Abstract
The ascomycete Botrytis porri causes clove rot and leaf blight of garlic worldwide. We report here the biological and molecular features of a novel bipartite double-stranded RNA (dsRNA) mycovirus named Botrytis porri RNA virus 1 (BpRV1) from the hypovirulent strain GarlicBc-72 of B. porri. The BpRV1 genome comprises two dsRNAs, dsRNA-1 (6,215 bp) and dsRNA-2 (5,879 bp), which share sequence identities of 62 and 95% at the 3′- and 5′-terminal regions, respectively. Two open reading frames (ORFs), ORF I (dsRNA-1) and ORF II (dsRNA-2), were detected. The protein encoded by the 3′-proximal coding region of ORF I shows sequence identities of 19 to 23% with RNA-dependent RNA polymerases encoded by viruses in the families Totiviridae, Chrysoviridae, and Megabirnaviridae. However, the proteins encoded by the 5′-proximal coding region of ORF I and by the entire ORF II lack sequence similarities to any reported virus proteins. Phylogenetic analysis showed that BpRV1 belongs to a separate clade distinct from those of other known RNA mycoviruses. Purified virions of ∼35 nm in diameter encompass dsRNA-1 and dsRNA-2, and three structural proteins (SPs) of 70, 80, and 85 kDa, respectively. Peptide mass fingerprinting analysis revealed that the 80- and 85-kDa SPs are encoded by ORF I, while the 70-kDa SP is encoded by ORF II. Introducing BpRV1 purified virions into the virulent strain GarlicBc-38 of B. porri caused derivative 38T reduced mycelial growth and hypovirulence. These combined results suggest that BpRV1 is a novel bipartite dsRNA virus that possibly belongs to a new virus family.
INTRODUCTION
Mycoviruses (fungal viruses) have been recorded to be widespread in the major taxonomic groups of filamentous fungi, yeasts, and oomycetes (16, 39). Most mycoviruses have RNA genomes, which are either single-stranded (ss) or double-stranded (ds), and are now classified into 12 families (16). Members of four families (Chysoviridae, Partitiviridae, Reoviride, and Totiviridae) accommodate undivided (e.g., Totiviridae) or divided (e.g., Chysoviridae, Partitiviridae, and Reoviride) dsRNA genomes, which are encapsidated within capsid proteins with formation of rigid virus particles. Members of six families (Alphaflexiviridae, Barnaviride, Endornaviridae, Gammaflexiviridae, Hypoviridae, and Narnaviridae) accommodate ssRNA genomes, of which only two families (Alphaflexiviridae and Gammaflexiviridae) form virus particles, whereas members of the remaining four virus families are unencapsidated and do not form typical virions. Two families (Metaviridae and Pseudoviridae) accommodate RNA reverse-transcribing genomes. Meanwhile, an increasing number of novel mycoviruses have been reported recently (2, 9, 29, 53, 59). Chiba et al. (9) proposed to establish a new bipartite dsRNA virus family, Megabirnaviridae, to accommodate Rosellinia necatrix megabirnavirus 1 (RnMBV1) based on characterization of the dsRNA genome of RnMBV1. Besides RNA mycoviruses, Yu et al. (55) reported a geminivirus-related ssDNA mycovirus infecting Sclerotinia sclerotiorum (Lib.) de Bary, the causal agent of stem rot of oilseed rape (Brassica napus).
In many cases, mycovirus infection does not cause any visible abnormal symptoms for host fungi (cryptic infections). However, infection by some mycoviruses can cause abnormal symptoms in their host fungi, including reduced mycelial growth and debilitated virulence (e.g., hypovirulence) (37). Successful control of chestnut blight caused by virulent strains of Cryphonectria (Endothia) parasitica with hypovirulent strains of this pathogen in Europe (1, 19, 30) inspired numerous researchers to study the phenomenon of mycovirus-mediated hypovirulence in plant pathogenic fungi. Thus far, many mycoviruses belonging to Hypoviridae, Narnaviridae, Reoviridae, Megabirnaviridae, and some unassigned virus taxa have been identified to cause (or to be associated with) fungal hypovirulence (16, 38, 39).
Allium crops, including garlic (Allium sativum), garlic chives (A. tuberosum), bulb onions (A. cepa), and green onions (A. fistulosum), are important vegetables grown in China, as well as in many other countries. Gray mold disease caused by Botrytis spp., including B. porri, is one of the most important ecological factors limiting both the yield and the quality of these Allium crops (33, 57, 58). Previous studies indicated that B. porri can cause garlic clove rot (12), garlic leaf blight (57), and leek leaf rot (3). Several RNA mycoviruses have been reported in B. cinerea (5, 6, 21, 22, 23, 50, 52), a close relative of B. porri and the causal agent of gray mold disease on numerous plants. The genomes of some mycoviruses infecting B. cinerea have been characterized (22, 23, 52). However, no mycoviruses infecting other species of Botrytis, including B. porri, have been reported thus far.
A strain of B. porri, designated GarlicBc-72, was isolated from a diseased leaf of garlic grown in Zhushan County, Hubei Province, China (57). We found that it grew slowly on culture media and was hypovirulent to garlic leaves compared to other strains of B. porri. Two dsRNA segments approximately 6.2 and 6.5 kb in size, respectively, were detected in mycelia of strain GarlicBc-72, but were not detected in virulent strains of B. porri. These results suggested that strain GarlicBc-72 might be infected with an RNA mycovirus. Therefore, a study was conducted to fulfill the following three objectives: (i) to establish the cause-effect relationship between presence of the two dsRNA segments and hypovirulence in strain GarlicBc-72, (ii) to obtain and analyze the full-length cDNA sequences generated from the two dsRNA segments, and (iii) to purify the virus particles from strain GarlicBc-72 and to characterize their nucleic acids and the structural proteins at the molecular level.
MATERIALS AND METHODS
Fungal strains and isolates and biological characterization.
Fifteen strains or isolates of B. porri were used in the present study (Table 1). For simplicity, the names of strains GarlicBc-16, GarlicBc-38, GarlicBc-72, OnionBc-95, and GP72SC35 of B. porri were abbreviated here as Bc-16, Bc-38, Bc-72, Bc-95, and SC35, respectively. Maintenance of the stock cultures and establishment of working cultures of these strains of B. porri were carried out as described in our previous study (51). Mycelial agar plugs (6 mm in diameter) removed from the colony margin of a 3- to 5-day-old culture of each strain or isolate were placed on potato dextrose agar (PDA) in petri dishes (9 cm in diameter) at one plug per dish. The dishes were incubated at 20°C for determination of the mycelial growth rate and for observation of the colony morphology. Pathogenicity of each strain was determined by inoculating detached leaves of garlic (A. sativum cultivar Feng Suan No. 1) with agar plugs of actively growing mycelia according to the procedures of Zhang et al. (57, 58).
Table 1.
Origin of strains or isolates of Botrytis porri used in this study
| Strain or isolate | Origin (host, place, isolation yr) | Pathogenicity | Source or reference |
|---|---|---|---|
| GarlicBc-72 | Allium sativum, Zhushan, China, 2008 | Hypovirulent | This study |
| OnionBc-95 | Allium fistulosum, Qianjiang, China, 2008 | Virulent | 58 |
| GarlicBc-38 | Allium sativum, Laifeng, China, 2007 | Virulent | 58 |
| GarlicBc-16 | Allium sativum, Zhushan, China, 2007 | Virulent | 57 |
| GP72SC35 | A single-conidium isolate of GarlicBc-72, 2009 | Virulent | This study |
| 72-35-1 | GP72SC35 in a pairing culture of GP72SC35 and GarlicBc-72, 2009 | Hypovirulent | This study |
| 72-35-2 | GP72SC35 in a pairing culture of GP72SC35 and GarlicBc-72, 2009 | Hypovirulent | This study |
| 72-35-3 | GP72SC35 in a pairing culture of GP72SC35 and GarlicBc-72, 2009 | Hypovirulent | This study |
| 72-38-1 | GarlicBc-38-1 in a pairing culture of GarlicBc-38-1 and GarlicBc-72, 2009 | Virulent | This study |
| 72-38-2 | GarlicBc-38-1 in a pairing culture of GarlicBc-38-1 and GarlicBc-72, 2009 | Virulent | This study |
| 72-38-3 | GarlicBc-38-1 in a pairing culture of GarlicBc-38-1 and GarlicBc-72, 2009 | Virulent | This study |
| 72-95-1 | OnionBc-95 in a pairing culture of OnionBc-95 and GarlicBc-72, 2009 | Virulent | This study |
| 72-95-2 | OnionBc-95 in a pairing culture of OnionBc-95 and GarlicBc-72, 2009 | Virulent | This study |
| 72-95-3 | OnionBc-95 in a pairing culture of OnionBc-95 and GarlicBc-72, 2009 | Virulent | This study |
| 38T | GarlicBc-38-1 infected by BpRV1, 2010 | Hypovirulent | This study |
Vertical transmission of hypovirulence-associated dsRNAs.
Vertical transmission refers to transmission of hypovirulence-associated dsRNAs from mycelia to asexual or sexual spores (38). This experiment was done by determining the pathogenicity and the presence of dsRNAs in single-conidium (SC) isolates from the dsRNA-containing hypovirulent strain Bc-72. For obtaining conidia, strain Bc-72 was incubated at 20°C on PDA in petri dishes (9 cm in diameter) for 17 days. Colonies showing conidial production were selected. Conidia in one of the dishes were washed off by flooding the colony with ∼3 ml of sterile-distilled water (SDW), followed by scraping the surface of the colony with a sterilized glass rod. The conidial mixtures from the selected dishes were pooled and filtered through four-layer cheesecloth to remove mycelial fragments. The resulting conidial suspension was diluted to 104 conidia/ml with SDW. An aliquot of 100 μl of conidial suspension was pipetted onto a PDA dish, and the conidial suspension drop was carefully spread on the PDA plate using a sterilized glass rod. Individual conidia on the PDA surface were picked up using a sterilized fine glass needle under a light microscope and transferred to fresh PDA plate at one conidium per plate. The plates were incubated at 20°C for 5 days, the resulting cultures were identified, and those belonging to B. porri were individually tested for the mycelial growth rates on PDA (20°C), pathogenicity on detached garlic leaves (20°C, 72 h), and the presence of the two dsRNA segments in mycelia using previously described methods (51, 57, 58). The hypovirulent strain Bc-72 and the virulent strain Bc-16 of B. porri were used as controls in this experiment.
Horizontal transmission of hypovirulence-associated dsRNAs.
