Abstract
Herpesvirus entry into cells requires coordinated interactions among several viral glycoproteins. The final membrane fusion step of entry is executed by glycoprotein B (gB), a class III viral fusion protein that is conserved across all herpesviruses. Fusion proteins are metastable proteins that mediate fusion by inserting into a target membrane and refolding from a prefusion to postfusion conformation to bring the viral and cell membranes together. Although the structure of gB has been solved in a conformation that likely represents its postfusion form, its prefusion structure and the details of how it refolds to execute fusion are unknown. The postfusion gB structure contains a trimeric coiled-coil at its core and a long C-terminal arm within the ectodomain packs against this coil in an antiparallel manner. This coil-arm complex is reminiscent of the six-helix bundle that provides the energy for fusion in class I fusogens. To determine the role of the coil-arm complex, we individually mutated residues in the herpes simplex virus 1 gB coil-arm complex to alanine and assessed the contribution of each residue to cell-cell and virus-cell fusion. Several coil mutations resulted in a loss of cell surface expression, indicating that the coil residues are important for proper processing of gB. Three mutations in the arm region (I671A, H681A, and F683A) reduced fusion without affecting expression. Combining these three arm mutations drastically reduced the ability of gB to execute fusion; however, fusion function could be restored by adding known hyperfusogenic mutations to the arm mutant. We propose that the formation of the coil-arm complex drives the gB transition to a postfusion conformation and the coil-arm complex performs a function similar to that of the six-helix bundle in class I fusion. Furthermore, we suggest that these specific mutations in the arm may energetically favor the prefusion state of gB.
INTRODUCTION
Unlike most enveloped viruses, which use a single protein to mediate binding to and fusion with a target cell, herpesvirus entry requires the coordinated action of multiple entry glycoproteins (reviewed in references 11 and 24). Glycoprotein D (gD) is the main receptor-binding protein, and engagement with receptor triggers fusion mediated by the gH/gL heterodimer and gB, proteins that together represent the conserved fusion machinery of herpesviruses. The herpesvirus gB crystal structures (3, 25) and subsequent mutational analyses (2, 22) indicate that gB is the fusion protein for herpesviruses.
Fusion proteins are metastable proteins that execute the final membrane merger step of virus entry by inserting into a target membrane and refolding to bring the viral and cell membranes into proximity. Crystal structures of multiple viral fusion proteins have been solved, and the proteins can be classified into three categories (23, 58). Class I fusogens are homotrimers that are rich in α-helices. Most class I fusion proteins contain a hydrophobic fusion peptide and proteolytic processing N terminal to this peptide activates the fusogenic potential of the protein. Upon triggering by low pH and/or receptor binding, the fusion peptides insert into the target membrane, and the protein refolds into a stable postfusion conformation. During transition to the postfusion form, a C-terminal region of the ectodomain adjacent to the transmembrane (TM) region (called heptad repeat B [HRB]) packs against an N-terminal trimeric α-helical coiled-coil adjacent to the fusion peptide (called heptad repeat A [HRA]) in an antiparallel manner, thus bringing the two membrane-inserted portions of the protein together. In the postfusion conformation, HRA and HRB form an energetically stable “six-helix bundle,” which may provide the energy to drive membrane fusion (34, 39, 53). There is variability among class I fusion proteins. In some class I fusion proteins, such as the paramyxovirus F protein (61) and human immunodeficiency virus (HIV) gp41 Env (57), the six-helix bundle is long and extends through the region adjacent to the membrane. In contrast, the six-helix bundles of influenza virus hemagglutinin (HA) (7) and human T-cell leukemia virus (HTLV) gp21 Env (33) are relatively short and distal from the membrane. At the membrane proximal C-terminal end of the six-helix bundles of HA and gp21 Env, a primarily nonhelical strand (termed a leash) packs against the helical N-terminal coiled-coil (46). The spacing of the heptad repeats also differs among class I fusion proteins. For example, while only a few residues separate the heptad repeats of HA, the paramyxovirus F heptad repeats are separated by over 250 residues. In addition, instead of fusion peptides, the avian sarcoma/leukosis virus (13), filovirus (30, 35), and arenavirus (29, 32) fusion proteins have internal fusion loops lacking a free N terminus positioned at the ends of their six-helix bundles. Class II fusogens differ from class I fusogens in that they have a higher content of β-sheets, they transition from hetero- or homodimers into trimers during refolding, and they lack six-helix bundles. In addition, class II fusogens contain internal fusion loops that lie within domains comprised of mainly β-sheets (23, 58). Class III fusogens share features of both class I and class II fusogens, including an α-helical coiled-coil core and trimeric arrangement like class I and internal fusion loops within a predominantly β-sheet domain like class II. gB is a class III fusogen (Fig. 1A).
Fig 1.
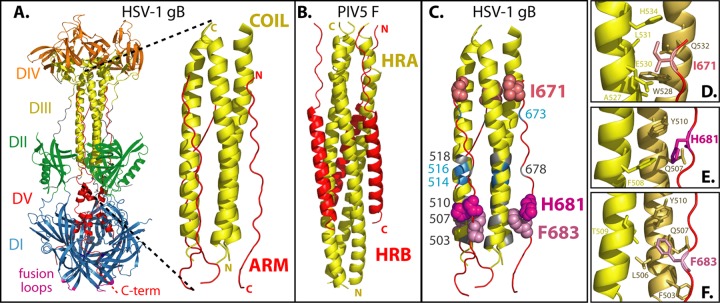
(A) HSV-1 gB ectodomain (PDB ID 2GUM). The structure of HSV-1 gB is shown and colored by domain. Domain I (blue) contains hydrophobic fusion loops (magenta) that insert into the cell membrane. Domain V (red) is comprised of a C-terminal arm that packs against a coiled-coil core formed by domain III (yellow) and proceeds through domain I toward the TM region at the C terminus of the ectodomain. This creates a hairpin-like organization, with the fusion loops and TM domain at the same end of the trimer. A close-up of the anti-parallel coil-arm complex is shown. (B) For comparison, the six-helix bundle from the paramyxovirus class I fusogen PIV5 F (PDB ID 1SVF) (5) is shown, comprised of three HRA (yellow) and HRB (red) peptides. (C) The gB coil-arm complex is colored to summarize the results. When the numbered residues were mutated to alanine, either gB was poorly surface expressed (gray), gB expression and fusion were at wild-type levels (blue), or gB was expressed at wild-type levels but fusion was reduced (shades of pink, space-filling residues). (D) gB arm residue I671 (red sticks) contacts residues A527, E530, L531, and H534 (yellow sticks) on one helix of the coil and residue W528 (brown sticks) on a second helix of the coil. (E) gB arm residue H681 contacts F508 (yellow sticks) on one helix of the coil and Q507 and Y510 (brown sticks) on a second helix of the coil. (F) gB arm residue F683 contacts residue T509 (yellow sticks) on one helix of the coil and residue F503, L506, Q507, and Y510 (brown sticks) on a second helix of the coil.
