Abstract
An attenuation of the HIV-1 replication capacity (RC) has been observed for immune-mediated escape mutations in Gag restricted by protective HLA alleles. However, the extent to which escape mutations affect other viral proteins during natural infection is not well understood. We generated recombinant viruses encoding plasma HIV-1 RNA integrase sequences from antiretroviral-naïve individuals with early (n = 88) and chronic (n = 304) infections and measured the in vitro RC of each. In contrast to data from previous studies of Gag, we observed little evidence that host HLA allele expression was associated with integrase RC. A modest negative correlation was observed between the number of HLA-B-associated integrase polymorphisms and RC in chronic infection (R = −0.2; P = 0.003); however, this effect was not driven by mutations restricted by protective HLA alleles. Notably, the integrase variants S119R, G163E, and I220L, which represent uncommon polymorphisms associated with HLA-C*05, -A*33, and -B*52, respectively, correlated with lower RC (all q < 0.2). We identified a novel C*05-restricted epitope (HTDNGSNF114–121) that likely contributes to the selection of the S119R variant, the polymorphism most significantly associated with lower RC in patient sequences. An NL4-3 mutant encoding the S119R polymorphism displayed a ∼35%-reduced function that was rescued by a single compensatory mutation of A91E. Together, these data indicate that substantial HLA-driven attenuation of integrase is not a general phenomenon during HIV-1 adaptation to host immunity. However, uncommon polymorphisms selected by HLA alleles that are not conventionally regarded to be protective may be associated with impaired protein function. Vulnerable epitopes in integrase might therefore be considered for future vaccine strategies.
INTRODUCTION
HIV-1 in vitro replication capacity (RC) can be impaired through the selection of human leukocyte antigen (HLA) class I-restricted CD8+ cytotoxic T-lymphocyte (CTL) escape mutations (20, 24, 25, 48). This phenomenon is most notable for mutations of the Gag protein (8, 41, 50, 59) that lie within CTL epitopes restricted by protective HLA class I alleles, such as HLA-B*13 (29), HLA-B*27 (55), HLA-B*51 (34), HLA-B*57 (6, 9), and HLA-B*81 (64). Using recombinant viral approaches, we have observed HLA-associated reductions in RC for patient-derived Gag-protease sequences (8). Notably, significant Gag RC defects were seen in the context of protective HLA class I alleles during acute/early infection but not during chronic infection, most likely due to the accumulation of compensatory mutations (8). Protective HLA allele-associated viral attenuation has also been demonstrated by using Gag sequences obtained from spontaneous HIV-1 controllers (42, 46), suggesting that immune-mediated fitness defects contribute to this rare phenotype (39, 43, 66).
The extent to which host immune pressure on regions outside Gag can affect RC in naturally occurring HIV-1 sequences remains incompletely known. A limited number of reports by our group and others have investigated the impact of CTL escape mutations in reverse transcriptase (RT)-integrase (16), Nef (60), and envelope (59). However, no studies to date have assessed this question using large, well-powered, population-based analyses. An improved understanding of HIV-1 adaptation to host immunity and its consequence for viral replicative fitness may reveal important epitopes that can assist in the development of vaccines designed to target the most vulnerable regions of HIV-1.
The HIV-1 integrase protein is a highly conserved 32-kDa enzyme that plays an essential role in the viral life cycle by catalyzing the 3′ DNA-processing and strand transfer reactions that result in the covalent ligation of the viral genome into the host cell chromosome (19, 37). It is therefore a critical target for new antiviral therapies (45, 49). Viral fitness defects resulting from mutations in the Pol gene following antiretroviral therapy are well documented (30, 51, 61), and the raltegravir resistance mutations N115H and Q148H in integrase were associated recently with reduced viral RC (30). Integrase is also a target for CTL responses, and it encodes a number of immunogenic epitopes restricted by protective HLA class I alleles, including B*27 (47), B*51 (38, 58), and B*57/B*58 (1, 53). T-cell responses against integrase are often detectable in individuals with reduced viral loads (44). Moreover, certain integrase epitopes are known to be targeted early following infection (3, 57) and exhibit strong and reproducible patterns of immune escape at the population level (15).
We have demonstrated previously that HLA-associated RC defects occur in RT-integrase sequences obtained from HIV-1 elite controllers (12); however, the integrase protein has not been evaluated systematically for evidence of immune-mediated attenuation or as a potential target for vaccine design. In order to examine the impact of HLA-associated immune pressure on HIV-1 integrase function, we investigated the in vitro RC of recombinant viruses expressing patient-derived sequences obtained from individuals during early and chronic infections. Our results demonstrate that mutations in integrase-derived CTL epitopes restricted by HLA class I alleles that are not typically considered to be protective can attenuate HIV-1 replication.
MATERIALS AND METHODS
Patients and samples.
Samples from antiretroviral-naïve individuals with early infection (n = 88; median time postinfection, 77 days [interquartile range {IQR}, 54 to 90]) were obtained from Acute Infection and Early Disease Research Program (AIEDRP) sites in the United States and Australia (n = 25) (12), a private medical clinic in Germany (n = 21) (12), and the Vancouver Injection Drug Users study (n = 42) (63). Of these, 9 individuals (10%) had positive HIV RNA (>5,000 copies/ml) or detectable serum p24 antigen but a negative HIV-1 enzyme immunoassay (EIA) result, 27 (31%) had a positive EIA result but a negative or indeterminate Western blot result, and 52 (59%) had a negative EIA result during the previous 6 months or a negative detuned EIA result (33) at enrollment. These categories are comparable to Fiebig stages 1/2, 3/4/5, and 6, respectively (21). Infection dates were estimated as follows (12). For HIV RNA-positive (RNA+)/EIA-negative (EIA−) patients, 4 weeks were subtracted from the date of the negative EIA. For patients with a positive EIA result but a negative/indeterminate Western blot result, 6 weeks were subtracted from the date of the positive EIA. For patients with a negative detuned EIA result, 4 months were subtracted from this date. For the remainder of the patients, infection dates were estimated as the midpoint between the last negative and the first positive EIA result. Clinical histories were incorporated into infection date estimates if available.
For the chronic infection cohort, samples (n = 304) were obtained from the baseline (antiretroviral-naïve) time point of the HOMER cohort, which is composed of individuals initiating their first highly active antiretroviral therapy (HAART) regimen in British Columbia, Canada (13, 15). Time since infection is unknown for these individuals. Clinical characteristics of patients with early and chronic infections are provided in Table 1.
Table 1.
Clinical characteristicsa
| Parameter | Median value (IQR) for group |
|
|---|---|---|
| Early cohort (n = 88) | Chronic cohort (n = 304) | |
| Plasma viral load (log10 copies/ml) | 5.26 (4.48–5.88)b | 5.16 (4.95–5.51) |
| CD4+ cell count (cells/mm3) | 522 (406–760)c | 285 (140–423) |
| CD8+ cell count (cells/mm3) | 881 (712–1153)d | NA |
| Estimated time since infection (days) | 77 (64–90) | NA |
All values reported as medians (IQR). NA, not available.
Clinical data were available for 43 individuals.
Clinical data were available for 37 individuals.
Clinical data were available for 36 individuals.
All patients were HLA class I typed by using sequence-based methods (12, 14). Ethical approval was obtained through the relevant institutional review boards.
Construction of recombinant viruses.
Recombinant viruses encoding patient-derived integrase sequences were generated on an NL4-3 background as described previously (8, 16), with the following modifications. HIV-1 integrase was RT-PCR amplified from plasma HIV RNA using sequence-specific, subtype B-optimized primers. Second-round PCR was performed by using PAGE-purified primers (forward primer 5′-AAA GGA AAA AGT CTA CCT GGC ATG GGT ACC AGC ACA CAA AGG AAT TGG AGG AAA TGA ACA AGT AGA TAA ATT GGT CAG TGC TGG AAT CAG GAA AGT ACT A-3′ and reverse primer 5′-ACT TAT TTT TGG ATT AGT ACT TTC ATA GTG ATG TCT ATA AAA CCA GTC CTT AGC TTT CCT TGA AAT ATA CAT ATG GTG TTT TAC TAA TCT TTT CCA TG-3′). These primers match the NL4-3 sequence directly upstream and downstream of integrase and are designed to facilitate the homologous recombination of the PCR product and the pNL4-3 backbone sequence (see below).