Horizontal transmission refers to transmission hypovirulence-associated dsRNAs from hypovirulent to virulent fungal strains through hyphal anastomosis (38). The experiment was done on PDA in petri dishes using the pairing culture technique (51, 52, 56). In the contact culture in each dish (9 cm in diameter), the dsRNA-containing hypovirulent strain Bc-72 served as the donor, whereas strains Bc-38, SC35, or Bc-95 served as recipients. After incubation of the contact cultures at 20°C for 8 days, three mycelial derivative isolates were obtained from three colonies of each recipient strain in the three contact cultures of Bc-72/Bc-38, Bc-72/SC35, or Bc-72/Bc-95 using the method described by Wu et al. (51). The isolates were designated 72-38-1, 72-38-2, and 72-38-3 for derivatives from strain Bc38, 72-35-1, 72-35-2, and 72-35-3 for derivatives from strain SC35, and 72-95-1, 72-95-2, and 72-95-3 for derivatives from strain Bc-95. All of these derivative isolates were individually tested for the mycelial growth rates on PDA (20°C), pathogenicity on detached garlic leaves (20°C, 72 h), and the presence of dsRNA in mycelia (51, 57, 58). The parental strains Bc-72, Bc-38, Bc-95, and SC35 were included as controls in this experiment.
TEM.
Subcellular characteristics of hyphal cells of strains Bc-16 (virulent) and Bc-72 (hypovirulent) were examined using transmission electron microscopy (TEM). A mycelial agar plug of each strain was inoculated in the center of a piece of sterilized cellophane film placed on PDA in a petri dish. The culture was incubated at 20°C for 3 days for strain Bc-16 and for 5 days for strain Bc-72. Small pieces (2 by 2 mm [length by width]) of the cellophane films colonized with mycelia of strains Bc-16 or Bc-72 were carefully cut off from the colony margin (∼3 mm in width) and the central area (∼10 mm from the margin area) using a razor blade. The mycelial specimens were fixed and dehydrated using conventional procedures. Ultrathin sections (50 to 60 nm in thickness) for the mycelial specimens were cut using a diamond knife, mounted on slotted and Formvar-coated grids, stained with 5% aqueous lead citrate and 5% uranyl acetate, and examined under TEM (Model Tecnai G2 20; FEI, Hillsboro, OR).
Extraction and purification of dsRNA from mycelia.
Mycelial agar plugs of each strain/isolate of B. porri were inoculated in potato dextrose broth in petri dishes (9 cm in diameter) at 1 agar plug per dish and 20 dishes per strain. The dishes were incubated at 20°C in the dark for 3 to 6 days. Mycelial mats in each dish were harvested and stored at −20°C until use. Extraction and purification of the dsRNAs from mycelia of each strain were performed according to the procedures of Morris and Dodds (34). Analysis of the dsRNA extracts and identification of the double-strandedness of RNAs were done with the methods described in our previous studies (51).
cDNA cloning, sequencing, and sequence analysis.
The dsRNAs extracted from strain Bc-72 of B. porri were fractioned by agarose gel electrophoresis, and the individual dsRNA bands (6.5 kb for dsRNA-1 and 6.2 kb for dsRNA-2) were cut off for further analysis. Cloning of the cDNAs for the dsRNAs from strain Bc-72 by random primer-mediated PCR, sequencing of the cDNAs, and analysis of the sequences were done using the procedures described in our previous studies (51, 52). To clone the terminal sequences of each dsRNA, the 3′ terminus of each strand of dsRNA-1 or dsRNA-2 was ligated to the 5′ terminus of the BPA adaptor (see Fig. S1 and Table S1 in the supplemental material) using T4 RNA ligase (TaKaRa Biotechnology Co., Ltd., Dalian, China) at 4 to 8°C for 18 h. The primer RCBPA was used for reverse transcription of dsRNA-1 or dsRNA-2 to cDNA. The primer pairs RCBPA/BP5-4 and RCBPA/BP3 were used for PCR amplification of the 5′-terminal sequence and the 3′-terminal sequence of dsRNA-1, respectively. Similarly, the primer pairs RCBPA/BG1 and RCBPA/BG2 were used to PCR amplify the 5′-terminal sequence and the 3′-terminal sequence of dsRNA-2, respectively. The PCR primers and their sequences mentioned above are listed in Table S1 in the supplemental material. The full-length cDNA sequence for dsRNA-1 or dsRNA-2 was obtained by assembling of the partial sequences in the different cDNA clones (see Fig. S1 in the supplemental material). Open reading frames (ORFs) in each full-length cDNA sequence were deduced using the ORF Finder program in the website of the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/gorf/gorf.html) with the standard codon usages. Database searches of the full-length cDNA sequences of dsRNA-1 and dsRNA-2 and deduced polypeptides from the two cDNA sequences were performed in the public database at NCBI using the programs of BLASTn and BLASTp, respectively. Motifs present in the deduced polypeptide sequences were searched in three databases, including the CDD database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), the Pfam database (http://pfam.sanger.ac.uk/) (15), and the PROSITE database (http://www.expasy.ch/). The deduced polypeptide sequences encoded by the two dsRNAs were also BLAST-searched in the MEROPS protease database (http://merops.sanger.ac.uk/) (40). Multiple alignments of the sequences of RNA-dependent RNA polymerase (RdRp) was accomplished using the CLUSTAL_X program (48). A phylogenetic tree was constructed using the neighbor-joining (NJ) method and tested with a bootstrap of 1,000 replicates to ascertain the reliability of a given branch pattern in the NJ tree. Potential secondary structures at the 3′- and 5′-terminal sequences of the positive strand of dsRNA-1 and dsRNA-2 were predicted using RNAstructure software (version 4.6) (32). The proline frequency on the two polypeptides deduced from ORF I on dsRNA-1 and ORF II on dsRNA-2 was calculated using Microsoft Office Excel 2007 as described by Spear et al. (46). The threshold value of 0.33 was used to indicate the minimum proline frequency required for generation a PPII-like helix (PxxPxxPxx) over a 9-amino-acid (aa) span.
Northern hybridization.
Northern hybridization was performed to verify the authenticity of the cDNA sequences generated from dsRNA-1 (6.5 kb) and dsRNA-2 (6.2 kb) in strain Bc-72 of B. porri. Meanwhile, it was also used to confirm the high nucleotide sequence similarity between the two dsRNAs at the 5′ terminus (Fig. 1A). The dsRNAs from strain Bc-72 were separated by agarose gel electrophoresis. They were then transferred from the agarose gel to an Immobilon-Ny+ membrane (Millipore, Bedford, MA) using the procedures described by Jiang and Ghabrial (25). Three DNA probes (423 bp for probes 1, 506 bp for probe 2, and 451 bp for probe 3) were designed on the basis of the full-length cDNA sequences of dsRNA-1 and dsRNA-2. They were generated by reverse-transcription PCR (RT-PCR) using dsRNA-1 or dsRNA-2 as a template with the specific primer pairs listed in Table S1 in the supplemental material. The probes were individually labeled with digoxigenin (DIG)-11-dUTP in the DIG High-Prime DNA labeling and detection starter kit I (Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Germany) and used to hybridize with the dsRNAs blotted on the nylon membrane. The hybridization signals were detected by enzymatic immunoassay using the reagents and the procedures recommended by the manufacturer (Roche Applied Science).
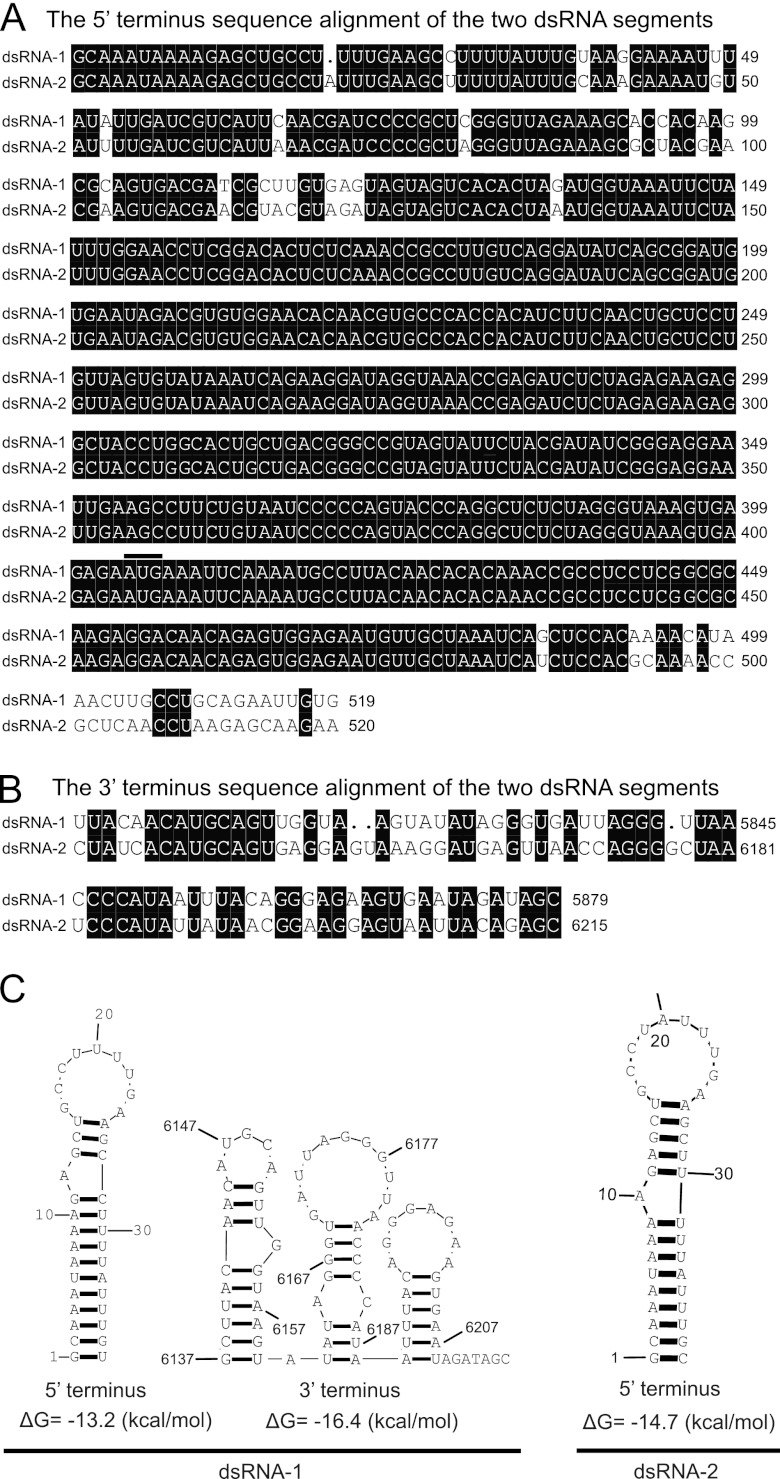
Fig 1.
Alignment of the 5′-terminal (A) and 3′-terminal (B) sequences of the coding strands of dsRNA-1 and dsRNA-2 of BpRV1. Black shading is used to highlight identical nucleotides. The “•” symbol represents a missing nucleotide. The start codon (AUG) at the 5′ terminus for both dsRNA-1 and dsRNA-2 is indicated with a black bar above the two nucleotide sequences. (C) Predicted secondary structures for the terminal regions of dsRNA-1 and dsRNA-2 of BpRV1.
Purification of viral particles.
