Abstract
Replication and assembly of hepatitis C virus (HCV) depend on the host's secretory and lipid-biosynthetic machinery. Viral replication occurs on endoplasmic reticulum (ER)-derived modified membranes, while viral assembly is thought to occur on lipid droplets (LDs). A physical association and coordination between the viral replication and assembly complexes are prerequisites for efficient viral production. Nonstructural protein 5A (NS5A), which localizes both to the ER and LDs, is an ideal candidate for this function. Here, the interaction of NS5A with host cell membranes and binding partners was characterized in living cells. The binding of NS5A to LDs is apparently irreversible, both in HCV-infected cells and when ectopically expressed. In HCV-infected cells, NS5A fluorescence was observed around the LDs and in perinuclear structures that were incorporated into a highly immobile platform superimposed over the ER membrane. Moreover, TBC1D20 and its cognate GTPase Rab1 are recruited by NS5A to LDs. The NS5A-TBC1D20 interaction was shown to be essential for the viral life cycle. In cells, expression of the Rab1 dominant negative (Rab1DN) GTPase mutant abolished steady-state LDs. In infected cells, Rab1DN induced the elimination of NS5A from viral replication sites. Our results demonstrate the significance of the localization of NS5A to LDs and support a model whereby its interaction with TBC1D20 and Rab1 affects lipid droplet metabolism to promote the viral life cycle.
INTRODUCTION
Millions of people worldwide are infected with hepatitis C virus (HCV). In most cases, HCV infection causes chronic hepatitis, which may progress to life-threatening liver cirrhosis and hepatocellular carcinoma (18). HCV is a positive single-strand RNA virus containing a 9.6-kb genome encoding a single ∼3,000-amino-acid (aa) polyprotein. This polyprotein is cleaved by cellular and viral proteases into the structural proteins (core, E1, and E2) that create the mature virion, as well as nonstructural (NS) proteins (P7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B), which are involved in replicating the viral genome (20). Replication occurs on distinct modified membrane domains derived from the endoplasmic reticulum (ER) (14). Assembly of HCV particles was shown to closely associate with lipid droplets (LDs) (5, 41, 53) that are thought to serve as a platform for viral assembly. This foretells an intimate physical association between the viral replication complex (RC) and LDs. To this end, the viral nonstructural proteins, essential for viral replication, were found to be recruited to the surface of LDs in infected cells (41). NS5A, a 56- to 58-kDa phosphoprotein (31, 44, 59), is an essential component of the viral replication complex (24). It is organized in three domains (I to III [61]). Domain I (aa 1 to 213) contains an N-terminal amphipathic helix (AH) (6, 17). Disruption of the amphipathic nature of this domain abolishes viral replication (17). Among the nonstructural proteins, NS5A harbors several properties rendering it a potential major link between the viral RC and assembly platform. NS5A was found to independently interact with LDs (27, 54), to have an RNA binding capacity (28), and to interact with the viral core protein (2, 38, 41). Both viral replication and assembly of HCV were found to depend on the host secretory and lipid-biosynthetic machinery. The function of NS5A in the aforementioned tasks is substantiated by its binding to a range of host proteins. Its interaction with several secretory trafficking proteins was shown to be essential for viral replication using small interfering RNA (siRNA)-mediated depletion (24). We have previously shown that interaction between HCV NS5A and the host protein, TBC1D20, is obligatory for efficient HCV replication (56). TBC1D20 is a Rab GTPase-activating protein (GAP) required for regulation of Rab1 (23, 55), a small GTPase that plays a key role in Golgi biogenesis, ER-to-Golgi trafficking (46, 51, 63, 67), and HCV replication (55).
Fatty acid synthesis (58) and components of the cholesterol and fatty acid-biosynthetic pathways are required for HCV RNA replication (32). Infectious virus particles are exclusively those that are exported from the cell coupled with lipoprotein complexes synthesized from LDs (1, 34, 45). NS5A is involved in this process as well, based on the findings that it interacts with apolipoprotein E, a lipoprotein enriched in viral infectious particles. This binding was shown to be essential for viral assembly (10). To elucidate the role of NS5A as a major link between the ER-derived replication sites and the LD assembly platform, its interaction with host ER and LD membranes and its intracellular dynamics were analyzed in living cells, including in the settings of infected cells. When expressed alone, NS5A-green fluorescent protein (GFP) localized to ER membranes and was highly concentrated on the surface of LDs. Fluorescence recovery after photobleaching (FRAP) was applied and demonstrated the tight interaction of NS5A with host membranes. In cells expressing JC1/GFP (a full-length genome with an in-frame GFP insertion into NS5A [52]), NS5A-decorated LDs were integrated into the viral perinuclear replication zone and were labeled by NS5A-GFP. FRAP analysis demonstrated complete immobilization of the entire NS5A-GFP population within the perinuclear replication platform. Furthermore, the host Rab1-GAP TBC1D20 and its cognate GTPase Rab1 colocalized with NS5A around LDs. Expression of Rab1DN resulted in the elimination of cellular LDs. Immunofluorescence analysis using anti-NS5A antibodies in infected cells demonstrated that Rab1DN removed NS5A from viral replication sites. These results emphasize the significance of the localization of NS5A on LDs and put forward a model in which the association of NS5A with TBC1D20 and Rab1 around LDs subverts the fate of LDs toward the progression of the viral life cycle.
MATERIALS AND METHODS
Materials.
Bodipy-558/568 dodecanoic acid (Bodipy) was purchased from Invitrogen (Carlsbad, CA). Oleic acid (OA) and fatty acid-free bovine serum albumin (BSA) were from Sigma (Rehovot, Israel). HCV genotype 1b NS5A cDNA was amplified from the Bart79I plasmid, which contains the HCV subgenomic replicon, as described previously (15). NS5A was then subcloned into pEGFP-N1 and pDsRed-N1 (Clontech, Palo Alto, CA). DHcRed-tagged GRASP65 (11) and TBC120-GFP (56) have been previously described. pEGFP-TBC1D20 R105A was made by introducing the R105A mutation using a QuikChange site-directed mutagenesis kit (Stratagene) as described previously (55). Rab1 was cloned into pEGFP-C1 (Clontech) so that the C terminus of GFP was fused in frame to the N terminus of Rab1. Dominant negative N121I and constitutively active Q67L mutations were introduced using the QuikChange site-directed mutagenesis kit. VSVGtsO45-AIA, a DXE motif-deficient mutant which is retained in the ER, has been previously described (12), and cDNA encoding this mutant was subcloned into pmCherry-C1 (Clontech). HCV genotype 1b core cDNA was amplified from Bart79I and cloned into pmCherry-C1 using the XhoI and SacII restriction sites. HCV genotype 2a NS5A cDNA was amplified from pFL-J6/JFH (33) and cloned into pEGFP-N1 using the HindIII and AgeI restriction sites. NS5A D3-GFP cDNA was amplified from the JC1/GFP coding plasmid (52) and cloned into pCDNA3.1(+) using the HindIII and EcoRI restriction sites.
Cell culture and transfections.
Huh7, Huh7.5, and COS7 cells were grown at 37°C in a 5% CO2–humidified atmosphere. Cell cultures were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) fetal calf serum and penicillin-streptomycin (Biological Industries, Bet-Haemek, Israel). A final concentration of 1% (vol/vol) nonessential amino acids was added to the Huh7 cells' culture media. Lipofectamine 2000 reagent (Invitrogen) or Fugene 6 reagent (Roche, Indianapolis, IN) was used for plasmid DNA transfections of subconfluent COS7 and Huh7 cells. Confocal laser scanning microscopy experiments were carried out from 18 to 24 h after transfection.
In vitro transcription and electroporation of HCV RNAs.
In vitro transcripts of the JC1/GFP (kindly provided by R. Bartenschlager, University of Heidelberg, Heidelberg, Germany) were generated by linearizing the plasmid using MluI as previously described (52, 64). The linearized DNA template, purified by phenol-chloroform extraction and ethanol precipitation, was resuspended at a final concentration of 1 μg/μl. The linearized DNA template was transcribed with T7 RNA polymerase using a MEGAscript T7 kit (Ambion, Austin, TX) according to the manufacturer's instructions. After transcription, synthesized RNA was treated with DNase I. The integrity of the RNA was analyzed by nondenaturing agarose gel electrophoresis, and the yield was determined spectrophotometrically. The RNA concentration was adjusted to 2 μg/μl, and the RNA was stored at −70°C until electroporation. For the electroporation, Huh7.5 cells were grown to 60 to 80% confluence, trypsinized, and washed twice in cold RNase-free phosphate-buffered saline (PBS; BioWhittaker Inc., Walkersville, MD). Cells were resuspended in cold PBS at a concentration of 1.5 × 107 cells/ml; 0.4 ml of the cell suspension was mixed with 10 μg in vitro-transcribed JC1/GFP RNA. The mixture was dispensed into a 2-mm-gap-width cuvette (BTX, San Diego, CA), and electroporation was performed using a BTX model 830 electroporator (820 V, five 99-μs pulses given at 220-ms intervals). Cells were left to recover for 15 min at 22°C and then mixed with 10 ml of prewarmed (37°C) growth medium. Cells were then seeded in six-well plates on coverslips and examined under a microscope at 24 and 48 h postelectroporation.