Plasmid pNL4-3Δintegrase was developed by inserting unique BstEII restriction sites at the 5′ and 3′ ends of the integrase gene (HXB2 nucleotide positions 4229 and 5093, respectively), using the QuikChange XL kit (Stratagene). This was followed by the deletion of the entire integrase coding sequence using BstEII digestion (New England BioLabs) and gel purification. Plasmid pNL4-3Δintegrase was religated by using T4 DNA ligase (New England BioLabs) and maintained in Stbl3 Escherichia coli cells (Invitrogen). To generate recombinant viruses, 10 μg of BstEII-linearized pNL4-3Δintegrase plus 50 μl of a second-round amplicon (approximately 5 μg) were mixed with 2.0 × 106 CEM-derived GXR25 reporter T cells (10) in 800 μl of Mega-Cell medium (Sigma) and transfected by electroporation using a Bio-Rad GenePulser II instrument in single cuvettes (exponential protocol of 300 V and 500 μF) or using a Bio-Rad MxCell_Pro instrument with 96-well electroporation plates (exponential protocol of 250 V and 2,000 μF). Following transfection, cells were rested for 15 min at room temperature, transferred into 25-cm2 flasks in 5 ml of R20+ medium (RPMI 1640 containing 20% fetal calf serum [FCS], 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin), and fed with 5 ml R10+ medium on day 5 and with replacement thereafter. Tat-driven green fluorescent protein (GFP) expression was monitored by flow cytometry (FACSCalibur [BD Biosciences] or Guava 8HT [Millipore]) starting on day 12, as described previously (10). Once the GFP-positive (GFP+) expression level reached >15% among viable cells, supernatants containing the recombinant viruses were harvested, and aliquots were stored at −80°C.
Replication capacity assays.
Viral titers and replication capacity (RC) assays were performed as described previously, using the GXR25 reporter T-cell line (8, 42). RC assays were initiated at a multiplicity of infection (MOI) of 0.003 and included one negative control (uninfected cells only) and one positive control (NL4-3 integrase reintroduced into the NL4-3Δintegrase backbone using identical methods) per 24-well plate. Assays typically featured four to six 24-well plates, each containing 22 recombinant viruses, one positive control, and one negative control. For each virus, the natural log slope of the percentage of GFP+ cells was calculated during the exponential phase of viral spread (days 3 to 6). This value was divided by the mean rate of spread of all NL4-3 controls to generate a normalized, quantitative measure of RC. RC values of <1.0 or >1.0 indicate rates of spread that were lower than or higher than those of NL4-3, respectively. Each virus was assayed in a minimum of two independent experiments, and average replication rates are reported. Previously reported quality control experiments confirmed that RC values of independently generated bulk (i.e., quasispecies-containing) recombinant viruses were highly concordant for a given patient sample and that the reintroduction of cloned NL4-3 sequences into an NL4-3 backbone did not alter viral fitness compared to that of the parental NL4-3 strain (8).
Sequence analysis.
Bulk plasma HIV-1 RNA integrase sequences from individuals early in infection were collected by using standard methods, as described previously (15). Briefly, HIV-1 RNA was extracted from 200 to 500 μl of plasma by using the QIAamp viral RNA kit (Qiagen) or the NucliSENS easyMag system (bioMérieux), amplified by nested RT-PCR using sequence-specific primers, and sequenced bidirectionally on an ABI 3730xl sequencer (Applied Biosystems). Chromatograms were analyzed by using Sequencher 4.9 (Gene Codes) or the custom software RECall (11). Nucleotide mixtures were called where the secondary peak height exceeded 25% of the dominant peak height. All sequences were confirmed as subtype B sequences by comparison to reference sequences (http://www.hiv.lanl.gov). Nucleotide alignments were performed by using a modified NAP algorithm (32). Bulk HIV-1 RNA sequences were previously collected by using comparable methods for the chronic infection cohort (15). In addition, HIV-1 RNAs from all recombinant viral stocks were extracted from culture supernatants, amplified by nested RT-PCR, and resequenced to confirm patient origin and to assess viral diversity. Maximum likelihood phylogenetic trees were generated by using PhyML (26) and visualized by using Figtree v.1.2.2 (http://tree.bio.ed.ac.uk/software/figtree). HIV-1 sequences from chronically infected individuals were previously deposited into the GenBank database (15).
Site-directed mutagenesis.
NL4-3 integrase was subcloned into a TOPO-TA vector (Invitrogen), and the mutations S119R and A91E were introduced either alone or in combination by site-directed mutagenesis using the QuikChange XL kit (Stratagene) and custom-designed primers. After verification by DNA sequencing, mutant integrase sequences were amplified from plasmid DNA, and recombinant viruses were generated and assayed as described above.
Identification of HLA-associated polymorphisms and covarying sites.
HLA-associated polymorphisms and their respective covarying polymorphic sites in integrase were defined based on a previously reported analysis of plasma HIV RNA sequences from the International HIV Adaptation Collaborative, comprised of 1,317 antiretroviral-naïve, chronically infected individuals from cohorts in Canada, the United States, and Australia (>97.5% subtype B) (15). All polymorphisms that were significant at a q value of <0.2 are shown Fig. 4.
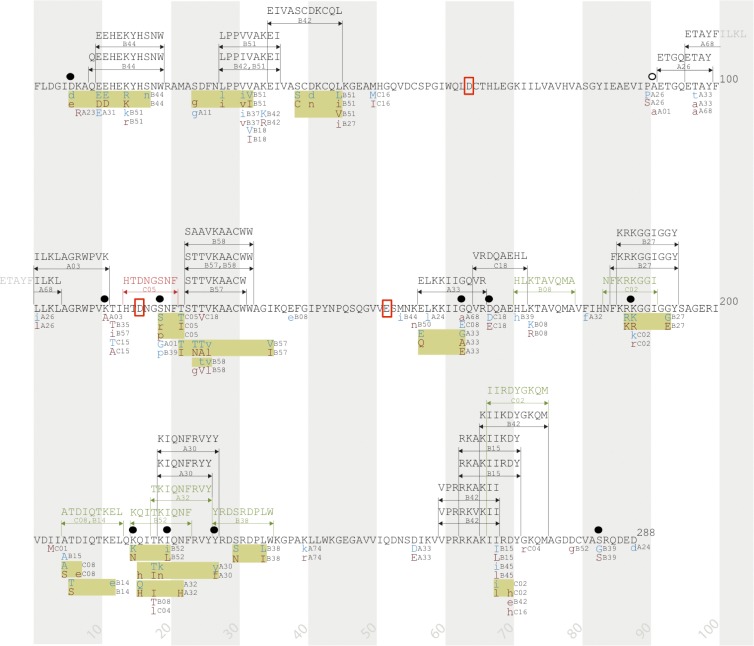
Fig 4.
Integrase immune escape map. HLA-associated polymorphisms, known and predicted CTL epitopes, and mutations associated with reduced viral replication capacity (RC) are indicated along the linear sequence of the HIV-1 integrase protein. HLA-associated polymorphisms (all q < 0.2) were defined based on an analysis of 1,317 antiretroviral-naïve, chronically infected individuals from cohorts in Canada, the United States, and Australia (>97.5% subtype B) (15). Shaded vertical bars separate blocks of 10 amino acids. Known CTL epitopes and restricting HLA class I alleles are indicated above the sequence with black arrows and text, as reported by the HIV Molecular Immunology Database (www.hiv.lanl.gov). Putative CTL epitopes and restricting HLA alleles are indicated above the sequence with green arrows and text, as predicted in this study. The C*05-restricted HTDNGSNF (HF8) epitope, validated in this study, is indicated above the sequence with red arrows and text. HLA-associated polymorphisms are shown below the protein sequence: “adapted” amino acids (enriched in the presence of the HLA allele) are displayed in red text, and “nonadapted” amino acids (enriched in the absence of the HLA allele) are displayed in blue text. Uppercase letters distinguish polymorphisms that survive correction for codon covariation. Polymorphisms associated with the same HLA allele that occur in proximity to one another are grouped together in yellow boxes. Amino acid residues associated with integrase RC are indicated by ●, and the compensatory site A91 is noted by ○. Red boxes highlight the essential catalytic-site amino acids D64, D116, and E152.
Prediction of novel HIV-1 epitopes and associations with RC.
To investigate HLA-associated polymorphisms linked to reduced RC that were not located within or near a previously reported CTL epitope, we examined gamma interferon (IFN-γ) enzyme-linked immunosorbent spot (ELISpot) assay responses from 372 HIV-positive (HIV+) individuals screened with overlapping subtype B consensus peptides 15 to 18 amino acids in length spanning the integrase protein (22; our unpublished data). Fisher's exact test was used to assess whether the expression of the HLA class I allele restricting the polymorphism of interest was significantly associated with the targeting of the respective overlapping peptide, evidence which would support the existence of a novel epitope within this peptide. Sequences of significantly reactive overlapping peptides were then scanned by using the bioinformatic epitope prediction algorithms NetCTL (36) (for HLA-B alleles) and Epipred (27a) (for HLA-B and -C alleles) to predict the most likely CTL epitope.
