Abstract
In infected cells, the chromatin structure of the adenovirus genome DNA plays critical roles in its genome functions. Previously, we reported that in early phases of infection, incoming viral DNA is associated with both viral core protein VII and cellular histones. Here we show that in late phases of infection, newly synthesized viral DNA is also associated with histones. We also found that the knockdown of CAF-1, a histone chaperone that functions in the replication-coupled deposition of histones, does not affect the level of histone H3 bound on viral chromatin, although CAF-1 is accumulated at viral DNA replication foci together with PCNA. Chromatin immunoprecipitation assays using epitope-tagged histone H3 demonstrated that histone variant H3.3, which is deposited onto the cellular genome in a replication-independent manner, is selectively associated with both incoming and newly synthesized viral DNAs. Microscopic analyses indicated that histones but not USF1, a transcription factor that regulates viral late gene expression, are excluded from viral DNA replication foci and that this is achieved by the oligomerization of the DNA binding protein (DBP). Taken together, these results suggest that histone deposition onto newly synthesized viral DNA is most likely uncoupled with viral DNA replication, and a possible role of DBP oligomerization in this replication-uncoupled histone deposition is discussed.
INTRODUCTION
In the cell nucleus, the genomic DNA is not naked but forms a chromatin structure with chromatin proteins. The fundamental unit of the chromatin structure is the nucleosome, which consists of a histone octamer (two copies each of histones H2A, H2B, H3, and H4) and DNA wrapping around the octamer. The deposition of histones and/or the remodeling of nucleosome arrays is a critical process for the expression of genome functions (2), since nucleosome packaging could be a barrier for trans-acting factors to access their cognate sites on DNA. Thus, the nucleosome structure must be strictly and dynamically regulated in connection with several events on chromatin, such as transcription, DNA replication, and DNA repair.
Currently, it is known that histone deposition is carried out mainly in two fashions, DNA replication-dependent and independent ones, and a role of histone variants in these deposition pathways has been elucidated (14). In mammalian somatic cells, there are three major histone H3 variants, H3.1, H3.2, and H3.3, and they have only slight differences in amino acid sequences (16). The canonical histone H3, histone H3.1, and the highly related variant H3.2 (which differs by only 1 amino acid [aa] from H3.1) are expressed exclusively during the S phase, while the expression of the variant H3.3, which differs by 4 and 5 aa from H3.2 and H3.1, respectively, is observed throughout the cell cycle. Thus, this variant is called a “replication-independent” one (11). Tagami et al. demonstrated previously that the canonical histone H3 (H3.1) interacts with the histone chaperone CAF-1 complex and is deposited onto DNA in a replication-dependent manner, while HIRA specifically binds to and deposits histone variant H3.3 onto DNA independently of DNA synthesis (43). CAF-1 is composed of three subunits, p150, p60, and p48, and is associated with the cellular DNA replication machinery through an interaction with PCNA, a sliding clamp for DNA polymerases, allowing the DNA replication-coupled deposition of histones (40, 41, 50). On the other hand, HIRA was identified as a DNA synthesis-independent histone chaperone in cell-free systems using Xenopus laevis egg extracts (32), and histone variant H3.3 was shown to mark transcriptionally active genomic regions (1). Furthermore, additional H3.3-specific chaperones were recently identified. Daxx is one of the components of promyelocytic leukemia (PML) nuclear bodies and was reported to deposit histone H3.3 onto specific genomic regions, such as telomeres and pericentric heterochromatin, together with an ATP-dependent chromatin remodeler, ATRX (10, 21). It was also reported that in Drosophila melanogaster cells, DEK is a coactivator of a nuclear receptor and functions as an H3.3-specific chaperone (37). Thus, the mechanistic evidences for histone deposition are accumulating in the case of cellular chromatin.
The regulatory events for the chromatin structure are not limited to the cellular genome, as some viruses also have chromatin and/or chromatin-like structures with their own genomes. The adenovirus (Ad) genome is a linear double-stranded DNA (dsDNA) of ∼36,000 bp in length. In the virion, the Ad genome forms a chromatin-like structure with viral basic core proteins, as revealed by electron microscopic analyses showing that viral core protein-DNA complexes purified from the virion show a “beads-on-a-string” structure (49). Among core proteins, protein VII is a major DNA binding protein that can introduce superhelical turns into DNA, as do cellular histones (4), and remains associated with viral DNA after nuclear import of the virus genome (7, 17). When viral DNA-core protein complexes purified from the virion are used as a template for cell-free DNA replication/transcription systems, the reactions occur at a much lower level than in the case of naked DNA, indicating that the viral chromatin-like structure must be remodeled to execute its genome functions (22, 23). Previously, we identified host cell-derived remodeling factors for Ad chromatin by biochemical analyses (19, 22, 24, 26) and demonstrated that TAF-I, one of these host factors, plays an important role in the regulation of viral early gene expression in infected cells through interactions with protein VII (15, 17, 18, 20, 27). Thus, it was indicated that the remodeling of Ad chromatin is a crucial process for its genome functions (13), as is the case for the cellular genome. In addition, using chromatin immunoprecipitation (ChIP) assays, we recently reported that in early phases of infection, cellular histones are incorporated into viral DNA-protein VII complexes and that a histone modification occurs depending upon the transcription states on viral chromatin, suggesting that cellular histones could be functional components of viral chromatin in infected cells (20).
As described above, although the viral chromatin structure and its regulation in early phases of infection are being clarified, it is quite unclear how the viral chromatin structure is regulated in late phases of infection. In particular, since the expression of viral late genes is dependent largely on its own DNA replication (45), the regulation of the chromatin structure during viral DNA replication could be a key step. Therefore, in this study, we sought to elucidate the regulatory mechanism of how the chromatin structure is formed on newly synthesized viral DNA through viral DNA replication, in particular with respect to histone deposition. We found that after the onset of viral DNA replication, cellular histones are also incorporated into viral chromatin. We also found that although CAF-1 is accumulated at the site of viral DNA replication, this factor seems not to be involved in histone deposition during viral DNA replication, since the knockdown of CAF-1 did not affect the binding level of histone H3 on viral chromatin, and that histone variant H3.3, which is deposited onto DNA in a DNA synthesis-independent manner, is specifically deposited onto viral DNA even after the onset of viral DNA replication. Microscopic analyses suggested that histones but not USF1, a transcription factor which was shown previously to bind to and regulate transcription from the viral major late promoter (MLP) (46), are excluded form the site of viral DNA replication, possibly by the oligomerization of Ad single-stranded DNA (ssDNA) binding protein (DBP), one of the viral DNA replication factors. Based on these results, we propose a model whereby, unlike cellular chromatin, histone deposition onto the newly synthesized viral DNA is not coupled with viral DNA replication. A feasible role of this uncoupled deposition mechanism mediated by DBP oligomerization in Ad genome functions is discussed.
MATERIALS AND METHODS
Cells and viruses.
The maintenance of HeLa cells and the purification and infection of human adenovirus type 5 (HAdV5) were carried out essentially as described previously (18, 20). Hydroxyurea (HU) was added at a final concentration of 2 mM immediately after infection when DNA replication was to be blocked. HeLa cells stably expressing enhanced green fluorescent protein (EGFP)-tagged histones H3.2 and H3.3 (a kind gift from M. Okuwaki, University of Tsukuba) were also maintained as described above. The transfection of expression plasmids was performed by using GeneJuice (Novagen) according to the manufacturer's protocol.
Antibodies.