The crystal structures of herpes simplex virus 1 (HSV-1) (25) and Epstein-Barr virus (EBV) gB (3) most likely represent postfusion conformations, although this has not been proven. The vesicular stomatitis virus G protein (VSV-G) is a class III fusogen that is pH dependent, and its structure has been solved in two forms that are proposed to represent a prefusion (crystallized at basic pH) and postfusion (crystallized at acidic pH) conformation (50, 51). Despite a lack of sequence homology with VSV-G, the HSV and EBV gB structures resemble the postfusion structure of VSV-G. The gB structures were solved using a soluble form of gB lacking a TM domain. Removal of the gB TM domain may have caused the protein to spontaneously adopt a postfusion conformation, as has been observed for other fusion proteins including class III baculovirus gp64 (31) and class I paramyxovirus F protein (61).
Similar to class I fusogens, the gB postfusion structure contains a trimeric α-helical coiled-coil at its core (25) (Fig. 1A). A long C-terminal arm within the ectodomain packs against this coiled-coil core in an antiparallel manner, reminiscent of the class I six-helix bundle (29) (Fig. 1B). The gB arm is nonhelical, and thus it more closely resembles the leash present in influenza virus HA and HTLV gp21 Env (7, 33, 46). The gB coil-arm complex consists of residues 500 to 541 in the coil and residues 670 to 690 in the arm. This hairpin-like arrangement most likely brings the C-terminal TM region into proximity with the fusion loops at the base of gB, further supporting the concept that this structure represents a postfusion gB conformation.
gB exhibits some notable differences from class I fusogens. In class I fusogens, the HRA coiled-coil is adjacent to the fusion peptide. In contrast, the gB coiled-coil lies over 200 residues downstream from its fusion loops, such that the coil and fusion loops are separated by the entirety of domain I (Fig. 1A). Similarly, in most class I fusogens, the HRB or leash region is proximal to the TM region. For gB, the arm that packs against the coil lies nearly 80 residues upstream of the TM domain.
We hypothesize that gB and other class III fusogens refold in a manner similar to class I fusogens and that the packing of the arm against the coiled-coil provides a driving force for gB refolding from a prefusion to a postfusion conformation. To investigate the role of the coil and arm regions in fusion, we mutated residues that make contacts within the coil-arm complex and assessed the effect of these mutations on the ability of gB to execute fusion.
MATERIALS AND METHODS
Cells, virus, antibodies, and plasmid constructs.
Chinese hamster ovary (CHO-K1) cells lack HSV-1 receptors and were grown in Ham F-12 medium supplemented with 10% fetal bovine serum (FBS). CHO cells stably expressing the gD receptor HVEM (CHO-HVEM) (40) or nectin-1 (CHO-nectin-1) (21) were grown in Ham F-12 with 10% FBS and 250 μg of G418/ml. Vero and Vero-B24 cells (i.e., Vero cells stably expressing HSV-1 gB [27], also called VgB) were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS. Vero-B24 cells were used to propagate the gB-negative HSV-1 (KOS) strain K082 (8). Confluent Vero-B24 cells were infected with K082 at 0.01 PFU/cell. After 3 days, the supernatants were harvested and titered on Vero-B24 cells. The antibodies used included anti-gB polyclonal serum R74 (26), the anti-gB monoclonal antibodies (MAbs) I-1-7, I-84-5, I-144-2, I-252, II-105, and II-125-4 (38, 45), I-59-6 and II-137-1 (45), H1397 and H336 (gifts from L. Pereira) (47), and B1 (a gift from J. Glorioso) (28) and the anti-VP5 MAb H1.4 (Biodesign International, Sato, ME). pT7EMCLuc plasmid encoding a firefly luciferase reporter gene under the control of the T7 promoter and pCAGT7 plasmid encoding T7 RNA polymerase (44) were used in the fusion assay. gD, gH, and gL were expressed from pCAGGS constructs (48). gB mutants were created by site-directed mutagenesis of the HSV-1 gB gene in a pSG5 vector using QuikChange (Agilent, Santa Clara, CA). The wild-type gB used here contains a few conservative amino acid changes compared to the published sequence for HSV-1 strain KOS gB (GenBank no. K01760.1), including T33S, P58A, T313S, Q443L, and R515H. The GenBank sequence was used to construct the mutagenic primers, and consequently the Q514A, H516A, and N518A mutants contain an H515R amino acid change that should not affect gB function.
Quantitative cell-cell fusion assay.
Effector CHO-K1 cells (4 × 104 cells/well) were seeded in 96-well plates and target CHO-HVEM or CHO-nectin-1 cells (6 × 105 cells/well) were seeded in six-well plates overnight. Effector cells were transfected in triplicate with plasmid DNA (total, 110 ng/well; 30 ng of gB or empty vector and 20 ng of each of HSV-1 gD, gH, gL, and T7 RNA polymerase) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) (0.35 µl/well) in Opti-MEM (50 μl/well, total volume). Target cells were transfected with 2.2 μg of pT7EMCLuc/well using 7 μl of Lipofectamine 2000/well (1 ml/well, total volume). After 3 h of transfection at 37°C, the cells were rinsed, and target cells were released using EDTA. Target cells were overlaid onto the effector cells at a 1:1 ratio and coincubated at 37°C for 15 h. The cells were rinsed and lysed in 50 μl of passive lysis buffer (Promega, Madison, WI)/well. Luciferase activity was measured by adding 50 μl of luciferase assay reagent (Promega)/well and detecting light output using a luminometer (Perkin-Elmer).
Cell-based enzyme-linked immunosorbent assay (CELISA).
To determine the level of gB expression on the surface of the cells used in the fusion assay (12, 20), a duplicate 96-well plate of transfected effector cells was fixed in 3% paraformaldehyde. The cells were rinsed and incubated in triplicate with anti-gB R74 polyclonal antibody (PAb) at 1:5,000, followed by goat anti-rabbit IgG coupled to horseradish peroxidase (HRP) at 1:1,000. Expression was determined by adding BioFx TMB one-component HRP microwell substrate (SurModics, Eden Prarie, MN) and measuring the absorbance at 370 nm. To probe gB conformation by CELISA, CHO-K1 cells seeded overnight in six-well plates (106 cells/well) were transfected with DNA encoding gB or empty vector (2.2 µg/well) using 7 μl Lipofectamine 2000/well (1 ml/well, total volume). The cells were replated into 96-well plates the next day, followed by incubation at 37°C overnight. The wells were blocked for 30 min at room temperature with PBS-BSA (phosphate-buffered saline containing 0.5 mM Mg2+, 0.9 mM Ca2+ [PBS+], and 3% [wt/vol] bovine serum albumin). PAb R74 diluted 1:5,000 or MAbs diluted 1:1,000 in PBS-BSA were added to cells in triplicate for 1 h at room temperature. The cells were rinsed and fixed in PBS+ with 2% formaldehyde and 0.2% glutaraldehyde. Cells were incubated with biotin-conjugated goat anti-rabbit IgG or goat anti-mouse IgG (Sigma) at 1:500 dilution for 1 h, followed by streptavidin-HRP (GE Healthcare) at a 1:10,000 dilution for 1 h. Expression was measured using HRP substrate as described above.
