Abstract
Influenza virus nucleoprotein (NP) is the major component of the viral ribonucleoprotein complex, which is crucial for the transcription and replication of the viral genome. We have determined the crystal structure of influenza B virus NP to a resolution of 3.2 Å. Influenza B NP contains a head, a body domain, and a tail loop. The electropositive groove between the head and body domains of influenza B NP is crucial for RNA binding. This groove also contains an extended flexible charged loop (amino acids [aa] 125 to 149), and two lysine clusters at the first half of this loop were shown to be crucial for binding RNA. Influenza B virus NP forms a crystallographic homotetramer by inserting the tail loop into the body domain of the neighboring NP molecule. A deeply buried salt bridge between R472 and E395 and a hydrophobic cluster at F468 are the major driving forces for the insertion. The analysis of the influenza B virus NP structure and function and comparisons with influenza A virus NP provide insights into the mechanisms of action and underpin efforts to design inhibitors for this class of proteins.
INTRODUCTION
Influenza viruses are RNA viruses and classified into three types: A, B, and C. While much attention has been paid to influenza A virus, the severity of influenza B virus cannot be underestimated. In the past 70 years, there have been 16 epidemics, resulting in excess morbidity and mortality, at least partially caused by influenza B virus (34). From July 2010 to June 2011, 25.0% of influenza positive specimens found globally were of influenza B virus (33). Influenza B virus also causes substantial mortality among pediatric patients. Among the 116 deaths associated with influenza infections occurring in the 2010-2011 flu season in the United States, 45 were due to influenza B virus (6). Therefore, it is important to find out how influenza B virus functions and compare it with influenza A virus, so that more effective means could be developed to combat the highly infectious influenza viruses in general.
The genome of influenza B virus comprises eight negative-sense RNA segments encoding 11 polypeptides (14). Among these proteins, nucleoprotein (NP) is the major component of the ribonucleoprotein complex (RNP), which consists of RNA, NP, and RNA polymerase and plays a vital role in the transcription and replication of the viral genome (reviewed in reference 25).
Influenza B virus NP (BNP) is a basic protein (pI > 9) with 560 amino acids and molecular mass of 62 kDa. The primary sequence of BNP has several distinctive features compared to influenza A virus NP (ANP): (i) BNP contains a significantly extended N-terminal region (amino acids [aa] 1 to 70); (ii) both nuclear localization signal 1 (NLS-1) and NLS-2 in ANP (31, 32) are absent in BNP; (iii) some known RNA-binding regions in ANP, especially the flexible basic loop of aa 74 to 88 (21), are not conserved in BNP; (iv) the linkers and tail loops for NP homo-oligomerization are not conserved; and (v) gaps are found when aligning the C termini of ANP and BNP. Recently, the crystal structures of ANP from H5N1 and H1N1 viruses have been determined by us and others (12, 21, 35). Here we report the crystal structure of BNP (Protein Data Bank [PDB] code: 3TJ0) at 3.2 Å and compare its structure and function with those of ANP for RNA binding and for forming oligomers.
MATERIALS AND METHODS
Biological materials.
The 293T cell line (ATCC, Manassas, VA) was cultivated in minimal essential medium (MEM) (Invitrogen, Carlsbad, CA) with 10% fetal calf serum (Invitrogen). Anti-BNP antibody (Santa Cruz Biotechnology, Santa Cruz, CA), anti-myc antibody (Cell Signaling Technology), anti-Flag antibody (Sigma-Aldrich, St. Louis, MO), and anti-beta-actin antibody (GenScript, Piscataway, NJ) were purchased commercially. Plasmids pCIPA, pCIPB1, and pCIPB2 expressing RNA polymerase subunits of influenza B/Panama/45/90 virus were described previously (15). Plasmid pPolI-Luc-RT for the generation of pPol-Luci-BNA-RT and pEGFP were kindly provided by L. L. M. Poon of the University of Hong Kong (18). The pPol-Luci-BNA-RT transcribes a viral RNA (vRNA)-like RNA, in which the noncoding sequences of influenza B NA segment flank the coding region of firefly luciferase. Plasmid pcDNA-BNP was generated by inserting the wild-type and mutant NP genes of B/HongKong/CUHK-24964/2004 into mammalian expression vector pcDNA3 (Invitrogen) for BNP expression in 293T cells. The genes of wild-type and mutant BNP were also cloned into pcDNA3.1/myc-His (Invitrogen) for myc-tagged BNP expression in mammalian cells. pCMV-Tag2B obtained from Agilent Technologies, Inc., Santa Clara, CA, was for Flag-tagged BNP expression in mammalian cells, and pRHisMBP obtained from K. B. Wong, the Chinese University of Hong Kong, was for the expression of maltose binding protein (MBP)-tagged BNP variants in Escherichia coli.
Expression and purification of BNP.
MBP-tagged BNP variants were expressed in Escherichia coli C41 (DE3). The cells were lysed in 20 mM sodium phosphate, 150 mM NaCl, pH 6.5. The lysate was passed through an amylose column (New England BioLabs, Ipswich, MA). The bound protein was eluted in buffer containing 20 mM maltose. The eluate was incubated with thrombin (100 U) (Sigma-Aldrich) and RNase A (300 U) (Sigma-Aldrich) at 4°C overnight to remove the MBP tag and RNA from BNP. It was then passed through a heparin high-performance (HP) column (GE Healthcare, Waukesha, WI). NP was eluted with a 0 to 1.5 M NaCl gradient in the same buffer. For the purpose of crystallization, the BNP protein was concentrated and buffer exchanged into 20 mM MOPS (morpholinepropanesulfonic acid), 100 mM NaCl, pH 7.0.
Crystallization and structure determination.
RNase-treated BNP in a concentration of 15 mg/ml was mixed in a 1:1 ratio with 1.4 M phosphate, pH 7.0. Crystals were grown in hanging drops at 16°C. Crystals were frozen in crystallization buffer with 20% glycerol and brought to the Shanghai Synchrotron Radiation Facility for data collection. The data were collected at 100 K using beamline BL17U (with X-rays at a wavelength of 0.97941 Å) and were processed and scaled with the HKL2000 suite (22). The structure was determined by molecular replacement using the tail loop-deleted influenza A H5N1 NP monomer (21) as the search model. Calculations and model building were carried out using Phenix (1) and Coot (10), respectively. The final model was analyzed by the program PROCHECK (16). Figures of protein structures were prepared with the program PyMOL (9).