Lipid droplet induction and labeling.
OA-BSA complexes were prepared as described previously (57). Fatty acid-free BSA was dissolved in PBS to a final concentration of 2 mM. OA dissolved in ethanol was added to a final concentration of 12 mM. For the induction of LDs, the cells were incubated with 400 μM OA for at least 3 h or with 200 μM OA overnight.
For labeling, Huh7 or COS7 cells at 60% confluence were incubated with 20 μg/ml Bodipy in cell culture medium for 20 min at 37°C, rinsed with fresh medium to remove excess stain, and further incubated for 60 min at 37°C in cell culture medium.
Live-cell microscopy.
Cells were imaged in DMEM without phenol red but with supplements, including 20 mM HEPES, pH 7.4. Transfection and imaging were carried out in a 35-mm glass-bottomed microwell dish (MatTek, Ashland, MA) or on glass coverslips. A Zeiss LSM Pascal confocal laser scanning microscope (Carl Zeiss MicroImaging, Jena, Germany) was equipped with an Axiovert 200 inverted microscope. Fluorescence emissions resulting from 488-nm excitation for GFP and 543-nm excitation for DsRed or mCherry were detected using filter sets supplied by the manufacturer. The confocal and time-lapse images were captured using a Plan-Apochromat ×63 1.4-numerical-aperture (NA) objective (Carl Zeiss MicroImaging). The temperature on the microscope stage was held stable during time-lapse sessions using an electronic temperature-controlled airstream incubator. Images and movies were generated and analyzed using the Zeiss LSM software and NIH Image and ImageJ software (W. Rasband, National Institutes of Health, Bethesda, MD).
Western blotting.
Proteins were extracted from cells using radio immunoprecipitation assay (RIPA) buffer supplemented with a protease inhibitor cocktail (Sigma) and separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gel was electroblotted to a nitrocellulose membrane, soaked for 1 h in a blocking solution (PBS containing 5% nonfat milk [Sigma]), and incubated for 1 to 2 h at room temperature with polyclonal rabbit anti-GFP antibody (Abcam, Cambridge, MA) or with an actin antibody (Sigma), followed by a corresponding pair of Alexa Fluor 680- or 800-coupled secondary antibodies (Li-Cor, Lincoln, NE). Proteins were visualized with a Li-Cor infrared imager (Odyssey), and the images were processed with Odyssey (version 1.2) infrared imaging software.
Confocal laser scanning microscopy, time-lapse imaging, FLIP and FRAP analyses, and image processing.
Long time-lapse image sequences and fluorescence loss in photobleaching (FLIP) and FRAP experiments were carried out using the autofocusing function integrated into the advanced time series macroset (Carl Zeiss MicroImaging). For quantitative FRAP measurements, a ×63 1.4-NA Plan-Apochromat objective was used. Photobleaching of GFP or mCherry was performed using four to six rapid scans with the laser at full power. Pre- and postbleach images were captured at 0.5- to 3-s intervals, using low laser intensity. Fluorescence recovery in the bleached region during the time series was quantified using LSM software (Carl Zeiss MicroImaging). For presentation purposes, confocal images were exported in TIFF, and their contrast and brightness were optimized in Adobe Photoshop software (San Jose, CA). The characteristic fluorescence recovery time (τ) values for membrane turnover were calculated from the photobleaching data using a simple exponential equation. Diffusion coefficient (D) values were calculated from the photobleaching data as described previously (11), using a fit to an approximate solution of the diffusion equation when the elongated rectangular bleach box has a width a, which is much smaller than its length. According to this solution, the fluorescence intensity in the bleached box at time t, Ft, can be presented as
where Mf is the mobile fraction and A serves as a second fitting parameter. Time-dependent changes in fluorescence intensity in the bleached box were normalized to prebleach and postbleach values of 1 and 0, respectively. Data were then fitted to the equation presented above using the Kalaidagraph program (Synergy Software, Essex Junction, VT). For quantitative FLIP measurements, cells were repeatedly imaged and bleached at intervals of 30 to 40 s in the same defined region.
FRAP beam-size analysis.
FRAP beam-size studies were conducted in Hanks' balanced salt solution (HBSS; Sigma) supplemented with 20 mM HEPES (pH 7.2) and 2% BSA fraction V on COS7 cells overexpressing NS5A-GFP at 24 h posttransfection. An argon ion laser beam (Innova 70C; Coherent) was focused through a fluorescence microscope (AxioImager.D1; Carl Zeiss MicroImaging) to a Gaussian spot of 0.77 ± 0.03 μm (Plan-Apochromat ×63 1.4-NA oil-immersion objective) or 1.17 ± 0.05 μm (C-Apochromat ×40 1.2-NA water-immersion objective). FRAP experiments were conducted with each objective (beam-size analysis [25]). The ratio between the bleach areas was 2.28 ± 0.17 (n = 59). After a brief measurement at monitoring intensity (488 nm, 1 μW), a 5-mW pulse (5 to 10 ms) bleached 60 to 75% of the fluorescence in the spot, and recovery was followed by the monitoring beam. τ and Mf were extracted from the FRAP curves by nonlinear regression analysis, fitting to a lateral diffusion process (25). The Mf values for NS5A-GFP were 80 to 85% in all cases. The significance of differences between τ values measured with the same beam size was evaluated by Student's t test. Bootstrap analysis was used to compare ratio measurements [τ(×40)/τ(×63) and ω2(×40)/ω2(×63), where ω is the Gaussian radius (see Results)] (13). The bootstrap analysis was performed as described previously (16, 22), using 1,000 bootstrap samples.
Immunofluorescence microscopy.
Cells cultured on coverslips were fixed with 4% formaldehyde, permeabilized with 0.2% saponin, and blocked with 3% fetal bovine serum (FBS), before immunolabeling with an NS5A monoclonal primary antibody (9E10; kindly provided by Charles M. Rice, The Rockefeller University). Secondary antibody was Alexa 555 goat anti-mouse antibody (Invitrogen). Images were acquired using a Zeiss Pascal confocal laser scanning microscope as described above.
RESULTS
Localization and dynamics of ectopically expressed NS5A.
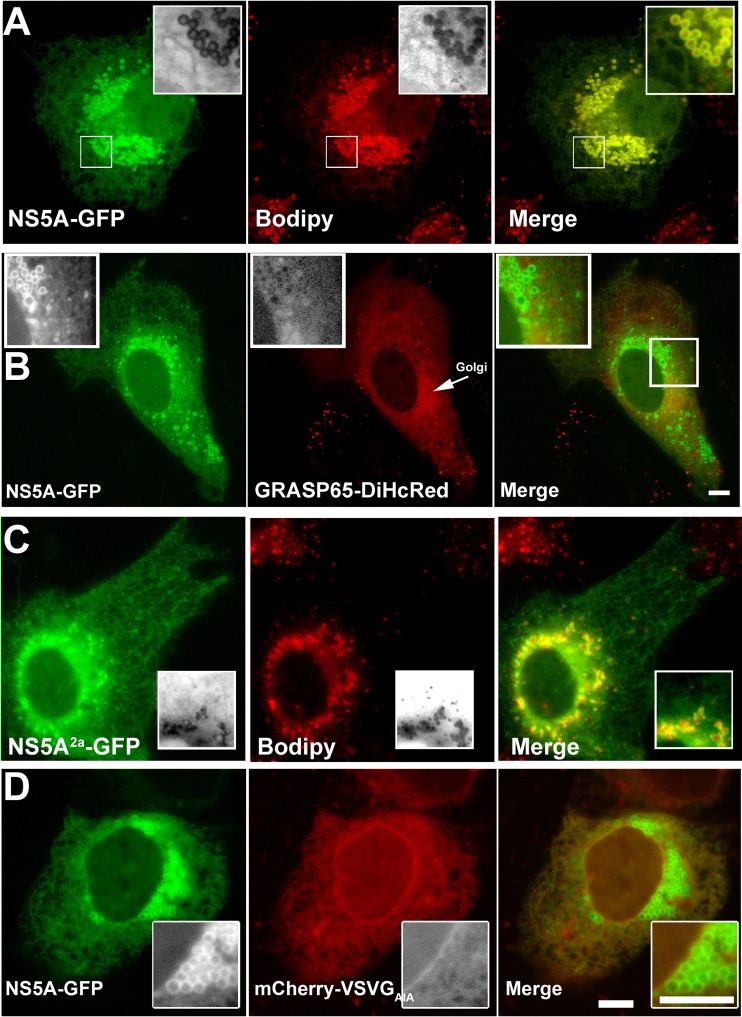
To study the intracellular distribution and dynamics of NS5A, GFP-tagged NS5A (NS5A-GFP) was expressed in Huh7 cells in the absence of other viral proteins. Figure 1A shows a typical Huh7 cell 24 h after transfection with NS5A-GFP. Prior to image capture, cells were labeled with Bodipy-558/568 dodecanoic acid (Bodipy), a marker of the LD core. NS5A-GFP is uniformly distributed throughout the ER membranes. In addition, significantly stronger NS5A fluorescence intensity was observed at the rims of LDs (Fig. 1A). Although membrane-associated cytosolic proteins typically contain a fraction that is soluble in the cytoplasm, NS5A was previously reported to act as an integral membrane protein in in vitro experiments and upon overexpression in osteosarcoma cell line (6). Only negligible amounts of NS5A were biochemically detected in the cytoplasm using fractionation. Comparably, in our hands, NS5A-GFP localized to ER and LD membranes without any apparent cytosolic fraction. This was also evident by the lack of colocalization with a typical membrane-associated protein Golgi reassembly stacking protein 65 (GRASP65) tagged with DiHcRed (tandem dimer red fluorescent protein) cotransfected with NS5A (Fig. 1B). GRASP65 is localized to Golgi membranes and a cytoplasmic fraction (Fig. 1B). Together, the lack of an apparent cytosolic fraction and the high fluorescence intensity of NS5A-GFP indicated that NS5A is tightly bound to both ER and LD membranes. To confirm that this tight binding is an intrinsic propensity of NS5A conserved among the various HCV genotypes, we further tested NS5A from genotype 2a (NS5A2a). NS5A2a-GFP showed an identical distribution in ER and LDs (Fig. 1C). It was previously shown that the NS5A AH is an LD-targeting domain (27); a different study suggested that the AH interacts with the host target membranes via an interacting receptor (9). To show that the NS5A localization is not cell type specific, we analyzed its distribution in COS7 cells. NS5A-GFP was coexpressed with an ER membrane marker that is not associated with LDs. We used a mutant version of the viral membrane cargo protein vesicular stomatitis virus G (VSVG) that lacks the cytosolic ER export motif (VSVGAIA-mCherry). Figure 1D shows the intracellular distribution of NS5A and VSVGAIA in COS7 cells after OA induction of lipid accumulation. COS7 cells were used because they have smaller amounts of LDs at steady state than Huh7 cells (see Fig. S1 in the supplemental material). In COS7 cells, while VSVG was restricted to the ER membranes, NS5A-GFP labeled both the ER and LD surface.