Antibodies used in this study are as follows: rabbit anti-histone H3 (catalog no. ab1791; Abcam), rabbit anti-histone H4 (catalog no. 04-858; Millipore), rabbit anti-histone H2A (catalog no. ab18255; Abcam), mouse anti-HIRA (catalog no. 04-1488; Millipore), mouse anti-FLAG M2 (catalog no. F3165; Sigma), rat anti-hemagglutinin (HA) (3F10; Roche), and mouse anti-β-actin (Sigma) antibodies. Rabbit anti-histone H2A-H2B, mouse anti-CAF-1 p150, and mouse anti-DBP antibodies were kindly provided by M. Okuwaki (University of Tsukuba), A. Verreault (University of Montreal), and W. C. Russel, respectively. Rat anti-protein VII antibody was described elsewhere previously (17).
Vector construction.
To construct the expression vectors for USF1, full-length DBP, and its deletion mutant (DBPΔC, which lacks the C-terminal 17 aa), cDNA fragments of USF1, DBP, and DBPΔC were amplified by PCR; digested with BamHI and EcoRI; and cloned in frame into the pCHA vector containing an HA epitope tag and the puromycin resistance gene (pCHA-puro vector; kindly provided by K. Kajitani, University of Tsukuba). The resulting vectors were designated pCHA-puro-USF1, pCHA-puro-DBP, and pCHA-puro-DBPΔC, respectively. Similarly, for the expression vector of PCNA, an amplified cDNA fragment was digested with BamHI and cloned into the pCHA-puro vector digested with BamHI and EcoRV (pCHA-puro-PCNA). The primers used here were as follows: 5′-GTTTAGGATCCCATATGAAGGGGCAGCAG-3′ and 5′-GGGCCGAATTCTTAGTTGCTGTCATTCTTG-3′ for USF1 cDNA, 5′-AAAGGATCCATGGCCAGTCGGG-3′ and 5′-GCGGAATTCTTAAAAATCAAAGGGGTTCTG-3′ for DBP cDNA, 5′-AAAGGATCCATGGCCAGTCGGG-3′ and 5′-CCCGAATTCTTAGTTGCGATACTGG-3′ for DBPΔC cDNA, and 5′-AAAGGATCCATGTTCGAGGCGC-3′ and 5′-ATCGTCGACCTAAGATCCTTCTTC-3′ for PCNA cDNA.
For the preparation of cells stably expressing HA-PCNA, HeLa cells were transfected with pCHA-puro-PCNA and cultured in the presence of 2 μg/ml puromycin for 2 weeks.
For the construction of the expression vector of histone H3.1, a cDNA fragment of histone H3.1 was amplified by PCR, digested with NcoI, and cloned into the pBS-FLAG vector (pBS-H3.1-FLAG). The DNA fragment containing cDNA of H3.1 and the C-terminal FLAG tag was then obtained from pBS-H3.1-FLAG by digestion with BamHI and EcoRI and cloned into the pcDNA3 vector (pcDNA3-H3.1-FLAG). The primers used here were as follows: 5′-AAAACCATGGCGCGTACTAAGCAG-3′ and 5′-TTATTCCATGGCCGCCCTCTCCCCA-3′.
The expression vectors for FLAG-tagged histones H3.2 and H3.3 (pcDNA3-H3.2-FLAG and pcDNA3-H3.3-FLAG, respectively) and HA-tagged DEK (pCHA-DEK) were generously provided by M. Okuwaki and S. Saito, respectively (University of Tsukuba).
Indirect immunofluorescence assays.
Indirect immunofluorescence (IF) assays were carried out essentially as previously described (18). The localization of the protein was visualized with secondary antibodies (anti-mouse IgG conjugated with Alexa Fluor 488, anti-mouse IgG conjugated with Alexa Fluor 568, and anti-rabbit IgG conjugated with Alexa Fluor 568; Invitrogen). DNA was visualized by staining with TO-PRO-3 iodide (Invitrogen). Labeled cells were observed by confocal laser scanning microscopy (LSM5 Exciter; Carl Zeiss), using argon laser (488 nm) and He/Ne laser (546 and 633 mm) lines.
ChIP, RT-PCR, siRNA-mediated knockdown, and Western blot assays.
ChIP, reverse transcription (RT)-PCR, small interfering RNA (siRNA)-mediated knockdown, and Western blot assays were carried out essentially as described previously (20). siRNA targeted for CAF-1 p150 was purchased commercially (Stealth siRNA; Invitrogen). Primers used for CAF-1 p150 mRNA are as follows: 5′-GGAGCAGGACAGTTGGAGTG-3′ and 5′-GACGAATGGCTGAGTACAGA-3′. Other primers for ChIP and RT-PCR assays were described elsewhere previously (20). In ChIP and RT-PCR assays using quantitative PCR (qPCR), the obtained cDNAs were quantitatively measured by qPCR, and mean values with standard deviations (SD) were obtained from three independent experiments.
RESULTS
Cellular histones are bound with viral chromatin in both early and late phases of infection.
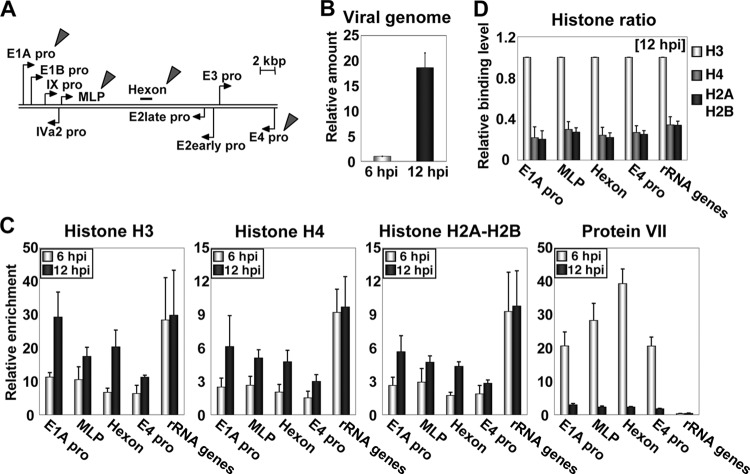
Previously, we reported that in early phases of infection (before the onset of viral DNA replication), viral chromatin is composed of both viral core protein VII and cellular histones and that this “chimeric” chromatin functions as the template for transcription (20). To examine whether histones are also bound with viral chromatin after the onset of viral DNA replication, we performed ChIP assays using antibodies against histones and protein VII (Fig. 1). Viral DNA replication starts at around 8 hpi (hours postinfection) under our conditions (17, 20). In order to reveal the viral chromatin state during or immediately after viral DNA replication, HeLa cells infected at an MOI (multiplicity of infection) of 100 were harvested at 6 and 12 hpi for ChIP assays. We chose five regions for ChIP assays, four viral genome regions (E1A pro, MLP, hexon, and E4 pro) (Fig. 1A) and one cellular genomic region (rRNA gene) as a control (20). Under this condition, the amount of viral DNA was increased by ∼20-fold through viral DNA replication (Fig. 1B). At 6 hpi, all core histones were bound with viral chromatin but at a low binding level compared with that of cellular chromatin (histones H3, H4, and H2A-H2B) (Fig. 1C). This was in good agreement with our previous observations (20). At 12 hpi, core histones were also found to be associated with viral chromatin. The binding level of histones on viral chromatin at 12 hpi was higher than that at 6 hpi but slightly lower than that on cellular chromatin. This is consistent with a previous report of electron microscopic analyses showing that viral genome DNA purified from infected cells at late phases of infection has nucleosome-like particles, which are less dense than those in cellular nucleosome arrays (3). In contrast, the binding level of protein VII was drastically decreased after the onset of viral DNA replication (Fig. 1C), suggesting that newly synthesized viral DNA is associated mainly with cellular histones. We do not exclude the possibility that protein VII remains associated with a small population of viral chromatin, because the binding level of protein VII on viral chromatin was still higher than that on cellular chromatin even at 12 hpi. The ratio among core histones bound on viral chromatin was almost the same as that among core histones bound on cellular chromatin both at 6 hpi (20; data not shown) and at 12 hpi (Fig. 1D), indicating that viral chromatin contains the canonical nucleosome structure.