Polymerase activity assay.
Plasmids pCIPA, pCIPB1, pCIPB2, pcDNA-BNP (wild type or NP mutants), and pPol-Luci-BNA-RT (0.125 μg each, except the amount of pcDNA-BNP was individually adjusted according to their expression levels) were cotransfected to 1.125 × 105 293T cells for RNP complex reconstitution. Plasmid for enhanced green fluorescent protein (EGFP) expression (0.06 μg) was also cotransfected for normalization (18). For the negative control, empty pcDNA plasmid instead of pcDNA-BNP was transfected to cells. At 48 h posttransfection, 293T cells were lysed by Steady-Glo assay reagent (Promega) for 5 min followed by luminescence measurement on a luminometer (Victor2 1420 multilabel counter; Wallac) according to the manufacturer's instructions.
Static light scattering.
Wild-type or variant BNP proteins were subject to static light scattering using a miniDAWN triangle (45°, 90°, and 135°) light scattering detector (Wyatt Technology, Santa Barbara, CA) connected to an Optilab DSP interferometric refractometer (Wyatt Technology Corporation, Santa Barbara, CA). This system was connected to a Superdex 200 column (GE Healthcare) controlled by an AKTAexplorer chromatography system (GE Healthcare). Before sample injection, the miniDAWN detector system was equilibrated with 100 mM sodium phosphate (pH 6.0) and 100 mM NaCl for at least 2 h to ensure a stable baseline signal. The flow rate was set to 0.5 ml/min, and the sample volume was 100 μl. The laser scattering (687 nm) and the refractive index (690 nm) of the respective protein solutions were recorded. Wyatt ASTRA software was used to evaluate all data obtained.
Coimmunoprecipitation.
Two micrograms of Flag-tagged and myc-tagged BNP plasmids were transfected into human kidney 293T cells in suspension. Since the presence of RNA may cause nonspecific interaction of BNP molecules without the tail loop insertion event, the G1 cluster in some of these myc- and Flag-tagged BNP constructs were mutated to alanine, for preventing the binding of RNA from interfering with the results interpretation. Coimmunoprecipitation was performed at 48 h posttransfection. Cells were lysed in 50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100, pH 7.6 (co-IP buffer). The lysate was centrifuged at 16,000 × g for 10 min at 4°C. The supernatant was treated with 150 U RNaseA (Sigma-Aldrich) and incubated at 4°C overnight with or without anti-Myc antibody (Cell Signaling Technology, Danvers, MA). The mixture was then incubated with protein A beads for 1.5 h at 4°C with shaking. The beads were centrifuged and washed with co-IP buffer three times before being boiled in SDS loading dye and analyzed by Western blotting.
Surface plasmon resonance (SPR).
A biotinylated 2′-O-methylated RNA oligonucleotide with the sequence 5′-UUU GUU ACA CAC ACA CAC GCU GUG-3′ was prepared in running buffer (10 mM Tris-HCl, 150 mM NaCl, 0.005% surfactant P20, 1 mM dithiothreitol [DTT], 5% glycerol, pH 8) and immobilized on an SA sensor chip (GE Healthcare) until the surface density reached 30 to 35 response units (RU), according to manufacturer's instructions (GE Healthcare). Kinetic measurements were carried out with a BIAcore 3000 system at 25°C. Concentration series of NP variants were injected onto the SA chip at 30 μl/min in running buffer. NaCl (2 M) was used to regenerate the chip surface. Data were analyzed with BIAevaluation v. 4.1 software, using the “1:1 Langmuir” model.
RESULTS
Crystal structure of influenza B nucleoprotein.
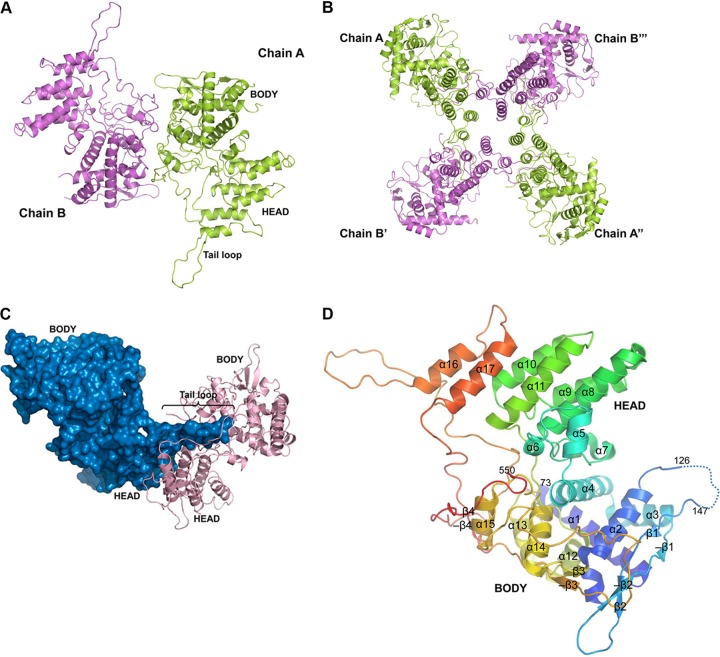
BNP was overexpressed in Escherichia coli and purified to more than 95% homogeneity. BNP formed oval-shaped crystals in orthorhombic space group I222. A diffraction data set of 3.2 Å resolution was collected and the structure of BNP was determined by molecular replacement, using the influenza A H5N1 NP monomer (PDB: 2Q06, chain A) as the search model, with the linkers and tail loop region (aa 395 to 437) excluded. The structure was refined to R/Rfree = 0.245/0.291 (Table 1). One asymmetric unit contains two BNP molecules, named chains A and B (Fig. 1A). In the structure, 458 residues (aa 73 to 126 and 147 to 550) were modeled for chain A, while 457 residues (aa 72 to 124 and 148 to 551) for chain B. BNP was crystallized in tetrameric form in this study (Fig. 1B), in contrast to the trimeric forms of ANP (12, 21, 35). Their differences and the biological implications of these will be further discussed below.
Table 1.