Virus complementation assay.
Confluent Vero cells in six-well plates were transfected with plasmid DNA encoding gB mutants (2 μg/well) using Lipofectamine 2000 (7 μl/well) in Opti-MEM (1 ml, total volume). After overnight incubation, the cells were rinsed and infected with K082 (5 × 106 PFU/well) for 2 h. The cells were rinsed with an acid wash (40 mM sodium citrate [pH 3], 10 mM KCl, 135 mM NaCl) to inactivate extracellular virus, and DMEM supplemented with 10% FBS was added (1 ml/well). After an overnight incubation at 37°C, virus was harvested from both cells and medium by exposing the plates to three freeze-thaw cycles and removing the cellular debris by pelleting in a microcentrifuge. To titer the complemented virus stocks, supernatants were serially diluted and added to Vero-B24 monolayers in triplicate for 3 h. The cells were overlaid with DMEM containing 5% methylcellulose and 5% heat-inactivated fetal calf serum (FCS). After 3 days, the cells were fixed with methanol and stained with Giemsa stain, and the plaques were counted.
Western blotting.
Vero cells transfected as described above were either infected as described above or mock infected. The cells were subjected to three rounds of freeze-thawing, and both the cells and the supernatants were harvested. Cellular debris was removed by pelleting in a microcentrifuge, and 900 μl of the resulting supernatants was pelleted at 55,000 × g for 2 h in a Ti50 rotor. The supernatants were removed, and the pellets were resuspended in loading buffer (2% sodium dodecyl sulfate [SDS], 175 mM β-mercaptoethanol) and separated by electrophoresis on 10% gels (Bio-Rad, Hercules, CA). The proteins were transferred to nitrocellulose membranes and probed for gB using R74 diluted 1:10,000 and VP5 using MAb H1.4 diluted 1:4,000, followed by the addition of IRDye 800CW goat anti-rabbit IgG (Li-Cor, Lincoln, NE) and IRDye 680LT donkey anti-mouse antibody at 1:10,000. The bands were visualized using Odyssey imaging (Li-Cor).
RESULTS
We predict that if the interaction between the gB C-terminal arm and the coiled-coil core drives the refolding of gB, mutation of residues at these sites should inhibit gB refolding and block fusion. Linker-insertion mutagenesis within the arm region of HSV-1 or HSV-2 gB has been shown previously to abrogate gB function (37, 38). Candidate residues within HSV-1 gB that contribute to the coil-arm interaction were identified by examining the gB structure (25). Residues were selected based on the number of side-chain contacts each residue made with the opposing region (Tables 1 and 2). Selected residues included F503, Q507, Y510, Q514, H516, and N518 in the coil region and I671, L673, L678, H681, and F683 in the arm region (Fig. 1C). Using site-directed mutagenesis, we individually mutated each of these residues in HSV-1 gB (strain KOS) to alanine to assess the contribution of each to gB function.
Table 1.
Selected candidate coil-arm interaction residues from the gB coil
| N-terminal gB coil residue | No. of contactsa with arm residues | No. of side-chain contactsb with arm residues | H-bond contacts with arm residues (no. of side chains) | No. of arm residues contacted |
|---|---|---|---|---|
| F503 | 13 | 12 | 3 | |
| Q507 | 16 | 13 | 2 | 3 |
| Y510 | 17 | 17 | 1 | 4 |
| Q514 | 13 | 13 | 1 | 3 |
| H516 | 16 | 16 | 3 | |
| N518 | 16 | 16 | 1 | 2 |
A contact is defined to occur when one atom lies within 4 Å of another atom, as determined using MacPyMol.
Side-chain contacts are defined as contacts involving atoms past the Cα of the selected amino acid residue.
Table 2.
Selected candidate coil-arm interaction residues from the gB arm
| C-terminal gB arm residue | No. of contactsa with coil residues | No. of side-chain contactsb with coil residues | H-bond contacts with coil residues (no. of main chains) | No. of coil residues contacted |
|---|---|---|---|---|
| I671 | 24 | 14 | 1 | 6 |
| L673 | 7 | 7 | 3 | |
| L678 | 28 | 12 | 1 | 5 |
| H681 | 12 | 7 | 1 | 3 |
| F683 | 18 | 13 | 1 | 5 |
A contact is defined to occur when one atom lies within 4 Å of another atom, as determined using MacPyMol.
Side-chain contacts are defined as contacts involving atoms past the Cα of the selected amino acid residue.
Mutations in the coil-arm complex affect gB expression and function in fusion.