Fig 1.
Viral chromatin structure in early and late phases of infection. (A) Structure of the Ad genome. Arrows represent promoters of viral genes. Target regions for ChIP assays are indicated by arrowheads. (B) Amounts of viral DNA. HeLa cells were infected with HAdV5 at an MOI of 100, and DNA samples were purified from infected cells at 6 and 12 hpi. The amount of viral DNA was quantitatively measured by qPCR using primers for the E1A promoter region. The amount of viral DNA is graphed as a ratio relative to that at 6 hpi. (C) ChIP assays. HeLa cells were infected with HAdV5 at an MOI of 100 and subjected to ChIP assays using infected cells at 6 and 12 hpi. Immunoprecipitation was carried out by using the indicated antibodies and an anti-FLAG antibody (as a negative control). The obtained DNAs were quantitatively measured by qPCR using the indicated primer sets. The binding levels of each protein were calculated as relative enrichment against that obtained in a negative control (anti-FLAG antibody). (D) Binding levels of core histones. Based on the results of ChIP assays, shown in panel C, the binding levels of histone H4 and H2A-H2B were normalized to that of histone H3.
CAF-1 and PCNA are not involved in histone deposition onto newly synthesized viral DNA.
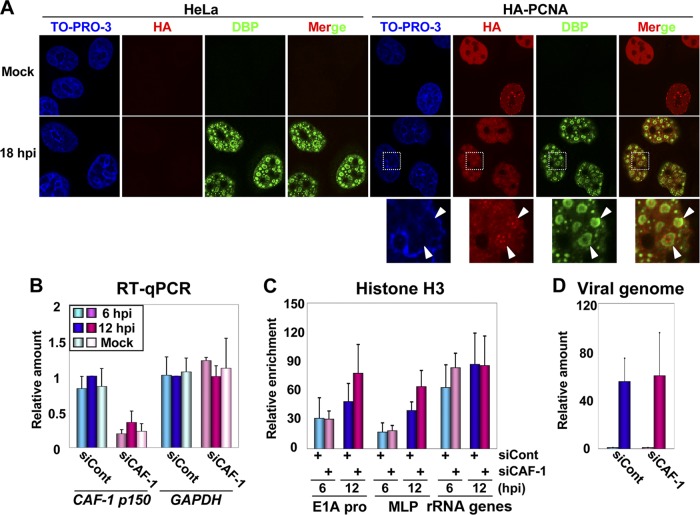
It is known that during the DNA replication of the cellular genome and some DNA virus genomes, histones are deposited by CAF-1, a replication-dependent histone chaperone (41, 43). CAF-1 is associated with the DNA replication machinery through interactions with PCNA, thereby enabling the replication-coupled deposition of histone H3-H4 complexes (40). Thus, it was worthwhile to examine whether CAF-1 and PCNA are also involved in the histone deposition onto newly synthesized Ad DNA, although there is no definitive evidence that these two are involved in Ad DNA replication. To test this, we first performed IF assays using cells stably expressing HA-tagged PCNA (HA-PCNA) to examine the relationship among viral DNA replication, PCNA, and CAF-1 (Fig. 2A). By using an antibody against DBP, an Ad ssDNA binding protein involved in viral DNA replication, the places for viral DNA replication, designated “viral DNA replication foci” (here referred to as “VDRF”), can be visualized (31). HeLa cells or cells stably expressing HA-PCNA were infected at an MOI of 50, and at 18 hpi, the cells were subjected to IF assays using anti-DBP and anti-HA antibodies. In mock-infected cells, HA-PCNA was localized throughout the nucleus and showed a punctate localization in some cell populations, as reported previously (29). At 18 hpi, VDRF was observed as a donut-like signal by using an anti-DBP antibody, and we found that HA-PCNA, showing a punctate localization, was accumulated inside VDRF. We also observed a similar localization pattern of CAF-1 inside VDRF (see Fig. 4A). These results suggest that PCNA and CAF-1 are recruited together to the site of viral DNA replication.
Fig 2.
Localization and role of PCNA and CAF-1 during viral DNA replication. (A) IF assays. HeLa cells and cells stably expressing HA-PCNA grown on coverslips were mock infected or infected with HAdV5 at an MOI of 50. At 18 hpi, the localization patterns of HA-PCNA and DBP were analyzed by IF using anti-HA and anti-DBP antibodies. DNA was visualized by TO-PRO-3 iodide staining. Merged images are also indicated. Higher-magnification images of the regions marked by squares are shown below. (B) RT-qPCR assays. HeLa cells were treated with siCont or siCAF-1 and then either mock infected or infected with HAdV5 at an MOI of 100, and total RNAs were purified at 6 and 12 hpi. cDNAs were synthesized with reverse transcription and subjected to qPCR using primer sets for CAF-1 p150 and GAPDH mRNAs. The mRNA levels relative to those of control cells at 12 hpi were graphed. (C) ChIP assays. siRNA-treated cells were infected with HAdV5 at an MOI of 100, and at 6 and 12 hpi, the cells were subjected to ChIP assays using anti-histone H3 and anti-FLAG antibodies. (D) Relative amounts of viral DNA. Viral DNA was purified from lysates for ChIP assays (C) and subjected to qPCR using the primer set for the E1A promoter. The DNA amounts at 12 hpi relative to those at 6 hpi are shown.
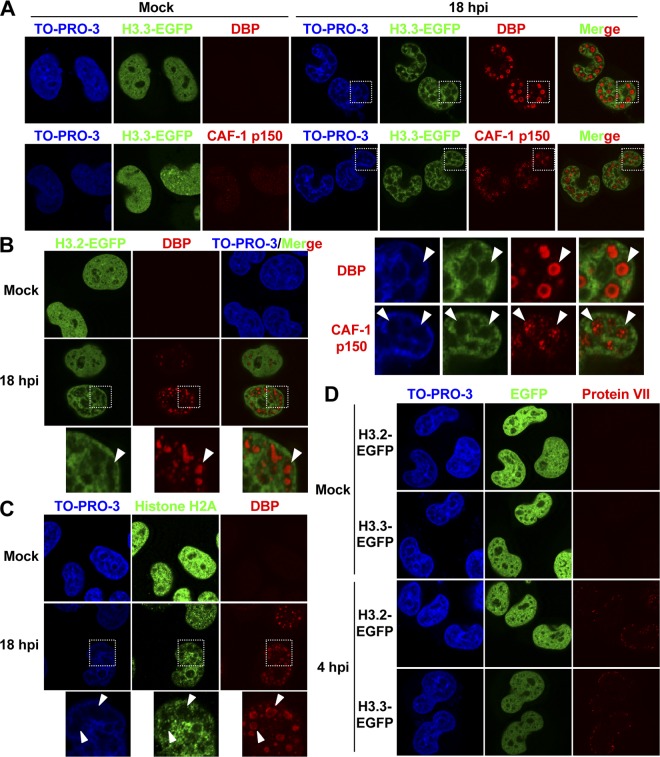
Fig 4.