Strain Bc-72 of B. porri was grown at 20°C for 6 days on sterilized cellophane films placed on PDA. Mycelia were harvested, and a sample of 10 g fresh mycelia was ground to fine powder in liquid nitrogen using a sterilized mortar and pestle. The powder was transferred to a 1,000 ml-blender containing 150 ml of extraction solution [0.1 M sodium phosphate and 3% (wt/vol) Triton X-100 (pH 7.0). The suspension was blended for 30 s and then transferred to a plastic tube (200 ml), followed by centrifugation at 10,000 × g for 20 min to remove the hyphal cell debris. The supernatant was then carefully pipetted out to another plastic tube, which was then subject to a 2-h ultracentrifugation at 119,000 × g under 4°C to precipitate all of the particles, including the virus particles in the supernatant. The pellet was resuspended in 4 ml of sodium phosphate buffer (0.1 M), and the resulting suspension was centrifuged at 16,000 × g for 20 min to precipitate large particles. The supernatant containing the virus particles was then overlaid on a centrifuge tube containing sucrose solutions with a concentration gradient ranging from 10 to 40% (wt/vol) and centrifuged at 70,000 × g under 4°C for 2 h. Fractions in the middle portion of the tube were individually measured for the presence of the virus particles by detection of the dsRNA segments using the agarose gel electrophoresis. The fraction containing virus particles was carefully collected and suspended in 100 μl of 0.05 M sodium phosphate buffer (pH 7.0). The virus particles were stained with phosphotungstic acid solution (20 g/liter [wt/vol], pH 7.4) and observed under TEM. dsRNA from viral particles was sequentially extracted with phenol, phenol-chloroform, and chloroform-isoamyl alcohol and fractionated by electrophoresis on a 1% (wt/vol) agarose gel. The SPs from viral particles were detected by electrophoresis on a 12% (wt/vol) polyacrylamide gel amended with 1% (wt/vol) sodium dodecyl sulfate (SDS).
Peptide mass fingerprinting (PMF).
The virus particle suspension, derived from B. porri strain Bc-72, was loaded on a 12% polyacrylamide gel amended with 1% SDS. After electrophoresis, the gel was stained with Coomassie brilliant blue G250. The protein bands on the gel were individually excised to make different gel samples. Each gel sample was sequentially destained with NH4HCO3 (100 mM) and 30% (wt/vol) acetonitrile (ACN). The gel samples were lyophilized, followed by rehydration in 5 μl of NH4HCO3 solution (50 mM) amended with 50 ng of trypsin (sequencing grade; Promega, Madison, WI). The mixtures in small tubes were bath incubated at 37°C overnight to digest the proteins. The resulting peptides from each gel sample were extracted three times with 1 ml of extraction buffer containing 0.1% (wt/vol) trifluoroacetic acid (TFA) and 60% ACN. The aqueous phase of the extracts for each sample was pooled and lyophilized. The lyophilized peptides mixture for each protein was dissolved in 0.1% TFA. The resulting solution was desalted and concentrated using ZipTip pipette tips (Millipore, Bedford, MA). The peptides were then dissolved in R-cyano-4-hydroxycinnamic acid (5 mg/ml) amended with 0.1% TFA and 50% ACN and analyzed by matrix-assisted laser desorption/ionization–time of flight mass spectrometry (MALDI-TOF-MS) and MALDI-TOF tandem MS on a 4800 Plus MALDI TOF/TOF analyzer (Applied Biosystems, Foster City, CA). The molecular weight of each peptide was compared to that deduced from the nucleotide sequences of ORF I on dsRNA-1 or ORF II on dsRNA-2 using the MASCOT program (Matrix Science, Ltd., London, United Kingdom).
Protoplast transfection.
The virus-free strain Bc-38 of B. porri was used as the recipient in this experiment. Hypovirulence-associated dsRNAs in strain Bc-72 could not be transmitted to from strain Bc-72 to strain Bc-38 in the horizontal transmission experiment of the present study. Protoplasts were prepared from actively growing mycelia of strain Bc-38 using the procedures described by Yelton et al. (54). The method described by Kanematsu et al. (27) was used to transfect protoplasts of strain Bc-38 with the purified virus particles from strain Bc-72 in the presence of polyethylene glycol (PEG) 4000. The transfected protoplast suspension was incubated without shaking at 20°C in the dark for 2 days and plated on YCGA medium (0.1% yeast extract, 0.1% casein hydrolysate, 0.5 M glucose, 1.5% agar) in petri dishes (9 cm in diameter). This was followed by incubation at 20°C for 3 days. Agar plugs containing mycelia generated from the protoplasts were individually transferred to PDA plates at one plug per plate. The dishes were incubated at 20°C in the dark for 12 days, and the resulting cultures were identified and screened for abnormal colonies (slow growth and formation of mycelial sectors). A representative virus-transfected isolate 38T was tested for mycelial growth rate (20°C) on PDA, pathogenicity on detached garlic leaves (20°C, 72 h), and the presence of dsRNAs in the mycelia. The parental strain Bc-38 was used as a control strain in these tests.
Statistical analysis.
Data on mycelial growth rate and leaf lesion length caused by strains of B. porri were analyzed using analysis of variance in SAS V8.0 (SAS Institute, Cary, NC). Treatment means on each of these two parameters for the tested strains or isolates were compared using the least-significant-difference test at α = 0.05. Significant differences between strains Bc-38 and 38T of B. porri in each of the two parameters were detected by using a Student t test at α = 0.01.
RESULTS
Virus particles.
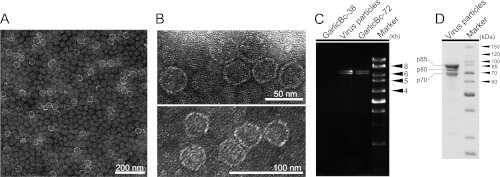
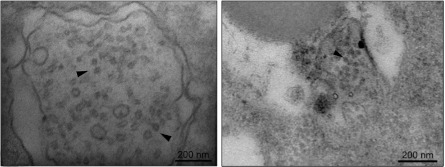
The virus particles purified from strain Bc-72 of B. porri were isometric and ∼35 nm in diameter (Fig. 2A and B). The virus particles accommodate two dsRNA segments with similar sizes to dsRNA-1 and dsRNA-2 extracted from mycelia of strain Bc-72 (Fig. 2C). SDS-PAGE of the purified particles revealed the presence of three major proteins with estimated molecular mass of 70, 80, and 85 kDa designated p70, p80, and p85, respectively (Fig. 2D). It is worth noting that the virion dsRNA segments showed brightness similar to that of the two dsRNAs extracted from the mycelia, suggesting that the two dsRNAs occur at the similar molar ratios in both cases.
Fig 2.
BpRV1 particles from strain GarlicBc-72 of B. porri. (A and B) TEM images (negative staining) of the virus particles of BpRV1. (C) Agarose gel electrophoresis of the dsRNAs extracted from purified virus particles of BpRV1 and from the mycelia of strains GarlicBc-72 and GarlicBc-38. Marker, 1-kb DNA ladder. Note the similar size for the dsRNAs from the purified virus particles and from the mycelia. (D) SDS-PAGE analysis of purified virus particles of BpRV1 showing three protein components (p85, p80, and p70).
Nucleotide sequences of dsRNA-1 and dsRNA-2.
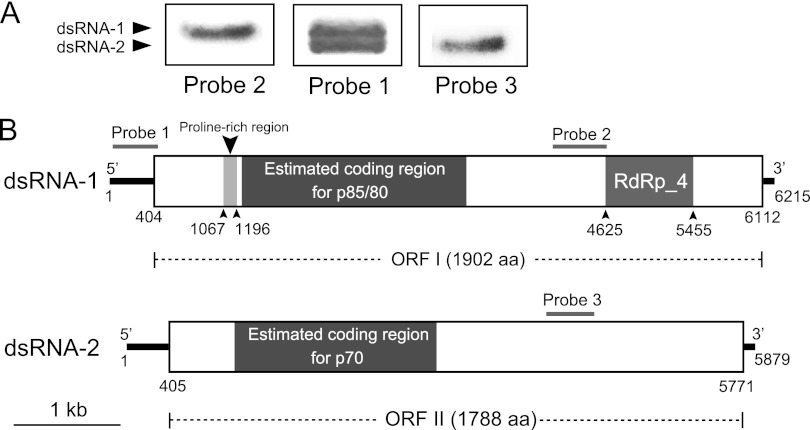
A total of 21 cDNA clones were obtained using random hexameric primers. Assembly sequences of these cDNA clones generated two contigs. Contig 1 comprised 13 clones belonging to dsRNA-1, and contig 2 comprised 8 clones belonging to dsRNA-2 (see Fig. S1 in the supplemental material). No other cDNA clones were found to make additional contigs, suggesting that no other dsRNAs in strain Bc-72 have the similar size to dsRNA-1 or dsRNA-2. The full-length cDNA sequences for dsRNA-1 (GenBank accession no. JF716350) and dsRNA-2 (GenBank accession no. JF716351) were 6,215 and 5,879 bp in length, respectively (Fig. 3). Interestingly, we observed that the nucleotide sequences of dsRNA-1 and dsRNA-2 at the 3′-termini (80-bp) and the 5′ termini (500-bp) shared sequence identities of 62 and 95%, respectively (Fig. 1A and B). Moreover, the extreme terminal sequences of dsRNAs are strictly conserved. The results of the Northern hybridization analysis confirmed the high nucleotide sequence identity at the 5′ termini of dsRNA-1 and dsRNA-2 (Fig. 3A). Probe 1 (423 bp) corresponding to sequences at the 5′ terminus of dsRNA-1 simultaneously hybridized with both dsRNA-1 and dsRNA-2. However, probe 2 (506 bp) corresponding to sequences at the middle region of dsRNA-1 hybridized only with dsRNA-1 but did not hybridize with dsRNA-2. Similarly, probe 3 (451 bp) corresponding to sequences at the middle region of dsRNA-2 hybridized only with dsRNA-2.
Fig 3.
Molecular characteristics of dsRNA-1 and dsRNA-2 of BpRV1. (A) Northern blot detection of dsRNA-1 and dsRNA-2 using digoxigenin-labeled probes 1, 2, and 3. The locations of these probes are indicated in panel B. (B) Schematic diagrams of the genetic organization of BpRV1 dsRNA-1 and dsRNA-2. The coding strand of dsRNA-1 is 6,215 bp long and comprises one large ORF, designated ORF I, which encodes a polyprotein of 1,902 aa. The coding strand of dsRNA-2 is 5,788 bp long and also comprises one large ORF, designated ORF II, which encodes a polypeptide of 1,788 aa. p70, p80, and p85 are the structural proteins of BpRV1 inferred from peptide fingerprinting analyses.
The 5′- and 3′-untranslated regions of dsRNA-1 and dsRNA-2 were examined for potential secondary structures using the RNA structure software (version 4.6) (32). The results showed that formation of a stem-loop structure was detected at the 3′- and 5′-terminal regions of dsRNA-1 and at the 5′-terminal region of dsRNA-2 but was not detected at the 3′ terminal region of dsRNA-2 (Fig. 1C). The results also showed that both the 3′- and 5′-terminal regions of dsRNA-1 and dsRNA-2 lack the inverted complementarity required for the formation of any panhandle structures.
