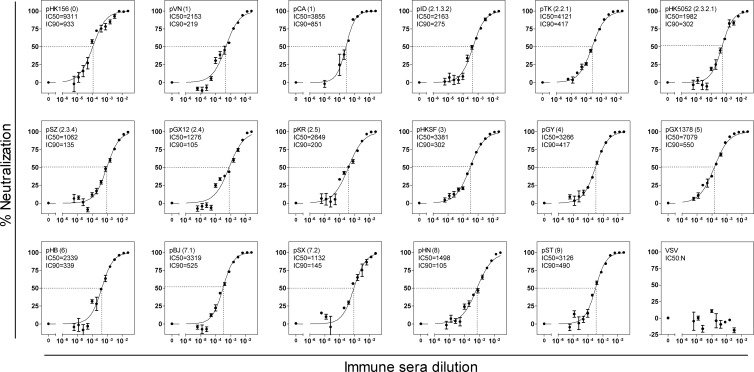
Based on the cross-reactivity of neutralizing antibody responses and antigenic cartography, we selected three DNA plasmids pHK156 (clade 0), pHK5052 (subclade 2.3.2.1), and pSX (subclade 7.2) to formulate a triclade DNA vaccine that would elicit neutralizing antibody responses against all H5 clades and subclades. To evaluate its immunogenicity, female BALB/c mice were i.m. immunized three times with either triclade DNA vaccine or empty vector. Immune sera were analyzed for their ability to neutralize H5N1 pseudotypes covering all H5 clades and subclades. Remarkably, immune sera elicited with the triclade DNA vaccine neutralized all H5N1 pseudotypes with a great degree of potency (Fig. 2). The IC50 ranged from 1,062 against pSZ (subclade 2.3.4) to 9,311 against pHK156 (clade 0), and the IC90 ranged from 105 against pGX (subclade 2.4) and pHN (clade 8) to 933 against pHK156 (clade 0), respectively. In contrast, the empty vector immunized sera did not exhibit any neutralizing activity (Zhou et al., data not shown).
Fig 2.
Titration of neutralizing antibody responses of immune sera elicited with the triclades DNA vaccine against a panel of H5N1 pseudotypes listed in Table 1. The IC50 and IC90 were defined as the reciprocal dilutions of the immune sera that resulted in 50% (dash line) and 90% inhibition, respectively. The data were collected from triplicate experiments and are presented as means ± the standard errors of the mean (SEM). N, not detected. VSV (vesicular stomatitis virus) was used here as a negative control. The virus is represented by strain and (sub)clade in parenthesis and the abbreviation of the H5N1 strain (listed in Table 1).
HA peptide-specific CD8 T-cell responses elicited with the triclade DNA vaccine.
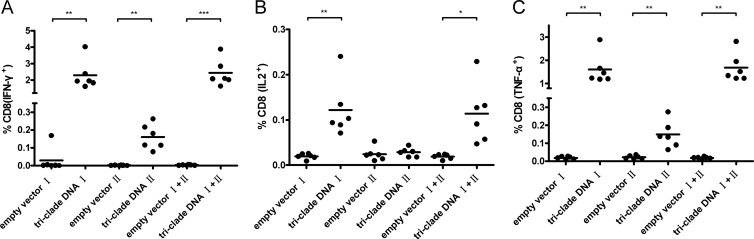
Previously, Deng et al. (11) reported two H-2Kd-restricted HA peptides. One peptide (peptide I, IYSTVASSL) is conserved among various HA subtypes, and the other peptide (peptide II, LYEKVRLQL) is conserved among various H5 strains. Therefore, to test whether the triclade DNA vaccine might also elicit an HA peptide-specific T-cell response, splenocytes from triclade DNA- and empty vector-immunized BALB/c mice were assayed against the two HA peptides using intracellular cytokine staining for IFN-γ, IL-2, and TNF-α. Compared to CD8 T cells from empty-vector-immunized mice, CD8 T cells from triclade DNA-immunized mice exhibited statistically significant peptide-specific responses against peptide I, peptide II, or both, except for peptide II-induced, IL-2-secreted CD8 T cells (Fig. 3). On average, 2.2, 0.13, and 1.6% of the CD8 T cells from triclade DNA-immunized mice secreted IFN-γ, IL-2, or TNF-α, respectively, in response to peptide I; 0.16, 0.03, and 0.15% of the CD8 T cells from triclade DNA-immunized mice secreted IFN-γ, IL-2, or TNF-α, respectively, in response to peptide II; and 2.3, 0.12, and 1.8% of the CD8 T cells from triclade DNA-immunized mice secreted IFN-γ, IL-2, or TNF-α, respectively, in response to both peptides. Thus, we conclude that the triclade DNA vaccine also elicits HA peptide-specific CD8 T-cell responses.