Localization of histones in late phases of infection. (A) IF analyses using cells stably expressing histone H3.3-EGFP. HeLa cells stably expressing histone H3.3-EGFP grown on coverslips were mock infected or infected at an MOI of 50, and at 18 hpi, the cells were subjected to IF assays using anti-DBP (top) and anti-CAF-1 p150 (bottom) antibodies. Higher-magnification images of the regions marked by squares are shown below. (B) Localization of histone H3.2 in late phases of infection. HeLa cells stably expressing histone H3.2-EGFP were mock infected or infected with HAdV5 at an MOI of 50, and at 18 hpi, the cells were subjected to IF analyses using an anti-DBP antibody. Higher-magnification images of the regions marked by squares are shown. (C) Localization of endogenous histone H2A in late phases of infection. HeLa cells were mock infected or infected with HAdV5 at an MOI of 50, and at 18 hpi, the cells were subjected to IF assays using anti-histone H2A and anti-DBP antibodies. Higher-magnification images of the regions marked by squares are shown below. (D) Histone localization in early phases of infection. HeLa cells stably expressing histone H3.2-EGFP and H3.3-EGFP were mock infected or infected with HAdV5 at an MOI of 250, and at 4 hpi, the cells were subjected to IF analyses using an anti-protein VII antibody.
Next, to investigate a role of CAF-1 in histone deposition onto viral DNA, the siRNA-mediated knockdown of CAF-1 p150, the largest subunit of the CAF-1 complex, was carried out (Fig. 2B to D). HeLa cells were treated with control siRNA (siCont) or siRNA for CAF-1 p150 (siCAF-1), infected with HAdV5 at an MOI of 100, and harvested at 6 and 12 hpi. First, we examined the knockdown efficiency of CAF-1 p150 by RT-qPCR assays (Fig. 2B). The mRNA level of CAF-1 p150 in siRNA-treated cells was about 20% of that in control cells, although at 12 hpi, the level was slightly increased, possibly due to the S-phase-like environment induced by Ad infection (30). In contrast, the mRNA level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was almost unaffected by siRNA treatment and Ad infection. Under this condition, the binding level of histone H3 on viral chromatin was examined by ChIP assays (Fig. 2C). The binding level of H3 on viral chromatin was not decreased by the CAF-1 knockdown and, rather, slightly increased (but not statistically significantly) at 12 hpi (E1A pro and MLP) (Fig. 2C). Note that the binding level of H3 on cellular chromatin was also unaffected by the CAF-1 knockdown (rRNA gene) (Fig. 2C, and see Discussion). In addition, we could not observe any effect of the CAF-1 knockdown on viral DNA replication levels (Fig. 2D). Taken together, these results suggest that it is not likely that CAF-1 is involved in the histone deposition onto viral chromatin during viral DNA replication, although CAF-1 was accumulated at VDRF together with PCNA.
Replication-independent histone H3.3 is selectively incorporated into viral chromatin.
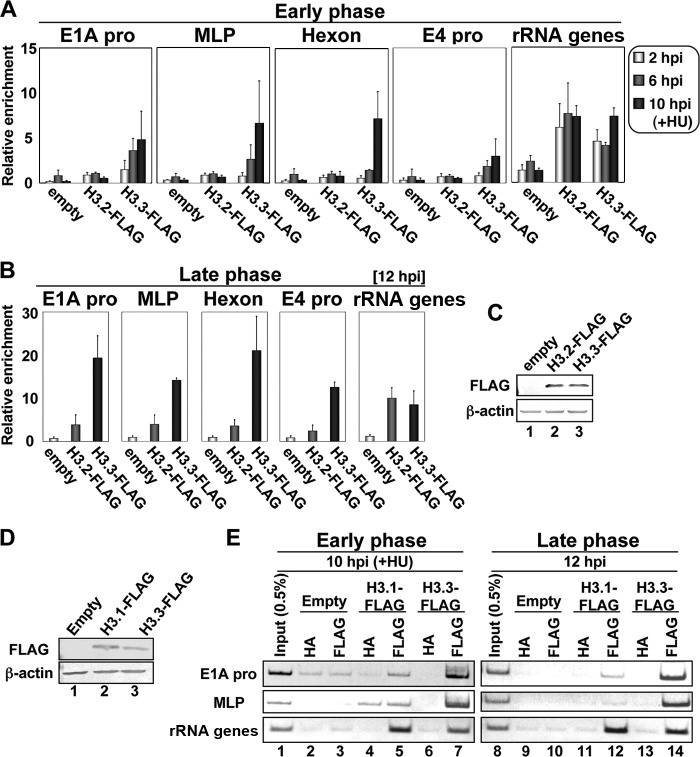
It is known that among histone H3 variants, histones H3.1 and H3.2 are deposited onto DNA by CAF-1 during DNA replication, while H3.3 is deposited independently of DNA replication (11, 14). If CAF-1 is not involved in histone deposition during viral DNA replication, histone H3.3, rather than H3.1 and H3.2, could be incorporated into newly synthesized viral DNA. Therefore, to examine this possibility, we performed ChIP assays using FLAG-tagged histones H3.2 and H3.3 (Fig. 3). HeLa cells were transfected with expression vectors for H3.2 and H3.3, and at 24 hpt (hours posttransfection), the cells were infected at an MOI of 100. We first studied cells at early phases of infection (before the onset of viral DNA replication) (Fig. 3A). Infected cells were harvested at 2 and 6 hpi as well as at 10 hpi in the presence of HU to block viral DNA replication (20) and subjected to ChIP assays with an anti-FLAG antibody. As shown in Fig. 3A, the exclusive binding of H3.3 on viral chromatin was observed at all the regions that we tested, with a gradual increase as infection proceeded. This finding is consistent with data from a recent report of a helper-dependent Ad vector (HdAd) indicating that histone H3.3 is specifically deposited onto HdAd DNA by HIRA, an H3.3-specific histone chaperone (34). Since HdAd alone does not undergo its DNA replication, the chromatin state of HdAd may reflect that of wild-type Ad in early phases of infection (13, 34).
Fig 3.
Incorporation of histone H3 variants into viral chromatin. (A and B) ChIP assays with FLAG-tagged histone H3 variants. HeLa cells were transfected with the pcDNA3 empty vector, pcDNA3-H3.2-FLAG, or pcDNA3-H3.3-FLAG and infected with HAdV5 at an MOI of 100 at 24 hpt (hours posttransfection). At 2, 6, and 10 hpi (A) or at 12 hpi (B), ChIP assays were carried out by using anti-FLAG and anti-HA (as a negative control) antibodies. Note that at 10 hpi, HU was added to block viral DNA replication. The results were graphed as relative enrichments. (C) Western blot analyses. At 24 hpt, lysates were prepared from cells transfected with the pcDNA3 empty vector (lane 1), pcDNA3-H3.2-FLAG (lane 2), and pcDNA3-H3.3-FLAG (lane 3) and subjected to 15% SDS-PAGE, followed by Western blot analyses using anti-FLAG (top) and anti-β-actin (bottom) antibodies. (D) Western blot analyses. HeLa cells were transfected with the pcDNA3 empty vector (lane 1), pcDNA3-H3.1-FLAG (lane 2), or pcDNA3-H3.3-FLAG (lane 3), and at 24 hpt, lysates were prepared and subjected to 15% SDS-PAGE, followed by Western blot analyses using anti-FLAG (top) and anti-β-actin (bottom) antibodies. (E) ChIP assays. HeLa cells transfected with the pcDNA3 empty vector (lanes 2, 3, 9, and 10), pcDNA3-H3.1-FLAG (lanes 4, 5, 11, and 12), or pcDNA3-H3.3-FLAG (lanes 6, 7, 13, and 14) were infected with HAdV5 at an MOI of 100. At 10 hpi (left, lanes 1 to 7) or at 12 hpi (right, lanes 8 to 14), ChIP assays were carried out by using anti-FLAG (lanes 3, 5, 7, 10, 12, and 14) and anti-HA (as a negative control) (lanes 2, 4, 6, 9, 11, and 13) antibodies. At 10 hpi, HU was added to block viral DNA replication. The immunoprecipitated DNAs were amplified by semiquantitative PCR using the indicated primer sets. PCR products were separated on a 7% polyacrylamide gel and visualized by staining with ethidium bromide (EtBr). Input DNAs (lanes 1 and 8) were purified from 0.5% of lysates of cells transfected with the empty vector.