Analysis of the putative polypeptides.
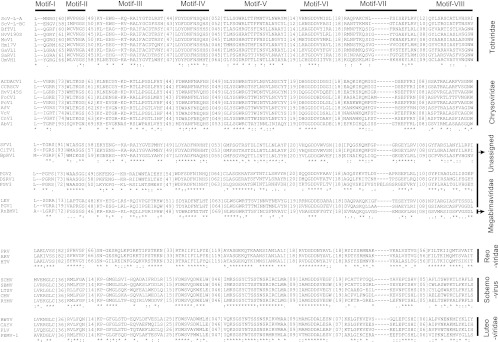
Both strands of the full-length cDNAs of dsRNA-1 and dsRNA-2 were examined for ORFs. Two ORFs—ORF I and ORF II—were found on the positive strand of dsRNA-1 (positions 404 to 6112) and dsRNA-2 (positions 405 to 5771), respectively (Fig. 3B). ORF I was deduced to encode a polypeptide with 1,902 aa. It has a proline-rich region at positions 221 to 264 (see Fig. S2 in the supplemental material), corresponding to the nucleotides (nt) 1067 to 1196 (Fig. 3B). It also contains RNA-dependent RNA polymerase (RdRp)_4 surperfamily domain at nt 4625 to 5455 (Fig. 3). The predicted RdRp sequence includes the eight conserved motifs (I to VIII) similar to the RdRp sequences of viruses in the families Totiviridae, Chrysoviridae, Megabirnaviridae, and a few unassigned virus taxa (Fig. 4). Therefore, dsRNA-1 and dsRNA-2 are proposed as the genome segments of a mycovirus. We designated this mycovirus as Botrytis porri RNA virus 1 (BpRV1). The results from BLASTp analysis showed that the RdRp sequence has low levels of sequence similarity (<25%) to the RdRp sequences of viruses in the families Totiviridae, Chrysoviridae, and Megabirnaviridae, and as well as to those of a few unassigned virus taxa (Table 2). With the exception of the RdRp sequence, no proteins homologous to the remaining part of ORF I-encoded polypeptide were identified in the database of the NCBI. Moreover, the polypeptide of ORF I was searched for homologous proteases in the MEROPS protease database. However, no hits with E values less than 0.01 were detected.
Fig 4.
Multiple alignment of RdRp sequences of BpRV1 and other RNA viruses showing the structures of the eight motifs (I to VIII). The alignment was made using CLUSTAL_ X. “*” indicates identical amino acid residues; “:” and “.” indicate high and low chemically similar amino acid residues, respectively. Abbreviations: UmVH1, Ustilago maydis virus H1 (GenBank accession no. NC_003823); ScV-l-A, Saccharomyces cerevisiae virus L-A (J04692); ScV-l-BC, Saccharomyces cerevisiae virus L-BC (La) (U01060); HvV145S, Helminthosporium victoriae virus 145S (AF297176); PcV, Penicillium chrysogenum virus (AF296339); FoCV1, Fusarium oxysporum chrysovirus 1 (EF152346); AfV, Aspergillus fumigatus chrysovirus (FN178512); CnV1, Cryphonectria nitschkei chrysovirus 1 (GQ290650); ACDACV, Amasya cherry disease-associated chrysovirus (NC_009947); VcV, Verticillium chrysogenum virus (HM004067); AbV1, Agaricus bisporus virus 1 (X94361); CCRSCV, Cherry chlorotic rusty spot-associated chrysovirus (AJ781397); Hm17V, Helicobasidium mompa no. 17 dsRNA virus (AB085814); SsRV1, Sphaeropsis sapinea RNA virus 1 (NC_001963); MoV1, Magnaporthe oryzae virus 1 (AB176964); GaVL1, Gremmeniella abetina RNA virus L1 (AF337175); BfTV1, Botryotinia fuckeliana totivirus 1 (AM491608); HvV190S, Helminthosporium victoriae virus 190S (U41345); RnBMV1, Rosellinia necatrix megabirnavirus 1 (AB512282); SFV1, Spissistilus festinus virus 1 (GU979419); CiTV1, Cirulifer tenellus virus 1 (GU979420); PGV1, Phlebiopsis gigantea mycovirus dsRNA 1 (AM111096); FgV3, Fusarium graminearum dsRNA mycovirus 3 (NC_013469); DsRV1, Diplodia scrobiculata RNA virus 1 (NC_013699); PGV2, Phlebiopsis gigantea mycovirus dsRNA 2 (AM111097); LEV, Lentinula edodes mycovirus HKB (AB429554); PRV, Porcine rotavirus (Q85036); HTV, Human rotavirus (Q91E95); ARV, Avian rotavirus PO-13 (O55590); CMV, Cocksfoot mottle virus (Q66150); RYMV, Rice yellow mottle virus (Q86519); SCMV, Southern cowpea mosaic virus (P21405); SBMV, Southern bean mosaic virus (O72157); LTSV, Lucerne transient streak virus (Q83093); BWYV, Beet western yellows virus (P09507); CAYV, Cucurbit aphid-borne yellows virus (Q65969); PLV, Potato leafroll virus (P11623); PEMV-1, Pea enation mosaic virus 1 (P29154).
Table 2.
Summary of results of a BLASTP search for RdRp encoded by ORF I on dsRNA-1 of Botrytis porri RNA virus 1 (BpRV1)
| Virusa | RdRp size (aa) | RdRp_4 conserved region |
BLASTP |
GenBank accession no. | ||||
|---|---|---|---|---|---|---|---|---|
| Motif (I-VIII)b | Length (aa) | % Identity | Overlap | Bit score | E value | |||
| Totivirus (Totiviridae) | ||||||||
| UmVH1c | 1,820 | 1142–1530 | 389 | 23 | 132/506 | 154 | 1e–34 | NC_003823 |
| ScV-L-A | 731 | 169–520 | 352 | 19 | 67/254 | 73.2 | 4e–10 | J04692 |
| ScV-L-BC | 863 | 306–654 | 349 | 20 | 98/424 | 67.0 | 3e–08 | U01060 |
| Chrysovirus (Chrysoviridae) | ||||||||
| HvV145S | 1,086 | 426–805 | 380 | 23 | 164/616 | 121 | 1e–24 | AF297176 |
| PcV | 1,117 | 453–832 | 380 | 23 | 145/563 | 115 | 1e–22 | AF296339 |
| FoCV1 | 858 | 360–738 | 379 | 21 | 133/527 | 99.8 | 4e–18 | EF152346 |
| AfV | 1,114 | 451–829 | 379 | 21 | 135/561 | 97.1 | 3e–17 | FN178512 |
| CnV1 | 962 | 442–822 | 381 | 20 | 99/412 | 89.4 | 5e–15 | GQ290650 |
| ACDACV | 1,087 | 427–806 | 380 | 21 | 121/474 | 91.7 | 1e–15 | NC_009947 |
| VcV | 1,108 | 443–823 | 381 | 20 | 122/513 | 87.8 | 1e–14 | HM004067 |
| AbV1 | 1,078 | 372–785 | 414 | 20 | 62/238 | 67.8 | 2e–08 | X94361 |
| CCRSCV | 1,087 | 427–806 | 380 | 21 | 64/282 | 60.5 | 3e–06 | AJ781397 |
| Victorivirus (Totiviridae) | ||||||||
| Hm17V | 845 | 243–594 | 352 | 21 | 115/469 | 87.0 | 3e–14 | AB085814 |
| SsRV1 | 838 | 241–589 | 349 | 22 | 74/256 | 79.0 | 8e–12 | NC_001963 |
| MoV1 | 845 | 227–575 | 353 | 20 | 98/407 | 78.2 | 1e–11 | AB176964 |
| GaVL1 | 825 | 221–573 | 353 | 20 | 103/427 | 68.6 | 1e–08 | AF337175 |
| BfTV1 | 838 | 242–585 | 344 | 21 | 57/219 | 67.4 | 3e–08 | AM491608 |
| HvV190S | 835 | 241–585 | 345 | 21 | 64/241 | 59.7 | 5e–06 | U41345 |
| Megabirnavirus (Megabirnaviridae) | ||||||||
| RnBMV1 | 1,111 | 378–765 | 388 | 21 | 95/389 | 95.9 | 6e–17 | AB512282 |
| Unassigned | ||||||||
| SFV1 | 1,326 | 537–941 | 405 | 25 | 125/468 | 139 | 7e–30 | GU979419 |
| CiTV1 | 1,338 | 550–953 | 404 | 23 | 145/591 | 108 | 6e–21 | GU979420 |
| PGV1 | 1,414 | 669–1061 | 393 | 20 | 79/287 | 96.3 | 5e–17 | AM111096 |
| FgV3 | 1,311 | 383–774 | 392 | 19 | 97/367 | 93.6 | 3e–16 | NC_013469 |
| DsRV1 | 1,110 | 227–620 | 394 | 20 | 89/367 | 67.4 | 2e–08 | NC_013699 |
| PGV2 | 1,153 | 230–624 | 395 | 18 | 87/374 | 66.2 | 5e–08 | AM111097 |
| LEV | 1,245 | 491–878 | 388 | 18 | 64/287 | 52.8 | 6e–04 | AB429554 |
See the legend to Fig. 4 for definitions of the virus abbreviations.
Numbers refer to the amino acid position.
The cap-pol fusion protein of this virus was used for analysis.
ORF II on dsRNA-2 was deduced to encode a polypeptide with 1,788 aa (Fig. 3B). The polypeptide sequence was detected to contain numerous phosphorylation/glycosylation sites. No conserved proteins homologous to this polypeptide were found in the database of NCBI. Moreover, no hits indicating the presence of any known homologous proteases on this polypeptide were detected in the MEROPS protease database.
The three proteins (p70, p80, and p85) detected in the purified virus particles of BpRV1 were individually cleaved with trypsin, and the resulting peptide fragments for each protein were subjected to PMF analysis. The results showed that p70, p80, and p85 generated a total of 13, 23, and 22 peptide fragments, respectively (see Tables S2, S3, and S4 in the supplemental material). Of these peptide fragments, 11 from p70 matched the partial peptide sequence encoded by ORF II at amino acids 200 to 850, accounting for 30.7% of the entire coverage (651 aa). Four peptide fragments matched the deduced sequences HSVNLWSYRPTSAQSR (aa 319 to 334), AVNTGIGYTSPR (aa 337 to 348), TDLSIVPTYYTYQTDIR (aa 505 to 521), and YLEETFTTGR (aa 714 to 723), with ion scores higher than 46 (>95% confidence) (see Table S4 in the supplemental material). Twenty-one peptide fragments from p80 and eighteen peptide fragments from p85 matched the partial peptide sequence encoded by ORF I at aa 200 to 1050, accounting for 41.9 and 30.7% of the entire coverage (851 aa), respectively. Seven peptide fragments from these two proteins matched the deduced sequences, including DTIAYNVPFGMSVYNR (aa 407 to 422), FLLPVEDGDDAPIRK (aa 501 to 515), ITTYVTAGPGAPDLAVHSTIDLYVPEQR (aa 552 to 579), FTLCPLNGSELIEER (aa 580 to 594), YLTAHNLWQQFPVMR (aa 616 to 630), QILQQEYCTER (aa 807 to 817), and VAQENVGAVTYYQHEILPSESVSDTPVVR (aa 829 to 857), with ion scores higher than 46. Therefore, p80 might be derived from p85 via a posttranslational modification in a manner comparable to that of the related capsid proteins of the victorivirus Helminthosporium victoriae virus 190S (HvV190S) (24).