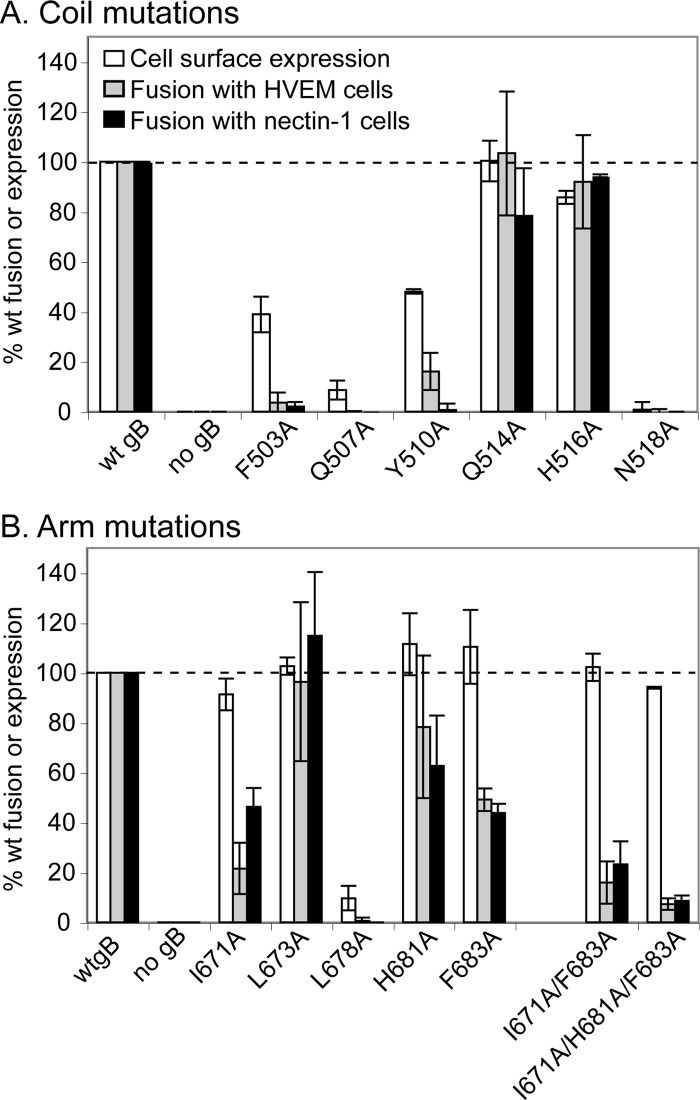
We used a quantitative cell-cell fusion assay to evaluate the function of the mutants. Effector CHO-K1 cells were transfected with constructs encoding wild-type or mutant gB and wild-type gD, gH, gL, and T7 RNA polymerase. CELISA was used to assess cell surface expression of the gB constructs. Most of the mutations within the coiled-coil core (F503A, Q507A, Y510A, and N518A) resulted in proteins with poor surface expression (Fig. 2A). This was not surprising since these residues are buried in the postfusion structure and may be critical for proper protein folding (25). Only one mutation in the arm region, L678A, impaired cell surface expression (Fig. 2B). Mutations in the remaining two coil residues (Q514A and H516A) and four arm residues (I671A, L673A, H681A, and F683A) resulted in wild-type levels of cell surface expression, suggesting the proteins were properly folded enough to be transported to the cell surface.
Fig 2.
Expression of and cell-cell fusion mediated by coil (A) and arm (B) gB mutants. CHO effector cells were transfected with plasmids encoding T7 RNA polymerase, gD, gH, gL, and a gB mutant. CHO-HVEM or CHO-nectin-1 target cells were transfected with a plasmid encoding luciferase under a T7 promoter. Cells were cocultured for 15 h, and the luciferase activity was recorded as a measure of cell-cell fusion. Surface expression was determined by CELISA using anti-gB PAb R74. Wild-type gB levels of expression and fusion were set to 100% (dashed line). The data from three independent experiments were averaged, and standard deviations are shown.
A luciferase reporter gene assay was used to assess cell-cell fusion (Fig. 2). The effector cells described above were overlaid with target CHO cells that express a single gD entry receptor (HVEM or nectin-1) and were transfected with a plasmid encoding luciferase under the control of the T7 promoter. Fusion of the effector and target cells results in luciferase expression. As expected, all of the constructs with poor surface expression (F503A, Q507A, Y510A, N518A, and L678A) yielded poor cell-cell fusion. The two coil mutants with wild-type expression levels, Q514A and H516A, and one of the arm mutants with a wild-type expression level, L673A, executed wild-type levels of fusion. The rest of the arm mutations (I671A, H681A, and F683A) resulted in wild-type expression levels but reduced the ability of gB to execute cell-cell fusion, using either HVEM- or nectin-1-expressing target cells (Fig. 2B). Mutants I671A, H681A, and F683A mediated fusion with HVEM-expressing cells at 22, 78, and 49% of wild-type levels, respectively, and fusion with nectin-1-expressing cells at 46, 63, and 44% of wild-type levels.
Thus, residues in the arm region located at both ends of the coil-arm complex contribute to gB function. I671 at the N terminus of the arm makes 24 contacts (14 side chain) with 6 coil residues on two helices (Fig. 1D and Table 2). At the C terminus of the arm, F683 makes 18 contacts (13 side chain) with 5 coil residues on two helices and H681 makes 12 contacts (7 side chain) with 3 coil residues on two helices (Fig. 1E and F). F683 and H681 contact one another and also share contact residues in the coil. Consistent with the functional importance of F683, this residue is conserved across herpesviruses, including HSV, EBV, and cytomegalovirus.
We hypothesized that combining the three arm mutations that diminish fusion would have an additive effect on fusion inhibition. As predicted, the combinations of I671A/F683A and I671A/H681A/F683A resulted in stepwise reductions in cell-cell fusion with both target cell types without compromising cell surface expression (Fig. 2B). Mutants I671A/F683 and I671A/H681A/F683A mediated fusion with HVEM-expressing cells at 16 and 7% of wild-type levels, respectively, and fusion with nectin-1-expressing cells at 23 and 9% of wild-type levels.
The diminished fusion caused by the gB arm mutations can be rescued by adding hyperfusogenic mutations to gB.
We have demonstrated that residues within the arm of gB are critical for gB function. In the postfusion structure, the intimate contacts of these arm residues with the coil may facilitate the transition of gB to its postfusion conformation. Mutation of these arm residues may destabilize the postfusion coil-arm complex and thus stabilize the gB prefusion conformation. In addition, these arm residues may form stabilizing contacts in the currently unknown prefusion structure.
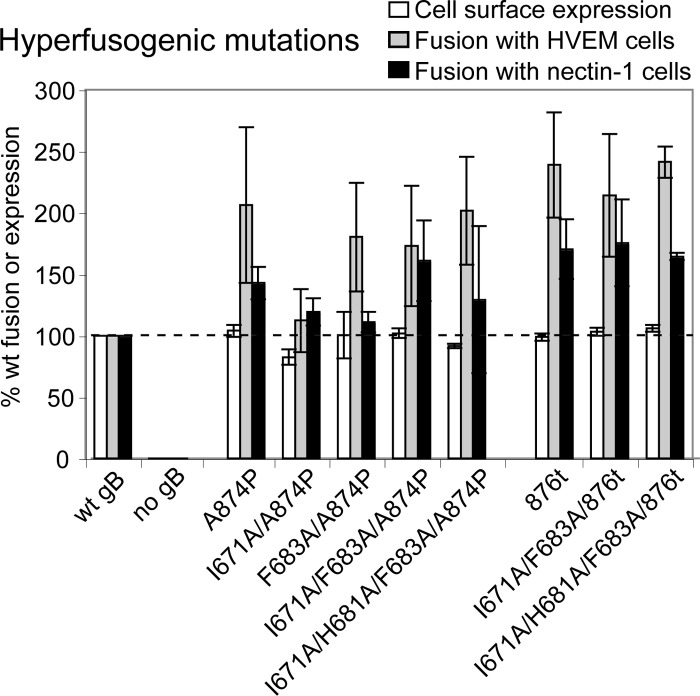
We hypothesized that the reduced fusion caused by the arm mutations could be rescued by the addition of hyperfusogenic mutations. The ability to rescue gB function with additional mutations would support the concept that the arm mutations do not inhibit fusion simply by causing gB to misfold. We added known hyperfusogenic mutations to the cytoplasmic tails of wild-type and mutant gB proteins to determine whether altering the energetics of the gB arm mutants would reverse their fusion deficits. We tested the mutants for cell surface expression and fusion using the luciferase reporter gene assay as described above. As expected, a point mutation (A874P) in the cytoplasmic tail of wild-type gB resulted in increased levels of fusion with both HVEM and nectin-1 expressing target cells without affecting cell surface expression (18). Truncation of the final 28 residues of the wild-type gB tail (876t) similarly increased cell-cell fusion without affecting cell surface expression (4, 18). When either of these hyperfusogenic mutations were added to the gB arm mutations, they independently rescued the ability of the gB arm mutants to mediate cell-cell fusion without altering cell surface expression (Fig. 3). Single mutations within the gB arm were rescued as well as the double- and triple-mutation combinations. These data are consistent with the concept that the arm mutations (I671A/H681A/F683A) inhibit cell-cell fusion by trapping gB in a prefusion conformation.