Next, we performed ChIP assays at 12 hpi to examine which H3 variant is deposited onto newly synthesized viral DNA (Fig. 3B). We found that histone H3.3 but not H3.2 was associated with viral chromatin at this time point, as observed in the early phases of infection. The expression levels of both H3 variants were comparable (Fig. 3C), and both variants were associated with cellular chromatin with a similar binding level (rRNA gene) (Fig. 3A and B). These findings strongly suggest that this result was not due to some technical issues. Furthermore, we obtained the same results by using FLAG-tagged histone H3.1 instead of H3.2 (Fig. 3D and E). Thus, these results suggest that replication-independent histone variant H3.3 is selectively deposited onto not only incoming but also newly synthesized viral DNA in infected cells.
Histones but not the transcription factor USF1 are excluded from the site of viral DNA replication.
To further investigate the fluctuation in histone levels during viral DNA replication, we performed IF analyses using HeLa cells stably expressing EGFP-tagged histone H3.3 (Fig. 4A). Cells were infected at an MOI of 50 and were subjected to IF assays using an anti-DBP antibody at 18 hpi (Fig. 4A, top). VDRF was observed at 18 hpi, as described above, and H3.3-EGFP was found to be excluded from VDRF. Similar results were obtained by using cells stably expressing EGFP-tagged histone H3.2 (Fig. 4B). This was due to neither exogenous expression nor the EGFP tag of H3 variants, as we observed a similar exclusion of the endogenous histone using anti-histone H2A antibody (Fig. 4C). This localization pattern was specific for the late phases of infection, since the localization of EGFP-tagged H3 variants was not changed in early phases of infection (Fig. 4D). We also performed IF analyses using anti-CAF-1 p150 antibody and observed that CAF-1 was accumulated at a “histoneless” region, that is, VDRF (Fig. 4A, bottom), as was observed for HA-PCNA (Fig. 2A). In summary, these results indicated that histones are localized reciprocally to VDRF (and CAF-1/PCNA) in late phases of infection.
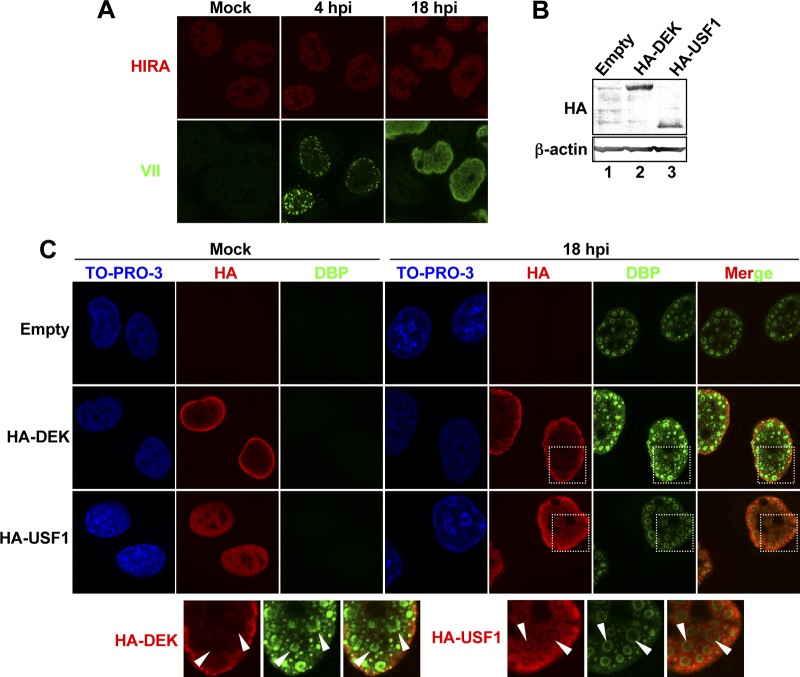
To gain more insights into the accessibility of other nuclear proteins to VDRF, we again performed IF analyses (Fig. 5). First, IF analyses were carried out by using antibody against HIRA, an H3.3-specific histone chaperone, and it was observed that the localization of HIRA was not drastically changed in both the early and late phases of infection (Fig. 5A). We also performed IF analyses using cells transiently transfected with expression vectors for HA-tagged DEK and USF1 (Fig. 5B and C). DEK is a cellular chromatin protein with potential histone chaperone activity (37), and USF1 is an E-box binding transcription factor and was reported previously to bind to and regulate transcription from the MLP region (46). HeLa cells were transfected with expression vectors, and at 24 hpt, the cells were subjected to Western blot analyses (Fig. 5B) or infected at an MOI of 50. At 18 hpi, the localization of DBP, HA-DEK, and HA-USF1 was visualized by IF analyses using anti-DBP and anti-HA antibodies (Fig. 5C). In mock-infected cells, both HA-tagged proteins showed a nuclear localization, and in the case of HA-DEK, a strong signal was observed at the nuclear periphery. At 18 hpi, HA-DEK appeared to be excluded from VDRF (Fig. 5C). However, in contrast to HA-DEK, we observed that HA-USF1 could be localized inside VDRF (Fig. 5C). Taken together, our IF analyses suggest that VDRF may allow the selective access of cellular nuclear proteins, and at least one of the transcription factors, USF1, is able to access VDRF.
Fig 5.
Localization of nuclear proteins in late phases of infection. (A) IF analyses using anti-HIRA antibody. HeLa cells were mock infected or infected with HAdV5 at an MOI of 250 (for 4 hpi) or 50 (for 18 hpi) and subjected to IF analyses using anti-protein VII and anti-HIRA antibodies. (B) Western blot analyses. Lysates were prepared from HeLa cells transfected with the pCHA-puro empty vector (lane 1), pCHA-DEK (lane 2), or pCHA-puro-USF1 (lane 3) at 24 hpt and were subjected to 10% SDS-PAGE, followed by Western blot analyses using anti-HA (top) and anti-β-actin (bottom) antibodies. (C) Localization of HA-DEK and HA-USF1. HeLa cells were transfected with the pCHA-puro empty vector, pCHA-DEK, or pCHA-puro-USF1. At 24 hpt, cells were mock infected or infected with HAdV5 at an MOI of 50, and at 18 hpi, the cells were subjected to IF assays using anti-HA and anti-DBP antibodies.
Oligomerization of DBP is critical for histone exclusion from VDRF.