Gamma interferon ELISpot assays and C*05 epitope mapping.
Epitope mapping and HLA restriction were conducted as previously described (23). Thirty-one integrase peptides overlapping the region of the HLA C*05-associated polymorphism at position 119 were used for IFN-γ ELISpot assays at 20 μg/ml to stimulate peripheral blood mononuclear cells (PBMC) (100,000 cells/well) from patient F702 (HLA-A*02, -A*30, -B*53, -B*58, -C*04, and -C*05). The magnitude of each response was determined as the number of spot-forming cells (SFC) per million input cells and was measured by using a CTL-ImmunoSpot automatic ELISpot reader (CTL, Bonn, Germany). The threshold for positive responses was defined as the highest of the following three criteria: a minimum of 5 spots per well, the mean number of SFC of the negative-control wells plus three times its standard deviation, or three times the mean of negative-control wells. Further fine mapping was performed by titrating peptides that elicited a positive response in serial 10-fold dilutions in ELISpot assays, with peptide concentrations ranging from 20 μg/ml to 0.2 ng/ml.
Statistical analysis.
The Mann-Whitney U test was used to compare differences in replication capacities between groups (e.g., the presence versus the absence of HLA alleles or the presence versus the absence of specific viral polymorphisms). The relationship between RC and the number of HLA-associated escape mutations or clinical parameters (CD4 and plasma viral load [pVL]) was assessed by using Spearman's correlation. In exploratory analyses of integrase polymorphisms associated with RC, multiple tests were addressed by using a q value approach (56).
Nucleotide sequence accession numbers.
Accession numbers for the acute/early sequences are JQ080085 to JQ080172.
RESULTS
Construction of recombinant viruses from integrase samples from early and chronic infections.
In order to assess the potential impact of HLA-mediated immune pressure on integrase activity, we developed a recombinant viral assay to measure the in vitro replication capacity (RC) of patient-derived plasma HIV-1 integrase RNA sequences. Integrase recombinant viruses were generated from 88 early and 304 chronic HIV-1 subtype B-infected antiretroviral-naïve individuals (Table 1), using methods designed to capture in vivo quasispecies diversity (8, 42). Recombinant viral stocks were resequenced to confirm patient origin and to assess sequence diversity. As expected, bulk plasma HIV RNA and recombinant virus sequences from early infection displayed limited diversity: ∼70% of plasma and recombinant viruses were apparently clonal (i.e., they displayed no mixtures at the amino acid level). Plasma and recombinant viral sequences from chronic infection displayed higher levels of diversity: 23% and 33%, respectively, were clonal at the amino acid level. Recombinant viruses were highly concordant with the input PCR product: a median of 0 (IQR, 0 to 0) (out of 288) full amino acid differences in integrase were observed between plasma and recombinant viral sequences.
In vitro RC was determined for each recombinant virus by using a GFP-reporter T-cell assay (8, 42), and the results were normalized to those of the parental NL4-3 strain such that RC values of 1.0 signify replication equivalent to that of NL4-3, whereas values of <1 or >1 indicate a lower or higher RC than that of NL4-3, respectively. Median RC values were comparable between early and chronic cohorts (medians of 0.99 [IQR, 0.92 to 1.04] for early viruses and 1.01 [IQR, 0.96 to 1.04] for chronic viruses; P = 0.1 by Mann-Whitney U test), indicating that on average, the in vitro function of patient integrase sequences was comparable to that of NL4-3 regardless of the time since infection (Fig. 1). Median RCs of early and chronic viruses encoding clonal integrase sequences (n = 58 and n = 99, respectively) versus those of viruses containing at least one amino acid mixture (n = 30 and n = 205, respectively) were also comparable (P > 0.1 by Mann-Whitney U test) (data not shown), demonstrating that results were not confounded by differences in the quasispecies diversity of recombinant viral stocks.
Fig 1.
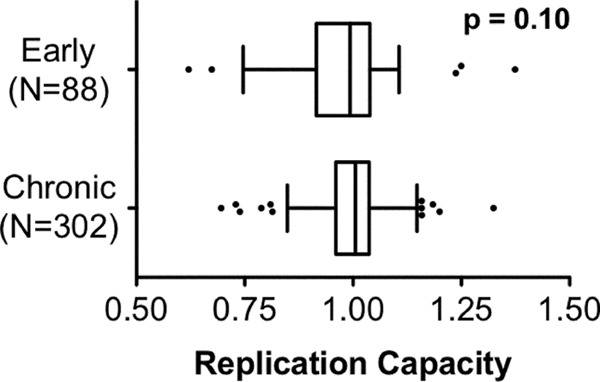
Integrase replication capacities of early and chronic samples. NL4-3 normalized replication capacities (RC) of recombinant viruses expressing integrase from 88 patients with early infection and 304 patients with chronic infection are indicated with Tukey box plots. Box boundaries display the medians and interquartile ranges, whiskers display maximum and minimum values excluding outliers, and outliers (defined as greater than 1.5 times the interquartile distances) are shown as points. No significant difference in integrase RC was observed between cohorts (P = 0.1 by Mann-Whitney U test).
Modest relationship between HLA class I-associated pressure and integrase replication capacity.
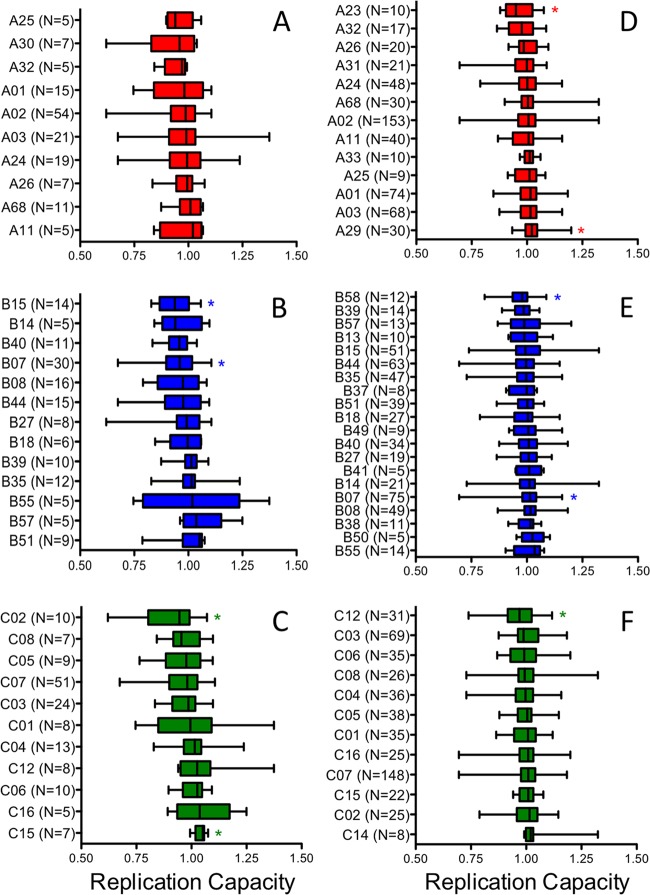
To assess the potential consequence of HLA-mediated selection pressure on integrase function during natural infection, recombinant viral RC values were stratified according to host HLA-A, -B, and -C allele expression genotypes and analyzed in both early and chronic infection cohorts (Fig. 2). In a systematic pairwise analysis of viral RC in early samples, host expression of HLA-B*07, -B*15, and -C*02 were each independently associated with modestly lower RC values than those for individuals not expressing these alleles, while C*15 was associated with modestly higher RC values than those for individuals not expressing this allele (all P < 0.05 by Mann-Whitney U test); however, these associations did not remain significant after correction for multiple comparisons (all q > 0.4). Integrase RC did not correlate significantly with the estimated time since infection either overall (n = 88; Spearman R = 0.15; P = 0.16) or in analyses restricted to protective alleles only (B*27, B*51, B*57, and/or B*58) (n = 25; Spearman R = −0.3; P = 0.16). In a systematic pairwise analysis of chronic viral RC for each HLA, the host expressions of A*23, B*58, and C*12 were associated with modestly lower RC values, while A*29 and B*07 were associated with modestly higher RC values (all P < 0.05 by Mann-Whitney U test); however, these associations were not significant after correction for multiple comparisons (all q > 0.35). We observed no significant associations between integrase RC and plasma viral loads (pVL) (R = 0.03; P = 0.6) or CD4 counts (R = −0.03; P = 0.7) for samples from chronic infections. Recombinant viruses encoding Gag-protease had been constructed previously for a subset of chronic patients (n = 284) (8); however, no correlation between integrase and Gag-protease-mediated RC was observed (R = 0.03; P = 0.6).