Data collection and refinement statistics (molecular replacement)
| Parameter | Value for BNPb |
|---|---|
| Data collection statistics | |
| Space group | I 2 2 2 |
| Cell dimensions | |
| a, b, c (Å) | 106.95, 123.34, 198.08 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Resolution (Å) | 104.73–3.23 (3.35–3.23)a |
| Rsym or Rmerge | 0.113 (0.387) |
| I /σI | 13.8 (2.9) |
| Completeness (%) | 93.2 (95.1) |
| Redundancy | 7.2 (7.3) |
| Refinement statistics | |
| Resolution range (Å) | 47.055–3.233 |
| No. of reflections | 19,862 |
| Rwork/Rfree | 0.245/0.291 |
| No. of atoms | 7022 |
| B-factors | 77.10 |
| RMSD | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.729 |
| Ramachandran plot (%) | |
| Most favored | 88.5 |
| Additional allowed | 9.0 |
| Generously allowed | 2.5 |
| Disallowed | 0.0 |
Data from a single crystal.
Values in parentheses are for highest-resolution shell.
Fig 1.
Crystal structure of BNP. (A) Two molecules in the asymmetric unit of the BNP crystal structure. (B) BNP forms a crystallographic tetramer, with the tail loop inserting in the order of A-B′-A″-B‴-A. (C) BNP (surface representation in blue) inserts its tail loop into the neighboring molecule (pink). (D) Structural organization of BNP (chain A). α-Helices and β-strands are numbered. Antiparallel β-strands are denoted with “−”.
BNP folds into a head and a body domain, both of which are largely helical. The polypeptide chain goes back and forth several times between the two domains, which could impose a constraint on the relative movement of the two domains. The electron densities for the first 71 residues of BNP were not visible, indicating that this N-terminal region of the protein is highly flexible. The head domain consists of aa 212 to 323 and 496 to 509, while the body domain is formed by aa 72 to 208, 336 to 404, and 521 to 551. Like what is seen in ANP, a special feature of BNP is a long tail loop (aa 459 to 486) which inserts into a neighboring molecule, facilitating the formation of BNP homo-oligomers (Fig. 1C). These structural features are linked together by several loop regions (aa 209 to 211, 324 to 335, 405 to 458, 487 to 495, and 510 to 520) (Fig. 1D).
The overall folds of influenza A and B NP are similar, yet a few regions have high root-mean-square deviation (RMSD) of Cα, and some regions could not be aligned together. For example, the tail loop is oriented differently in the two structures. The least-aligned regions are mostly flexible surface loops. One of these loops (aa 263 to 271) in BNP was found to be a cytotoxic-T-lymphocyte epitope (26).
The role of the basic groove and the flexible charged loop in RNA binding.
Purified influenza virus NP could bind single-stranded RNA, but with no or little sequence specificity (4). Purified monomeric ANP mutants were found to have significantly reduced RNA-binding affinity. On the other hand, purified trimeric and tetrameric ANP, which better represent the oligomeric NP in physiological states, bear high affinity toward RNA (5, 7).
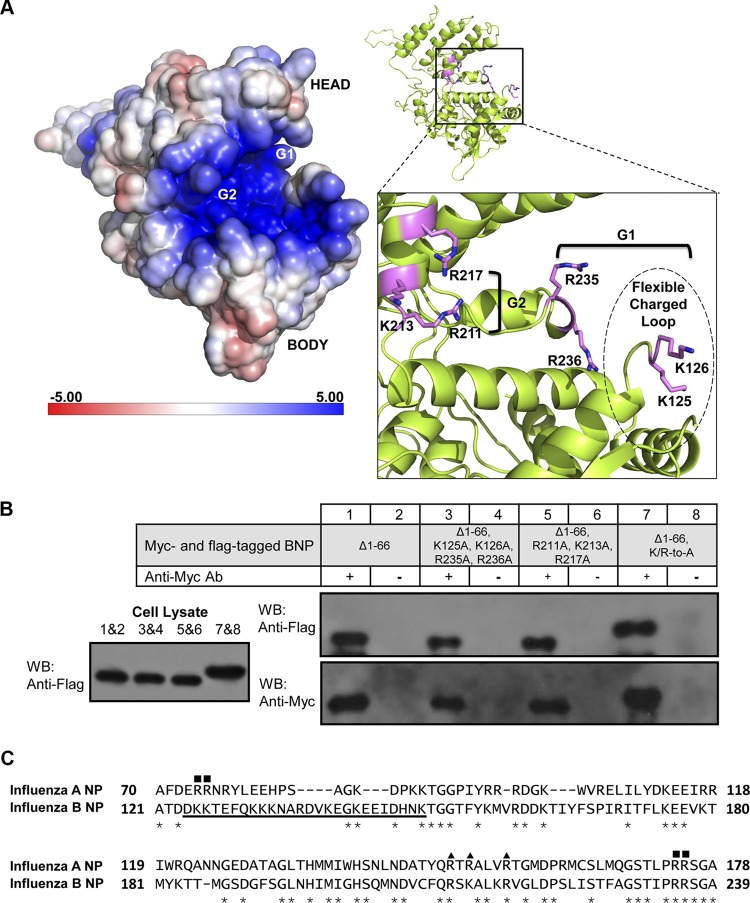
In the BNP structure, an electropositive groove is found between the head and body domains of BNP, which is likely to interact with RNA (Fig. 2A). Two clusters of positively charged residues are identified in this groove, including clusters G1 (aa K125, K126, R235, R236) and G2 (aa R211, K213, R217). Charged-to-alanine mutations on these two clusters reduced the polymerase activity to less than 10% of the wild type (Table 2). Surface plasmon resonance (SPR) also showed that the protein variants were unable to interact with RNA (Table 2). It is noted that the homo-oligomerization of the variants was not affected by the mutations (Fig. 2B). The findings are consistent with the phenotypes of the corresponding ANP mutants (reference 21 and data not shown).
Fig 2.
RNA binding of BNP. (A) Electrostatic potential representation of BNP. The groove between the head and body domains is highly electropositive. Residues in the G1 and G2 clusters are shown as stick models. The estimated position of the flexible charged loop is indicated in the broken-line circle. (B) Coimmunoprecipitation of BNP RNA-binding mutants. The three protein variants which showed no binding toward RNA in the SPR experiment could homo-oligomerize. The DNA sequences of the mutants were confirmed by sequencing. Ab, antibody; WB, Western blot. (C) Sequence alignment of ANP and BNP shows that BNP has a much lengthened charged loop (underlined), while G1 (■) and G2 (▲) clusters are mostly conserved. Stars indicate identical sites.
Table 2.