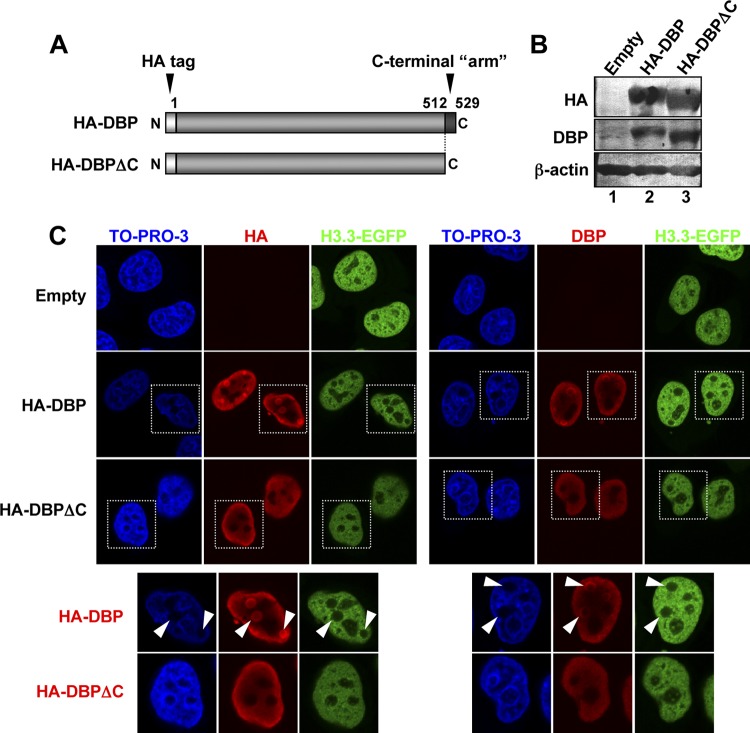
To investigate the mechanism of histone exclusion from VDRF, we hypothesized that DBP may play a role, since an abundant amount of DBP was associated with Ad DNA in VDRF. The crystal structure of DBP revealed that this protein has a 17-aa extension at its C terminus (Fig. 6A), and this C-terminal “arm” hooks onto the next DBP molecule, resulting in the oligomerization of DBP (47). It was also reported previously that the oligomerization of DBP mediated by the C-terminal arm enables the ATP-independent unwinding of dsDNA, and thus, full-length DBP but not the deletion mutant that lacks the C-terminal arm (DBPΔC) could support viral DNA replication in vitro (9). Therefore, to examine a role of DBP and its oligomerization in histone localization, HeLa cells expressing histone H3.3-EGFP were transfected with the expression vectors for HA-tagged full-length DBP or DBPΔC, and at 36 hpt, the cells were subjected to Western blotting and IF assays using anti-HA and anti-DBP antibodies (Fig. 6B and C). The expression levels of both DBP proteins were almost the same, as indicated by Western blotting (Fig. 6B). By IF analyses, we observed that full-length DBP forms foci like VDRF in the absence of any viral proteins/DNA, and histone H3.3-EGFP was excluded from these foci, as observed for infected cells (HA-DBP) (Fig. 6C). In sharp contrast, DBPΔC was localized throughout the nucleus and did not form such foci (Fig. 6C). Taken together, these results suggest that the oligomerization of DBP has a critical role in histone exclusion from VDRF.
Fig 6.
Role of DBP oligomerization in histone localization. (A) Schematic diagrams of full-length DBP and the C-terminally deleted mutant DBPΔC. DBP of HAdV5 consists of 529 aa, and the C-terminal 17 aa (aa 513 to 529) function as an “arm” for oligomerization. DBPΔC lacks the C-terminal 17 aa. (B) Western blot analyses. HeLa cells stably expressing histone H3.3-EGFP were transfected with the pCHA-puro empty vector (lane 1), pCHA-puro-DBP (lane 2), or pCHA-puro-DBPΔC (lane 3), and lysates prepared at 36 hpt were subjected to 10% SDS-PAGE, followed by Western blot analyses using anti-HA (top), anti-DBP (middle), and anti-β-actin (bottom) antibodies. (C) IF analyses. At 36 hpt, cells grown on coverslips were subjected to IF analyses using anti-HA (left) and anti-DBP (right) antibodies (B). Higher-magnification images of the regions marked by squares are shown below.
DISCUSSION
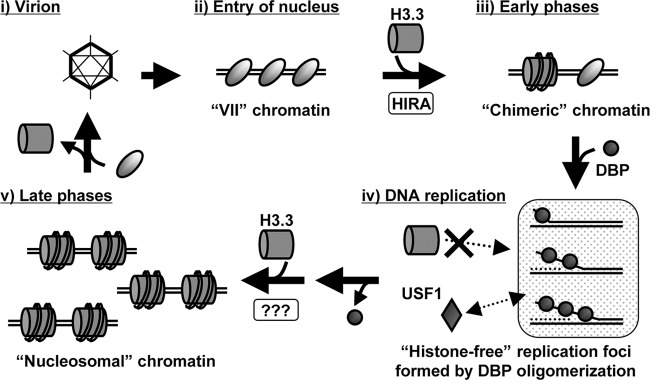
In this study, we showed that replication-independent histone variant H3.3 is deposited onto both incoming and newly synthesized Ad DNA (Fig. 3). These results, together with the results from knockdown experiments with CAF-1 (Fig. 2) and microscopic analyses (Fig. 4 to 6), indicated that the histone deposition onto the replicated virus genome is most likely uncoupled with viral DNA replication. Based on these results, together with data from our previous work (20), we hypothesize a model with respect to the fluctuation of the viral chromatin structure during the infection cycle (Fig. 7). In virions, viral DNA is tightly packed with viral core proteins (13). After entry into the cell, cellular histones are incorporated into incoming viral DNA-protein VII complexes in the nucleus, and viral chromatin composed of both protein VII and histones functions as the template for viral early gene expression (20). In this process, histone H3.3 is specifically deposited onto viral DNA, possibly by a histone chaperone, HIRA (34). As infection proceeds and viral DNA replication is then initiated, the oligomerization of DBP establishes a “histone-free” environment for viral DNA replication. Newly synthesized viral DNA is then associated with histone H3.3 in a replication-uncoupling fashion and might be acting as the template for viral late gene expression outside VDRF (31). In later phases of infection (∼24 hpi), both histones and newly synthesized core proteins VII and V are associated with viral DNA, which likely reflects processes during progeny virion assembly (35). Since histones are not included in virions, histones must be removed and replaced with newly synthesized core proteins for progeny virions. Although the packaging mechanism of progeny viral DNA during virion assembly remains unclear, we have reported the involvement of nucleolar protein B23/nucleophosmin in the regulation of the viral chromatin structure during progeny virion assembly (35, 36).
Fig 7.
Hypothetical model for the viral chromatin structure during the infection cycle. For details, see Discussion.
The mechanistic details of the histone deposition after viral DNA replication still remain unclear. First, what factor(s) is involved in histone deposition at late phases of infection? HIRA is a potential candidate for this process as well as in early phases of infection (34). However, we did not perform knockdown experiments for HIRA, since even if we could observe some effect of an HIRA knockdown on viral chromatin in late phases of infection, we could hardly distinguish whether the knockdown affects the chromatin structure of progeny viral DNA directly or whether the effect is derived indirectly from earlier events on incoming viral chromatin. IF analyses showed that the localization of HIRA was not drastically changed during the infection cycle (Fig. 5A). Recent reports indicated that Daxx, a component of PML bodies, is also an H3.3-specific histone chaperone (10, 21). However, Daxx seems not to function in the H3.3 deposition onto viral DNA, because during Ad infection, some components of PML bodies, including Daxx, are relocalized by the viral protein E4orf3, possibly for the inactivation of the components (6, 42). Indeed, it was shown previously that the Daxx-mediated antiviral response is antagonized by E4orf3 (48). It was also revealed that Daxx negatively functions and undergoes E1B-55K- and proteasome-dependent degradation during Ad infection (39). Furthermore, most recently, Schreiner et al. reported that during or immediately after the nuclear import of the incoming virus genome, protein VI, one of the capsid proteins, binds to and counteracts Daxx, at least partly by displacing it from PML bodies (38). Those reports strongly suggest that Daxx is inactivated entirely throughout the infection cycle by viral proteins. DEK was also recently reported to be a chaperone for histone H3.3 in Drosophila cells (37), but it is unknown whether human DEK also functions as a variant-specific chaperone or not. Our IF analyses indicated that exogenously expressed DEK is excluded from VDRF (Fig. 5B and C). Further studies are needed to elucidate the functions of these factors in late phases of infection.