Fig 3.
Expression and fusion of gB mutants carrying hyperfusogenic mutations. Effector cells were transfected with plasmids carrying T7 RNA polymerase, gD, gH, gL, and a gB mutant. HVEM- or nectin-1-expressing target cells were transfected with a plasmid carrying luciferase under a T7 promoter. The cells were cocultured for 15 h, and the luciferase activity was recorded as a measure of cell-cell fusion. Surface expression was determined by CELISA using anti-gB PAb R74. Wild-type gB levels of expression and fusion were set to 100% (dashed line). The data from three independent experiments were averaged, and standard deviations are shown.
Mutations in the gB arm inhibit virus entry.
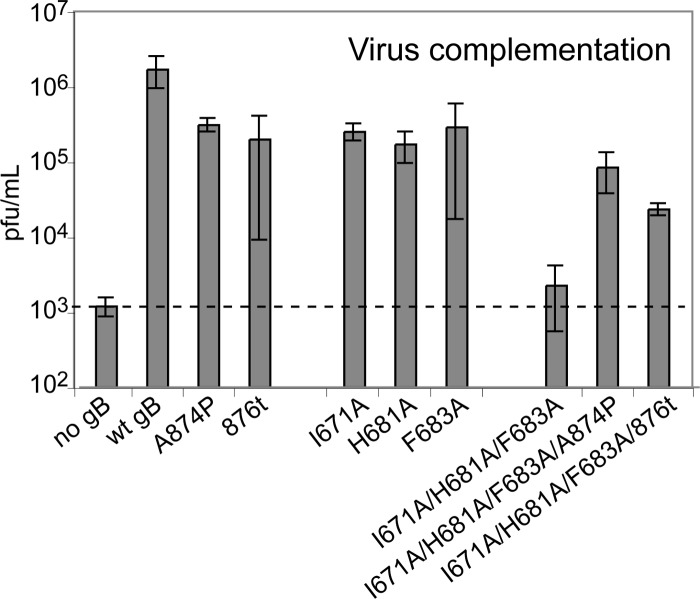
To determine whether the cell-cell fusion defects exhibited by the gB arm mutants are apparent also in the context of virus-cell fusion, gB-null virus was phenotypically complemented with the gB mutants, and the complemented viruses were titered on Vero cells stably expressing gB to allow for plaque formation. Individual gB mutations of I671A, H681A, or F683A resulted in an ∼1-log decrease in virus titer (Fig. 4). When the three gB arm mutations—I671A, H681A, and F683A–were combined, the complemented virus titer decreased by 3 logs and became indistinguishable from background. Thus, mutations in the gB arm region affect gB function during virus-cell as well as cell-cell fusion.
Fig 4.
Mutant gB function during virus entry (virus complementation). Vero cells were transfected overnight with gB mutant plasmids and then infected with gB-null virus that had been phenotypically complemented with wild-type gB. After 24 h, virus complemented with mutant gB proteins was harvested by freeze-thawing the cells. Infectious titers were determined by titration of the complemented stocks on a cell line that constitutively expresses wild-type gB. The data from three independent experiments were averaged, and the standard deviations are shown.
When the hyperfusogenic gB mutations A874P or 876t were added to wild-type gB, the complemented virus titers decreased by ∼1 log. This is consistent with a previously reported modest decrease in titer for virus complemented with the 876t gB mutant (4) or with some other (although not all) gB tail mutants that enhance cell-cell fusion (16, 52). When the hyperfusogenic gB mutations were added to the I671A/H681A/F683A gB triple-arm mutations, the complemented virus titers were enhanced by 1 or 2 logs. Thus, in both virus-cell and cell-cell fusion, the hyperfusogenic mutations were able to rescue the fusion deficiency imparted by the I671A/H681A/F683A gB mutations, a finding consistent with the notion that the gB triple-arm mutant is energetically trapped in a prefusion conformation.
We attempted to assess the incorporation of gB into virions by pelleting the complemented virus preparations by ultracentrifugation and analyzing the pelleted proteins by SDS-PAGE and Western blotting. Both gB and the HSV-1 capsid protein VP5 were present in all of the virus preparations; however, similar amounts of gB were found in samples derived from mock-infected cells transfected with gB (data not shown). The release of gB from cells in the absence of infection has been reported previously (37), and this complication prevents us from assessing the virion incorporation of our gB mutants. Although the consistency between the cell-cell and virus-cell fusion results suggests that the entry defect seen for the triple-arm mutant is due to a defect in fusion, we cannot rule out the possibility that insufficient virion incorporation of this mutant contributes to its lower virus titer.
gB mutants are properly folded and transported to the cell surface.
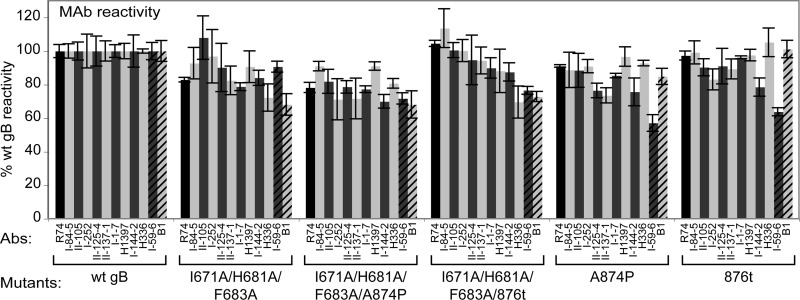
To examine the conformational state of the gB mutants, the binding of a panel of 11 MAbs to gB on the cell surface was assessed by CELISA (Fig. 5). None of the gB mutants tested demonstrated a consistent decrease in MAb binding, suggesting that none of the gB mutants have global alterations in gB folding, processing, or transport to the surface. Excluding the MAbs I-59-6 and B1 (discussed below), all of the MAbs bound every gB mutant with >70% reactivity compared to wild-type gB.