RNA-binding affinity and polymerase activity of BNP mutants
| Target region | BNP | Polymerase activity assaya |
SPRb (mean Kd [nM]) | |
|---|---|---|---|---|
| Relative luciferase activity at 48 h posttransfection (%) | P | |||
| / | Wild type | 99.00 ± 3.42 | 12.8 ± 0.3 | |
| / | Negative controlc | 0.56 ± 0.04 | ** | — |
| G1 | K125A, K126A, R235A, R236A | 2.06 ± 0.58 | ** | ND |
| G2 | R211A, K213A, R217A | 2.58 ± 0.44 | ** | ND |
| 125KKTEFQKKKNARDVKEGKEEIDHNK149 | Δ125–146 | 2.14 ± 0.60 | ** | 195 ± 9.5 |
| 125KKTAFQKKKNARAVKAGKAAIAHNK149 | D/E-to-A | 47.77 ± 3.23 | ** | — |
| 125AATEFQAAANAADVAEGAEEIDHNA149 | K/R-to-A | 2.92 ± 1.39 | ** | ND |
| 125KKTEFQKKKNAADVAEGAEEIDHNA149 | R136A, K139A, K142A, K149A | 108.21 ± 1.58 | — | |
| 125AATEFQAAANARDVKEGKEEIDHNK149 | K125A, K126A, K131A, K132A, K133A | 5.85 ± 0.84 | ** | — |
| 125AATEFQKKKNARDVKEGKEEIDHNK149 | K125A, K126A | 101.17 ± 3.46 | — | |
| 125KKTEFQAAANARDVKEGKEEIDHNK149 | K131A, K132A, K133A | 81.21 ± 5.03 | * | — |
The residues with charge-to-alanine mutation are underlined. The relative luciferase activity reflects the ability of BNP mutants to support polymerase activity. Mutants which exhibited <10% activity of the wild type are defined as having very low or no polymerase activity. Mutants with 11 to 50% activity are defined as partially active. Mutants whose activities are 51 to 100% are active. *, P < 0.01; **, P < 0.001.
The dissociation constant (Kd) was measured by SPR. N-terminally (aa 1 to 66)-deleted BNP variants were used to prevent unwanted interference of the chip surface. It is noted that the N terminus did not interact with RNA (data not shown), and it has been previously shown that the N terminus is not required for the expression and replication of a model RNA (25). ND, the RNA-binding affinity is too weak to be determined; —, not attempted.
Transfection was carried out without the BNP plasmid.
Sequence alignment between ANP and BNP, however, identifies a much-extended flexible charged loop in BNP between aa 125 and aa 149. It contains nine basic and six acidic residues (Fig. 2C). Only aa 125, 126, and 147 to 149 were visible in the density map, showing that this loop is highly flexible. Deletion of this loop in BNP resulted in a 14-fold decrease in the RNA-binding affinity, which is significantly more dramatic than the 6.4-fold decrease in the flexible loop-deleted ANP equivalent (21), implying that this loop in BNP contributes more to RNA binding. While the loop-deleted BNP mutant rendered the polymerase inactive, mutation of the negative charges in the loop (D/E to A) did not cause significant reduction in polymerase activity (Table 2). However, when the positively charged residues in the loop were mutated to alanine, the K/R-to-A mutant lost most of the polymerase activity (Table 2). SPR data of the K/R-to-A mutant also showed no RNA binding, with affinity lower than that of the loop-deleted mutant (Table 2). This is possibly due to the charge-charge repulsion of the remaining acidic residues in the loop against the negatively charged RNA immobilized on the chip. Nevertheless, the positive charges in the loop are essential for RNA-binding affinity, as well as polymerase activity in the context of viral RNP. To further map the crucial basic residues on the flexible charged loop, we constructed another four multiple-point charged-to-alanine mutants. Residues R136, K139, K142, and K149 at the second half of the loop were found to be nonessential, as mutations of these did not reduce polymerase activity (Table 2). In contrast, five-point mutations on the first half of the loop (K125A, K126A, K131A, K132A, and K133A forming two clusters) reduced polymerase activity to 5.85% of the wild type, and the protein variant was unable to bind RNA in the SPR measurement (Table 2). However, mutating individual clusters ([K125A, K126A] or [K131A, K132A, K133A]) did not cause reduction in polymerase activities (Table 2). These data indicate that the two lysine clusters are important to the viral RNP activity and that either lysine cluster is sufficient for the flexible basic loop to bind RNA.
BNP homo-oligomerizes by tail loop insertion.
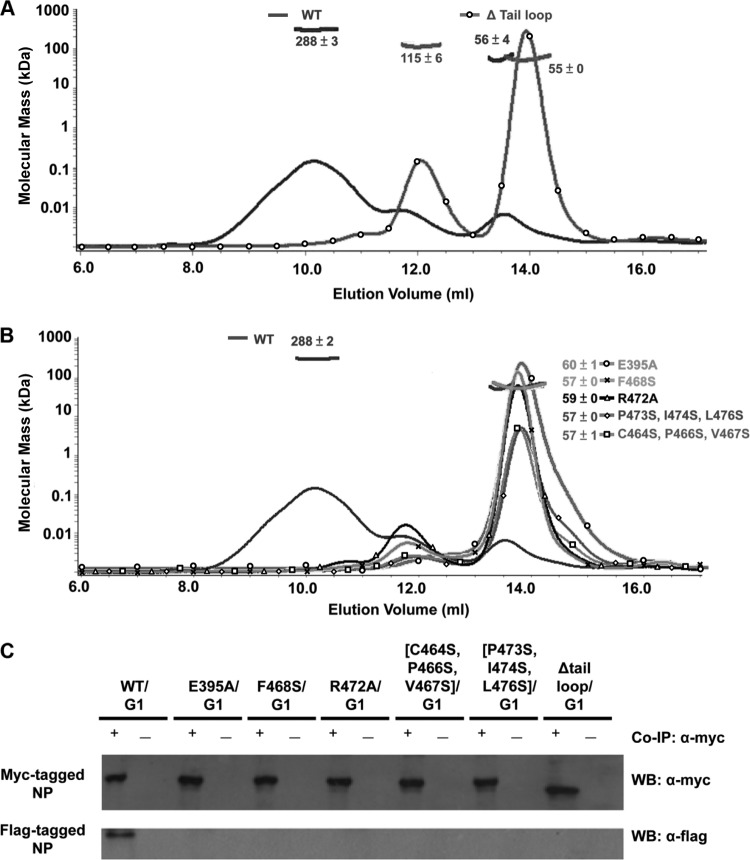
In the tetrameric crystal structure of BNP, each tetramer is composed of two A chains and two B chains (Fig. 1B). This is largely facilitated by the tail loop insertion of one molecule into the next, following the following order: from chain A to chain B′, to chain A″, to chain B‴, and back to chain A. The use of such an extended loop near the C terminus for homo-oligomerization is a common strategy adopted by NP molecules of different viruses, including influenza A virus (12, 21, 35), rabies virus (2), and vesicular stomatitis virus (13). The BNP tail loop (aa 458 to 484) (Fig. 3A) inserts deeply into the body groove of a neighboring BNP molecule (Fig. 1C). The insertion is crucial for the function of the RNP, as deletion of the tail loop resulted in a dramatic reduction (3.52% of the wild type) in polymerase activity (Table 3). Static light scattering showed that the tail loop-deleted BNP exists mainly in monomers, in contrast to the tetramer population of wild-type BNP in solution (Fig. 4A). The small proportion of dimer observed is probably due to the relatively weak interaction between the body domains of two BNP molecules, as seen in the crystal asymmetric unit (Fig. 1A). As coimmunoprecipitation showed that myc- and Flag-tagged tail loop deleted BNPs do not interact with each other (Fig. 4C), it is the tail loop insertion that provides the major driving force for oligomerization.