Fig 1.
Intracellular localization of NS5A expressed in living cells. (A) Colabeling of NS5A-GFP with the LD core marker Bodipy. Huh7 cells were transfected with NS5A-GFP (green); at 24 h posttransfection, the cells were labeled for 20 min with 20 μg/ml Bodipy (red). (Insets) A cluster of LDs. (B) Comparison of the intracellular localization of NS5A-GFP (green) with the host Golgi-binding peripheral protein GRASP65-DiHcRed (red) cotransfected into Huh7 cells. (Insets) Enlarged LDs. Bar = 5 μm. (C) Colabeling of NS5A from genotype 2a (NS5A-GFP2a) with the LD marker Bodipy. Huh7 cells were transfected with NS5A-GFP2a (green); at 24 h posttransfection, the cells were labeled for 20 min with 20 μg/ml Bodipy (red). (Insets) Enlarged LDs. Bar = 5 μm. (D) Intracellular localization of NS5A in OA-treated COS7 cells. COS7 cells were cotransfected with NS5A-GFP (green) and the ER membrane marker VSVGAIA-mCherry (red) that lacks the ER export motif. Cells were pretreated with 400 μM OA complexed to BSA for 4 h. (Insets) A cluster of LDs. Bar = 5 μm.
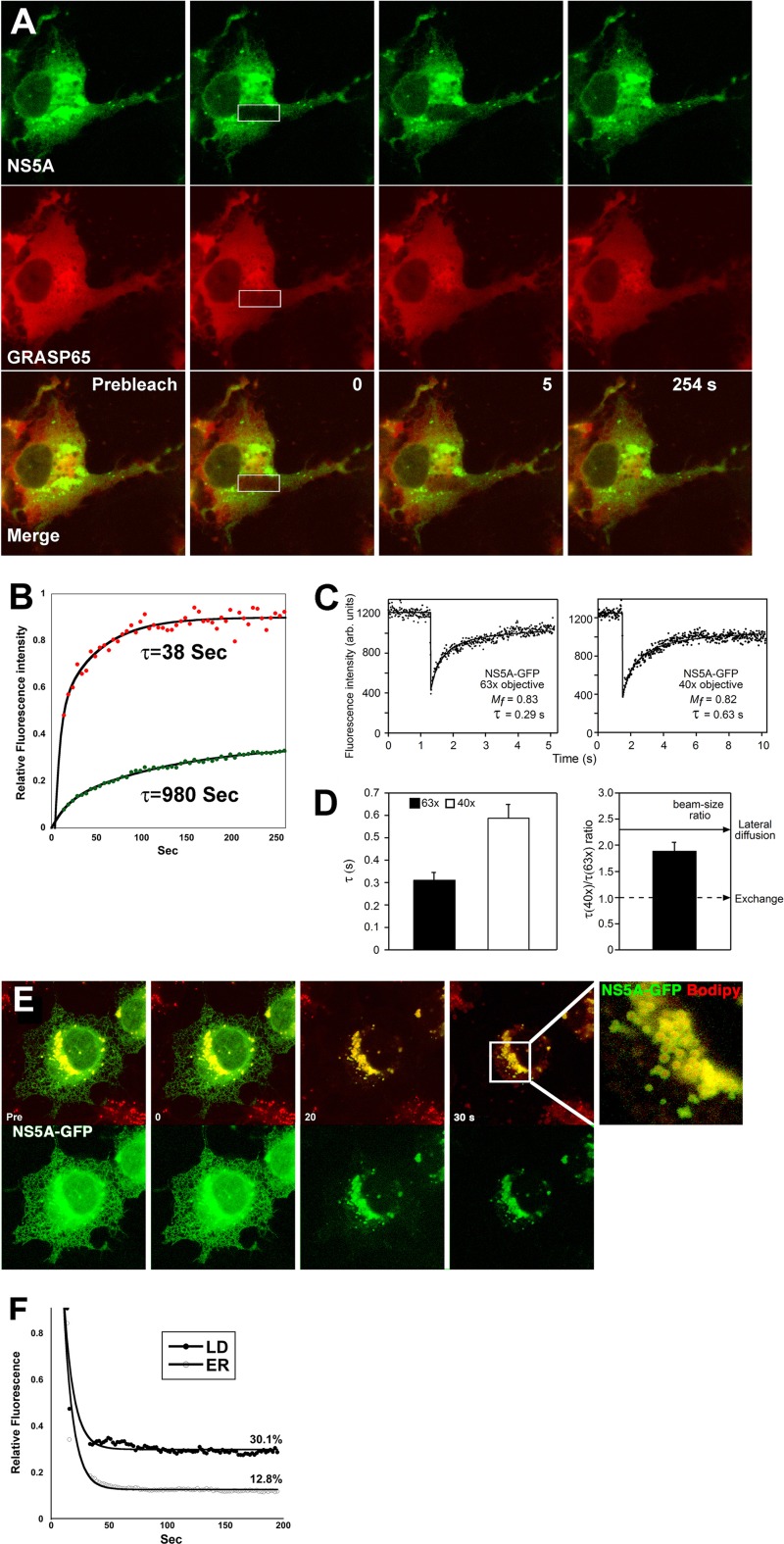
To substantiate the strong association of NS5A to LD membranes, FRAP was used to compare the membrane turnover of NS5A with that of DiHcRed-GRASP65. As shown in Fig. 2A and Movie S1 in the supplemental material, a rectangle over a cluster of NS5A-decorated LDs and cytosolic GRASP65 was photobleached. GRASP65 recovered rapidly, with a time scale of 38 s, while very little recovery was observed for NS5A during the 4.2-min-long experiment (τ = 980 s; Fig. 2B). In contrast, a similar FRAP of ER-associated NS5A-GFP resulted in complete recovery in our hands (see Fig. S2 in the supplemental material) as well as in another study (30). We hypothesized that since no cytosolic fraction was apparent, the recovery after photobleaching of NS5A on ER membranes is unlikely to be the result of NS5A turnover (exchange of membrane-bound with cytosolic fraction) but rather is indicative of lateral diffusion. To validate this hypothesis, FRAP beam-size analysis was performed (25) (Fig. 2C and D). In this method, FRAP is performed using two different Gaussian laser beam sizes by means of the ×63 (smaller ω) or ×40 (larger ω) objectives. τ represents the characteristic fluorescence recovery time, which may occur by a process other than diffusion and thus does not have to be equal to τD, where D is the lateral diffusion coefficient. If recovery takes place by dynamic exchange between membrane-bound and cytosolic pools, τ reflects the chemical relaxation time due to exchange, which is equal on all surface regions regardless of whether they are illuminated by the beam and therefore does not depend on the beam size. Thus, a τ(×40)/τ(×63) ratio of 1 will indicate FRAP by exchange between membrane-associated and cytoplasmic pools. If recovery occurs by lateral diffusion, where τ is equal to τD, it will be proportional to the area illuminated by the beam (τ = τD = ω2/4D). Therefore, for FRAP by lateral diffusion, the expected τ(×40)/τ(×63) ratio equals the beam-size ratio (2.28). Intermediate τ ratios will suggest mixed recovery (16). To this end, cells were transfected with NS5A-GFP and FRAP analysis was performed using the two objectives (Fig. 2C). The calculated τ values obtained are shown in Fig. 2D. The average measured beam-size ratio was 2.28 ± 0.17, while the τ ratio of NS5A-GFP was 1.9 ± 0.16 (n = 59; Fig. 2D). Bootstrap statistical analysis (see Materials and Methods) showed that the τ ratio of NS5A-GFP (1.9 ± 0.16, Fig. 2D) is not significantly different from the 2.28 beam-size ratio (P > 0.08), corresponding to FRAP by lateral diffusion. These data demonstrate that in the measured time resolution, the exchange rate is much slower than lateral diffusion. Thus, NS5A is strongly associated with host ER and LD membranes.