Fig 3.
Cellular immune responses elicited with the triclade DNA vaccine against two HA-specific peptides. Intracellular cytokine staining was performed to analyze the CD8 T-cell responses against two HA-specific peptides. The percentages of activated CD8+ T cells that produce IFN-γ (A), IL-2 (B), and TNF-α (C) are shown. Splenocytes from mice (n = 6 per group) immunized with empty vector or mice (n = 6 per group) immunized with the triclade DNA were assessed, and immune responses were measured 10 days after the final boost. “I” stands for peptide I (IYSTVASSL); “II” stands for peptide II (LYEKVRLQL). The mean value is shown with the horizon bar. *, P = 0.01 to 0.05; **, P = 0.001 to 0.01; ***, P < 0.001.
Immunity elicited with the triclade DNA vaccine protected mice from HPAI H5N1 challenge.
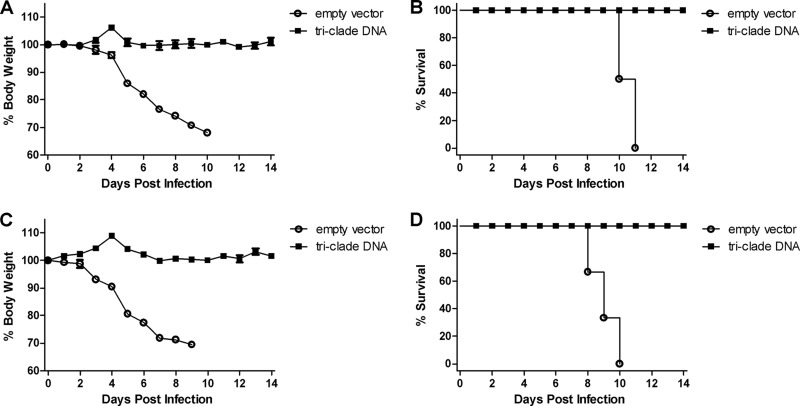
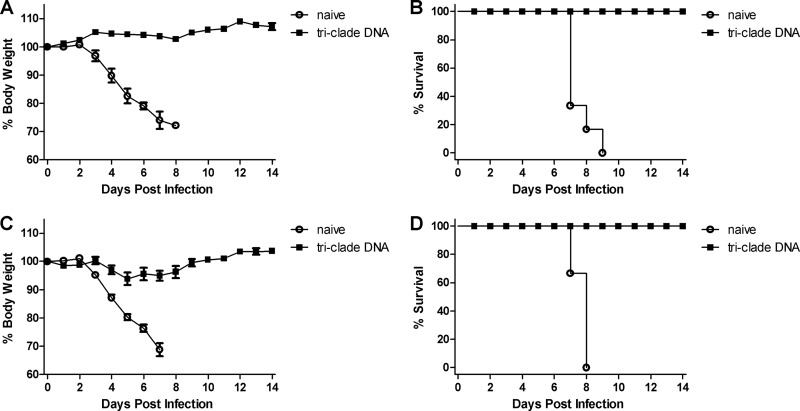
To evaluate whether the triclade DNA vaccine would confer protection against HPAI H5N1 challenge, female BALB/c mice were immunized i.m. three times with triclade DNA vaccine or empty vector, and challenged with two heterologous HPAI H5N1 strains: 100 MLD50 of A/Cambodia/P0322095/05 (clade 1, Fig. 4A and B) and 100 MLD50 of A/Shenzhen/406H/06 (clade 2.3.4, Fig. 4C and D). After the challenge, body weight (Fig. 4A and C) and survival (Fig. 4B and D) were monitored for 14 days. Beginning at day 4, mice in empty-vector immunization group became sick, as evidenced by a rough coat, decreased reactivity, passivity during handling, rolled-up posture, labored breathing, and rapidly lost weight. All mice died between days 8 and 11. In contrast, in the triclade DNA vaccine group after challenge, no mice had any sign of illness or weight loss and all survived. Thus, we conclude that indeed triclade DNA vaccine confers protection against heterologous HPAI H5N1 viruses in mice.