In this study, we could not observe a role of CAF-1 in histone deposition onto viral DNA, while the accumulation of CAF-1 at VDRF was observed (Fig. 2 and 4A). The CAF-1 knockdown did not affect the binding levels of histone H3 on viral chromatin (Fig. 2C). Although we could not exclude the possibility that the knockdown efficiency of CAF-1 is not sufficient under the conditions employed here, we concluded that the function of CAF-1 is largely inhibited under our conditions: we observed that siCAF-1-treated cells exhibit aberrant cell shapes (data not shown) and that the knockdown affected viral gene expression (see below). Second, histone H3.3 is selectively incorporated into viral chromatin (Fig. 3), while CAF-1 generally functions as a chaperone for H3.1 and H3.2 (43). Third, although CAF-1 was reported previously to be able to be associated with H3.3 under some specific conditions (10, 21), we could not observe any interaction between CAF-1 and H3.3 during Ad infection, at least under our experimental conditions (data not shown). In addition to their roles during DNA replication, CAF-1 and PCNA were also reported previously to be involved in the DNA damage response pathway (29). Carson et al. reported that the DNA damage response pathway is only partially activated during Ad infection and that some related factors, such as ATRIP and TopBP1, are accumulated at VDRF (5). Therefore, CAF-1 (and PCNA) might localize at VDRF in the course of this limited DNA damage response. Recently, it was reported that FANCD2, one of the factors involved in the DNA damage response, is accumulated at VDRF and that the loss of this protein results in lower expression levels of viral late, but not early, genes (8). Similarly, we observed that the CAF-1 knockdown affected mRNA levels of viral late genes without any effect on viral DNA replication (our unpublished observations). Thus, factors related to the DNA damage response, such as FANCD2 and CAF-1, might be required for viral late gene expression, although the underlying mechanisms are unknown. Under our conditions, the CAF-1 knockdown did not affect the binding level of histone H3 on cellular chromatin (rRNA gene) (Fig. 2C). This is consistent with a previous report that the loss of CAF-1 impairs the replication-coupled deposition of histones but that the formation of nucleosome arrays on genomic DNA was still observed in the absence of CAF-1 (44). In addition, a recent report demonstrated that a defect of histone H3.1 deposition by CAF-1 depletion could be rescued by HIRA-mediated H3.3 deposition (33). Thus, in the case of cellular chromatin, an alternative histone deposition pathway(s) could rescue the loss of CAF-1 function.
It remains to be clarified what is the biological/virological significance of histone deposition uncoupled with viral DNA replication. On cellular chromatin, a replication-dependent histone chaperone, CAF-1, is associated with the DNA replication machinery and deposits histone H3.1-H4 (and H3.2-H4) complexes during DNA replication (14, 40, 43). This DNA replication-coupled system of histone deposition is thought to also be utilized by some DNA viruses. For instance, the DNA replication of simian virus 40 (SV40) is dependent largely on the cellular replication machinery, and indeed, CAF-1 was originally identified by using cell-free DNA replication systems of SV40 (41). For cytomegalovirus infection, it was reported previously that cellular histones, CAF-1, and PCNA accumulated at viral replication compartments (25). In the case of herpes simplex virus 1, it was shown that histone H3.3 is first deposited onto incoming viral DNA by HIRA and that H3.1 then becomes associated with viral DNA accompanied by viral DNA replication (28). It was suggested that this functional link between DNA replication and histone deposition enables the transfer of “epigenetic memory,” such as histone modifications, to the daughter DNA strands (43). Thus, some DNA viruses might take advantage of this system for late gene expression, which generally occurs after viral DNA replication. On the other hand, Ad seems to utilize another strategy, that is, the uncoupling mechanism, as shown here. Like other DNA viruses, Ad late genes are expressed only after the onset of viral DNA replication. Thomas and Mathews demonstrated previously that Ad late gene expression requires its DNA replication in cis (45), although the molecular mechanism remains to be determined. This report leads us to hypothesize that the regulation of the viral chromatin structure during DNA replication could be an important process for late gene expression. In general, the histone/nucleosome structure on DNA restricts the access of trans-acting factors, such as transcription factors. In this view, DBP is an attractive candidate for the key regulatory factor for the DNA replication-dependent expression of viral late genes. By oligomerization, DBP is able to not only support viral DNA replication but also establish the “histone-free” environment, which could be an opportunity window for transcription factors to access the viral DNA for the activation of viral late genes. Our IF analyses showed that the transcription factor USF1, which binds to the MLP region after viral DNA replication (46), is not excluded from VDRF (Fig. 5B and C), supporting this notion. Furthermore, this is in agreement with a previous report that DBP enhances the binding of USF1 to the MLP region in vitro (51). Overall, we speculate that the uncoupling of histone deposition with viral DNA replication is mediated by DBP oligomerization, at least partly, and plays a role in the DNA replication-dependent activation of viral late gene expression.
The expression of certain cellular genes, such as the HoxB gene, has been shown to require DNA replication (12). However, the regulation mechanism of “DNA replication-dependent gene expression” remains to be determined. As Ad has late genes, the expressions of which are DNA replication dependent (45), this virus could be a good model for analyses of such regulations. Therefore, this study might give a clue to an understanding of the functional relationship between DNA replication and transcription on cellular and/or viral chromatin.
ACKNOWLEDGMENTS
We thank M. Okuwaki, S. Saito, A. Verreault, W. C. Russel, and K. Kajitani for their kind gifts.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to K.N.) and the University of Tsukuba research infrastructure support program (to T.K.).