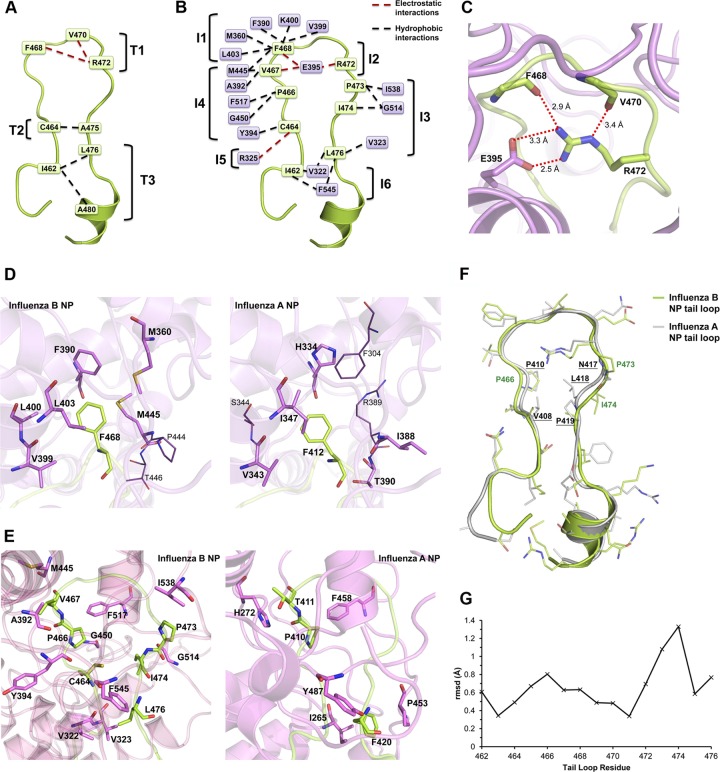
Fig 3.
Tail loop interaction of BNP. (A) Intrachain interactions of the tail loop. Electrostatic and hydrophobic interactions are in red and black dotted lines, respectively. (B) Interchain interactions between the tail loop and the neighboring BNP. Tail loop residues are in pale green boxes, while residues in neighboring BNP are in purple boxes. (C) The E395-R472 salt bridge between the tail loop (green) and the neighboring BNP (purple). The position of the R472 side chain is oriented by the hydrogen bond interaction with the main chain O of F468 and V470. (D) BNP F468 and ANP F412 make distinct interactions with the neighboring molecule. Residues in sticks interact with F468/F412, while residues in lines do not. M360, L400, and M445 of BNP interact with F468, but the equivalent F304, S344, and R38 of ANP do not. (E) Residues C464, P466, V467, P473, I474, and L476 in the stem region of the BNP tail loop (green) make novel contacts with the neighboring molecule (pink). These interactions are not observed in ANP. The stem region of the ANP tail loop forms a zig-zag pattern of intrachain hydrophobic interaction instead. (F) Structural alignment of the tail loops in ANP (gray) and BNP (green). I474 of BNP faces toward the neighboring molecule but not toward P466, which is different from L418 in ANP. (G) Graph of RMSD of individual residues when the stem and tip regions of the tail loops in ANP and BNP are aligned. Residues P473 and I474 are not well aligned.
Table 3.
Polymerase activity of BNP homo-oligomerization mutants
| Cluster | NP mutant disrupting the interactions in the cluster | Relative luciferase activity at 48 h posttransfection (%) | Pc |
|---|---|---|---|
| Wild type | 99.00 ± 3.42 | ||
| Negative control | 0.56 ± 0.04 | ** | |
| Δtail loop (aa 458–484)a | 3.52 ± 0.91 | ** | |
| Myc-tagged wild typea | 100.00 ± 12.4 | ||
| Myc-tagged Δtail loopa | 4.44 ± 0.86 | * | |
| T1 | R472Ab | 7.66 ± 1.34 | ** |
| T2 | C464Sb | 120.11 ± 7.90 | * |
| A475S | 124.98 ± 18.26 | ||
| T3 | I462Sb | 71.73 ± 1.79 | * |
| L476Sb | 77.77 ± 9.22 | ||
| A480S | 108.35 ± 5.21 | ||
| I1 | F468S | 3.07 ± 1.49 | ** |
| I2 | E395A | 4.14 ± 1.05 | ** |
| R472Ab | 7.66 ± 1.34 | ** | |
| I3 | P473S | 125.84 ± 23.03 | |
| I474S | 106.02 ± 2.12 | ||
| L476Sb | 77.77 ± 9.22 | ||
| [P473S, I474S] | 27.90 ± 4.91 | ** | |
| [P473S, L476S] | 49.11 ± 3.08 | ** | |
| [I474S, L476S] | 20.77 ± 2.41 | ** | |
| [P473S, I474S, L476S] | 9.15 ± 2.27 | ** | |
| Myc-tagged [P473S, I474S, L476S]a | 2.78 ± 1.39 | * | |
| I4 | C464Sb | 120.11 ± 7.90 | * |
| P466S | 79.13 ± 6.58 | ||
| V467S | 113.45 ± 8.08 | ||
| [464S, P466S] | 53.71 ± 3.85 | ** | |
| [C464S, V467S] | 87.04 ± 2.71 | ||
| [P466S, V467S] | 63.79 ± 1.64 | ** | |
| [C464S, P466S, V467S] | 8.87 ± 1.24 | ** | |
| I5 | R325A | 41.32 ± 0.79 | ** |
| I6 | I462Sb | 71.73 ± 1.79 | * |
Mutants which could not be detected with the anti-BNP commercial antibodies were also cloned into myc-tagged vector, and their polymerase activities were compared to those of the myc-tagged wild-type BNP.