Fig 4.
Triclade DNA vaccination confers protection against high lethal challenge by heterologous H5N1 viruses. BALB/c mice were immunized three times with triclade DNA or with an empty vector. The immunized mice were i.n. challenged with 100 MLD50 (50 TCID50) of A/Cambodia/P0322095/05 (A and B) and 100 MLD50 (5,623 TCID50) of A/Shenzhen/406H/2006 (C and D). After virus challenge, body weight (A and C) and survival (B and D) were recorded for 14 days. Body weight is presented as the mean ± the SEM.
Immune sera elicited with the triclade DNA vaccine confer protection against HPAI H5N1.
To determine the contribution of immune sera to the protection, we immunized female BALB/c mice with the triclade DNA vaccine. Immune sera were collected at 14 days after the last immunization and combined. For passive immunization, female BALB/c mice were i.p. injected with 700 μl of immune sera. Control mice were injected with the same amount of sera collected from naive mice. After 24 h, the mice were challenged i.n. with 100 MLD50 of A/Cambodia/P0322095/05 (clade 1, Fig. 5A and B) and 100 MLD50 of A/Shenzhen/406H/06 (clade 2.3.4, Fig. 5C and D) strains. After the challenge, body weight (Fig. 5A and C) and survival (Fig. 5B and D) were monitored for 14 days. Mice that had been passively immunized with sera from naive mice became sick on day 3 after the challenge, developed all of the symptoms mentioned above, lost weight rapidly, and died between days 7 and 9. In contrast, mice that had been passively immunized with sera from the triclade DNA vaccine group did not exhibit any sign of illness nor weight loss, survived after challenge with A/Cambodia/P0322095/05 (Fig. 5A and B), and slightly lost weight between days 4 and 9, and of the animals all survived after challenge with A/Shenzhen/406H/06 (Fig. 5C and D). Taken together, these results indicate that although the triclade DNA vaccine elicits both broadly neutralizing antibody and HA peptide-specific CD8 T-cell responses, broadly neutralizing antibody responses alone could confer protection from a lethal-dose challenge of heterologous HPAI H5N1 viruses in mice.
Fig 5.
Passive immunization of immune sera elicited with the triclade DNA vaccine confers protection against lethal challenge by heterologous H5N1 viruses. To assess the protective effect of immunized sera, mice first received sera from naive mice or immune sera elicited with the triclade DNA vaccine and, 24 h later, were i.n. challenged with 100 MLD50 (50 TCID50) of A/Cambodia/P0322095/05 (A and B) and 100 MLD50 (5,623 TCID50) of A/Shenzhen/406H/2006 (C and D). After virus challenge, body weight (A and C) and survival (B and D) were recorded for 14 days. Body weight is presented as the mean ± the SEM.
DISCUSSION
As the major viral protein to elicit neutralizing antibody responses, HA is also the most variable influenza virus protein both genetically and antigenically. This variability of HA poses a great scientific challenge and economic burdens in influenza vaccine development. For H5N1 viruses, during the past 15 years viruses have evolved into genetically distinct clades and subclades. To cover this diversity, the WHO has already created 20 seed recombinant vaccine strains (38), and many new approaches that have applied both reverse and analytic vaccinology have been evaluated (2, 7, 13, 15, 18, 26, 27, 30, 36, 37). For reverse vaccinology, H5 HA sequence diversity was analyzed, and phylogenetic trees were constructed. Vaccine candidates based on consensus, ancestral, mosaic, and multiclades of H5 HA sequences have been designed (7, 13, 27, 30). For analytic vaccinology, several broadly neutralizing antibodies against HA in influenza vaccinated and/or infected humans have been isolated, and the structural basis of antibody recognition and neutralization has been recently elucidated (8, 9, 16, 24, 31, 33, 43). Vaccine candidates based on conserved neutralizing epitopes were designed (2, 15, 26, 36). For example, Wang et al. (36) designed a peptide-based vaccine candidate based on conserved peptide sequence at amino acid residues between positions 76 and 130 of the HA2 region and demonstrated that this peptide could elicit antibody responses against both H5 HA and HA from other subtypes.