Fig 2.
Limited evidence for HLA-associated attenuation of integrase during natural HIV-1 infection. The viral replication capacities (RC) of early integrase samples (A to C) and of chronic integrase samples (D to F) were stratified according to the HLA-A (A and D), HLA-B (B and E), and HLA-C (C and F) allele expression of the host. Results are displayed as box plots, arranged by median RC from lowest (top) to highest (bottom). Box boundaries display the medians and interquartile ranges, and whiskers display maximum and minimum values. HLA associations with RC are indicated by asterisks (P < 0.05 by Mann-Whitney U test).
Limited impact of known HLA-associated mutations on integrase replication capacity.
To explore the relationship between known HLA-associated mutations and the integrase RC, we employed a previously reported list of HLA-associated HIV-1 subtype B polymorphisms, derived through statistical analyses of a cohort of 1,317 antiretroviral-naïve individuals with chronic infection (15). For each recombinant viral sequence, we determined the total number of HLA-associated polymorphisms according to the patient's six HLA class I alleles. In samples from early infection, we observed no correlations between the total number of HLA-A-, -B-, or -C-associated polymorphisms specific for the patient's HLA and RC (data not shown). For samples from chronic infection, we observed a modest yet statistically significant negative correlation between the total number of HLA-B-associated integrase polymorphisms and viral RC (R = −0.2; P = 0.003), while no correlation was seen for either HLA-A- or HLA-C-associated mutations (data not shown). Further analyses of the HLA-B locus revealed no significant relationships between RC and the number of integrase mutations associated with any single allele (data not shown), suggesting an additive negative effect of mutations selected by multiple HLA-B alleles on protein function.
Identification of integrase amino acids associated with viral replication capacity.
We next performed an exploratory codon-by-codon analysis of integrase sequences in the chronic and the combined (early plus chronic) data sets in order to identify polymorphisms that were significantly associated with viral RC. Note that this investigation did not consider host HLA allele expression, and therefore, it may identify transmitted polymorphisms as well as de novo CTL escape mutations. We observed that the integrase polymorphisms K111Q, S119R, G163E, D167E, K188R, K215X, I220L, and Y227X were each linked with lower RC (all P < 0.05) (Table 2), while the D6E and S283G polymorphisms were each linked with higher viral RC (P < 0.05). To assess the potential additive effects of these 10 polymorphisms on integrase function, we stratified viral RC according to the number of mutations encoded by each patient's sequence (0 to 5) and observed a statistically significant inverse correlation (Spearman R = −0.35; P < 0.0001) (Fig. 3).
Table 2.
Exploratory analysis of integrase polymorphisms and viral RCa (P < 0.05)
| Cohort | Integrase codon | aa | No. of patients with aa | No. of patients without aa | Median RC with aa | Median RC without aa | P value | q value | HLA associationb | HLA-restricted epitope(s) | Response frequency (%)c |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chronic | S119 | R | 12 | 292 | 0.94 | 1.01 | 0.0001 | 0.02 | C*05 | HTDNGSNF (confirmed) | 18 |
| Chronic | D167 | E | 5 | 299 | 0.94 | 1.01 | 0.028 | 0.45 | C*18 | VRDQAEHL | ND |
| Chronic | K188 | K | 298 | 6 | 1.01 | 0.93 | 0.006 | 0.35 | Consensus | ||
| Chronic | K188 | R | 6 | 298 | 0.93 | 1.01 | 0.014 | 0.35 | B*27 and C*02 | KRKGGIGGY (B*27), NFKRKGGI (C*02) (predicted) | 59, 62 |
| Chronic | K215 | K | 298 | 6 | 1.01 | 0.93 | 0.021 | 0.44 | Consensus | ||
| Chronic | I220 | I | 292 | 12 | 1.01 | 0.95 | 0.012 | 0.35 | Consensus | ||
| Chronic | I220 | L | 8 | 296 | 0.95 | 1.01 | 0.013 | 0.35 | B*52 | KQITKIQNF, KIQNFRVYY (predicted) | ND |
| Chronic | Y227 | Y | 299 | 5 | 1.01 | 0.93 | 0.032 | 0.45 | Consensus | ||
| Chronic | S283 | G | 17 | 287 | 1.03 | 1.00 | 0.032 | 0.45 | None | ||
| Merged | D6 | D | 376 | 16 | 1.00 | 1.04 | 0.021 | 0.34 | Consensus | ||
| Merged | D6 | E | 16 | 376 | 1.04 | 1.00 | 0.021 | 0.34 | B*44 | QEEHEKYHSNW | 13 |
| Merged | K111 | Q | 5 | 387 | 0.93 | 1.00 | 0.026 | 0.35 | None | ||
| Merged | S119 | R | 12 | 380 | 0.94 | 1.01 | 0.001 | 0.13 | C*05 | HTDNGSNF (confirmed) | 18 |
| Merged | G163 | E | 26 | 366 | 0.95 | 1.01 | 0.003 | 0.14 | A*33 | ELKKIIGQVR | 20 |
| Merged | G163 | G | 368 | 24 | 1.01 | 0.95 | 0.004 | 0.14 | Consensus | ||
| Merged | D167 | D | 386 | 6 | 1.00 | 0.94 | 0.010 | 0.25 | Consensus | ||
| Merged | K188 | K | 384 | 8 | 1.00 | 0.93 | 0.014 | 0.30 | Consensus | ||
| Merged | K188 | R | 8 | 384 | 0.93 | 1.00 | 0.024 | 0.35 | B*27 and C*02 | KRKGGIGGY (B*27), NFKRKGGI (C*02) (predicted) | 59, 62 |
| Merged | K215 | K | 386 | 6 | 1.00 | 0.93 | 0.033 | 0.41 | Consensus | ||
| Merged | I220 | I | 376 | 16 | 1.01 | 0.95 | 0.005 | 0.14 | Consensus | ||
| Merged | I220 | L | 13 | 379 | 0.95 | 1.01 | 0.002 | 0.13 | B*52 | KQITKIQNF, KIQNFRVYY (predicted) | ND |
The analysis is restricted to polymorphisms with a frequency greater than or equal to 5. Codon associations that are significant (q values of <0.2) are indicated using boldface type. Abbreviations: aa, amino acid; RC, replication capacity; ND, not determined.
Based on data from reference 15.
Frequency of positive IFN-γ ELISpot responses to overlapping 18-mer peptides in individuals expressing the indicated HLA.
Fig 3.
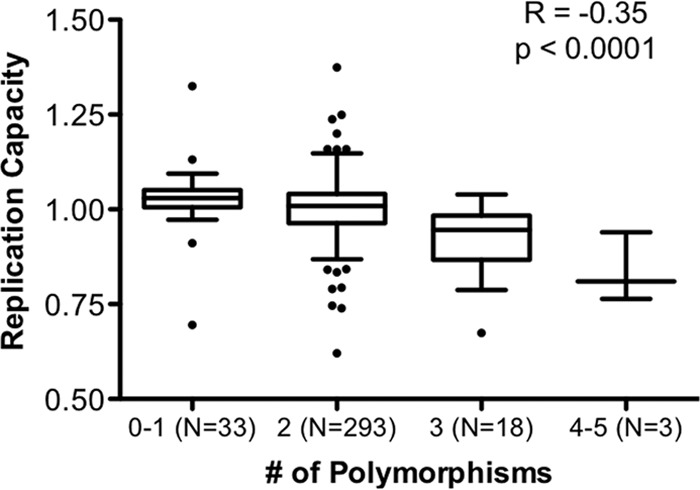
Additive effect of integrase polymorphisms on viral replication capacity. The viral replication capacities (RC) were stratified according to the total number of polymorphisms (E6D, K111Q, S119R, G163E, D167E, K188R, K215X, I220L, Y227X, and G283S) (all P < 0.05) (Table 2) encoded by each patient-derived sequence. A significant inverse association between viral RC and the number of polymorphisms was observed (Spearman R = −0.35; P < 0.0001).