Mutants which are in bold appear in more than one cluster.
*, P < 0.01; **, P < 0.001.
Fig 4.
Static light scattering and coimmunoprecipitation of the homo-oligomerization mutants. (A) Static light scattering of wild-type (WT) and tail loop-deleted BNP. Curves are the refractive interference signal. Values are native molecular masses of BNP in the population of the peaks. (B) Static light scattering of the polymerase activity-defective BNP variants, compared to the wild type. (C) Coimmunoprecipitation of BNP oligomerization mutants. BNP without RNA-binding activity (WT/G1) could self-interact and served as the positive control. All BNP oligomerization mutants with defective polymerase activity could not homo-oligomerize.
The crystal structure of BNP reveals an extensive network of intrachain (clusters T1 to T3) and interchain (clusters I1 to I6) interactions at the tail loop region, which we classified into different clusters according to their proximity and nature of interactions (Fig. 3A and B). To understand the role of these clusters toward BNP homo-oligomerization, the involved hydrophilic and hydrophobic residues were mutated to alanine and serine, respectively, for the elimination of the interactions concerned. The expression levels of these mutants were normalized (data not shown) before investigating their effect on polymerase activity and oligomeric states.
Although most of the tail loop interactions are hydrophobic in nature, a salt bridge between E395 and R472 (cluster I2) is found deeply buried within (Fig. 3C). Mutating either residue to alanine led to dramatic decrease in polymerase activities (4.1% and 7.6% of the wild type, respectively) (Table 3). Both the E395A and R472A protein variants resulted in the loss of homo-oligomerization ability in vitro (Fig. 4B) and in vivo (Fig. 4C). The main chains of F468 and V470 also form hydrogen bonds with the side chain of R472 (cluster T1), which possibly helps to orient R472 for the formation of the interchain salt bridge with E395.
The tip region of the tail loop not only comprises a salt bridge but also contains an extensive network of hydrophobic contacts (cluster I1). The tail loop residue F468 interacts with M360, F390, V399, L400, and L403 of the neighboring BNP (Fig. 3D). Mutation of F468S led to a dramatic decrease in polymerase activity (3.1% of the wild type). Static light scattering and coimmunoprecipitation also demonstrated that the F468S mutant could not form homo-oligomers (Fig. 4B and C). Mutating F412 (the BNP F468 equivalent) in ANP also had similar effects (8, 17). Although this residue is conserved in ANP and BNP, there are significant differences in its interactions (Fig. 3D). Only one out of the five residues that interact with it is homologous (V399 in BNP and I/L/V343 in ANP). Even though the interactions are conserved, F390 and L403 in BNP become H/N334 and I/V347 in ANP, respectively. The ANP equivalents of BNP, M360 and L400, do not play any role in the interaction.
The hydrophobic residues in the stem of the BNP tail loop also interact with the neighboring BNP molecule (Fig. 3E), including residues in cluster I3 (P473, I474, and L476) and cluster I4 (C464, P466, and V467). Single- and double-point mutations in these clusters did not cause a major effect on polymerase activities (Table 3). However, when all three residues were mutated to serine (Myc-tagged [P473S, I474S, L476S] and [C464S, P466S, V467S]), polymerase activities dropped significantly (2.8% and 8.9% of the wild type) (Table 3). These two mutants could not form BNP oligomers either (Fig. 4B and C). No major structural changes were detected for these two triple-point mutants using circular dichroism spectroscopy (data not shown). Many of the contacts in clusters I3 and I4 are nonconserved between ANP and BNP (Fig. 3E). Structural alignment between the tail loops (not the full-length protein) of ANP and BNP (Fig. 3F) shows that the RMSD of residues P473 and I474 is higher than that of the rest of the loop (Fig. 3G). Residue P473 in BNP (N417 in ANP) restricts the phi-psi dihedral angles. The conserved I474 (L418 in ANP) faces the neighboring BNP molecule instead of having van der Waals contacts with the conserved P466 (P410 in ANP) (Fig. 3F). The crucial zig-zag pattern of hydrophobic interactions at the stem for maintaining the tail loop conformation in ANP (7) is thus not observed in BNP.
The base of the tail loop features clusters T3, I5, and I6. While cluster T3 is not essential, a hydrogen bond between R325 and C464 (cluster I5) plays a certain role in tail loop insertion, as the polymerase activity of the R325A mutant dropped to 41% of the wild type (Table 3). Regarding cluster I6, a similar interaction pair is found in ANP (I406 to Y487), and mutation of I406S renders the polymerase partially active (7), A BNP I462S mutation did not cause major reduction in polymerase activity (Table 3), which showed that cluster I6 is likely to have limited functional importance.
DISCUSSION
BNP is indispensable for the transcription and replication of the influenza B virus RNA genome. This report reveals the structure of influenza B virus NP and analyses the functional importance of amino acid residues involved in RNA binding and homo-oligomerization.