Fig 2.
Analysis of the binding dynamics of NS5A-GFP to ER and LD membranes. (A) Dual FRAP analysis of NS5A-GFP (green) and GRASP65-DiHcRed (red) in Huh7 cells. A rectangle over an area containing a cluster of LDs in a cell coexpressing NS5A-GFP and DiHcRed-GRASP65 was photobleached using high-power Ar 488-nm and He Ne 543-nm lasers. Images were captured for approximately 4 min. (B) Quantitative analysis of the experiment whose results are shown in panel A. The average fluorescence intensity of NS5A-GFP (green filled circles) and DiHcRed-GRASP65 (red filled circles) in the bleach box is plotted against time and fitted to the equation described in Materials and Methods. τ values are the inverse of the k of the exponential. R2 values were 0.92 and 0.99 for NS5A-GFP and GRASP65, respectively. (C) FRAP beam-size analysis of ER membrane-associated NS5A-GFP. Typical FRAP curves of NS5A-GFP using ×63 and ×40 objectives at 37°C. Solid lines, best fit of a nonlinear regression analysis (Materials and Methods). Note the different time scales between the two objectives. (D) FRAP beam-size analysis. Bars are the mean ± standard error of the mean of 35 to 45 measurements. The τ values (left) and ratios (right) are shown. (E) Comparison between the ER and LD binding of NS5A using saponin treatment. COS7 cells expressing NS5A-GFP (green and bottom) were labeled with Bodipy (red) after 4 h of OA treatment. Saponin was added to a final concentration of 0.2%, and images were captured for several minutes. (Inset) Magnified NS5A-GFP-decorated LDs. (F) Quantitative analysis of the experiment described in panel E. The changes in relative residual fluorescence intensity of NS5A-GFP in two regions of interest over LDs and the ER are plotted against time upon addition of the detergent. Data were fitted to an exponential-decay equation. The results shown are typical results from three independent experiments.
To further substantiate the differences between NS5A's binding to the ER and LDs, the effect of saponin on NS5A-GFP-expressing cells was analyzed. Saponin is a detergent that perforates membranes via cholesterol sequestration. Compared to other detergents, saponin treatment is considered mild and does not dissociate the ER membranes (see Fig. S3 in the supplemental material). Saponin treatment (0.2%, wt/vol) of NS5A-GFP-expressing cells labeled with Bodipy resulted in partial dissociation of both LD- and ER-bound fractions of NS5A-GFP. Figures 2E and F demonstrate that while the ER fraction of NS5A-GFP is almost completely eliminated (12.8% remains bound), an approximately 3-fold larger fraction of LD-bound NS5A is apparent (30.1% remains bound). This observation demonstrates the stronger binding of NS5A to LDs than to ER membranes.
Targeting of NS5A to LDs.
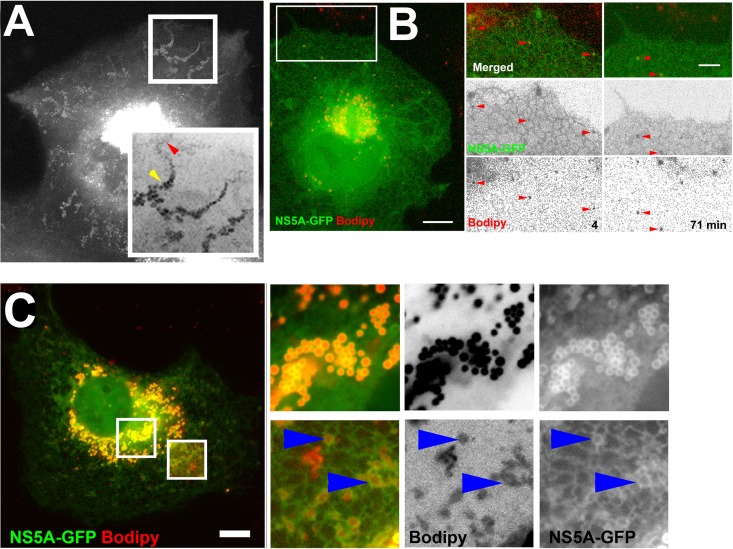
LDs bud from the ER membrane to form independent organelles that are limited by a monolayer of phospholipids and LD-associated proteins (37). There are several models describing LD biogenesis, as reviewed elsewhere (37, 43, 48). Common to all these models is the fact that the mechanism for targeting and binding of LD-associated proteins is unclear. Our finding that NS5A strongly binds LDs with very slow turnover raises a similar question regarding its association with these organelles. This issue is pertinent at the onset of viral infection, when the very low concentrations of NS5A face the ER surface, which is considerably larger than that of steady-state LDs. In one current model of LD formation, neutral lipids accumulate between the leaflets of the ER membrane and subsequently bud as nascent LDs (6). Thus, we hypothesize that ER-bound NS5A-GFP has an affinity to the specialized membrane domains where these lipids accumulate. To this end, a time-lapse experiment of OA-induced cells expressing NS5A-GFP was carried out. Figure 3A is a brightest-pixel projection of a 2-h time-lapse showing the dynamics of nascent NS5A-positive structures as they appear in the cell periphery. These structures slowly migrate toward the cell center as their diameter increases. Figure 3B and the corresponding Movie S2 in the supplemental material show a time-lapse sequence demonstrating that nascent NS5A-positive structures are colabeled with Bodipy, indicating that they are LDs. Next, we asked if NS5A is seen to be recruited to potential ER sites of neutral lipid accumulations. Toward this hypothesis, cells expressing NS5A-GFP were labeled with Bodipy following incubation in the presence of OA. The insets shown in Fig. 3C focus on two distinct populations of NS5A-labeled structures; the perinuclear region mostly contains strongly NS5A- and Bodipy-colabeled LDs (Fig. 3C, top). At the periphery of the cell, less defined structures with weak Bodipy labeling were detected (Fig. 3C, bottom). The membranes defining these structures contained a higher intensity of NS5A-GFP labeling than the surrounding ER. These data are consistent with the hypothesis that NS5A may partition to membrane domains where LDs are potentially assembled.
Fig 3.
Targeting of NS5A-GFP to LDs. (A) Time-lapse analysis of OA-induced NS5A-decorated LDs. COS7 cells were transfected with NS5A-GFP. At 24 h posttransfection, cells were treated with 400 μM OA-BSA complex and images were captured at 30-s intervals for 2.8 h. Shown is a brightest-pixel projection of the entire sequence. The enlarged inverted inset shows the transport-coupled maturation of LDs. The red arrowhead points to the LD at an early time point. The yellow arrowhead points to a mature LD at a late time point. (B) Bodipy labeling during NS5A-decorated LD biogenesis. Cells expressing NS5A-GFP were treated with 400 μM OA after labeling with Bodipy. An image of the entire cell on the left-end side was captured at 71 min after OA addition. Bar = 10 μm. The area marked by the white rectangle is enlarged in the panels on the right. Included are two time points in the time-lapse movie in Movie S2 in the supplemental material. Bar = 5 μm. (C) Localization of NS5A-GFP to OA-induced, Bodipy-labeled LDs. Huh7 cells were transfected with NS5A-GFP (green). At 24 h posttransfection, cells were incubated with 400 μM OA-BSA complex for 4 h and labeled for 20 min with 20 μg/ml Bodipy (red). Insets are magnified (3×), and inverted separate channels of Bodipy and GFP are shown. Top left and bottom right insets show NS5A-labeled strictures at the perinuclear region and cell periphery, respectively. The brightness and contrast of the lower right inset were adjusted to compensate for the weak signal. Blue arrowheads indicate the area within the ER where Bodipy labeling coincides with increased NS5A-GFP labeling. Bar = 5 μm.
Localization and dynamics of NS5A in HCV-infected cells.