Notably, 8 (out of 10) of these variants represent polymorphisms that are significantly associated with host HLA expression based on population-level studies (15) (Table 2 and Fig. 4), suggesting that immune pressure on integrase can contribute to viral RC in some cases. Further data exploration revealed that polymorphisms located at previously identified HLA-associated sites tended to be more costly to integrase RC than polymorphisms located at non-HLA-associated sites (P = 0.018) (data not shown). In addition, known epitope-specific CTL responses were reported previously to overlap several of these mutations, including D6E (B*44-QW11) (15, 54), G163E (A*33-ER10) (15, 35), D167E (C*18-VL8) (5, 15, 38), and K188R (B*27-KY9) (47). However, all of these polymorphisms are observed infrequently (<7% frequency at the population level) (15), suggesting that these epitopes are rarely targeted by CTL and/or that these polymorphisms represent uncommon pathways of CTL escape.
Of the polymorphisms identified here, the S119R (C*05), G163E (A*33), and I220L (B*52) polymorphisms remained significant after correction for multiple comparisons (all q < 0.2) (Table 2). The stratification of viral RC according to the number of these mutations that were encoded by patient sequences (0, 1, or 2) demonstrated an inverse correlation, indicating that these three polymorphisms could contribute additively to integrase function (Spearman R = −0.31; P < 0.0001) (data not shown). As noted above, the G163E polymorphism is likely to be driven by the known CTL response directed against the A*33-ER10 epitope (15, 35). Despite strong statistical associations linking C*05 with the S119R polymorphism and B*52 with the I220L polymorphism (15), no CTL epitopes restricted by these alleles have been defined in these regions. To investigate the existence of novel epitopes, we analyzed IFN-γ ELISpot responses to overlapping 18-mer subtype B consensus integrase peptides in 372 chronically infected individuals (22; our unpublished data). We applied Fisher's exact test to assess whether the expression of each HLA was significantly associated with host responses to a peptide(s) containing the position of interest, and we used the bioinformatics tools Epipred and NetCTL (36) to predict the most likely epitope sequences. In our data, the expression of C*05 (n = 33) was significantly associated with IFN-γ reactivity to two overlapping peptides in this region: AGRWPVKTIHTDNGSNF105–121 (odds ratio [OR], 6.4; P = 0.003) and TIHTDNGSNFTSTTVKAA112–129 (OR, 7.2; P = 0.01). Bioinformatics analysis suggested that the most likely C*05-restricted epitope sequence in the overlapping region was IHTDNGSNF (HXB2 positions 113 to 121; Epipred score, −4.69). Insufficient ELISpot data were available for B*52+ individuals; however, bioinformatics analysis suggested that KQITKIQNF (HXB2 positions 215 to 223; Epipred score, −3.26; NetCTL value, 104 nM) or KIQNFRVYY (HXB2 positions 219 to 227; Epipred score, −3.22; NetCTL value, 423 nM) was the most likely B*52-restricted epitope in this region. The location of each epitope and the frequency of positive IFN-γ ELISpot responses to long peptides are indicated in Fig. 4 and Table 2, respectively.
Confirmation of a novel C*05 epitope spanning integrase codon S119.
Very few C*05-restricted epitopes have been described. As a result, epitope prediction algorithms for this allele are limited. To confirm our prediction that a novel C*05 response may drive viral escape at integrase position S119, we used IFN-γ ELISpot assays to test a panel of shorter peptides spanning codons 105 to 129 in PBMC from a C*05-expressing individual who displayed reactivity in this region. We observed a loss of IFN-γ reactivity in PBMC for all peptides that did not have the sequence HTDNGSNF114–121, indicating that this 8-mer peptide was the minimal C*05-restricted epitope (Fig. 5). Peptide titration assays demonstrated that the half-maximal stimulatory dose (SD50) for HTDNGSNF114–121 (∼10 ng/ml) was substantially lower than that for other peptides spanning this sequence, including the predicted sequence IHTDNGSNF113–121, illustrating that this 8-mer peptide is the optimal C*05 epitope in this region of integrase.
Fig 5.
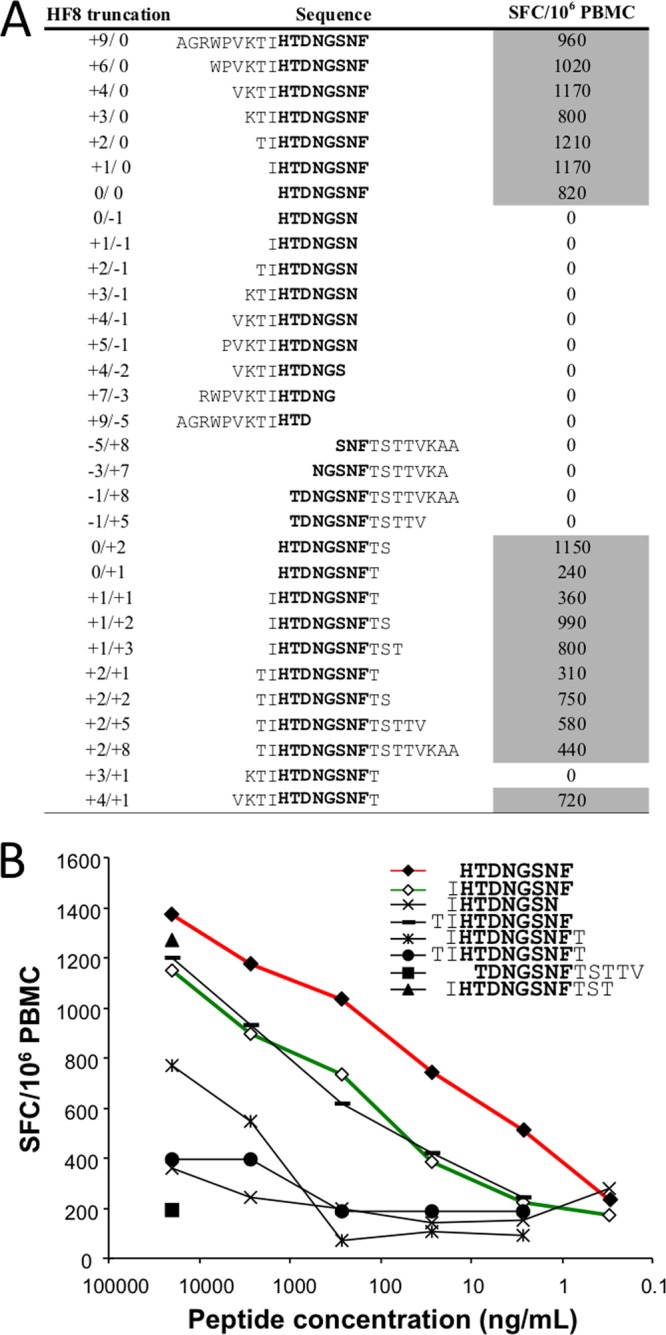
Detection of a novel C*05 epitope at integrase positions 114 to 121. (A) ELISpot assays were used to test PBMC from C*05+ patient F702. IFN-γ spot-forming cells (SFC) per 106 PBMC after the subtraction of negative-control values are shown on the right. Positive responses (shaded) were determined as described in Materials and Methods. Aligned sequences indicate IFN-γ reactivity against the minimal peptide HTDNGSNF114–121 (HF8) (shown in boldface type). (B) Serial dilutions of peptides spanning codons 112 to 126 were used to stimulate PBMC from patient F702. Responses to the predicted peptide (IHTDNSNF) are indicated by the green line (open diamonds). The HF8 peptide (red line and closed diamonds) displayed the lowest SD50 value, and it is therefore proposed to be the optimal epitope presented by C*05.
Validation of impaired replication by the S119R polymorphism and its compensation by the A91E mutation.
The C*05-restricted polymorphism S119R was most significantly associated with lower RC in chronic patient sequences (P = 0.0001; q = 0.02) (Table 2). To validate this result, we generated an NL4-3 site-directed mutant encoding the S119R polymorphism and tested its function. In vitro assays revealed that the S119R virus exhibited a ∼35% lower RC than did the parental NL4-3 strain (Fig. 6), demonstrating that this mutation alone could substantially diminish integrase function.
Fig 6.