In the present study, we took another approach, i.e., designing vaccine candidates based on a comprehensive serologic study. To accomplish this, we constructed panels of DNA plasmids, pseudotypes, and corresponding immune sera of 17 representative strains covering all reported H5 clades and subclades. We then performed an across-the-board pseudotype-based neutralization assay and determined the cross-reactivity of neutralizing antibody responses, as well as antigenic clusters, among all H5 clades and subclades. Based on the serologic data, we then designed a triclade DNA vaccine and demonstrated for the first time that immune sera elicited with this triclade DNA vaccine can neutralize representative strains from all H5 clades and subclades and conferred immune protection against a high-lethal-dose challenge of two heterologous H5N1 viruses (see Fig. 1, 2, 4, and 5). Here, we should point out three caveats. First, although strains within a given clade or subclade are expected to be antigenically related to some extent, very few studies have thoroughly investigated the antigenicity among sublineages of H5N1 strains. Therefore, although in the present study we show that immune sera elicited with the triclade vaccine were able to neutralize representative strains of all H5 clades and subclades and confer the protection against a high-lethal-dose challenge of heterologous HPAI H5N1 viruses, additional tests against more H5N1 strains from different sublineages of various H5 clades and subclades are needed in order to get the full picture of coverage by this triclade vaccine. Second, although in the present study we demonstrated that a triclade vaccine that consists of HA from (sub)clades 0, 2.3.2.1, and 7.2 elicits neutralizing antibody responses against representative strains from all H5 clades and subclades, this does not imply that only this combination can elicit broadly neutralizing antibody responses against all H5 clades and subclades. Multiple-clade immunogens based on other combinations may also be effective. Third, although DNA vaccines have certain advantages over conventional inactivated and live attenuated influenza vaccines, in the present study we chose an HA-based DNA vaccine simply because an HA-based DNA vaccine would allow us to look exclusively into anti-HA immune responses without any complications caused by immune responses elicited by other viral proteins. In addition, we immunized mice three times with relatively high DNA doses (200 μg per immunization) to maximize the neutralizing antibody responses. Therefore, other vaccine modalities, particularly those based on currently licensed vaccine platform with the same triclade HA combination and a more practical immunization schedule, should be examined.
Previously, using mouse monoclonal antibodies and HI assay, Wu et al. (44) grouped H5N1 viruses into four antigenically distinct clusters (A to D). Cluster A contained subclade 2.1 and 2.4 viruses, as well as A/Hong Kong/213/03 (clade 1), cluster B contained clades 1, 4, 5, 7, and 9, cluster C contained subclades 2.2, 2.3.2, and 2.3.3, and cluster D contained subclades 2.3.2 and 2.3.4. Similar findings were described in another study using HI assays using ferret or chicken polyclonal sera (13, 35). These findings suggested a link between genetic and antigenic distances, but they also highlighted the antigenic complexity of clade 2 strains. In the present study we did compare the cartography of IC50 values generated from an across-the-board pseudotype-based neutralization assay. We found that (sub)clades 0, 1, 3, 4, 5, 6, 7.1, and 9 were grouped into antigenic cluster 1, that (sub)clades 2.1.3.2, 2.3.4, 2.4, 2.5, and 8 were grouped into another with subclade 2.2.1 loosely connected to it, and that each of subclades 2.3.2.1 and 7.2 was by itself. Thus, to a certain extent, the clusters defined by our present study based on antigenicity and those reported earlier (13, 35, 44) are similar, although here we included six additional (sub)clades—(sub)clades 0, 2.5, 3, 6, 7.2, and 8—in the analyses.
To better understand uniqueness of immunogenicity and antigenicity of (sub)clades 2.3.2.1 and 7.2, we used CBS Prediction Servers (http://www.cbs.dtu.dk/services/) to predict glycosylations among representative H5 HA from various clades and subclades. The software program predicts neither C-linked mannosylation nor O-linked glycosylation among H5 HAs. For N glycosylation, subclade 2.3.2.1 tends to be more likely to be N-linked glycosylated at the amino acid position 181 than other clades and subclades at the same position, and subclade 7.2 is very likely to have two additional N-linked glycosylations at positions 88 and 178. Therefore, we speculate that the uniqueness in immunogenicity and antigenicity of subclades 2.3.2.1 and 7.2 may be due to their differences in N-linked glycosylation pattern.