Fig 5.
MAb reactivity with gB mutants. CHO cells were transfected with plasmids encoding the gB mutants listed and stained for surface expression of gB using PAb R74 (black bar) or one of 11 MAbs. For each antibody, data were normalized by setting reactivity with wild-type gB at 100%. MAbs I-59-6 and B1 (stripes) are highlighted.
Interestingly, the differential reactivity of the I-59-6 and B1 MAbs with the gB mutants may reveal modest conformational differences among the mutants. The gB triple-arm I671A/H681A/F683A mutant shows high reactivity with I-59-6 (91% of wild-type gB levels) and lower reactivity with B1 (68%). Conversely, the hyperfusogenic gB mutants A874P and 876t both show low reactivity with I-59-6 (57 and 64%) and high reactivity with B1 (85 and 101%). Adding the triple-arm gB mutations to the hyperfusogenic gB mutations causes the I-59-6 reactivity to increase (72 and 77%) and the B1 reactivity to decrease (68 and 73%).
DISCUSSION
gB is a class III fusogen, and the mechanism by which it refolds from a prefusion to postfusion form to mediate fusion is not yet understood. Using site-directed mutagenesis, we have demonstrated that the coil-arm complex of gB is important for its function. Several mutations to the coil disrupted gB expression, suggesting that this region is critical for proper folding of the protein. Previous studies have shown that gB is sensitive to mutagenesis and mutations frequently result in gB misfolding or aberrant processing (37, 38, 43). In contrast, three mutations to the arm region—I671A, H681A, and F683A—resulted in wild-type levels of surface expression and MAb reactivity but decreased ability to mediate both cell-cell and virus-cell fusion. We conclude that these three arm residues are critical for gB fusion execution.
The gB coil-arm complex resembles the six-helix bundle present in the postfusion structures of class I fusion proteins (23, 29, 58). The coil-arm complex and six-helix bundles are both trimeric and their C-terminal regions (arm or HRB) pack on the outside and antiparallel to their N-terminal regions (coil or HRA) (Fig. 1A and B). The gB coil-arm complex most closely resembles the six-helix bundle regions of the influenza virus HA and HTLV gp21 Env because the C-terminal portions that pack against those coils are primarily nonhelical leashes (7, 33). Similar to our observations for the gB arm, mutations of residues within the HA leash inhibit fusion (46). The class I six-helix bundle is energetically stable, and its formation during the transition from a prefusion to postfusion conformation has been suggested to provide the energy to execute membrane fusion (5, 34, 39). Our results suggest that the coil-arm complex of gB may play the same role as the six-helix bundle during herpesvirus entry, despite the fact that the coil-arm complex is positioned relatively distantly from the membrane-embedded fusion loops and transmembrane domain of the protein (Fig. 1A). We predict that the alanine substitutions in the arm weaken its interaction with the coil, and thus the energy released when the arm-coil complex forms is reduced. In addition, lowering the affinity of the arm for the coil may reduce the frequency at which gB achieves its postfusion conformation.
Consistent with this model for how the mutations in the arm inhibit fusion, the addition of known hyperfusogenic mutations to the gB triple-arm mutant rescued its fusion function in both cell-cell and virus-cell fusion. This suggests that the arm mutations did not cause global disruption in gB folding, processing, or transport. The hyperfusogenic mutations in the gB cytoplasmic tail may decrease the activation energy required to convert gB to its postfusion conformation and thus allow gB to execute fusion despite the presence of the triple arm mutations. Although the mechanism by which gB cytoplasmic tail mutations enhance fusion is unknown, a previous study hypothesized that the gB tail negatively regulates fusion by interacting with the membrane (10). Residues 869 to 882 were predicted to form a helix that stably interacts with membranes. A truncation at residue 876 or a proline substitution at residue 874 may enhance fusion by disrupting this helix and its membrane association. The cytoplasmic regions of fusion proteins from other viruses have been shown to similarly negatively regulate fusion, including those of HIV (60) and paramyxovirus (56). For the paramyxovirus fusion protein, the suppression of fusion mediated by a long cytoplasmic tail can be overcome by adding a hyperfusogenic mutation to the ectodomain (56); thus, as we have shown for our gB mutants, mutations in the tail and ectodomain can compensate for one another.
Since the hyperfusogenic gB tail mutations modestly decrease the complemented virus titers (Fig. 4), it may initially seem counterintuitive that adding these tail mutations to the triple-arm gB mutant enhances the virus titer. We believe the explanation comes down to energetics. The hyperfusogenic mutations may destabilize the triple-arm mutant enough to rescue its function and restore virus entry. The virus carrying the hyperfusogenic gB mutants may exhibit a decreased titer because a portion of the gB is so destabilized that it adopts a postfusion conformation prematurely and is thus inactive for fusion. A caveat to these experiments is that we were unable to assess virion incorporation for the various gB mutants.
Peptides corresponding to the HRA and HRB sequences of several class I fusogens inhibit fusion by blocking six-helix bundle formation (39, 53), and peptides corresponding to the C-terminal leash of HA and gp21 Env also inhibit fusion (36, 49). We originally focused our mutations on the C-terminal end of the arm because when a library of peptides spanning the gB ectodomain was tested for inhibition, peptides containing residues from this end of the arm were reported to inhibit entry better than peptides derived from the N-terminal end of the arm (1). In congruence, peptides derived from the N-terminal end of the coil (the region that contacts the C terminus of the arm) also were reported to inhibit entry better than peptides corresponding to the C terminus of the coil (1, 19). As we predicted, we found two residues, H681 and F683, at the C terminus of the arm that contribute to gB function. We were somewhat surprised to find that residue I671 near the N terminus of the arm is also critical for gB function. Thus, residues at both ends of the coil-arm complex are important. I671 contacts five coil residues (Fig. 1D and Table 2), and we predict that some of these coil residues contribute to gB function, most notably W528 and Q532, which both make 11 side-chain contacts with two arm residues.
Results from a previous random linker-insertion mutagenesis study of HSV-1 gB support the concept that the arm is important for gB function (38). Insertions of five amino acids after residues 671, 673, or 690 in the arm inhibited fusion; however, these insertions likely had a more drastic effect on gB folding than the point mutations described here, because surface expression of the insertion mutants was decreased by 53, 43, and 85%, respectively, and MAb reactivity and processing for the 673 insertion mutant was reduced.