Phylogenetic analysis.
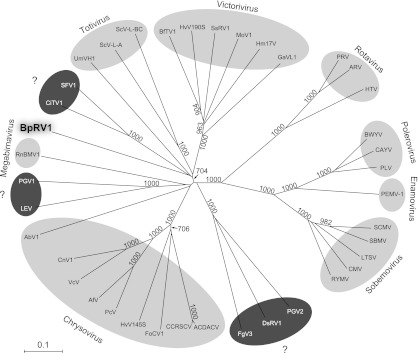
A phylogenetic tree based on RdRp sequences of BpRV1 and 38 selected RNA viruses was generated using the NJ method (Fig. 5). Since no members of the reported bipartite dsRNA viral families Birnaviridae, Partitiviridae, and Picobirnaviridae were identified through BLAST searching (Table 2), these viruses were not selected to establish the phylogenetic tree. Phylogenetic analysis revealed that BpRV1 appears to be more closely related to members of the genera Totivirius and Victorivirus (both in the family Totiviridae) than to members of the families Chrysoviridae and Megabirnaviridae. BpRV1 clustered with two unassigned insect viruses, Spissistilus festinus virus 1 (SFV1) and Cirulifer tenellus virus 1 (CiTV1), forming the BpRV1-SFV1-CiTV1 clade, which was more closely related to Ustilago maydis virus H1 (UmVH1) than to ScV-l-A and ScV-LBC, forming the BpRV1-SFV1-CiTV1-UmVH1 clade. However, the bootstrap support for generation of the two clades was weak, i.e., 408 and 343 (1,000 replicates) for support of the clades of BpRV1-SFV1-CiTV1 and BpRV1-SFV1-CiTV1-UMVH1, respectively.
Fig 5.
Phylogenetic analysis of BpRV1 and 38 selected RNA viruses presented in an NJ tree inferred from the RdRp sequences. Animal viruses (rotaviruses) and plant viruses (poleroviruses, enamoviruses, and sobemoviruses) with their RdRps belonging to the RdRp_4 superfamily were included as outgroups. Numbers at the nodes are bootstrap values out of 1,000 replicates. Bootstrap values lower than 700 are not shown in the figure. The taxonomic status of the viruses labeled with the question mark (?) has not been determined. See the legend to Fig. 4 for definitions of the virus abbreviations.
Hypovirulence, abnormal colony morphology, and dsRNAs.
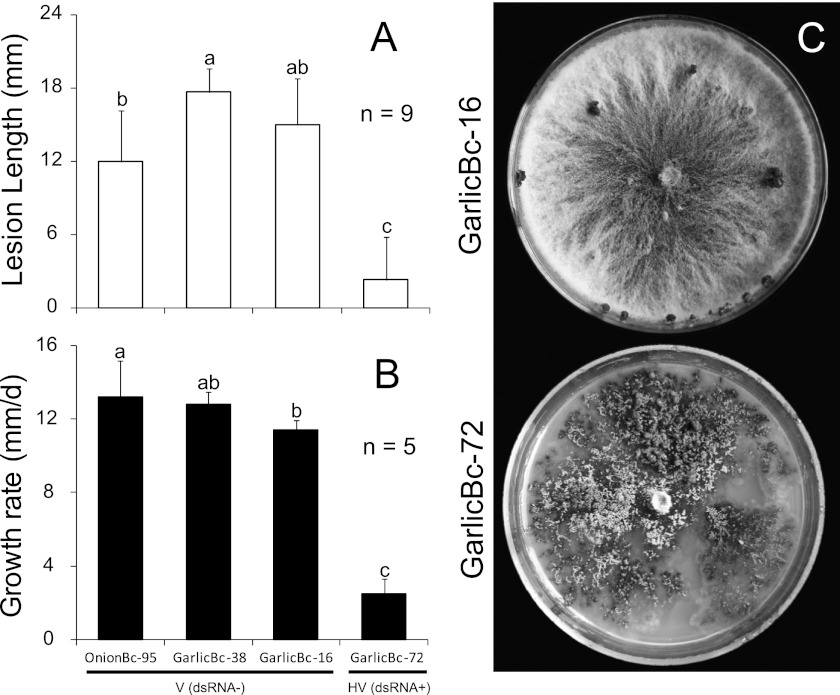
Strains Bc-16, Bc-38, and Bc-95 of B. porri, which lack detectable dsRNAs, caused large necrotic lesions on garlic leaves (20°C, 72 h) with average lesion lengths of 12 to 17 mm (Fig. 6A). On PDA, they grew at radial growth rates ranging from 11.4 to 13.2 mm/day under 20°C (Fig. 6B) and formed whitish-gray colonies and black cerebriform sclerotia after incubation for 17 days (Fig. 6C). In contrast, strain Bc-72 of B. porri harboring two dsRNA segments caused small lesions on garlic leaves with an average lesion length of 2.3 mm (Fig. 6A). On PDA, it grew at a radial growth rate of 2.3 mm/day under 20°C (Fig. 6B) and produced grayish-brown colonies with the formation of numerous mycelial sectors at the colony margin (Fig. 6C). No sclerotia were formed on the colonies of strain Bc-72 after incubation for 17 days (Fig. 6C).
Fig 6.
Leaf lesion length on garlic, mycelial growth rate on PDA, and the colony morphology of different strains of B. porri. (A) Average lesion length caused by strains GarlicBc-16, GarlicBc-38, GarlicBc-72, and OnionBc-95 on detached garlic leaves (20°C, 72 h). (B) Average radial growth rates of these strains of B. porri on PDA at 20°C. (C) Cultures of strains GarlicBc-16 and GarlicBc-72 on PDA (20°C, 17 days). In histograms A and B, the results are expressed as arithmetic means ± the standard errors of the means. Bars in each histogram labeled with the same letters are not significantly different (P > 0.05) according to the least-significant-difference test. dsRNA−, dsRNA negative; dsRNA+, dsRNA positive; V, virulent; HV, hypovirulent.
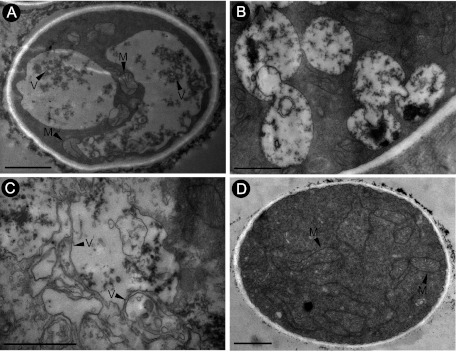
Hyphae of strains Bc-16 and Bc-72 were sampled both from the colony margin area and from the colony center area. Ultrathin hyphal sections for each strain were observed under TEM. The cytoplasm of hyphal compartments in strain Bc-72 showed a degeneration syndrome manifested by formation of abundant vacuole-like membranous structures and small membranous vesicles (Fig. 7A, B, and C). In most cases, the membranous vacuoles/vesicles aggregated and merged. In contrast, the cytoplasm of hyphal compartments of comparable age from strain Bc-16 was densely stained and evenly distributed without formation of abundant and aggregated membranous vacuoles or vesicles (Fig. 7D). Moreover, virus-like particles with a size similar to that of the virus particles of BpRV1 were observed in the hyphal cells of Bc-72 (Fig. 8).
Fig 7.
TEM images showing the cytoplasmic characteristics of the hypovirulent strain GarlicBc-72 and the virulent strain of B. porri. The mycelial samples examined were collected from the colony margin area of each strain. (A, B, and C) Three hyphal cells of strain GarlicBc-72 show a degeneration sign characterized by formation of abundant vacuole-like membranous structures and small membranous vesicles. (D) Hyphal cell of strain GarlicBc-16 with dense and evenly distributed cytoplasm. V, vacuoles or vesicles; M, mitochondrion. Scale bar, 1 μm.
Fig 8.
TEM images of hyphal cells of the hypovirulent strain GarlicBc-72 containing virus-like particles inside the vesicles (arrowheads).
Vertical transmission of hypovirulence-associated dsRNAs.
Thirty-five SC isolates derived from strain Bc-72 were obtained. Thirty-four SC isolates (GP72SC01 to GP72SC34) were determined to be BpRV1 positive, and only one SC isolate named GP72SC35 (shortened as SC35) was determined to be BpRV1 negative (Fig. 9). On PDA, all of the BpRV1-postive SC isolates grew at the average mycelial growth rates ranging from 1.0 to 6.4 mm/day, which is significantly lower (P < 0.05) than for the BpRV1-free strain Bc-16 and isolate SC35 (9.9 mm/day). On garlic leaves, they produced no lesions or small lesions, with the average length ranging from 0 to 3.6 mm, which is significantly shorter (P < 0.05) than the average lesion lengths for strain Bc-16 (15.1 mm) and isolate SC35 (11.7 mm). We also found that the average lesion length caused by the BpRV1-positive SC isolates were not significantly correlated with the average mycelial growth rates of these isolates (r2 = 10−7, P > 0.01). Therefore, BpRV1 in strain Bc72 can be vertically transmitted from mycelia to conidia, and the hypovirulence phenotype appears to be qualitatively correlated with the transmission of BpRV1.
Fig 9.
Lesion length on detached garlic leaves (20°C, 72 h) (A) and mycelial growth rates on PDA at 20°C (B) for strains GarlicBc-16 and GarlicBc-72 of B. porri and 35 SC isolates (GP72SC01 to GP72SC35) of strain GarlicBc-72. The results are expressed as arithmetic means ± the standard errors of the means. In each histogram, bars labeled with the same letters are not significantly different (P > 0.05) according to the least-significant-difference test. The symbols “+” and “−” indicate the presence and absence of dsRNA-1/dsRNA-2, respectively. The dsRNAs were extracted from the mycelia of each strain or isolate and were detected by agarose gel electrophoresis.
Horizontal transmission of hypovirulence-associated dsRNAs.
The results of the horizontal transmission experiment showed that in the single cultures, strains Bc-38, Bc-95, and SC35 grew rapidly on PDA and colonized the entire dishes after incubation for 8 days (20°C), whereas strain Bc-72 grew slowly and formed small abnormal colonies (Fig. 10). In the contact cultures of Bc-72/Bc-38 and Bc-72/Bc-95, strains Bc-38 and Bc-95 still grew rapidly and colonized the entire dishes after 8 days. In the contact cultures of Bc-72/SC35, strain SC35 grew rapidly in the first 3 days, followed by reduced growth and formation of mycelial sectors at the colony margin (Fig. 10). However, strain SC35 still failed to colonize the entire dishes in the contact cultures of Bc-72/SC-35 after incubation for 8 days.