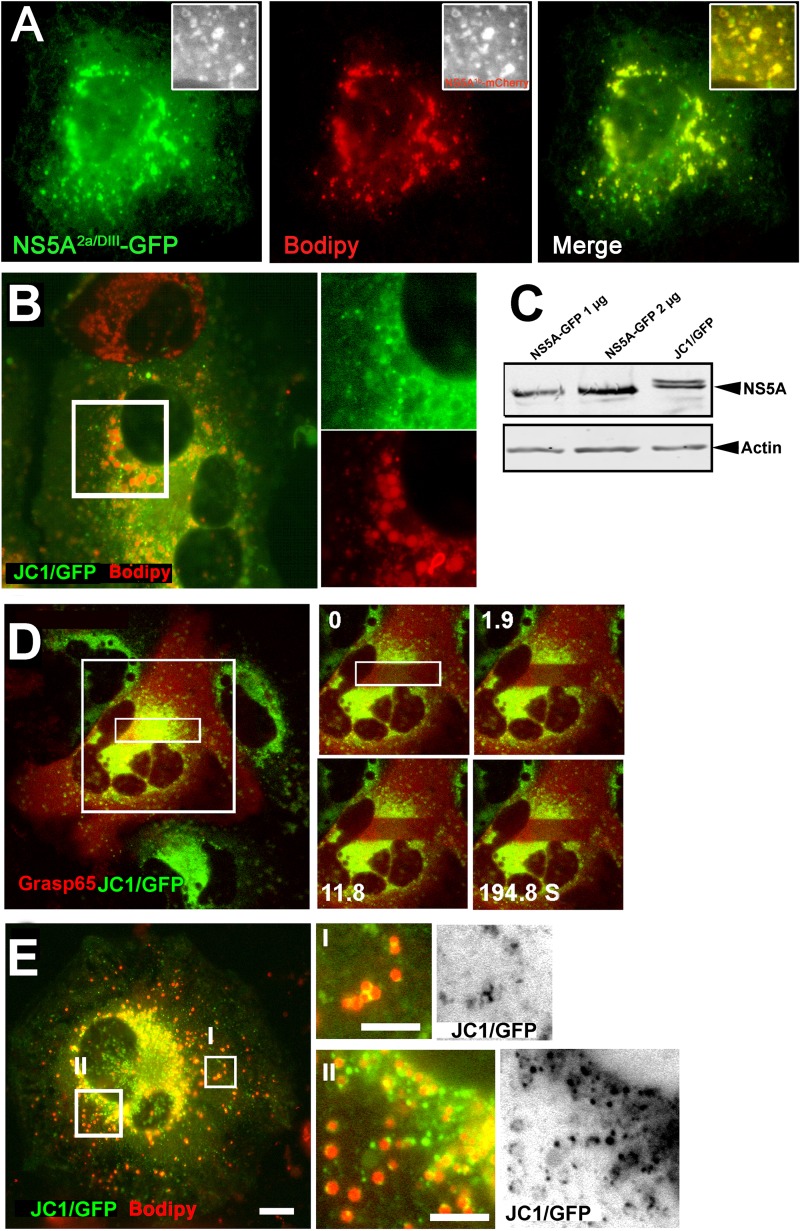
To study the intracellular distribution and dynamics of NS5A in the context of all other viral proteins, we used live cells containing the JC1/GFP full-length HCV genome. This genome replicates to a level comparable to that of the wild-type virus, and it is capable of producing infectious virions (52). We set out to visualize NS5A in Huh7.5 cells actively replicating and producing viral particles, as well as to establish its relationship with LDs. Primarily, we verified that the intracellular distribution of the ectopically expressed NS5A2a with GFP inserted in domain III (NS5A2a/DIII-GFP) is similar to that of NS5A from genotypes 1b and 2a used in our previous experiments. Figure 4A as well as FRAP experiments (not shown) demonstrated that NS5A2a/DIII-GFP is identically localized to the ER and LDs and displays similar dynamics. In the JC1/GFP-infected cells, NS5A colocalized with the ER membrane marker VSVGAIA (see Fig. S4 in the supplemental material). Steady-state LDs labeled with Bodipy were found to tightly associate with the perinuclear region that contained a large number of NS5A-labeled foci (Fig. 4B). NS5A with an insertion within domain III expressed from a subgenomic genome was previously shown to be incorporated into functional HCV replication complexes (42, 68). Here, NS5A was localized to the surface of the LDs as well. Under these conditions, the labeling of the LD surface was weaker than that with the ectopically expressed NS5A (Fig. 4B). The mechanism for the differences in NS5A distribution in these systems is unclear. It is plausible that the presence of additional LD-binding viral proteins facilitates the change in NS5A distribution. We excluded core from facilitating this redistribution, as coexpression of core with NS5A had no effect on its ER and LD localization (see Fig. S5 in the supplemental material). The cause of these changes cannot be attributed to differences in expression, since NS5A-GFP and JC1/GFP showed comparable expression levels (Fig. 4C). In agreement with previous work (30), photobleaching inside the perinuclear region, which contains a large number of presumed replication complexes, demonstrated that NS5A was immobilized (Fig. 4D). Thus, we propose that the firm interactions of NS5A with host ER and LDs shown in previous figures play a part in the formation and maintenance of this unique immobilized structure. To study the association of NS5A with nascent LDs, lipid accumulation was induced with OA. As observed in Fig. 4E, the majority of LDs were incorporated into the perinuclear region containing the NS5A-labeled foci (Fig. 4EII), while a significant proportion of the LDs localized throughout the periphery of the cell (Fig. 4EI). The NS5A-GFP fraction that was not incorporated into the perinuclear region localized to a distinct spot on the rims of almost each LD or to the interface among clustered LDs (Fig. 4EI and II). The data demonstrate that the unique distribution of NS5A in HCV-infected cells is influenced by other viral components.
Fig 4.
Intracellular distribution of NS5A in JC1/GFP-containing cells. (A) NS5A from genotype 2a containing a GFP inserted within domain III (green, NS5A2a/DIII-GFP) is localized to the ER and to Bodipy-labeled (red) LDs. (Insets) Colocalization of NS5A2a/DIII-GFP with NS5A from genotype 1b (NS5A1b-mCherry). (B) Colocalization of GFP-tagged NS5A and steady-state LDs in full-length JC1/GFP genome-containing cells. Cells were labeled with Bodipy (red) 48 h after electroporation of the JC1/GFP (green) in vitro-transcribed RNA. The inset over the perinuclear replication zone is enlarged to show the LDs assimilated into the replication zone. (C) Western blot analysis comparing expression levels of ectopic NS5A-GFP and NS5A expressed from the full-length JC1/GFP genome. NS5A-GFP (1 or 2 μg, as indicated) was transfected into Huh7 cells. At 48 h posttransfection, cells were lysed and analyzed by Western blotting using a primary anti-GFP antibody. JC1/GFP-expressing cells were lysed and analyzed as described in the Materials and Methods section. (D) FRAP of NS5A-GFP and mCherry-GRASP65 in full-length JC1/GFP genome-containing cells. Cells were electroporated with the JC1/GFP RNA and at 48 h posttransfection were transfected with mCherry-tagged GRASP65. After an additional 24 h, a rectangle shown in the image on the left-hand side was photobleached using high-power Ar 488-nm and He Ne 543-nm lasers and images were captured for approximately 200 s. (E) Colocalization of the GFP-tagged NS5A and OA-induced LDs in JC1/GFP cells. Cells containing the JC1/GFP (green and inverted images on the right-hand side) genome were treated with OA prior to labeling with Bodipy (red). (Inset I) Cell periphery containing potentially forming LDs; (inset II) perinuclear region NS5A-labeled LDs are incorporated into the replication zone. Bar = 5 μm.
NS5A at the virus-host interface.