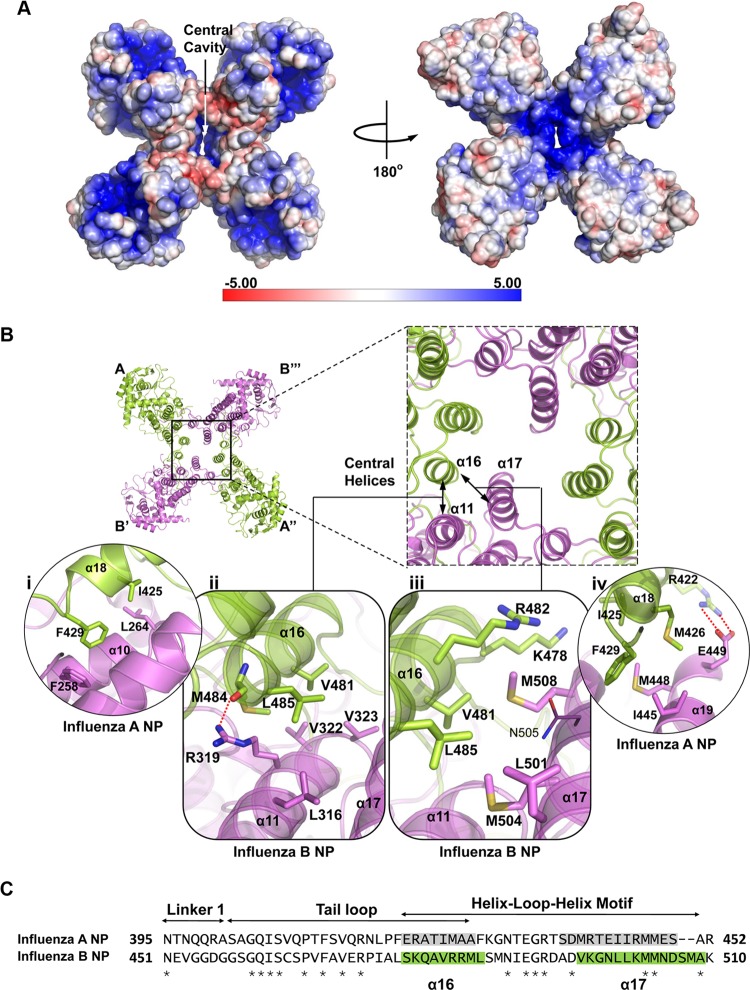
So far, all NP crystal structures from single-stranded negative sense RNA viruses have a two-domain fold (2, 11, 13, 23, 24, 27, 30, 35). For BNP, the proposed RNA-binding groove is found at the periphery of the tetrameric BNP (Fig. 5A), which is similar to the docked 9-mer ANP model after electron microscopy (EM) 3-dimensional (3D) reconstruction (8). On the contrary, the RNA of rabies and vesicular stomatitis viruses is completely sequestered in the NP polymer ring and the opening to access the RNA faces the center of the ring, as revealed in their crystal structures (2, 13). Besides, one NP molecule of rabies and vesicular stomatitis viruses bind 9 nucleotides of RNA (2, 13), while one NP molecule of influenza A virus binds 24 to 27 RNA nucleotides in vivo (3, 19).
Fig 5.
Tetramer formation of BNP. (A) Electrostatic potential representation of BNP tetramer. The RNA-binding groove is located at the periphery of the tetramer, at the highly positively charged region. An obvious space is observed in the middle of the tetramer. (B) Interactions of the central helices of ANP and BNP. (i) Hydrophobic interactions between α10 and α18 in ANP. (ii) In BNP, M484 of the tail loop forms electrostatic interactions with R319 of the neighboring BNP molecule. V481 of the tail loop also forms hydrophobic interactions with V322 and V323 of the neighboring BNP. M484 and L485 form another van der Waals contacts with R319. (iii) K478 and N505 do not form salt bridges in BNP. Instead, M508, which is not found in ANP, interacts with K478, V481, and R482. (iv) The R422-E449 salt bridge is prominent in ANP. (C) Sequence alignment of influenza A and B shows that the regions for homo-oligomerization are less conserved (29% identical). Shaded regions are the helices α16 and α17 of the helix-loop-helix motif. α17 is especially misaligned between the two NPs and suggests that the motif is flexible in nature. Stars indicate identical sites.
NP of single-stranded negative-sense RNA viruses not only encapsidates RNA in different manners, they also adopt various strategies to form homo-oligomers. Lassa virus NP uses its N-terminal domain to interact with the C-terminal domain of the neighboring molecule (23). The extreme N terminus of Rift Valley fever virus NP forms an alpha-helical arm for inserting into the neighboring molecule (11). Human respiratory syncytial virus and Borna disease virus NP use their extreme N and C termini to interact with the neighboring NP, but without tight contacts (27, 30). Rabies and vesicular stomatitis virus NPs use both an extended loop and the extreme N terminus for the interactions (2, 13). While influenza NPs also utilize the extended tail loop for homo-oligomer formation, whether the N-terminal region takes part in the process like the other viral NPs is yet to be determined.
Nine ANP monomers are organized in a ring-like structure in a reconstituted mini-RNP (19). Based on the flexibility between the tail loop and the rest of ANP structure observed, we have attempted to construct a 9-mer model (21). The newly determined BNP structure has provided some support to this model. Despite the fact that different interactions at the central helices (α11, α16, and α17) are found between BNP tetramer and ANP trimer, several hydrophobic interactions are likely to be maintained. Residue L485 in BNP helix α16 interacts with the neighboring molecule through residue L316 in helix α11 and residues L501 and M504 of helix α17 (Fig. 5Bii and iii). Accordingly, the equivalent ANP residue F429 interacts with F258, I445, and M448 (Fig. 5Bi and iv). These conserved interactions in ANP trimer and BNP tetramer are likely to be kept in NP higher oligomers. The loop region in helix-loop-helix motif (aa 458 to 509), for connecting the tail loop to the main body of NP may vary in structural organization upon the formation of different oligomers, as we have observed that A494 and D495 of BNP are part of the loop, while their corresponding residues in ANP S438 and D439 are part of the helix (Fig. 5C). Therefore, the structural features observed for the formation of BNP tetramer, together with the trimeric ANP, provide insights into how NP forms higher oligomers.
The N-terminal regions of both ANP and BNP are not visible in the crystal structure. BNP, however, has a much-extended N-terminal region than ANP, as revealed by their sequence comparisons. This N terminus is nonconserved among the >300 BNP sequences in the NCBI database. Twenty-three out of the 70 residues (32.9%) are polymorphic (defined as bearing more than one amino acid variation in that particular residue), and the percentage of polymorphism is much higher than of the rest of the BNP (8.8%). While the N terminus of ANP has been well characterized for carrying an unconventional nuclear localization signal (31) and a binding site for the cellular splicing factor BAT1/UAP56 (20), no particular functions were found to associate with this region of BNP (29).
Structural comparisons of ANP and BNP tail loop interactions reveal a strikingly conserved R472-E395 salt bridge (R416-E339 in ANP). Primary sequence analysis also showed that these two residues are conserved among different strains of influenza A virus and different lineages of influenza B virus, which indicate the essentiality of the interaction. The charged side chains of the two residues neutralize one another, and the salt bridge is deeply buried in the hydrophobic insertion groove. The identification of ligands which prevent the formation of the salt bridge will be of particular interest to combat both types of influenza viruses. Intriguingly, an inhibitor which targets the salt bridge of ANP has been recently identified (28). It will be interesting to find out whether this inhibitor also targets the BNP salt bridge.