HSV gB likely adopts multiple conformations within infected cells. Although HSV entry into some cells occurs at the plasma membrane, entry into other cell types occurs via endocytosis (41, 42), and low pH has been shown to alter the gB conformation in a reversible manner (9, 14, 15, 55). In addition, soluble forms of gB produced from different expression systems exhibit different properties (15). The prefusion structure of gB is currently unknown; however, our results predict that the coil-arm complex would not be formed in the gB prefusion conformation. A model of prefusion gB based on the prefusion VSV-G structure was created previously (3). Although the arm region is C terminal to this model and not shown, the N terminus of the coil is predicted to be nonhelical in the prefusion state, which is consistent with the absence of a coil-arm complex.
We propose the arm mutations that block fusion may energetically trap gB in a prefusion state. Purification of a soluble form of gB carrying these arm mutations may serve as a starting point for crystallization of gB in its prefusion form. Previously, random mutagenesis of gB identified insertion mutants that were deficient for fusion (38); however, these mutants adopted a postfusion conformation when expressed as soluble proteins (54), leading to the conclusion that the insertion mutations interfered with gB function in a manner other than by preventing conversion to postfusion. Point mutations introduced into the arm region may be less disruptive to the overall folding of gB than the previously examined insertion mutations and thus may provide a good possibility for preserving the prefusion gB structure.
The triple-arm mutant retained wild-type reactivity with multiple MAbs, regardless of whether the hyperfusogenic tail mutations were added. A comparison of the prefusion and postfusion structures of VSV-G reveals that intact domains move with respect to one another, meaning that many of the antigenic structures may remain unchanged after the conversion. gB may undergo a similar conformational change with some of its domains shifting position but remaining relatively unchanged individually. This would make the prefusion and postfusion forms indistinguishable by most conformational MAbs. Indeed, a previous study demonstrated that a large panel of anti-gB MAbs can react with both purified soluble gB (postfusion) and cell surface-expressed gB, which should be at least partially prefusion (6).
Interestingly, the binding of MAbs I-59-6 and B1 was moderately affected by the mutations. I-59-6 bound better to the arm mutant, possibly indicating that it recognizes a prefusion conformation, and B1 bound better to the hyperfusogenic mutants, possibly indicating that it recognizes a postfusion conformation. The specific epitopes recognized by these MAbs need further characterization. gB conformations may exist in a mixed population on the cell surface and mutations to the arm that inhibit fusion (or mutations to the tail that enhance fusion) may alter the equilibrium between the prefusion and postfusion conformations.
Understanding the contribution of specific coil-arm residues to fusion execution should aid in the design of small peptide inhibitors of herpesvirus fusion, analogous to the peptide inhibitors that correspond to the HRA and HRB or leash segments of multiple class I fusogens and efficiently block fusion. The present study suggests that effective gB inhibitors should include residues at both ends of the coil-arm complex. Effective peptide inhibitors will be useful both for studying gB execution of fusion by trapping refolding intermediates and for potential clinical applications.
ACKNOWLEDGMENTS
We thank Nanette Susmarski for providing cell culture and Ted Jardetzky, Cindy Rowe, and Qing Fan for critical reading of the manuscript. We also thank all of the members of the Longnecker laboratory for their help and support. We thank Lenore Pereira, Joseph Glorioso, and Gregory Smith for antibodies.
This study was supported by NIH grants CA021776 and AI067048 to R.L.
Footnotes
Published ahead of print 4 April 2012
REFERENCES
- 1. Akkarawongsa R, Pocaro NE, Case G, Kolb AW, Brandt CR. 2009. Multiple peptides homologous to herpes simplex virus type 1 glycoprotein B inhibit viral infection. Antimicrob. Agents Chemother. 53:987–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Backovic M, Jardetzky TS, Longnecker R. 2007. Hydrophobic residues that form putative fusion loops of Epstein-Barr virus glycoprotein B are critical for fusion activity. J. Virol. 81:9596–9600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Backovic M, Longnecker R, Jardetzky TS. 2009. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 106:2880–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baghian A, Huang L, Newman S, Jayachandra S, Kousoulas KG. 1993. Truncation of the carboxy-terminal 28 amino acids of glycoprotein B specified by herpes simplex virus type 1 mutant amb1511-7 causes extensive cell fusion. J. Virol. 67:2396–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baker KA, Dutch RE, Lamb RA, Jardetzky TS. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309–319 [DOI] [PubMed] [Google Scholar]
- 6. Bender FC, et al. 2007. Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J. Virol. 81:3827–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bullough PA, Hughson FM, Skehel JJ, Wiley DC. 1994. Structure of influenza hemagglutinin at the pH of membrane fusion. Nature 371:37–43 [DOI] [PubMed] [Google Scholar]
- 8. Cai WZ, Person S, Warner SC, Zhou JH, DeLuca NA. 1987. Linker-insertion nonsense and restriction-site deletion mutations of the gB glycoprotein gene of herpes simplex virus type 1. J. Virol. 61:714–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cairns TM, et al. 2011. Capturing the herpes simplex virus core fusion complex (gB-gH/gL) in an acidic environment. J. Virol. 85:6175–6184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chowdary TK, Heldwein EE. 2010. Syncytial phenotype of C-terminally truncated herpes simplex virus type 1 gB is associated with diminished membrane interactions. J. Virol. 84:4923–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. 2011. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat. Rev. Microbiol. 9:369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Connolly SA, et al. 2003. Structure-based mutagenesis of herpes simplex virus glycoprotein D defines three critical regions at the gD-HveA/HVEM binding interface. J. Virol. 77:8127–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delos SE, White JM. 2000. Critical role for the cysteines flanking the internal fusion peptide of avian sarcoma/leukosis virus envelope glycoprotein. J. Virol 74:9738–9741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dollery SJ, Delboy MG, Nicola AV. 2010. Low pH-induced conformational change in herpes simplex virus glycoprotein B. J. Virol. 84:3759–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dollery SJ, Wright CC, Johnson DC, Nicola AV. 2011. Low-pH-dependent changes in the conformational and oligomeric state of the prefusion form of herpes simplex virus glycoprotein B are separable from fusion activity. J. Virol. 85:9964–9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fan Z, et al. 2002. Truncation of herpes simplex virus type 2 glycoprotein B increases its cell surface expression and activity in cell-cell fusion, but these properties are unrelated. J. Virol. 76:9271–9283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reference deleted.