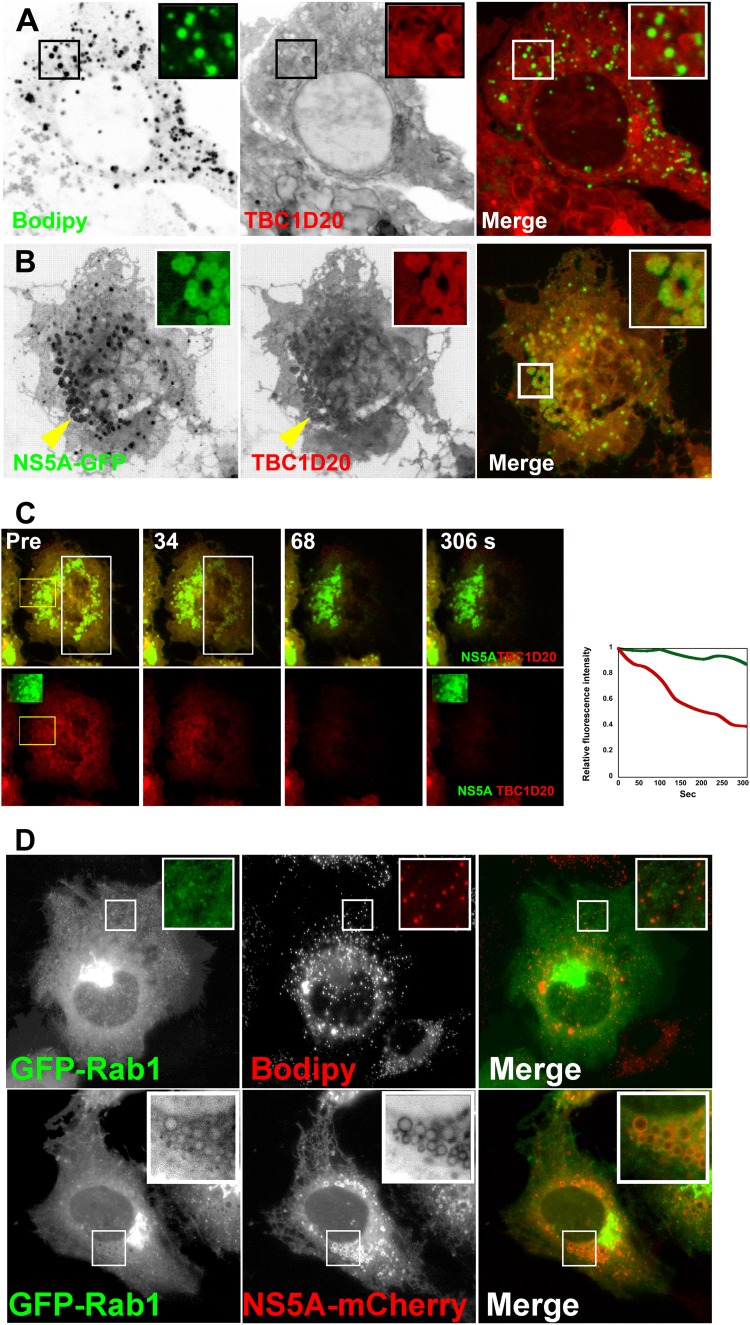
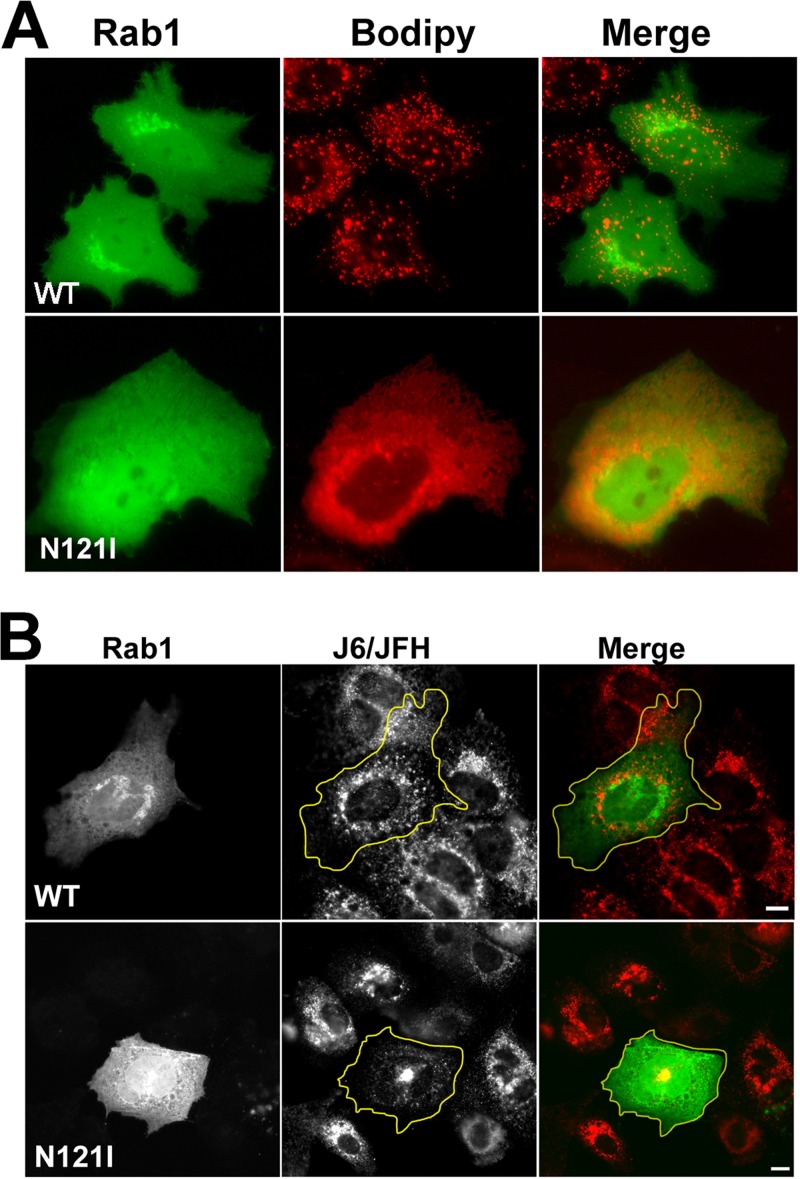
To this point, we characterized NS5A's interaction with LDs. Our previous studies showed that the ER resident Rab1-GAP TBC1D20 interacts with NS5A. This interaction is essential for the viral life cycle (55, 56). Thus, we asked if this interaction is also associated with LDs. We and others (23) failed to produce antibodies which specifically detect endogenous TBC1D20, possibly due to low expression levels. Therefore, overexpressed TBC1D20 was used in the following experiments. Figure 5 demonstrates cells overexpressing TBC1D20 in the absence or presence of NS5A. At steady state, TBC1D20 was evenly distributed in the ER membrane and did not colocalize with Bodipy-labeled LDs (Fig. 5A). In the presence of NS5A, TBC1D20 is concentrated in the vicinity of the LDs (Fig. 5B). The labeling of LDs by TBC1D20 seemed to be less confined than that of NS5A. This and the fact that TBC1D20 is an integral transmembrane protein suggest its actual localization to ER membranes surrounding LDs. To confirm this interpretation, the dynamics of the association of these two proteins with LDs were studied using FLIP. In FLIP, a region of the cell is repeatedly photobleached while fluorescence is monitored over time in a separate region of interest. The purpose of FLIP is to demonstrate membrane connectivity or continuity for a given tagged protein. The FLIP results are shown in Fig. 5C and Movie S3 in the supplemental material. Bleaching of a rectangle over the ER (white rectangle in Fig. 5C) did not significantly affect NS5A fluorescence (inset at the bottom and green line in the graph of Fig. 5C), while TBC1D20 fluorescence was depleted (yellow square at the bottom and red line in the graph in Fig. 5C). This experiment demonstrates that TBC1D20 is able to freely exchange within the ER membranes. Thus, we show that in the presence of NS5A, TBC1D20 is concentrated in the proximal ER membranes that surround LDs. Since TBC1D20 is the GAP of Rab1, we asked whether Rab1 is also associated with LDs. Rab1 is a small GTPase that plays a key role in ER-to-Golgi trafficking and Golgi biogenesis (46, 51, 63, 67). Immunofluorescence analysis of endogenous Rab1 showed that Rab1 localizes to the Golgi and the ER. Its association with NS5A and Bodipy-labeled LDs was variable and thus inconclusive (not shown). However, when coexpressed with NS5A-mCherry, GFP-Rab1 redistributed to the surface of LDs (Fig. 5D). Toward understanding the role of Rab1, we asked whether its activity is associated with LD metabolism. Cells were transfected with wild type or Rab1DN (N121I), and LDs were visualized using Bodipy staining. Figure 6A demonstrates that Rab1DN expression eliminated all steady-state LDs. In the presence of Rab1DN the Bodipy stain strongly labeled ER membranes, suggesting that lipid synthesis was not affected. Overexpression of wild-type Rab1 had no apparent effect on LDs. These data lend support to the premise that Rab1 is essential for LD metabolism. To determine the role of Rab1 during viral infection, Rab1 was transfected into cells with established transfection of the J6/JFH genome (Fig. 6B). Cells were fixed and permeabilized, and the localization of NS5A was visualized using immunofluorescence with a specific antibody. In infected cells, NS5A typically displays a speckle-like pattern thought to represent the membrane rearrangements accommodating the viral replication complexes (14, 19). In the presence of Rab1DN, the typical speckled NS5A pattern was lost. Instead, a perinuclear accumulation of NS5A was observed. We hypothesized that this structure represents a pool of NS5A undergoing degradation (29). In contrast, expression of wild-type Rab1 did not alter NS5A's staining pattern. These data demonstrate that Rab1 GTPase activity is essential to the integrity of viral replication complexes.
Fig 5.