Fig 10.
Transmission of BpRV1 from the hypovirulent strain GarlicBc-72 to virulent strain GP72SC35 of B. porri on PDA. All of the dishes were incubated at 20°C for 8 days. In the single cultures (top row), strains GP72SC35 and OnionBc-95 colonized the entire dishes, whereas strain GarlicBc-72 formed a small colony. In the contact culture of OnionBc-95/GarlicBc-72 (bottom row), OnionBc-95 colonized the entire dish, indicating unsuccessful transmission, since contact of the hyphae of GarlicBc-72 with the hyphae of GarlicBc-95 had no effect on the mycelial growth of GarlicBc-95. However, in the contact culture of GP72SC35/GarlicBc-72, GP72SC35 colonized only a part of the dish, with formation of mycelial sector along the colony margin, indicating successful transmission, since the contact of the hyphae of GarlicBc-72 with the hyphae of GP72SC35 produced a dramatic effect on mycelial growth of GP72SC35. The symbol “*” indicates the place to remove a mycelial agar plug for generation of a derivative isolate for GP72SC35 or OnionBc-95 transfected by BpRV1.
Three mycelial derivative isolates of strain SC35 (72-35-1, 72-35-2, and 72-35-3) were obtained from three recipient colonies of SC35 in the three contact cultures of Bc-72/SC35 (Fig. 10). They were similar to strain Bc-72 in mycelial growth on PDA, with average growth rates of 2.2 to 3.4 mm/day, and in pathogenicity on garlic leaves, with average leaf lesion lengths of 1.1 to 2.4 mm (see Fig. S3 in the supplemental material). However, these derivatives differed greatly from their parental strain SC35, which grew at 10.2 mm/day on PDA and caused lesions of 12.3 mm in length on garlic leaves. Both dsRNA-1 and dsRNA-2 were detected in 72-35-1, 72-35-2, and 72-35-3, as well as in strain Bc-72, but were not detected in strain SC35. Therefore, BpRV1 in strain Bc-72 can be horizontally transmitted to strain SC35 through hyphal contact, and the hypovirulence phenotype appears qualitatively correlated with virus transmission.
Three mycelial derivative isolates of strain Bc-38 (72-38-1, 72-38-2, and 72-38-3) were obtained from the three recipient colonies of strain Bc-38 in the contact cultures of Bc-72/Bc-38 (data not shown). Three mycelial derivatives of strain Bc-95 (72-95-1, 72-95-2, and 72-95-3) were obtained from the three recipient colonies of strain Bc-95 in the contact cultures of Bc-72/Bc-95 (Fig. 10). All of these mycelial derivative isolates were similar to the corresponding parental strains Bc-38 and Bc-95, both in mycelial growth rate on PDA (20°C) and in leaf lesion length on garlic leaves (see Fig. S3 in the supplemental material). No dsRNA was detected in these derivative isolates, as well as in their parental strains Bc-38 and Bc-95. Therefore, BpRV1 in strain Bc-72 cannot be horizontally transmitted to strains Bc-38 or Bc-95 through hyphal contact.
Virulence attenuation as a consequence of transfection with purified virions.
The contact culture experiments aiming at transmission of the hypovirulence-associated dsRNAs from strain Bc-72 to strain Bc-38 (BpRV1 free) was unsuccessful (see Fig. S3 in the supplemental material). As an alternative, we tried to transfect protoplasts of strain Bc-38 with the purified BpRV1 particles in the presence of PEG 4000. A derivative isolate 38T was obtained from the generated protoplasts of strain Bc-38. It grew at the average radial rate of 4.3 mm/day on PDA at 20°C with formation of abnormal whitish compacted colonies (Fig. 11A) and was hypovirulent on garlic leaves with an average lesion length of 3.3 mm (20°C, 72 h) (Fig. 11). Two dsRNA segments present in strain Bc-72 were consistently detected in the mycelia of 38T (data not shown). The identity of the dsRNAs in 38T as the genome of BpRV1 was confirmed using RT-PCR with the BpRV1-specific primers BVP1F/BVP1R (see Table S1 in the supplemental material).
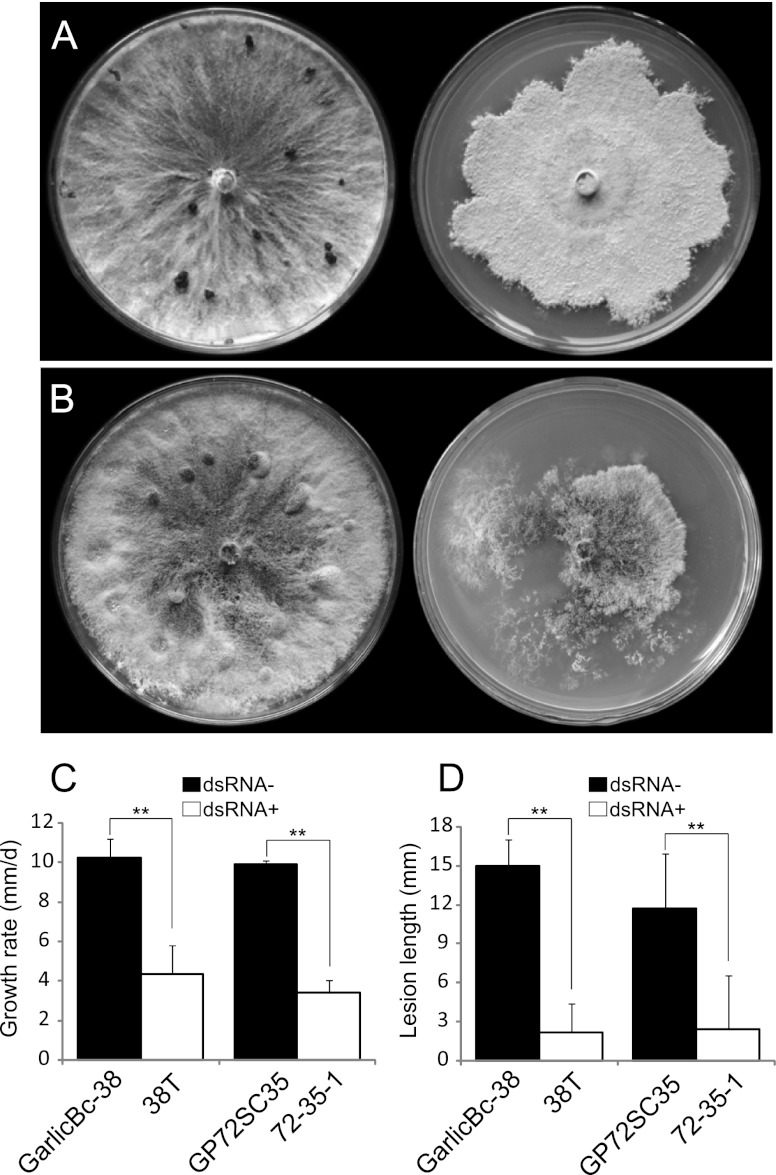
Fig 11.
Phenotypic effects of BpRV1 on fungal colony morphology, mycelial growth rate, and virulence in strains GarlicBc-38 and GP72SC35 of B. porri. (A) Comparison of the colony morphology between the BpRV1-free strain GarlicBc-38 (left) and the BpRV1-transfected isolate 38T (right) (20°C, 17 days). (B) Comparison of the colony morphology between BpRV1-free isolate GP72SC35 (left) and the BpRV1-infected isolate 72-35-1 (right) (20°C, 17 days). (C) Mycelial growth rates of strains GP72SC35, 72-35-1, GarlicBc-38, and 38T on PDA (20°C). (D) Lesion length caused by strains GP72SC35, 72-35-1, GarlicBc-38, and 38T on garlic leaves (20°C, 72 h). The results in each histogram are expressed as arithmetic means ± the standard errors of the means. “**” indicates a significant difference (P < 0.01) between strains GarlicBc-38 and 38T and between strains GP72SC35 and 72-35-1 in mycelial growth rate or lesion length according to the Student t test.
DISCUSSION
The present study reveals that hypovirulence of strain Bc-72 of B. porri is caused by infection with a novel bipartite dsRNA mycovirus (BpRV1). To our knowledge, this is the first report about the co-occurrence of a dsRNA mycovirus and hypovirulence in B. porri. This study provided three lines of experimental evidence supporting the conclusion that BpRV1 is the causal agent of hypovirulence in B. porri. The first evidence is based on the vertical transmission experiment. Thirty-four BpRV1-infected SC isolates of strain Bc-72 were hypovirulent on garlic leaves, whereas the BpRV1-free SC isolate SC35 became virulent on garlic leaves (Fig. 9). The second evidence is derived from the horizontal transmission experiment, in which BpRV1 was transmitted to the virus-cured strain SC35 through hyphal contact (Fig. 10). The three derivative isolates of strain SC35 showed reduced mycelial growth rates on PDA and debilitated virulence on garlic leaves (see Fig. S3 in the supplemental material). The third evidence is from the transfection experiment, in which the purified BpRV1 particles were successfully introduced into the protoplasts of the BpRV1-free virulent strain Bc-38 of B. porri. The transfected derivative isolate 38T showed debilitation symptoms, including reduced mycelial growth on PDA and hypovirulence on garlic leaves (Fig. 11A).
It is of interest that the 34 BpRV1-infected hypovirulent SC isolates of strain Bc-72 varied greatly both in mycelial growth rates (1.0 to 6.4 mm/day) on PDA and in leaf lesion length (0 to 3.6 mm) on garlic leaves. We interpret this kind of variation as interisolate variation, which might be caused by different levels of virus accumulation in different SC isolates. We also observed that there was no significant correlation (P > 0.01) between the two parameters. This result might be due to the unstable cultural characteristics for some SC isolates. We call this kind of variation intraisolate variation, which might be caused by the uneven distribution of BpRV1 in hyphal cytoplasm. We observed that the subcultures of even the same SC isolate differed greatly in colony morphology on PDA, with the colonies of some subcultures showing a normal appearance (slightly attenuated) and the colonies of some subcultures showing a very abnormal appearance (severely attenuated) (M. D. Wu and G. Q. Li, unpublished data). The intraisolate variability may mask or obscure the correlative relationship between the mycelial growth rates and the leaf lesion length values. We also observed that the hypovirulent isolate 38T infected by BpRV1 (protoplast infection) showed dramatic differences from the hypovirulent strain GarlicBc-72 and its hypovirulent derivatives, such as 72-35-1 (Fig. 11) in colony morphology on PDA, although the mycelial growth of all of these hypovirulent isolates was impaired. The difference between the hypovirulent strains 38T and 72-35-1 in colony morphology might be caused by different genetic backgrounds.