Through the study of the structure and functional analysis of BNP, we have identified crucial residues for RNA binding and tail loop insertion. The only conserved interchain interaction of R472-E395 in influenza A and B NP strengthens the idea that ligands targeting this salt bridge would be a universal drug for combating both A and B types of influenza viruses. Comparison of the ANP trimer and BNP tetramer structures suggests the structural reorganization of the helix-loop-helix motif and the maintenance of some conserved hydrophobic contacts in higher-order NP oligomer formation.
ACKNOWLEDGMENTS
We thank Wendy Barclay at Imperial College, University of London, for providing pCIPA, pCIPB1, and pCIPB2 plasmids and Ervin Fodor at University of Oxford for critical reading of the manuscript.
This research was supported by a General Research Fund grant (CUHK473810) from the Research Grants Council of Hong Kong (P.-C.S.) and the Claudia Adams Barr Program in Cancer Research (J.W.). Equipment used was obtained through a Special Equipment Grant (SEG CUHK08) from the University Grants Council of Hong Kong. The diffraction data were collected at the Shanghai Synchrotron Radiation Facility (Shanghai, China).
Footnotes
Published ahead of print 11 April 2012
REFERENCES
- 1. Adams PD, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Albertini AA, et al. 2006. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313:360–363 [DOI] [PubMed] [Google Scholar]
- 3. Area E, et al. 2004. 3D structure of the influenza virus polymerase complex: localization of subunit domains. Proc. Natl. Acad. Sci. U. S. A. 101:308–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baudin F, Bach C, Cusack S, Ruigrok RW. 1994. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 13:3158–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boulo S, et al. 2011. Human importin alpha and RNA do not compete for binding to influenza A virus nucleoprotein. Virology 409:84–90 [DOI] [PubMed] [Google Scholar]
- 6. Centers for Disease Control and Prevention 2011. FluView. http://www.cdc.gov/flu/ Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 7. Chan WH, et al. 2010. Functional analysis of the influenza virus H5N1 nucleoprotein tail loop reveals amino acids that are crucial for oligomerization and ribonucleoprotein activities. J. Virol. 84:7337–7345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coloma R, et al. 2009. The structure of a biologically active influenza virus ribonucleoprotein complex. PLoS Pathog. 5:e1000491 doi:10.1371/journal.ppat.1000491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. DeLano WL. 2002. The PYMOL user's manual. DeLano Scientific, San Carlos, CA [Google Scholar]
- 10. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ferron F, et al. 2011. The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes. PLoS Pathog. 7:e1002030 doi:10.1371/journal.ppat.1002030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gerritz SW, et al. 2011. Inhibition of influenza virus replication via small molecules that induce the formation of higher-order nucleoprotein oligomers. Proc. Natl. Acad. Sci. U. S. A. 108:15366–15371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Green TJ, Zhang X, Wertz GW, Luo M. 2006. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313:357–360 [DOI] [PubMed] [Google Scholar]
- 14. Hatta M, Kawaoka Y. 2003. The NB protein of influenza B virus is not necessary for virus replication in vitro. J. Virol. 77:6050–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jackson D, Cadman A, Zurcher T, Barclay WS. 2002. A reverse genetics approach for recovery of recombinant influenza B viruses entirely from cDNA. J. Virol. 76:11744–11747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Laskowski RA, Macarthur MW, Moss DS, Thornton JM. 1993. Procheck - a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 17. Li Z, et al. 2009. Mutational analysis of conserved amino acids in the influenza A virus nucleoprotein. J. Virol. 83:4153–4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li OT, et al. 2009. Full factorial analysis of mammalian and avian influenza polymerase subunits suggests a role of an efficient polymerase for virus adaptation. PLoS One 4:e5658 doi:10.1371/journal.pone.0005658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martin-Benito J, et al. 2001. Three-dimensional reconstruction of a recombinant influenza virus ribonucleoprotein particle. EMBO Rep. 2:313–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Momose F, et al. 2001. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J. Virol. 75:1899–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ng AK, et al. 2008. Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design. FASEB J. 22:3638–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 23. Qi X, et al. 2010. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 468:779–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raymond DD, Piper ME, Gerrard SR, Smith JL. 2010. Structure of the Rift Valley fever virus nucleocapsid protein reveals another architecture for RNA encapsidation. Proc. Natl. Acad. Sci. U. S. A. 107:11769–11774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Resa-Infante P, Jorba N, Coloma R, Ortin J. 2011. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol. 8:207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robbins PA, Rota PA, Shapiro SZ. 1997. A broad cytotoxic T lymphocyte response to influenza type B virus presented by multiple HLA molecules. Int. Immunol. 9:815–823 [DOI] [PubMed] [Google Scholar]
- 27. Rudolph MG, et al. 2003. Crystal structure of the borna disease virus nucleoprotein. Structure 11:1219–1226 [DOI] [PubMed] [Google Scholar]
- 28. Shen YF, et al. 2011. E339.R416 salt bridge of nucleoprotein as a feasible target for influenza virus inhibitors. Proc. Natl. Acad. Sci. U. S. A. 108:16515–16520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stevens MP, Barclay WS. 1998. The N-terminal extension of the influenza B virus nucleoprotein is not required for nuclear accumulation or the expression and replication of a model RNA. J. Virol. 72:5307–5312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tawar RG, et al. 2009. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 326:1279–1283 [DOI] [PubMed] [Google Scholar]
- 31. Wang P, Palese P, O'Neill RE. 1997. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza a virus nucleoprotein NP is a nonconventional nuclear localization signal. J. Virol. 71:1850–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weber F, Kochs G, Gruber S, Haller O. 1998. A classical bipartite nuclear localization signal on Thogoto and influenza A virus nucleoproteins. Virology 250:9–18 [DOI] [PubMed] [Google Scholar]
- 33. World Health Organization 2011. FluNet. http://www.who.int/influenza/gisrs_laboratory/flunet/en/ World Health Organization, Geneva, Switzerland [Google Scholar]
- 34. Wright PF, Webster RG. 2001. Orthomyxoviruses, p 1534–1577 In Knipe DM, et al. (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 35. Ye Q, Krug RM, Tao YJ. 2006. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 444:1078–1082 [DOI] [PubMed] [Google Scholar]