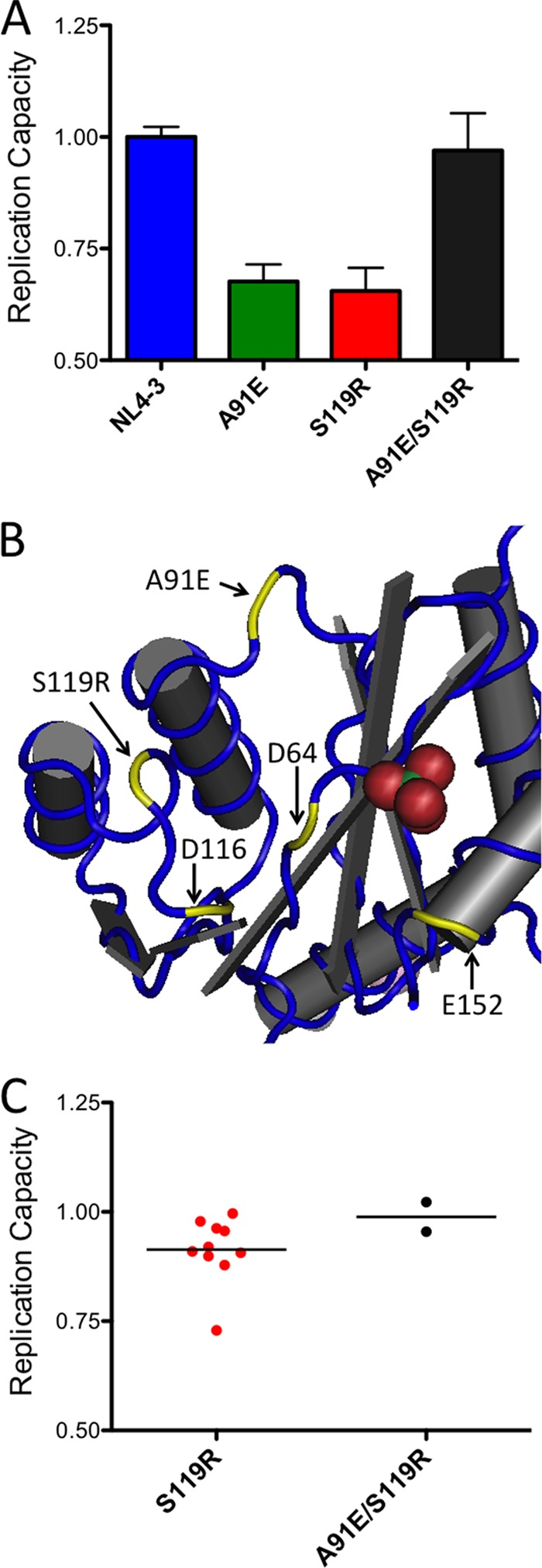
Confirmation of integrase S119R-mediated attenuation and its compensation by the A91E variant. (A) NL4-3 site-directed mutants encoding integrase mutations A91E and S119R, either alone or together, were constructed, and their in vitro replication capacities (RC) were assessed as described in Materials and Methods. Viral RC was impaired by either the A91E or S119R mutation alone, but RC was restored to wild-type levels when these mutations were introduced together. (B) The positions of residues A91 and S119 are indicated in the three-dimensional structure of the integrase protein (Protein Data Bank [PDB] accession number 1K6Y [62]). The three essential catalytic-site residues, D64, D116, and E152, are also highlighted. (C) In vitro RC results for integrase recombinant viruses encoding the S119R mutation only (n = 10) versus results for viruses encoding the S119R and A91E mutations (n = 2) indicate higher RC when these mutations are observed together in patient-derived sequences.
The in vitro RC defect displayed by the S119R site-directed mutant was notably greater than the average ∼7% lower RC observed for patient isolates that encoded this polymorphism. We therefore hypothesized that the S119R mutation could be compensated for by a secondary mutation(s) present in clinical sequences. Putative S119R compensatory mutations were identified by using previously reported data for covarying amino acids in HIV-1 integrase, generated using the same large cohort used to identify HLA-associated integrase polymorphisms at the population level (15). The residue most strongly covarying with the S119R mutation in patient sequences was the upstream A91E variant (P = 3.48 × 10−15; q = 1.26 × 10−11) (data not shown), which lies ∼12.5 Å away from codon 119 on an adjacent loop in the three-dimensional integrase protein structure (Fig. 6B).
We constructed additional site-directed mutants expressing the A91E variant alone as well as the A91E variant in combination with the S119R variant. The mutant expressing the A91E mutation alone exhibited a similar ∼35% lower RC than that of the parental NL4-3 strain; however, the combination of both the A91E and S119R mutations restored integrase RC to wild-type levels (Fig. 6A). Consistent with these results, the stratification of patient-derived S119R-containing integrase recombinant viruses into those expressing the S119R mutation only (n = 10) versus those expressing both the S119R and A91E mutations (n = 2) revealed higher RC when these two mutations were present together (Fig. 6C). The A91E mutation alone was not observed for any patient samples in our study. Together, these data demonstrate that reduced integrase function was observed for the C*05-associated S119R mutation and that this defect can be compensated for by a naturally occurring secondary mutation, A91E.
DISCUSSION
Despite the presence of immunogenic CTL epitopes in integrase restricted by protective HLA class I alleles and the reproducible patterns of immune escape in large-scale studies of infected individuals, our results indicate that the immune-driven attenuation of HIV-1 integrase function is not a general phenomenon during natural infection. In contrast to previous studies of recombinant viruses expressing Gag-protease (8, 31, 64), the range of integrase RC values observed in this study was relatively narrow, indicating that patient-derived sequences were in general highly functional. Nevertheless, we observed that polymorphisms at HLA-associated sites were more costly than those at non-HLA-associated sites, identified several HLA-associated polymorphisms that correlated with reduced RC, and confirmed a novel C*05-restricted epitope that may contribute to this effect. Together, these data suggest that immune pressure can select for sequence variants that compromise integrase function and reduce viral fitness.
Significant reductions in RC were observed in the context of certain HLA-associated mutations, of which the S119R, G163E, and I220L mutations were most prominent. These polymorphisms are selected by HLA class I alleles that are not considered to be protective, including C*05, A*33, and B*52, respectively. A previous study by our group featuring RT-integrase sequences derived from HIV-1 elite controllers identified an association between integrase codon 265 and lower RC (16); however, this was not observed in the present study. Notably, these attenuating mutations were not restricted by protective HLA alleles. Rather, polymorphisms that were significantly associated with RC were targeted by nonprotective alleles that have not been widely studied in the context of vaccine design. Since the majority of individuals in a population will not express a protective HLA class I allele, the development of a broadly effective CTL-based vaccine will likely require the design of immunogens that can stimulate host responses restricted by more common alleles (44). Our observations are therefore encouraging, since they support the notion that nonprotective HLA may be harnessed to more effectively target vulnerable regions of HIV-1, including sites in the integrase gene.
The integrase polymorphisms S119R and I220L are likely to lie within novel CTL epitopes restricted by HLA-C*05 and -B*52, respectively. Using functional (IFN-γ ELISpot) and computational approaches, we identified putative candidate epitope sequences at both positions. We validated one prediction by mapping a new optimal epitope (HTDNGSNF114–121) in a C*05-expressing individual which spans position S119. This result highlights the utility of combining sequence-based analyses of population-level immune escape with large-scale functional analyses to guide vaccine discovery for novel HIV-1 epitopes, particularly for viral proteins outside Gag and for HLA alleles that are not typically considered to be protective (2, 4).
Using site-directed mutagenesis, we confirmed that the most significant association identified using patient sequences, the S119R mutation, displayed a ∼35%-reduced RC when it was engineered alone into laboratory-adapted isolate NL4-3. We also identified a secondary mutation, A91E, which restored RC to wild-type levels in NL4-3 and which appeared to compensate for the S119R mutation in 2 out of 12 patient sequences. Covariation between integrase codons 91 and 119 was reported previously (52), although functional studies to test their interaction have not been performed previously. Notably, the impact of the S119R mutation on RC was substantially greater for NL4-3 than for patient-derived recombinant viruses, and the A91E mutation appeared to contribute to RC recovery in only a few cases. This finding strongly suggests that alternative mechanisms of S119R compensation exist; however, to date, we have not been able to identify additional pathways using our data.
Mutations at HIV-1 integrase codon 119 are known to affect integration site selection (27). A recent high-resolution structure of the prototype foamy virus (PFV) integrase bound to viral and target DNA demonstrated that residue A188 (which is homologous to HIV-1 integrase codon 119) formed a van der Waals interaction with a key cytosine in the minor groove of the target DNA sequence (40). The reduced viral RC that we observed with the S119R mutation is therefore consistent with a defect in integrase function at the step of target site recognition and strand transfer. Specifically, our data suggest that the substitution of a positively charged amino acid at this position (arginine) may be more costly to integrase function than the more conservative changes that have been analyzed previously, such as threonine, glycine, or alanine (27). More detailed studies will be required to confirm this hypothesis and to assess the mechanism by which the A91E mutation compensates for the S119R mutation. Similarly, additional work will be necessary to examine the other integrase mutations identified in this study, including assays to directly assess in vitro enzymatic activity. Codon K188, which is part of the KRK motif (residues 186 to 188), forms a salt bridge with D25 that stabilizes the multimer structure, and it may participate with K215 and I220 in dimerization and viral DNA binding (18, 37), but putative roles for codons D6, K111, G163, D167, Y227, and G283 have not been described. The generally conservative nature of amino acid substitutions observed at these sites suggests important structural contributions of these amino acids (52).