- 18. Foster TP, Melancon JM, Kousoulas KG. 2001. An alpha-helical domain within the carboxyl terminus of herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) is associated with cell fusion and resistance to heparin inhibition of cell fusion. Virology 287:18–29 [DOI] [PubMed] [Google Scholar]
- 19. Galdiero S, et al. 2008. The identification and characterization of fusogenic domains in herpesvirus glycoprotein B molecules. Chem. Biochem. 9:758–767 [DOI] [PubMed] [Google Scholar]
- 20. Geraghty RJ, Jogger CR, Spear PG. 2000. Cellular expression of alphaherpesvirus gD interferes with entry of homologous and heterologous alphaherpesviruses by blocking access to a shared gD receptor. Virology 268:147–158 [DOI] [PubMed] [Google Scholar]
- 21. Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618–1620 [DOI] [PubMed] [Google Scholar]
- 22. Hannah BP, et al. 2009. Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops. J. Virol. 83:6825–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harrison SC. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heldwein EE, Krummenacher C. 2008. Entry of herpesviruses into mammalian cells. Cell. Mol. Life Sci. 65:1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heldwein EE, et al. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217–220 [DOI] [PubMed] [Google Scholar]
- 26. Herold BC, Visalli RJ, Susmarski N, Brandt CR, Spear PG. 1994. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulfate and glycoprotein B. J. Gen. Virol. 75:1211–1222 [DOI] [PubMed] [Google Scholar]
- 27. Herold BC, WuDunn D, Soltys N, Spear PG. 1991. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J. Virol. 65:1090–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Highlander SL, Cai WH, Person S, Levine M, Glorioso JC. 1988. Monoclonal antibodies define a domain on herpes simplex virus glycoprotein B involved in virus penetration. J. Virol. 62:1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Igonet S, et al. 2011. X-ray structure of the arenavirus glycoprotein GP2 in its postfusion hairpin conformation. Proc. Natl. Acad. Sci. U. S. A. 108:19967–19972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ito H, Watanabe S, Sanchez A, Whitt MA, Kawaoka Y. 1999. Mutational analysis of the putative fusion domain of Ebola virus glycoprotein. J. Virol. 73:8907–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kadlec J, Loureiro S, Abrescia NG, Stuart DI, Jones IM. 2008. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 15:1024–1030 [DOI] [PubMed] [Google Scholar]
- 32. Klewitz C, Klenk HD, ter Meulen J. 2007. Amino acids from both N-terminal hydrophobic regions of the Lassa virus envelope glycoprotein GP-1 are critical for pH-dependent membrane fusion and infectivity. J. Gen. Virol. 88:2320–2328 [DOI] [PubMed] [Google Scholar]
- 33. Kobe B, Center RJ, Kemp BE, Poumbourios P. 1999. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc. Natl. Acad. Sci. U. S. A. 96:4319–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lamb RA, Jardetzky TS. 2007. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 17:427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee JE, et al. 2008. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454:177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee KK, et al. 2011. Capturing a fusion intermediate of influenza hemagglutinin with a cholesterol-conjugating peptide, a new antiviral strategy for influenza virus. J. Biol. Chem. 286:42141–42149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li W, Minova-Foster TJ, Norton DD, Muggeridge MI. 2006. Identification of functional domains in herpes simplex virus 2 glycoprotein B. J. Virol. 80:3792–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin E, Spear PG. 2007. Random linker-insertion mutagenesis to identify functional domains of herpes simplex virus type 1 glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 104:13140–13145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Melikyan GB, et al. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Montgomery RI, Warner MS, Lum BJ, Spear PG. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427–436 [DOI] [PubMed] [Google Scholar]
- 41. Nicola AV, Hou J, Major EO, Stras SE. 2005. Herpes simplex virus type 1 enters human epidermal keratinocytes, but not neurons, via a pH-dependent endocytic pathway. J. Virol. 79:7609–7616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nicola AV, McEvoy AM, Straus SE. 2003. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J. Virol. 77:5324–5332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Norton DD, Dwyer DS, Muggeridge MI. 1998. Use of a neural network secondary structure prediction to define targets to mutagenesis of herpes simplex virus glycoprotein B. Virus Res. 55:37–48 [DOI] [PubMed] [Google Scholar]
- 44. Okuma K, Nakamura M, Nakano S, Niho Y, Matsuura Y. 1999. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology 254:235–244 [DOI] [PubMed] [Google Scholar]
- 45. Para MF, Parish ML, Nobel AG, Spear PG. 1985. Potent neutralizing activity associated with anti-glycoprotein D specificity among monoclonal antibodies selected for binding to herpes simplex virions. J. Virol. 55:483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Park HE, Gruenke JA, White JM. 2003. Leash in the groove mechanism of membrane fusion. Nat. Struct. Biol. 10:1048–1053 [DOI] [PubMed] [Google Scholar]
- 47. Pereira L, Ali M, Kousoulas K, Huo B, Banks T. 1989. Domain structure of herpes simplex virus 1 glycoprotein B: neutralizing epitopes map in regions of continuous and discontinuous residues. Virology 172:11–24 [DOI] [PubMed] [Google Scholar]
- 48. Pertel PE, Fridberg A, Parish ML, Spear PG. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313–324 [DOI] [PubMed] [Google Scholar]
- 49. Pinon JD, Kelly SM, Price NC, Flanagan JU, Brightly DW. 2003. An antiviral peptide targets a coiled-coil domain of human T-cell leukemia virus envelope protein. J. Virol. 77:3281–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roche S, Bressanelli S, Rey FA, Gaudin Y. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313:187–191 [DOI] [PubMed] [Google Scholar]
- 51. Roche S, Rey FA, Gaudin Y, Bressanelli S. 2007. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 315:843–848 [DOI] [PubMed] [Google Scholar]
- 52. Ruel N, Zago A, Spear PG. 2006. Alanine substitution of conserved residues in the cytoplasmic tail of herpes simplex virus gB can enhance or abolish cell fusion activity and viral entry. Virology 346:229–237 [DOI] [PubMed] [Google Scholar]
- 53. Russell CJ, Jardetzky TS, Lamb RA. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Silverman JL, Sharma S, Cairns TM, Heldwein EE. 2010. Fusion-deficient insertion mutants of herpes simplex virus 1 glycoprotein B adopt the trimeric postfusion conformation. J. Virol. 84:2001–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stampfer SD, Lou H, Cohen GC, Eisenberg RJ, Heldwein EE. 2010. Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus 1. J. Virol. 84:12924–12933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Waning DL, Russell CJ, Jardetzky TS, Lamb RA. 2004. Activation of a paramyxovirus fusion protein is modulated by inside-out signaling from the cytoplasmic tail. Proc. Natl. Acad. Sci. U. S. A. 101:9217–9222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430 [DOI] [PubMed] [Google Scholar]
- 58. White JM, Delos SE, Brecher M, Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Reference deleted.
- 60. Wyss S, et al. 2005. Regulation of human immunodeficiency virus type 1 envelope glycoprotein fusion by a membrane-interactive domain on the gp41 cytoplasmic tail. J. Virol. 79:12231–12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. U. S. A. 102:9288–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]