In summary, in the present study we were able to provide the proof of concept that broadly neutralizing antibody responses against all H5 clades and subclades can indeed be elicited with a triclade DNA vaccine designed on the basis of a comprehensive serologic study. Therefore, such a triclade vaccine based on currently licensed vaccine platforms may be developed to effectively deal with the diversity of H5N1 viruses.
ACKNOWLEDGMENTS
We thank L. Naldini at the University of Turin Medical School, Turin, Italy, for the lentiviral transfer vector and Tong Zhang at Rutgers University and Jialiang Yang at Mississippi State University for their assistance with the antigenic cartography analyses.
This study was supported by research grants from the Chinese National Science Foundation (grant 31170871), the Chinese Science and Technology Ministry 973 Program Project (2006CB504308), National Science and Technology Major Projects (2008ZX10001-010, 2009ZX10004-105, and 2009ZX10004-016), the Shanghai Pasteur Foundation (SPHRF2007001), and the French Energy Company Areva to P.Z. Z.C. and X.-F.W. were supported by grant 1RC1AI086830 from the National Institutes of Health to X-F.W.
Footnotes
Published ahead of print 11 April 2012
REFERENCES
- 1. Barr IG, et al. 2010. Epidemiological, antigenic and genetic characteristics of seasonal influenza A(H1N1), A(H3N2), and B influenza viruses: basis for the WHO recommendation on the composition of influenza vaccines for use in the 2009–2010 northern hemisphere season. Vaccine 28:1156–1167 [DOI] [PubMed] [Google Scholar]
- 2. Bommakanti G, et al. 2010. Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc. Natl. Acad. Sci. U. S. A. 107:13701–13706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bright RA, et al. 2008. Cross-clade protective immune responses to influenza viruses with H5N1 HA and NA elicited by an influenza virus-like particle. PLoS One 3:e1501 doi:10.1371/journal.pone.0001501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buchy P, et al. 2007. Influenza A/H5N1 virus infection in humans in Cambodia. J. Clin. Virol. 39:164–168 [DOI] [PubMed] [Google Scholar]
- 5. Cai Z, Zhang T, Wan X-F. 2010. A computational framework for influenza antigenic cartography. PLoS Comput. Biol. 6:e1000949 doi:10.1371/journal.pcbi.1000949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carrat F, Flahault A. 2007. Influenza vaccine: the challenge of antigenic drift. Vaccine 25:6852–6862 [DOI] [PubMed] [Google Scholar]
- 7. Chen MW, et al. 2008. A consensus-hemagglutinin-based DNA vaccine that protects mice against divergent H5N1 influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 105:13538–13543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corti D, et al. 2010. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Invest. 120:1663–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Corti D, et al. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856 [DOI] [PubMed] [Google Scholar]
- 10. Crevar CJ, Ross TM. 2008. Elicitation of protective immune responses using a bivalent H5N1 VLP vaccine. Virol. J. 5:131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deng Y, Yewdell JW, Eisenlohr LC, Bennink JR. 1997. MHC affinity, peptide liberation, T cell repertoire, and immunodominance all contribute to the paucity of MHC class I-restricted peptides recognized by antiviral CTL. J. Immunol. 158:1507–1515 [PubMed] [Google Scholar]
- 12. Ding H, et al. 2011. Superior neutralizing antibody response and protection in mice vaccinated with heterologous DNA prime and virus like particle boost against HPAI H5N1 virus. PLoS One 6:e16563 doi:10.1371/journal.pone.0016563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ducatez MF, et al. 2011. Feasibility of reconstructed ancestral H5N1 influenza viruses for cross-clade protective vaccine development. Proc. Natl. Acad. Sci. U. S. A. 108:349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ehrlich HJ, et al. 2009. A cell culture (Vero)-derived H5N1 whole-virus vaccine induces cross-reactive memory responses. J. Infect. Dis. 200:1113–1118 [DOI] [PubMed] [Google Scholar]
- 15. Ekiert DC, et al. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ekiert DC, et al. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. FAO 2011. FAO ECTAD HPAI situation update no. 500. Food and Agriculture Association, New York, NY [Google Scholar]