Our analyses of integrase did not identify a consistent HLA allele or common HLA-associated polymorphism that contributed substantially to RC. Rather, we observed a small number of infrequent polymorphisms that appeared to compromise RC. Since our study of 392 recombinant viruses is well powered to assess frequent cases, our results indicate that immune selection pressure on integrase can result in a functional impairment that affects viral RC. However, the specific HLA-associated polymorphisms identified appear to be relatively rare escape mutations at the population level. For example, the S119R mutation is likely to represent the less common of two possible escape mutations in the C*05-restricted HTDNGSNF epitope described here. In a large cohort of infected individuals (15), the S119R mutation was observed for 13.9% of C*05-expressing individuals, while the more common S119P variant (which was not associated with lower RC in our study) was observed for 40.2% of persons expressing C*05. Such differences in the frequency of escape mutations can be attributed to a variation in host CTL pressure (i.e., antigen receptor clonotypes) or intrinsic barriers to incorporating changes in the context of the founder viral protein sequence (i.e., the presence of secondary mutations that reduce fitness costs). To best utilize functional information for vaccine design, it may be necessary not only to enhance the frequency of CTL responses in persons expressing the relevant HLA but also to devise novel strategies that channel HIV-1 evolution in favor of mutations that reduce viral fitness. Indeed, the design of vaccine immunogens that incorporate both the wild-type sequence as well as the preferred escape variant(s) represents one feasible approach to achieve this end (4, 7, 14).
Several other caveats and limitations of this study merit mention. First, our experimental design utilized recombinant viruses that focused on a single gene product, and as such, potential epistatic interactions with other viral proteins that could affect RC have not been taken into account (17, 28). Indeed, false-positive (and false-negative) results could arise due to the choice of NL4-3 as a backbone vector for this work, and studies using other viral strains and whole-virus isolates may be necessary to determine the full impact of the polymorphisms identified here on viral fitness. Second, the relative reductions in viral RC observed for patient-derived sequences were modest, particularly in comparison to our previous results for Gag. This might be attributed to the relative sequence conservation of integrase (65) and its essential enzymatic role in the HIV-1 replication cycle. Notably, data from a recent report (49) indicated that the three codons associated with RC in our study demonstrate a higher degree of polymorphism than other integrase codons, S119 (7.5% nonconsensus amino acids), G163 (1.8%), and I220 (1.8%), versus a median integrase value of 0% (IQR, 0 to 0.5) (P = 0.006) (not shown). Finally, we did not observe a significant correlation between RC and clinical measures of HIV-1 infection (pVL and CD4 count). This may be due in part to the integrase-focused experimental design, the relatively progressed status of our chronic cohort, the small number of polymorphisms identified that were associated with integrase function, and the ability of secondary mutations to compensate for impaired RC in some cases. Despite these limitations, we were able to uncover HLA-associated polymorphisms that may contribute to integrase function, we confirmed the functional impact of our top hit (S119R) and a compensatory mechanism (A91E) using site-directed mutants, and we identified a novel C*05-restricted CTL response that is likely to contribute to these observations.
In summary, the HLA-mediated impairment of HIV-1 integrase does not appear to be a general phenomenon during viral adaptation to host immune responses. Nevertheless, we identified uncommon immune-driven polymorphisms that were associated with reduced viral RC. Attenuating mutations were restricted by HLA class I alleles that are not conventionally regarded as being protective, including the S119R mutation, which is situated within a novel C*05 epitope. This study highlights the potential utility of population-based functional studies for immunogen discovery. Indeed, from a vaccine perspective, it is encouraging to uncover new examples of viral mutations selected by nonprotective HLA alleles that may be able to reduce HIV-1 fitness. The data presented here suggest that certain integrase epitopes could be considered for vaccine strategies designed to diminish HIV-1 pathogenesis.
ACKNOWLEDGMENTS
We gratefully acknowledge Pamela Rosato for technical assistance with site-directed mutagenesis and viral vector construction; Christopher Lachowski for DNA sequencing; and Conan Woods, Calvin Lai, and Caitlin Johnson for data support. We also thank Evan Wood, Thomas Kerr, and the AIDS Care Cohort To Evaluate Access to Survival Services (ACCESS) project team for providing plasma samples used in this study.
This work was supported by an operating grant from the Canadian Institutes for Health Research (CIHR) (MOP-93536), by a Jim Gray Seed grant from Microsoft Research (to M.A.B. and Z.L.B.), and by the National Institutes of Health (NIH) (grant RO1AI030914 to B.D.W.). The VIDUS and ACCESS projects were funded by the National Institute on Drug Abuse, NIH (grants RO1DA011591 and RO1DA021525). D.R.C. is a recipient of the Canada-HOPE fellowship from CIHR and Sanofi-Aventis. C.J.B. is supported by a Vanier Canada graduate scholarship from the CIHR. E.M. is supported by a master's scholarship from the Canadian Association of HIV Research and Abbott Virology. P.R.H. is the CIHR-GSK research chair in HIV/AIDS. M.A.B. holds a Canada research chair, tier 2, in viral pathogenesis and immunity. Z.L.B. is the recipient of a CIHR new investigator award.
The opinions expressed in this work do not necessarily reflect the official policies of project funders, nor does mention of trade names, commercial practices, or organizations imply endorsement by the U.S. or Canadian government.
Footnotes
Published ahead of print 11 April 2012
REFERENCES
- 1. Allen TM, et al. 2005. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J. Virol. 79:13239–13249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Almeida CA, et al. 2011. Translation of HLA-HIV associations to the cellular level: HIV adapts to inflate CD8 T cell responses against Nef and HLA-adapted variant epitopes. J. Immunol. 187:2502–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altfeld M, et al. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8(+) T cell response against HIV-1. PLoS Med. 3:e403 doi:10.1371/journal.pmed.0030403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhattacharya T, et al. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 315:1583–1586 [DOI] [PubMed] [Google Scholar]
- 5. Boutwell CL, Essex M. 2007. Identification of HLA class I-associated amino acid polymorphisms in the HIV-1C proteome. AIDS Res. Hum. Retroviruses 23:165–174 [DOI] [PubMed] [Google Scholar]
- 6. Boutwell CL, Rowley CF, Essex M. 2009. Reduced viral replication capacity of human immunodeficiency virus type 1 subtype C caused by cytotoxic-T-lymphocyte escape mutations in HLA-B57 epitopes of capsid protein. J. Virol. 83:2460–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brander C, Frahm N, Walker BD. 2006. The challenges of host and viral diversity in HIV vaccine design. Curr. Opin. Immunol. 18:430–437 [DOI] [PubMed] [Google Scholar]
- 8. Brockman MA, et al. 2010. Early selection in Gag by protective HLA alleles contributes to reduced HIV-1 replication capacity that may be largely compensated for in chronic infection. J. Virol. 84:11937–11949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brockman MA, et al. 2007. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J. Virol. 81:12608–12618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brockman MA, Tanzi GO, Walker BD, Allen TM. 2006. Use of a novel GFP reporter cell line to examine replication capacity of CXCR4- and CCR5-tropic HIV-1 by flow cytometry. J. Virol. Methods 131:134–142 [DOI] [PubMed] [Google Scholar]
- 11. Brooks JI, et al. 2009. Evaluation of an automated sequence analysis tool to standardize HIV genotyping results, abstr O061. Abstr. 18th Annu. Can. Conf. HIV/AIDS Res., Vancouver, British Columbia, Canada [Google Scholar]
- 12. Brumme ZL, et al. 2008. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J. Virol. 82:9216–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brumme ZL, et al. 2007. Effects of human leukocyte antigen class I genetic parameters on clinical outcomes and survival after initiation of highly active antiretroviral therapy. J. Infect. Dis. 195:1694–1704 [DOI] [PubMed] [Google Scholar]
- 14. Brumme ZL, et al. 2007. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 3:e94 doi:10.1371/journal.ppat.0030094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brumme ZL, et al. 2009. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS One 4:e6687 doi:10.1371/journal.pone.0006687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brumme ZL, et al. 2011. Reduced replication capacity of NL4-3 recombinant viruses encoding reverse transcriptase-integrase sequences from HIV-1 elite controllers. J. Acquir. Immune Defic. Syndr. 56:100–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Buzon MJ, et al. 2010. The HIV-1 integrase genotype strongly predicts raltegravir susceptibility but not viral fitness of primary virus isolates. AIDS 24:17–25 [DOI] [PubMed] [Google Scholar]