NS5A-GFP interacts with the host partners TBC1D20 and Rab1 at the ER-LD contact zone. (A) The intracellular localization of TBC1D20-GFP (center inverted image and red) in the absence of NS5A. LDs are labeled with Bodipy (green and inverted image on the left). Huh7 cells were transfected with TBC1D20-GFP (green and inverted image on the left) and labeled for 20 min with 20 μg/ml Bodipy. Images are confocal thin slices of living cells. Insets are magnified regions of interest containing LDs (black squares). (B) The intracellular localization of TBC1D20-GFP (center inverted image and red) in the presence of NS5A-GFP (green and inverted image on the left). Huh7 cells were cotransfected with NS5A-GFP (green and inverted image on the left) and TBC1D20-mCherry (red and inverted image in the center). Yellow arrowheads point to an area containing LDs enlarged in upper-right inset. Bar = 5 μm. (C) FLIP analysis to localize TBC1D20 to ER membranes. Huh7 cells were cotransfected with NS5A-GFP (green) and TBC1D20-mCherry (red). A rectangular region (white rectangle) over the ER was continuously photobleached with high laser power of Ar at 488 nm and He Ne at 543 nm. Fluorescence loss in the yellow rectangle was monitored. (Insets on bottom row) Prebleaching (left) and last image at the end of the experiment (right). The change in fluorescence intensity of NS5A-GFP (green line) and TBC1D20-mCherry (red line) within the yellow rectangle is plotted against time on the graph on the right. (D) NS5A recruits Rab1 to LDs. Huh7 cells were transfected with Rab1 (top) and labeled with Bodipy (red) or cotransfected with GFP-Rab1 and NS5A-mCherry (bottom). Images were captured at 24 h posttransfection. (Insets) Magnified LDs.
Fig 6.
Rab1 activity is essential for LD biogenesis and HCV replication. (A) The dominant negative Rab1 N121I mutant blocks LD biogenesis. Huh7 cells were transfected with wild type (WT; top) or dominant negative (N121I; bottom) GFP-Rab1 (green). At 24 h posttransfection, images of living cells were captured after labeling for 20 min with 20 μg/ml Bodipy (red). (B) Huh7.5 cells were transfected with the J6/JFH genome by electroporation of 10 μg in vitro-transcribed J6/JFH RNA. After 24 h, the infected cells were transfected with wild-type (top) or dominant negative (N121I, bottom) GFP-Rab1 (green). At 24 h posttransfection, cells were fixed, permeabilized, and analyzed by immunofluorescence using anti-NS5A-specific antibody (middle and red). The contour of the Rab-GFP expressing cells is outlined in yellow. Bars = 10 μm.
In summary, we demonstrated the strong binding of ectopically expressed NS5A to host ER and LD membranes, with a preference for the latter. While NS5A's intracellular localization is altered in cells containing actively replicating virus, its firm interaction with the host membranes, namely, those of the ER and LDs, remains consistent. Moreover, the perinuclear replication zone containing multiple NS5A-labeled replication complexes displayed robust immobility. Finally, LDs and associated ER domains were identified as the location of interaction of NS5A with its host factors, TBC1D20 and Rab1. Moreover, the data here provide evidence for Rab1 as a modulator linking LD homeostasis and HCV replication.
DISCUSSION
HCV replication and assembly occur at the interface of LD and ER membranes (41). However, the exact details of these processes are still obscure (see references 3, 40, and 47 for reviews). Here, we studied the relationship between NS5A and host ER and LD membranes, both alone and in the context of a full-length genome. When expressed alone, NS5A was evenly distributed and tightly bound to both host ER and LD membranes, with a stronger preference for the latter. The strong association of NS5A with LD membranes is supported by four independent observations: (i) the fluorescence intensity surrounding LDs is stronger than that surrounding the ER, (ii) LD-bound NS5A is more resistant to saponin treatment than ER, (iii) turnover of LD-bound NS5A-GFP is particularly slow, and (iv) recovery after photobleaching of ER-bound NS5A is mediated by lateral diffusion and not membrane turnover. A similar tight and irreversible interaction has been reported for the host LD-associated protein adipose triglyceride lipase (57). In our experiments, NS5A distribution was unaffected by induction of LD biogenesis using OA. These experimental conditions were used on the basis of our view that the presence or absence of OA corresponds at least to some extent to physiologically relevant lipid-biosynthetic states of hepatocytes.
LD biogenesis experiments and time-lapse microscopy revealed that NS5A is found to be concentrated on LDs at the earliest detectable time. Furthermore, we followed NS5A-decorated LDs for several hours from the point of their generation at the cell periphery until their maturation to large perinuclear LDs. The growth of LDs occurred at least in part by homeotypic fusions (not shown), suggesting that NS5A does not interfere with this process. We also demonstrated that in the ER membrane, NS5A partitions into distinct domains that are associated with Bodipy-positive putative neutral lipid accumulation sites. Based on these observations, we postulated that NS5A is localized to the LD surface membrane prior to its budding from the ER. It was recently shown that the ER-localized triglyceride-synthesizing enzyme diacylglycerol acyltransferase-1 (DGAT1) interacts with core and is required for its targeting to lipid droplets (26). A similar mechanism may be responsible for the observations reported in Fig. 3, where the partitioning and concentration of NS5A around forming LDs are mediated by similar interactions.
NS5A dynamics have been determined in cells containing subgenomic genotype 2a (30) and genotype 1b (68) genomes. Those studies showed the static nature of NS5A in what was shown to be replication complexes. Since the viral core protein plays a major role in viral assembly and LD binding (41), we performed these experiments using the JC1/GFP full-length genome. We demonstrated that the apparent discrepancy in the distribution of NS5A when expressed alone versus within the replicating genome (Fig. 4) does not arise from the different HCV genotypes or positions of the tag. We propose that an added complex array of competing interactions resulting from the presence of all HCV proteins and RNAs (41) culminates in the restriction and alteration of the inherent characteristic intracellular distribution of NS5A. Moreover, a recent study has shown that formation of new replication complexes is abolished by several small-molecule HCV inhibitors that shift the localization of NS5A from the replication complexes to LDs (60).
The viral replication platform is interlaced with LDs. We speculate that NS5A might be responsible for the recruitment of the LDs to this region. The most prominent feature of the dense replication region is its seemingly complete immobility, as revealed by failure to recover after photobleaching. Time-lapse images of Bodipy-labeled LDs demonstrated that they are fully confined to and immobilized within this perinuclear area. An analogous immobility of NS5A from subgenomic genotype 1b and genotype 2a genomes has been previously observed (30, 68). The strong association of NS5A with host membranes may provide, at least in part, a mechanism for this immobility. Thus, future experiments should address the link between immobility and viral subsistence.
High-resolution structural studies of NS5A have revealed that the protein forms dimers via contacts near its N terminus (62). The positioning of NS5A in the interface of peripheral LDs suggests that its self-dimerization may contribute to recruitment of LDs to the replication zone. Site-directed mutagenesis of NS5A altering its LD-binding or homodimerization propensity is under way.
The significance of NS5A's localization to LDs is underscored by the fact that LDs are also the site of its interaction with key host factors. Here, we found that NS5A recruits TBC1D20 and its cognate GTPase Rab1 to the vicinity of LDs. Using immunofluorescence, we were unable to consistently demonstrate localization of endogenous Rab1 to LDs, presumably due to the dynamic nature of this GTPase. When coexpressed with NS5A, Rab1 clearly associated with LDs. In accord with these findings, Rab1 was reported to associate with LDs using proteomic analysis (4, 8, 35, 65). Using a dominant negative mutant of Rab1, we demonstrated that Rab1 GTPase activity is a prerequisite for LD homeostasis as well as for maintenance of viral replication complexes. These results are supported by previously published data showing that siRNA-mediated depletion of Rab1 significantly decreased HCV RNA levels (55). Additional small GTPases associated with secretory transport were shown to be involved in LD metabolism as well: Rab18 was demonstrated to be recruited to LDs during lipolysis and to induce the apposition of LDs to the ER membranes (36, 49). Similarly, Arf1 has been shown to act on the LD surface to regulate droplet morphology and lipid utilization (21). Arf1 inhibition was shown to inhibit HCV replication and to shift the localization of NS3 and NS5A from the ER to the LD surface (39).
Infectious virus particles are exclusively those that are exported from the cell complexed with lipoproteins synthesized from LDs (1, 34, 45). The data herein provide evidence linking the LD binding of NS5A to a proposed role in perturbing LD metabolism. It is tempting to hypothesize that NS5A binds to newly formed LDs and together with other viral LD-binding proteins, such as core, facilitates their incorporation into the perinuclear replication zone. Its interaction with TBC1D20 and, consequently, Rab1 at these sites might serve to recruit these host mediators. Our results confirm the independent localization of NS5A to LDs and support recent viral assembly models in which LD localization of both core and NS5A is pivotal for the assembly process (see reference 3 for updated models). Our data further confirm a role for the host factors TBC1D20 and Rab1 in HCV replication.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the European Union Marie Curie Reintegration Grant (to E.H.S.) and the Israel Cancer Association grant (to E.H.S. and K.H.).