- 18. Forrest HL, et al. 2009. Single- and multiple-clade influenza A H5N1 vaccines induce cross protection in ferrets. Vaccine 27:4187–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gerhard W, et al. 1990. Identification of eight determinants in the hemagglutinin molecule of influenza virus A/PR/8/34 (H1N1) which are recognized by class II-restricted T cells from BALB/c mice. J. Virol. 65:364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Giles BM, Ross TM. 2011. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 29:3043–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kong WP, et al. 2003. Immunogenicity of multiple gene and clade human immunodeficiency virus type 1 DNA vaccines. J. Virol. 77:12764–12772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Y, et al. 2010. Continued evolution of H5N1 influenza viruses in wild birds, domestic poultry, and humans in China from 2004 to 2009. J. Virol. 84:8389–8397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nguyen T, et al. 2009. Characterization of a highly pathogenic avian influenza H5N1 virus sublineage in poultry seized at ports of entry into Vietnam. Virology 387:250–256 [DOI] [PubMed] [Google Scholar]
- 24. Okuno Y, Isegawa Y, Sasao F, Ueda S. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A Virus H1 and H2 strains. J. Virol. 67:2552–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oran AE, Robinson HL. 2003. DNA vaccines, combining form of antigen and method of delivery to raise a spectrum of IFN-γ and IL-4-producing CD4+ and CD8+ T Cells. J. Immunol. 171:1999–2005 [DOI] [PubMed] [Google Scholar]
- 26. Prabakaran M, et al. 2010. Neutralizing epitopes of influenza virus hemagglutinin: target for the development of a universal vaccine against H5N1 lineages. J. Virol. 84:11822–11830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rao S, et al. 2008. Multivalent HA DNA vaccination protects against highly pathogenic H5N1 avian influenza infection in chickens and mice. PLoS One 3:e2432 doi:10.1371/journal.pone.0002432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 27:493–497 [Google Scholar]
- 29. Smith GJ, et al. 2006. Emergence and predominance of an H5N1 influenza variant in China. Proc. Natl. Acad. Sci. U. S. A. 103:16936–16941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Steel J, et al. 2010. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio 1:e00018–10 doi:10.1128/mBio.00018-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sui J, et al. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16:265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tao P, et al. 2009. Virus-like particle vaccine comprised of the HA, NA, and M1 proteins of an avian isolated H5N1 influenza virus induces protective immunity against homologous and heterologous strains in mice. Viral Immunol. 22:273–281 [DOI] [PubMed] [Google Scholar]
- 33. Throsby M, et al. 2008. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 3:e3942 doi:10.1371/journal.pone.0003942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tsai C, et al. 2009. Measurement of neutralizing antibody responses against H5N1 clades in immunized mice and ferrets using pseudotypes expressing influenza hemagglutinin and neuraminidase. Vaccine 27:6777–6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wan XF, et al. 2011. Indications that live poultry markets are a major source of human H5N1 influenza virus infection in China. J. Virol. 85:13432–13438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang TT, et al. 2010. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc. Natl. Acad. Sci. U. S. A. 107:18979–18984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wei CJ, et al. 2010. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science 329:1060–1064 [DOI] [PubMed] [Google Scholar]
- 38. WHO 2012. Antigenic and genetic characteristics of influenza A(H5N1) and influenza A(H9N2) viruses and candidate vaccine viruses developed for potential use in human vaccines. World Health Organization, Geneva, Switzerland [Google Scholar]
- 39. WHO 2011. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. World Health Organization, Geneva, Switzerland: [PubMed] [Google Scholar]
- 40. WHO 2012. Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003–2012 World Health Organization, Geneva, Switzerland [Google Scholar]
- 41. WHO 2011. H5N1 avian influenza: timeline of major events. World Health Organization, Geneva, Switzerland [Google Scholar]
- 42. WHO 2008. Update on avian influenza A (H5N1) virus infection in humans. N. Engl. J. Med. 358:261–273 [DOI] [PubMed] [Google Scholar]
- 43. Wrammert J, et al. 2008. Rapid cloning of high affinity human monoclonal antibodies against influenza virus. Nature 453:667–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu WL, et al. 2008. Antigenic profile of avian H5N1 viruses in Asia from 2002 to 2007. J. Virol. 82:1798–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang ZY, et al. 2004. pH-dependent entry of Severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 78:5642–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang ZY, et al. 2007. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science 317:825–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou B, Zhong N, Guan Y. 2007. Treatment with convalescent plasma for influenza A (H5N1) infection. N. Engl. J. Med. 357:1450–1451 [DOI] [PubMed] [Google Scholar]