- 18. Chen JC, et al. 2000. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl. Acad. Sci. U. S. A. 97:8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cherepanov P, Maertens GN, Hare S. 2011. Structural insights into the retroviral DNA integration apparatus. Curr. Opin. Struct. Biol. 21:249–256 [DOI] [PubMed] [Google Scholar]
- 20. Chopera DR, et al. 2008. Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage. PLoS Pathog. 4:e1000033 doi:10.1371/journal.ppat.1000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fiebig EW, et al. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 17:1871–1879 [DOI] [PubMed] [Google Scholar]
- 22. Frahm N, et al. 2004. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J. Virol. 78:2187–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frahm N, et al. 2007. Extensive HLA class I allele promiscuity among viral CTL epitopes. Eur. J. Immunol. 37:2419–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Friedrich TC, et al. 2004. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med. 10:275–281 [DOI] [PubMed] [Google Scholar]
- 25. Goepfert PA, et al. 2008. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J. Exp. Med. 205:1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 27. Harper AL, Sudol M, Katzman M. 2003. An amino acid in the central catalytic domain of three retroviral integrases that affects target site selection in nonviral DNA. J. Virol. 77:3838–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a. Heckerman D, Kadie CM, Listgarten J. 2006. Leveraging information across HLA alleles/supertypes improves HLA-specific epitope prediction. RECOMB 2006, Venice, Italy, 2 to 5 April 2006 [DOI] [PubMed] [Google Scholar]
- 28. Hinkley T, et al. 2011. A systems analysis of mutational effects in HIV-1 protease and reverse transcriptase. Nat. Genet. 43:487–489 [DOI] [PubMed] [Google Scholar]
- 29. Honeyborne I, et al. 2007. Control of human immunodeficiency virus type 1 is associated with HLA-B*13 and targeting of multiple gag-specific CD8+ T-cell epitopes. J. Virol. 81:3667–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu Z, Kuritzkes DR. 2010. Effect of raltegravir resistance mutations in HIV-1 integrase on viral fitness. J. Acquir. Immune Defic. Syndr. 55:148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang KH, et al. 2011. Progression to AIDS in South Africa is associated with both reverting and compensatory viral mutations. PLoS One 6:e19018 doi:10.1371/journal.pone.0019018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang X, Zhang J. 1996. Methods for comparing a DNA sequence with a protein sequence. Comput. Appl. Biosci. 12:497–506 [DOI] [PubMed] [Google Scholar]
- 33. Janssen RS, et al. 1998. New testing strategy to detect early HIV-1 infection for use in incidence estimates and for clinical and prevention purposes. JAMA 280:42–48 [DOI] [PubMed] [Google Scholar]
- 34. Kawashima Y, et al. 2010. Long-term control of HIV-1 in hemophiliacs carrying slow-progressing allele HLA-B*5101. J. Virol. 84:7151–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kiepiela P, et al. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46–53 [DOI] [PubMed] [Google Scholar]
- 36. Larsen MV, et al. 2005. An integrative approach to CTL epitope prediction: a combined algorithm integrating MHC class I binding, TAP transport efficiency, and proteasomal cleavage predictions. Eur. J. Immunol. 35:2295–2303 [DOI] [PubMed] [Google Scholar]
- 37. Li X, Krishnan L, Cherepanov P, Engelman A. 2011. Structural biology of retroviral DNA integration. Virology 411:194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Y, et al. 2006. Selection on the human immunodeficiency virus type 1 proteome following primary infection. J. Virol. 80:9519–9529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lobritz MA, Lassen KG, Arts EJ. 2011. HIV-1 replicative fitness in elite controllers. Curr. Opin. HIV AIDS 6:214–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maertens GN, Hare S, Cherepanov P. 2010. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468:326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martinez-Picado J, et al. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 80:3617–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miura T, et al. 2009. HLA-associated alterations in replication capacity of chimeric NL4-3 viruses carrying gag-protease from elite controllers of human immunodeficiency virus type 1. J. Virol. 83:140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miura T, et al. 2010. Impaired replication capacity of acute/early viruses in persons who become HIV controllers. J. Virol. 84:7581–7591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mothe B, et al. 2011. Definition of the viral targets of protective HIV-1-specific T cell responses. J. Transl. Med. 9:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nguyen BY, et al. 2011. Raltegravir: the first HIV-1 integrase strand transfer inhibitor in the HIV armamentarium. Ann. N. Y. Acad. Sci. 1222:83–89 [DOI] [PubMed] [Google Scholar]
- 46. O'Connell KA, Hegarty RW, Siliciano RF, Blankson JN. 2011. Viral suppression of multiple escape mutants by de novo CD8(+) T cell responses in a human immunodeficiency virus-1 infected elite suppressor. Retrovirology 8:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Payne RP, et al. 2010. Efficacious early antiviral activity of HIV Gag- and Pol-specific HLA-B 2705-restricted CD8+ T cells. J. Virol. 84:10543–10557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peyerl FW, et al. 2004. Fitness costs limit viral escape from cytotoxic T lymphocytes at a structurally constrained epitope. J. Virol. 78:13901–13910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Powderly WG. 2010. Integrase inhibitors in the treatment of HIV-1 infection. J. Antimicrob. Chemother. 65:2485–2488 [DOI] [PubMed] [Google Scholar]
- 50. Prado JG, et al. 2009. Functional consequences of human immunodeficiency virus escape from an HLA-B*13-restricted CD8+ T-cell epitope in p1 Gag protein. J. Virol. 83:1018–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Quinones-Mateu ME, Moore-Dudley DM, Jegede O, Weber J, Arts EJ. 2008. Viral drug resistance and fitness. Adv. Pharmacol. 56:257–296 [DOI] [PubMed] [Google Scholar]
- 52. Rhee SY, et al. 2008. Natural variation of HIV-1 group M integrase: implications for a new class of antiretroviral inhibitors. Retrovirology 5:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rodriguez WR, et al. 2004. CD8+ T lymphocyte responses target functionally important regions of protease and integrase in HIV-1 infected subjects. J. Transl. Med. 2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rousseau CM, et al. 2008. HLA class-I driven evolution of human immunodeficiency virus type 1 subtype C proteome: immune escape and viral load. J. Virol. 82:6434–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schneidewind A, et al. 2007. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 81:12382–12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100:9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Streeck H, et al. 2009. Human immunodeficiency virus type 1-specific CD8+ T-cell responses during primary infection are major determinants of the viral set point and loss of CD4+ T cells. J. Virol. 83:7641–7648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tomiyama H, et al. 1999. Identification of multiple HIV-1 CTL epitopes presented by HLA-B*5101 molecules. Hum. Immunol. 60:177–186 [DOI] [PubMed] [Google Scholar]
- 59. Troyer RM, et al. 2009. Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoS Pathog. 5:e1000365 doi:10.1371/journal.ppat.1000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ueno T, et al. 2008. CTL-mediated selective pressure influences dynamic evolution and pathogenic functions of HIV-1 Nef. J. Immunol. 180:1107–1116 [DOI] [PubMed] [Google Scholar]
- 61. Wainberg MA. 2004. The impact of the M184V substitution on drug resistance and viral fitness. Expert Rev. Anti Infect. Ther. 2:147–151 [DOI] [PubMed] [Google Scholar]
- 62. Wang JY, Ling H, Yang W, Craigie R. 2001. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 20:7333–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wood E, et al. 2005. Baseline self-perceived risk of HIV infection independently predicts the rate of HIV seroconversion in a prospective cohort of injection drug users. Int. J. Epidemiol. 34:152–158 [DOI] [PubMed] [Google Scholar]
- 64. Wright J, et al. 2010. Gag-protease-mediated replication capacity in HIV-1 subtype C chronic infection: associations with HLA type and clinical parameters. J. Virol. 84:10820–10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yusim K, et al. 2002. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J. Virol. 76:8757–8768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zaunders J, Dyer WB, Churchill M. 2011. The Sydney Blood Bank Cohort: implications for viral fitness as a cause of elite control. Curr. Opin. HIV AIDS 6:151–156 [DOI] [PubMed] [Google Scholar]