Footnotes
Published ahead of print 11 April 2012
REFERENCES
- 1. Ahmad K, Henikoff S. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191–1200 [DOI] [PubMed] [Google Scholar]
- 2. Bell O, Tiwari VK, Thoma NH, Schubeler D. 2011. Determinants and dynamics of genome accessibility. Nat. Rev. Genet. 12:554–564 [DOI] [PubMed] [Google Scholar]
- 3. Beyer AL, Bouton AH, Hodge LD, Miller OL., Jr 1981. Visualization of the major late R strand transcription unit of adenovirus serotype 2. J. Mol. Biol. 147:269–295 [DOI] [PubMed] [Google Scholar]
- 4. Burg JL, Schweitzer J, Daniell E. 1983. Introduction of superhelical turns into DNA by adenoviral core proteins and chromatin assembly factors. J. Virol. 46:749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carson CT, et al. 2009. Mislocalization of the MRN complex prevents ATR signaling during adenovirus infection. EMBO J. 28:652–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carvalho T, et al. 1995. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chatterjee PK, Vayda ME, Flint SJ. 1986. Adenoviral protein VII packages intracellular viral DNA throughout the early phase of infection. EMBO J. 5:1633–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cherubini G, et al. 2011. The FANC pathway is activated by adenovirus infection and promotes viral replication-dependent recombination. Nucleic Acids Res. 39:5459–5473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dekker J, et al. 1997. Multimerization of the adenovirus DNA-binding protein is the driving force for ATP-independent DNA unwinding during strand displacement synthesis. EMBO J. 16:1455–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. 2010. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24:1253–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elsaesser SJ, Goldberg AD, Allis CD. 2010. New functions for an old variant: no substitute for histone H3.3. Curr. Opin. Genet. Dev. 20:110–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fisher D, Mechali M. 2003. Vertebrate HoxB gene expression requires DNA replication. EMBO J. 22:3737–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Giberson AN, Davidson AR, Parks RJ. 2012. Chromatin structure of adenovirus DNA throughout infection. Nucleic Acids Res. 40:2369–2376 doi:10.1093/nar/gkr1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Groth A, Rocha W, Verreault A, Almouzni G. 2007. Chromatin challenges during DNA replication and repair. Cell 128:721–733 [DOI] [PubMed] [Google Scholar]
- 15. Gyurcsik B, Haruki H, Takahashi T, Mihara H, Nagata K. 2006. Binding modes of the precursor of adenovirus major core protein VII to DNA and template activating factor I: implication for the mechanism of remodeling of the adenovirus chromatin. Biochemistry 45:303–313 [DOI] [PubMed] [Google Scholar]
- 16. Hake SB, Allis CD. 2006. Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis.” Proc. Natl. Acad. Sci. U. S. A. 103:6428–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haruki H, Gyurcsik B, Okuwaki M, Nagata K. 2003. Ternary complex formation between DNA-adenovirus core protein VII and TAF-Ibeta/SET, an acidic molecular chaperone. FEBS Lett. 555:521–527 [DOI] [PubMed] [Google Scholar]
- 18. Haruki H, Okuwaki M, Miyagishi M, Taira K, Nagata K. 2006. Involvement of template-activating factor I/SET in transcription of adenovirus early genes as a positive-acting factor. J. Virol. 80:794–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kawase H, et al. 1996. NAP-I is a functional homologue of TAF-I that is required for replication and transcription of the adenovirus genome in a chromatin-like structure. Genes Cells 1:1045–1056 [DOI] [PubMed] [Google Scholar]
- 20. Komatsu T, Haruki H, Nagata K. 2011. Cellular and viral chromatin proteins are positive factors in the regulation of adenovirus gene expression. Nucleic Acids Res. 39:889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. 2010. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. U. S. A. 107:14075–14080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsumoto K, Nagata K, Ui M, Hanaoka F. 1993. Template activating factor I, a novel host factor required to stimulate the adenovirus core DNA replication. J. Biol. Chem. 268:10582–10587 [PubMed] [Google Scholar]
- 23. Matsumoto K, et al. 1995. Stimulation of DNA transcription by the replication factor from the adenovirus genome in a chromatin-like structure. J. Biol. Chem. 270:9645–9650 [DOI] [PubMed] [Google Scholar]
- 24. Nagata K, et al. 1995. Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc. Natl. Acad. Sci. U. S. A. 92:4279–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nitzsche A, Paulus C, Nevels M. 2008. Temporal dynamics of cytomegalovirus chromatin assembly in productively infected human cells. J. Virol. 82:11167–11180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okuwaki M, Iwamatsu A, Tsujimoto M, Nagata K. 2001. Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J. Mol. Biol. 311:41–55 [DOI] [PubMed] [Google Scholar]
- 27. Okuwaki M, Nagata K. 1998. Template activating factor-I remodels the chromatin structure and stimulates transcription from the chromatin template. J. Biol. Chem. 273:34511–34518 [DOI] [PubMed] [Google Scholar]
- 28. Placek BJ, et al. 2009. The histone variant H3.3 regulates gene expression during lytic infection with herpes simplex virus type 1. J. Virol. 83:1416–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Polo SE, Roche D, Almouzni G. 2006. New histone incorporation marks sites of UV repair in human cells. Cell 127:481–493 [DOI] [PubMed] [Google Scholar]
- 30. Polo SE, et al. 2004. Chromatin assembly factor-1, a marker of clinical value to distinguish quiescent from proliferating cells. Cancer Res. 64:2371–2381 [DOI] [PubMed] [Google Scholar]
- 31. Pombo A, Ferreira J, Bridge E, Carmo-Fonseca M. 1994. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 13:5075–5085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ray-Gallet D, et al. 2002. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9:1091–1100 [DOI] [PubMed] [Google Scholar]
- 33. Ray-Gallet D, et al. 2011. Dynamics of histone h3 deposition in vivo reveal a nucleosome gap-filling mechanism for h3.3 to maintain chromatin integrity. Mol. Cell 44:928–941 [DOI] [PubMed] [Google Scholar]
- 34. Ross PJ, et al. 2011. Assembly of helper-dependent adenovirus DNA into chromatin promotes efficient gene expression. J. Virol. 85:3950–3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Samad MA, Komatsu T, Okuwaki M, Nagata K. 15 February 2012, posting date B23/nucleophosmin is involved in regulation of adenovirus chromatin structure at late infection stages, but not in its replication and transcription. J. Gen. Virol. [Epub ahead of print.] doi:10.1099/vir.0.036665-0 [DOI] [PubMed] [Google Scholar]
- 36. Samad MA, Okuwaki M, Haruki H, Nagata K. 2007. Physical and functional interaction between a nucleolar protein nucleophosmin/B23 and adenovirus basic core proteins. FEBS Lett. 581:3283–3288 [DOI] [PubMed] [Google Scholar]
- 37. Sawatsubashi S, et al. 2010. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 24:159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schreiner S, et al. 2012. Transcriptional activation of the adenoviral genome is mediated by capsid protein VI. PLoS Pathog. 8:e1002549 doi:10.1371/journal.ppat.1002549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schreiner S, et al. 2010. Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J. Virol. 84:7029–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shibahara K, Stillman B. 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96:575–585 [DOI] [PubMed] [Google Scholar]
- 41. Smith S, Stillman B. 1989. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58:15–25 [DOI] [PubMed] [Google Scholar]
- 42. Stracker TH, et al. 2005. Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J. Virol. 79:6664–6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51–61 [DOI] [PubMed] [Google Scholar]
- 44. Takami Y, Ono T, Fukagawa T, Shibahara K, Nakayama T. 2007. Essential role of chromatin assembly factor-1-mediated rapid nucleosome assembly for DNA replication and cell division in vertebrate cells. Mol. Biol. Cell 18:129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thomas GP, Mathews MB. 1980. DNA replication and the early to late transition in adenovirus infection. Cell 22:523–533 [DOI] [PubMed] [Google Scholar]
- 46. Toth M, Doerfler W, Shenk T. 1992. Adenovirus DNA replication facilitates binding of the MLTF/USF transcription factor to the viral major late promoter within infected cells. Nucleic Acids Res. 20:5143–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tucker PA, et al. 1994. Crystal structure of the adenovirus DNA binding protein reveals a hook-on model for cooperative DNA binding. EMBO J. 13:2994–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ullman AJ, Hearing P. 2008. Cellular proteins PML and Daxx mediate an innate antiviral defense antagonized by the adenovirus E4 ORF3 protein. J. Virol. 82:7325–7335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vayda ME, Rogers AE, Flint SJ. 1983. The structure of nucleoprotein cores released from adenovirions. Nucleic Acids Res. 11:441–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Verreault A, Kaufman PD, Kobayashi R, Stillman B. 1996. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87:95–104 [DOI] [PubMed] [Google Scholar]
- 51. Zijderveld DC, d'Adda di Fagagna F, Giacca M, Timmers HT, van der Vliet PC. 1994. Stimulation of the adenovirus major late promoter in vitro by transcription factor USF is enhanced by the adenovirus DNA binding protein. J. Virol. 68:8288–8295 [DOI] [PMC free article] [PubMed] [Google Scholar]