The results of the horizontal transmission experiment showed that the transmission of BpRV1 was successful from strain Bc-72 to virulent strain SC35 of B. porri but was unsuccessful from Bc-72 to two other virulent strains (Bc-38 and Bc-95) of B. porri. Therefore, the strain-specific transmission manner appears to be similar to the horizontal transmission of Botrytis cinerea mitovirus 1 (BcMV1) (51, 52). The unsuccessful horizontal transmission of BpRV1 might be due to the vegetative incompatibility between the hypovirulent strain Bc-72 and the virulent strains Bc-38 and Bc-95. Strain SC35, an SC isolate of strain Bc-72, is isogenic to strain Bc-72, and they may have the same or similar alleles at the vegetative incompatibility (vic) loci, conferring a compatible interaction between hyphal cells of strains Bc-72 and SC35. In contrast, strains Bc-38, Bc-72, and Bc-95 were collected from different geographic locations in China (Table 1). They might have different alleles at the vic loci, thus perhaps conferring an incompatible interaction between hyphal cells and failure of BpRV1 transmission.
Previous studies showed that it is often difficult to establish the etiology of a mycovirus as an agent for hypovirulence in phytopathogenic fungi due to the lack of a proper infection method to complete the Koch's postulates (9). Two approaches have been successfully used to solve this problem. The first approach is to prepare virus-derived cDNA transcripts (7) or to construct infectious viral cDNA clones (10, 28). Both the synthetic cDNA transcripts and the infectious viral cDNA clones can initiate mycovirus infection in host fungi. This approach has been successfully applied in a hypovirus-C. parasitica system to ascertain the determinant role of hypovirus in hypovirulence of C. parasitica (7, 10, 28). Another approach is to transfect cells or protoplasts of host fungi with purified virus particles. A PEG-mediated transfection method with virus particles has been developed for a mycovirus in B. cinerea (6), for mycoreviruses in Cryphonectria parasitica (20) and Rosellinia necatrix (43), for a megabirnavirus (9) and a partitivirus (42) in R. necatrix, and for an ssDNA mycovirus in Sclerotinia sclerotiorum (55). In the present study, we successfully introduced the purified BpRV1 particles into virus-free strain Bc-38 (Fig. 11). This provides an additional example showing the potential of using the PEG-mediated mycovirus transfection technique to study the etiology of mycoviruses for fungal hypovirulence. This technique might even be useful for determining the experimental host range of BpRV1 in the future.
Sequence analysis indicates that dsRNA-1 and dsRNA-2 in strain Bc-72 comprise the genome of a bipartite virus. Only dsRNA-1 encodes an RdRp, implying that the two dsRNAs are not derived from two independent mycoviruses. Previous studies showed that many multipartite RNA viruses have conserved terminal regions (9, 25, 49). This phenomenon was observed in the two dsRNAs of BpRV1, which has conserved terminal regions (Fig. 1). This result suggests that dsRNA-2 of BpRV1 may depend on the RdRp encoded by ORF I on dsRNA-1 of BpRV1 for replication. We observed that the remaining regions on the two dsRNAs of BpRV1 have low sequence similarity (<40%), implying that dsRNA-2 is not a defective RNA of dsRNA-1. In addition, the genome size of BpRV1 appears different from that of other reported bipartite dsRNA viruses, including those in the Birnaviridae (hosts: vertebrates and invertebrates), Partitiviridae (hosts: fungi and plants), Picobirnaviridae (hosts: vertebrates), and Megabirnaviridae (hosts: fungi). The genome size of BpRV1 is ∼12 kb, smaller than that of Megabirnaviridae (∼17 kb) (9) but larger than those of Partitiviridae (2.8 to 4.6 kb) (16), Picobirnaviridae (∼4 kb) (9), and Birnaviridae (∼6 kb) (9).
The RdRp of BpRV1 contains eight conserved motifs, which appear similar to the conserved motifs of the viral RdRps in Totiviridae, Chrysoviridae, Megabirnaviridae, and a few taxonomically unassigned viruses, including SFV1 and CiTV1 (Fig. 4) but are different from the RdRp motifs of other bipartite viruses (Birnaviridae, Partitiviridae, and Picobirnaviridae). However, the RdRp sequence of BpRV1 shows low sequence similarity levels (<25%) to the RdRp sequences of the mycoviruses in Totiviridae, Chrysoviridae, and Megabirnaviridae (Table 2). In the phylogenetic tree inferred from the viral RdRp sequences (Fig. 5), BpRV1 formed a separate clade distinct from those for Totiviridae, Chrysoviridae, Megabirnaviridae, and a few taxonomically unassigned virus taxa. It has a closer phylogenetic relationship to SFV1, CiTV1, and UmVH1 than to RnMBV1. However, BpRV1 with two dsRNA segments differs from these three viruses in the number of dsRNA segments since genomes of SFV1, CiTV1, and UmVH1 are monosegmented (26, 46). Although BpRV1 and RnMBV1 have bipartite dsRNA genomes and their RdRps share sequence similarities, BpRV1 differs from RnMBV1 in the expression strategy for the generation of RdRp and the structural proteins. For BpRV1, the RdRp of BpRV1 is more likely expressed as a fusion protein containing a part of the structural proteins (p80/p85) (Fig. 3). The other structural protein component (p70) is more likely encoded by ORF II. However, RnMBV1 has two ORFs on dsRNA-1: ORF 1 for capsid protein and ORF 2 for RdRp (9).
Taking together the information about the number of dsRNA segments, the RNA genome size, the genetic organization and the RdRp sequences, we believe that BpRV1 is a novel bipartite dsRNA mycovirus, which cannot be assigned to any known mycovirus families. Furthermore, BpRV1 has been detected in a strain of Botrytis squamosa Walker, the causal agent of onion leaf blight (M. D. Wu, unpublished data). The BpRV1-infected strain of B. squamosa was collected from Tongshan County of Hubei Province, China, ∼500 km away from Zhushan County, where strain Bc-72 of B. porri was originally collected. Moreover, a bipartite dsRNA mycovirus infecting Sclerotinia sclerotiorum, the causal agent of sclerotinia stem rot of oilseed rape, was recently identified in China. The sequences of the two dsRNAs were identical to dsRNA-1 and dsRNA-2 of BpRV1 by 91 and 97%, respectively (L. J. Liu and D. H. Jiang, unpublished data). These results suggest that, besides B. porri, BpRV1 can infect B. squamosa and S. sclerotiorum. It might be widely distributed in regions where B. porri, B. squamosa, and S. sclerotiorum are found.
BpRV1 particles are ∼35 nm in diameter. The size is similar to that of the families Chrysoviridae (30 to 40 nm), Partitiviridae (30 to 40 nm), and Totiviridae (30 to 40 nm) but is smaller than that of RnMBV1 in Megabirnaviridae (∼50 nm) and Mycoreovirus (∼80 nm). Whether or not the two dsRNAs of BpRV1 are encapsidated in separate virus particles or within the same virus particles remains unknown. The dsRNA segments for the genomes of the viruses in Chrysoviridae and Partitiviridae are separately encapsidated (17, 18, 41). In addition, the two dsRNA segments for RnMBV1 were thought to be separately encapsidated based on the finding that the unequal molar ratios between dsRNA-1 and dsRNA-2 extracted directly from host mycelia and the purified virus particles (9). However, no significant molar ratio variation was observed for BpRV1 dsRNA-1 and dsRNA-2 (Fig. 2C). To address the question about the particle assembly in BpRV1, we calculated the gross volumes of virus particles of dsRNA viruses in all recognized genera based on diameter of virus particles (see Table S5 in the supplemental material). The values of the dsRNA length encapsidated within one volume unit of a virus particle were then inferred to indicate the dsRNA assembly density. For almost all dsRNA viruses, the density of dsRNA in one particle is within the range of 0.03 to 0.28 bp/nm3. For BpRV1, the values for the dsRNA density are 0.28 bp/nm3 under separate encapsidation and 0.53 bp/nm3 under combined encapsidation. From this point of view, the two dsRNAs of BpRV1 are likely to be separately encapsidated, since the virus particles might not be spacious enough for simultaneous packaging of the two dsRNAs.
The results of the PMF analysis of the three proteins (p70, p80, and p85) in BpRV1 indicate that there may be two structural proteins (SPs) for BpRV1 (see Fig. S4 and Tables S2, S3, and S4 in the supplemental material). The protein component p70 is encoded by ORF II on dsRNA-2, whereas p80 and p85 are encoded by ORF I at a comparable region on dsRNA-1. There are two possibilities for generation of p80 and p85. The first possibility is that p80 might be derived from p85 via posttranslational modification in a manner comparable to that of the related capsid proteins of HvV190S (24). Another possibility is that p80 and p85 are generated via different events in processing the ORF I-encoded polyprotein. In the PMF analysis, we also found that several peptide sequences matched the C-terminal regions, rather than the N-terminal regions, on the ORF I- and ORF II-encoded polypeptides (see Fig. S4 in the supplemental material). These are probably random matches, since the ion scores for these peptide sequences are lower than 46 (see Tables S2, S3, and S4 in the supplemental material).
The genomes of RNA viruses are susceptible to chemical or physical damage from environmental factors. They also encounter immune response or antiviral defense of their hosts. To overcome these challenges, many RNA viruses have developed a series of self-defense mechanisms (4). In the interaction between hypoviruses and Cryphonectria parasitica, the hypovirus-encoded papain-like protease p29 acts as an inhibitor of the RNA silencing-induced antiviral defense by C. parasitica, thus enhancing viral RNA accumulation and vertical transmission (44, 45, 47). Moreover, hypovirus infection of C. parasitica causes the proliferation of vesicle formation within hyphal cells (11, 35, 36), and these vesicles were determined to be sites for viral replication (13, 14). The replication of RNA viruses in these membranous lipid vesicles might also offer protection from nucleases (31).
In addition, Spear et al. (46) proposed that the proline-alanine-rich protein (PArp) encoded by SFV1 and CiTV1 may act as a scaffold protein to interact with some membrane components in insect cells, since many animal viruses can utilize a protein scaffold containing proline-rich “late” domain motifs to hijack host proteins involved in the vacuolar protein sorting pathway, resulting in the formation of multivesicular bodies (8). In the present study, the formation of abundant vacuole-like membranous structures and small membranous vesicles in hyphal cells infected with BpRV1 was observed (Fig. 7) and a proline-rich region (PrR) was detected on a putative polypeptide encoded by ORF I on dsRNA-1. Therefore, further studies to elucidate the function of PrR-containing polypeptide in the virus accumulation of BpRV1 and to understand the expression strategy of dsRNA-1 and dsRNA-2 are warranted.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by the Natural Science Foundation of China (grant 31070122).
We greatly appreciate the assistance of S. A. Ghabrial of the University of Kentucky for critical readings of the manuscript and providing useful literature on mycovirus taxonomy. We also thank N. Suzuki at Okayama University (Japan) for providing a protocol for the transfection experiment with the purified virus particles of BpRV1 and N. Hong of Huazhong Agricultural University (China) for her advice to purify virus particles of BpRV1.