We thank Charles M. Rice (The Rockefeller University, New York, NY), Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan), and Ralf Bartenschlager (University of Heidelberg, Heidelberg, Germany) for providing reagents. Y.I.H. is an incumbent of the Zalman Weinberg Chair in Cell Biology. Special thanks go to Eran Bachrach (Tel Aviv University) for helpful discussions.
Footnotes
Published ahead of print 4 April 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Andre P, et al. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 76:6919–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Appel N, et al. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035 doi:10.1371/journal.ppat.1000035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartenschlager R, Penin F, Lohmann V, Andre P. 2011. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 19:95–103 [DOI] [PubMed] [Google Scholar]
- 4. Bartz R, et al. 2007. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J. Proteome Res. 6:3256–3265 [DOI] [PubMed] [Google Scholar]
- 5. Boulant S, Targett-Adams P, McLauchlan J. 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 88:2204–2213 [DOI] [PubMed] [Google Scholar]
- 6. Brass V, et al. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130–8139 [DOI] [PubMed] [Google Scholar]
- 7. Reference deleted.
- 8. Cermelli S, Guo Y, Gross SP, Welte MA. 2006. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16:1783–1795 [DOI] [PubMed] [Google Scholar]
- 9. Cho NJ, Cheong KH, Lee C, Frank CW, Glenn JS. 2007. Binding dynamics of hepatitis C virus' NS5A amphipathic peptide to cell and model membranes. J. Virol. 81:6682–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cun W, Jiang J, Luo G. 2010. The C-terminal a-helix domain of apolipoprotein E is required for interaction with nonstructural protein 5A and assembly of hepatitis C virus. J. Virol. 84:11532–11541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dukhovny A, Papadopulos A, Hirschberg K. 2008. Quantitative live-cell analysis of microtubule-uncoupled cargo-protein sorting in the ER. J. Cell Sci. 121:865–876 [DOI] [PubMed] [Google Scholar]
- 12. Dukhovny A, Yaffe Y, Shepshelovitch J, Hirschberg K. 2009. The length of cargo-protein transmembrane segments drives secretory transport by facilitating cargo concentration in export domains. J. Cell Sci. 122:1759–1767 [DOI] [PubMed] [Google Scholar]
- 13. Efron B, Tibshirani R. 1993. Estimates of bias, p 124–130 In Cox DR, Hinkley DV, Reid N, Rubin D B, Silverman BW. (ed), An introduction to bootstrap. Chapman & Hall, London, United Kingdom [Google Scholar]
- 14. Egger D, et al. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Einav S, Elazar M, Danieli T, Glenn JS. 2004. A nucleotide binding motif in hepatitis C virus (HCV) NS4B mediates HCV RNA replication. J. Virol. 78:11288–11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eisenberg S, et al. 2011. Raft protein clustering alters N-Ras membrane interactions and activation pattern. Mol. Cell. Biol. 31:3938–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elazar M, et al. 2003. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J. Virol. 77:6055–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ghany MG, Strader DB, Thomas DL, Seeff LB. 2009. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology 49:1335–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gosert R, et al. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo Y, et al. 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453:657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gutman O, Walliser C, Piechulek T, Gierschik P, Henis YI. 2010. Differential regulation of phospholipase C-b2 activity and membrane interaction by Gaq, Gb1g2, and Rac2. J. Biol. Chem. 285:3905–3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haas AK, et al. 2007. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J. Cell Sci. 120:2997–3010 [DOI] [PubMed] [Google Scholar]
- 24. He Y, Staschke KA, Tan SL. 2006. HCV NS5A: a multifunctional regulator of cellular pathways and virus replication. In Tan SL. (ed), Hepatitis C viruses: genomes and molecular biology. Horizon Bioscience, Norfolk, United Kingdom: [PubMed] [Google Scholar]
- 25. Henis YI, Rotblat B, Kloog Y. 2006. FRAP beam-size analysis to measure palmitoylation-dependent membrane association dynamics and microdomain partitioning of Ras proteins. Methods 40:183–190 [DOI] [PubMed] [Google Scholar]
- 26. Herker E, et al. 2010. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat. Med. 16:1295–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hinson ER, Cresswell P. 2009. The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic alpha-helix. Proc. Natl. Acad. Sci. U. S. A. 106:20452–20457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang L, et al. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 280:36417–36428 [DOI] [PubMed] [Google Scholar]
- 29. Johnston JA, Ward CL, Kopito RR. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143:1883–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jones DM, Gretton SN, McLauchlan J, Targett-Adams P. 2007. Mobility analysis of an NS5A-GFP fusion protein in cells actively replicating hepatitis C virus subgenomic RNA. J. Gen. Virol. 88:470–475 [DOI] [PubMed] [Google Scholar]
- 31. Kaneko T, et al. 1994. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem. Biophys. Res. Commun. 205:320. [DOI] [PubMed] [Google Scholar]
- 32. Kapadia SB, Chisari FV. 2005. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl. Acad. Sci. U. S. A. 102:2561–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lindenbach BD, et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 34. Lindenbach BD, et al. 2006. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl. Acad. Sci. U. S. A. 103:3805–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu P, et al. 2004. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 279:3787–3792 [DOI] [PubMed] [Google Scholar]
- 36. Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG. 2005. Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J. Biol. Chem. 280:42325–42335 [DOI] [PubMed] [Google Scholar]
- 37. Martin S, Parton RG. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7:373–378 [DOI] [PubMed] [Google Scholar]
- 38. Masaki T, et al. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matto M, et al. 2011. Role for ADP ribosylation factor 1 in the regulation of hepatitis C virus replication. J. Virol. 85:946–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McLauchlan J. 2009. Lipid droplets and hepatitis C virus infection. Biochim. Biophys. Acta 1791:552–559 [DOI] [PubMed] [Google Scholar]
- 41. Miyanari Y, et al. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097 [DOI] [PubMed] [Google Scholar]
- 42. Moradpour D, et al. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murphy DJ. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40:325–438 [DOI] [PubMed] [Google Scholar]
- 44. Neddermann P, Clementi A, De Francesco R. 1999. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B, and NS5A encoded on the same polyprotein. J. Virol. 73:9984–9991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nielsen SU, et al. 2006. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J. Virol. 80:2418–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nuoffer C, Davidson HW, Matteson J, Meinkoth J, Balch WE. 1994. A GDP-bound of rab1 inhibits protein export from the endoplasmic reticulum and transport between Golgi compartments. J. Cell Biol. 125:225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ogawa K, et al. 2009. Hepatitis C virus utilizes lipid droplet for production of infectious virus. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85:217–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ohsaki Y, et al. 2009. Biogenesis of cytoplasmic lipid droplets: from the lipid ester globule in the membrane to the visible structure. Biochim. Biophys. Acta 1791:399–407 [DOI] [PubMed] [Google Scholar]
- 49. Ozeki S, et al. 2005. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci. 118:2601–2611 [DOI] [PubMed] [Google Scholar]
- 50. Reference deleted.
- 51. Plutner H, et al. 1991. Rab1b regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. J. Cell Biol. 115:31–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schaller T, et al. 2007. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J. Virol. 81:4591–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282:37158–37169 [DOI] [PubMed] [Google Scholar]
- 54. Shi ST, et al. 2002. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology 292:198–210 [DOI] [PubMed] [Google Scholar]
- 55. Sklan EH, et al. 2007. TBC1D20 is a Rab1 GTPase-activating protein that mediates hepatitis C virus replication. J. Biol. Chem. 282:36354–36361 [DOI] [PubMed] [Google Scholar]
- 56. Sklan EH, et al. 2007. A Rab-GAP TBC domain protein binds hepatitis C virus NS5A and mediates viral replication. J. Virol. 81:11096–11105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Soni KG, et al. 2009. Coatomer-dependent protein delivery to lipid droplets. J. Cell Sci. 122:1834–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Su AI, et al. 2002. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 99:15669–15674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tanji Y, Kaneko T, Satoh S, Shimotohno K. 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J. Virol. 69:3980–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Targett-Adams P, et al. 2011. Small molecules targeting hepatitis C virus-encoded NS5A cause subcellular redistribution of their target: insights into compound mode of action. J. Virol. 85:6353–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tellinghuisen TL, Marcotrigiano J, Gorbalenya AE, Rice CM. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 279:48576–48587 [DOI] [PubMed] [Google Scholar]
- 62. Tellinghuisen TL, Marcotrigiano J, Rice CM. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tisdale EJ, Bourne JR, Khosravi-Far R, Der CJ, Balch WE. 1992. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J. Cell Biol. 119:749–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tscherne DM, et al. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80:1734–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Umlauf E, et al. 2004. Association of stomatin with lipid bodies. J. Biol. Chem. 279:23699–23709 [DOI] [PubMed] [Google Scholar]
- 66. Reference deleted.
- 67. Wilson BS, et al. 1994. A Rab1 mutant affecting guanine nucleotide exchange promotes disassembly of the Golgi apparatus. J. Cell Biol. 125:557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wolk B, Buchele B, Moradpour D, Rice CM. 2008. A dynamic view of hepatitis C virus replication complexes. J. Virol. 82:10519–10531 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.