Footnotes
Published ahead of print 11 April 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Anagnostakis SL. 1982. Biological control of chestnut blight. Science 215:466–471 [DOI] [PubMed] [Google Scholar]
- 2. Aoki N, et al. 2009. A novel mycovirus associated with four double-stranded RNAs affects host fungal growth in Alternaria alternata. Virus Res. 140:179–187 [DOI] [PubMed] [Google Scholar]
- 3. Asiedu SK, Raghavan GSV, Gariepy Y, Reeleder R. 1986. Botrytis porri on leek in Canada. Plant Dis. 70:259 [Google Scholar]
- 4. Barr JN, Fearns R. 2010. How RNA viruses maintain their genome integrity. J. Gen. Virol. 91:1373–1387 [DOI] [PubMed] [Google Scholar]
- 5. Castro M, et al. 1999. A new double-stranded RNA mycovirus from Botrytis cinerea. FEMS Microbiol. Lett. 175:95–99 [DOI] [PubMed] [Google Scholar]
- 6. Castro M, Kramer K, Valdivia L, Ortiz S, Castillo A. 2003. A double-stranded RNA mycovirus confers hypovirulence-associated traits to Botrytis cinerea. FEMS Microbiol. Lett. 228:87–91 [DOI] [PubMed] [Google Scholar]
- 7. Chen B, Choi G, Nuss DL. 1994. Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 264:1762–1764 [DOI] [PubMed] [Google Scholar]
- 8. Chen BJ, Lamb RA. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiba S, et al. 2009. A novel bipartite double-stranded RNA mycovirus from the white root rot fungus Rosellinia necatrix: molecular and biological characterization, taxonomic considerations, and potential for biological control. J. Virol. 83:12801–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Choi GH, Nuss DL. 1992. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 257:800–803 [DOI] [PubMed] [Google Scholar]
- 11. Dodds JA. 1980. Association of type 1 viral-like double stranded RNA with club shaped particles in hypovirulent strains of Endothia parasitica. Virology 107:1–12 [DOI] [PubMed] [Google Scholar]
- 12. Dugan FM, Hellier BC, Lupien L. 2007. Pathogenic fungi in garlic seed cloves from the United States and China, and efficacy of fungicides against pathogens in garlic germplasm in Washington State. J. Phytopathol. 155:437–445 [Google Scholar]
- 13. Fahima T, Kazmierczak P, Hansen DR, Pfeiffer P, van Alfen NK. 1993. Membrane-associated replication of an unencapsidated double-strand RNA of the fungus, Cryphonectria parasitica. Virology 195:81–89 [DOI] [PubMed] [Google Scholar]
- 14. Fahima T, Wu Y, Zhang L, van Alfen NK. 1994. Identification of the putative RNA polymerase of Cryphonectria hypovirus in a solubilized replication complex. J. Virol. 68:6116–6119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Finn RD, et al. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ghabrial SA, Suzuki N. 2009. Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 47:353–384 [DOI] [PubMed] [Google Scholar]
- 17. Ghabrial SA, Castón JR. 2011. Chrysoviridae, p 509–513 In Fauquet CM, et al. (ed), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier, London, United Kingdom [Google Scholar]
- 18. Ghabrial SA, et al. 2011. Partitiviridae, p 523–534 In Fauquet CM, et al. (ed), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier, London, United Kingdom [Google Scholar]
- 19. Heiniger U, Rigling D. 1994. Biological control of chestnut blight in Europe. Annu. Rev. Phytopathol. 32:581–599 [Google Scholar]
- 20. Hillman BI, Supyani S, Kondo H, Suzuki N. 2004. A reovirus of the fungus Cryphonectria parasitica that is infectious as particles and related to the Coltivirus genus of animal pathogens. J. Virol. 78:892–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Howitt RLJ, Beever RE, Pearson MN, Forster RLS. 1995. Presence of double-stranded RNA and virus-like particles in Botrytis cinerea. Mycol. Res. 99:1472–1478 [Google Scholar]
- 22. Howitt RLJ, Beever RE, Pearson MN, Forster RL. 2001. Genome characterization of Botrytis virus F, a flexuous rod-shaped mycovirus resembling plant “potex-like” viruses. J. Gen. Virol. 82:67–78 [DOI] [PubMed] [Google Scholar]
- 23. Howitt RLJ, Beever RE, Pearson MN, Forster RL. 2005. Genome characterization of a flexuous rod-shaped mycovirus, Botrytis virus X, reveals high amino acid identity to genes from plant “potex-like” viruses. Arch. Virol. 151:563–579 [DOI] [PubMed] [Google Scholar]
- 24. Huang S, Ghabrial SA. 1996. Organization and expression of the double-stranded RNA genome of Helminthosporium victoriae 190S virus, a totivirus infecting a plant pathogenic filamentous fungus. Proc. Natl. Acad. Sci. of U. S. A. 93:12541–12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang DH, Ghabrial SA. 2004. Molecular characterization of Penicillium chrysogenum virus: reconsideration of the taxonomy of the genus Chrysovirus. J. Gen. Virol. 85:2111–2121 [DOI] [PubMed] [Google Scholar]
- 26. Kang JG, Wu JC, Bruenn JA, Park CM. 2001. The H1 double-stranded RNA genome of Ustilago maydis virus-H1 encodes a polyprotein that contains structural motifs for capsid polypeptide, papain-like protease, and RNA-dependent RNA polymerase. Virus Res. 76:183–189 [DOI] [PubMed] [Google Scholar]
- 27. Kanematsu S, et al. 2004. A reovirus causes hypovirulence of Rosellinia necatrix. Phytopathology 94:561–568 [DOI] [PubMed] [Google Scholar]
- 28. Lin HY, et al. 2007. Genome sequence, full-length infectious cDNA clone, and mapping of viral double-stranded RNA accumulation determinant of hypovirus CHV1-EP721. J. Virol. 81:1813–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu H, et al. 2009. A novel mycovirus that is related to the human pathogen hepatitis E virus and Rubi-like viruses. J. Virol. 83:1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. MacDonald W, Fulbright D. 1991. Biological control of chestnut blight: use and limitations of transmissible hypovirulence. Plant Dis. 75:656–661 [Google Scholar]
- 31. Mackenzie J. 2005. Wrapping things up about virus RNA replication. Traffic 6:967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mathews DH, et al. 2004. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U. S. A. 101:7287–7292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McDonald MR, et al. 2004. Management of diseases of onions and garlic, p 149–200 In Naqvi SAMH. (ed), Diseases of fruits and vegetables, vol II Kluwer, Dordrecht, Netherlands [Google Scholar]
- 34. Morris TJ, Dodds JA. 1979. Isolation and analysis of double-stranded RNA virus infected plant and fungal tissue. Phytopathology 69:854–858 [Google Scholar]
- 35. Newhouse JR, Hoch HC, MacDonald WL. 1983. The ultrastructure of Endothia parasitica comparison of a virulent with a hypovirulent isolate. Can. J. Bot. 61:389–399 [Google Scholar]
- 36. Newhouse JR, MacDonald WL, Hoch HC. 1990. Virus-like particles in hyphae and conidia of European hypovirulent dsRNA-containing strains of Cryphonectria parasitica. Can. J. Bot. 68:90–101 [Google Scholar]
- 37. Nuss DL, Koltin Y. 1990. Significance of dsRNA genetic elements in plant pathogenic fungi. Annu. Rev. Phytopathol. 28:37–58 [DOI] [PubMed] [Google Scholar]
- 38. Nuss DL. 2005. Hypovirulence: mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 3:632–642 [DOI] [PubMed] [Google Scholar]
- 39. Pearson MN, Beever RE, Boine B, Arthur K. 2009. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol. Plant Pathol. 10:115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rawlings ND, Morton FR, Barrett AJ. 2006. MEROPS: the peptidase database. Nucleic Acids Res. 34:270–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sanderlin RS, Ghabrial SA. 1978. Physicochemical properties of two distinct types of virus-like particles from Helminthosporium victoriae. Virology 87:142–151 [DOI] [PubMed] [Google Scholar]
- 42. Sasaki A, Kanematsu S, Onoue M, Oyama Y, Yoshida K. 2006. Infection of Rosellinia necatrix with purified viral particles of a member of Partitiviridae (RnPV1-W8). Arch. Virol. 151:697–707 [DOI] [PubMed] [Google Scholar]
- 43. Sasaki A, et al. 2007. Artificial infection of Rosellinia necatrix with purified viral particles of a member of the genus Mycoreovirus reveals its uneven distribution in single colonies. Phytopathology 97:278–286 [DOI] [PubMed] [Google Scholar]
- 44. Segers GC, van Wezel R, Zhang X, Hong Y, Nuss DL. 2006. Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot. Cells 5:896–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Segers GC, Zhang X, Deng F, Sun Q, Nuss DL. 2007. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. U. S. A. 104:12902–12906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Spear A, Sisterson MS, Yokomi R, Stenger DC. 2010. Plant-feeding insects harbor double-stranded RNA viruses encoding a novel proline-alanine rich protein and a polymerase distantly related to that of fungal viruses. Virology 404:304–311 [DOI] [PubMed] [Google Scholar]
- 47. Suzuki N, Maruyama K, Moriyama M, Nuss DL. 2003. Hypovirus papain-like protease p29 is an enhancer of viral dsRNA accumulation and vertical transmission. J. Virol. 77:11697–11707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aids by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Urayama S, et al. 2010. Mycoviruses related to chrysovirus affect vegetative growth in the rice blast fungus Magnaporthe oryzae. J. Gen. Virol. 91:3085–3094 [DOI] [PubMed] [Google Scholar]
- 50. Vilches S, Castillo A. 1997. A double-stranded RNA mycovirus in Botrytis cinerea. FEMS Microbiol. Lett. 155:125–130 [DOI] [PubMed] [Google Scholar]
- 51. Wu MD, et al. 2007. Hypovirulence and double-stranded RNA in Botrytis cinerea. Phytopathology 97:1590–1599 [DOI] [PubMed] [Google Scholar]
- 52. Wu MD, Zhang L, Li GQ, Jiang DH, Ghabrial SA. 2010. Genome characterization of a debilitation-associated mitovirus infecting the phytopathogenic fungus Botrytis cinerea. Virology 406:117–126 [DOI] [PubMed] [Google Scholar]
- 53. Xie JT, et al. 2011. A novel mycovirus closely related to hypoviruses that infects the plant pathogenic fungus Sclerotinia sclerotiorum.Virology 418:49–56 [DOI] [PubMed] [Google Scholar]
- 54. Yelton MM, Hamer JE, Timberlake WE. 1984. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc. Natl. Acad. Sci. U. S. A. 81:1470–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yu X, et al. 2010. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. Sci. U. S. A. 107:8387–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang X, Nuss DL. 2008. A host dicer is required for defective viral RNA production and recombinant virus vector RNA instability for a positive sense RNA virus. Proc. Natl. Acad. Sci. U. S. A. 105:16749–16754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang J, Li GQ, Jiang DH. 2009. First report of garlic leaf blight caused by Botrytis porri in China. Plant Dis. 93:1216. [DOI] [PubMed] [Google Scholar]
- 58. Zhang J, et al. 2010. Botrytis sinoallii: a new species of the grey mould pathogen on Allium crops in China. Mycoscience 51:421–431 [Google Scholar]
- 59. Zhang LY, et al. 2009. A novel virus that infecting hypovirulent strain XG36-1 of plant fungal pathogen Sclerotinia sclerotiorum. Virol. J. 6:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.