Abstract
Variants near the HLA-DP gene show the strongest genome-wide association with chronic hepatitis B virus (HBV) infection and HBV recovery/persistence in Asians. To test the effect of the HLA-DP region on outcomes to HBV infection, we sequenced the polymorphic HLA-DPB1 and DPA1 coding exons and the corresponding 3′ untranslated regions (3′UTRs) in 662 individuals of European-American and African-American ancestry. The genome-wide association study (GWAS) variant (rs9277535; 550A/G) in the 3′UTR of the HLA-DPB1 gene that associated most significantly with chronic hepatitis B and outcomes to HBV infection in Asians had a marginal effect on HBV recovery in our European- and African-American samples (odds ratio [OR] = 0.39, P = 0.01, combined ethnic groups). However, we identified a novel variant in the HLA-DPB1 3′UTR region, 496A/G (rs9277534), which associated very significantly with HBV recovery in both European and African-American populations (OR = 0.37, P = 0.0001, combined ethnic groups). The 496A/G variant distinguishes the most protective HLA-DPB1 allele (DPB1*04:01) from the most susceptible (DPB1*01:01), whereas 550A/G does not. 496A/G has a stronger effect than any individual HLA-DPB1 or DPA1 allele and any other HLA alleles that showed an association with HBV recovery in our European-American cohort. The 496GG genotype, which confers recessive susceptibility to HBV persistence, also associates in a recessive manner with significantly higher levels of HLA-DP surface protein and transcript level expression in healthy donors, suggesting that differences in expression of HLA-DP may increase the risk of persistent HBV infection.
INTRODUCTION
Hepatitis B virus (HBV) infection is a major global health concern that can result in liver cirrhosis and hepatocellular carcinoma. Upon HBV infection in adulthood, up to 95% of people have a robust immune response that leads to recovery from the infection and development of protective antibodies, but in 5 to 10% of the population, viral persistence occurs, leading to chronic infection. About 2 billion people worldwide have been infected with the virus and about 350 million live with chronic infection (11). Factors that determine viral recovery or persistence include age at infection, immunodeficiency states, gender, and host genetic variation (7, 29). Several reports have pointed to variation in the human leukocyte antigen (HLA) class I and II genes located within the major histocompatibility complex (MHC) as determinants of outcome after HBV infection and HBV disease pathogenesis in several global populations (23, 29). HLA genes encode molecules that are central to the host immune response, and variation in these genes likely affects outcome after infection with any given pathogen. Recently the importance of HLA genes was further highlighted by the first genome-wide association study (GWAS) in Asian patients with chronic hepatitis B, where HLA-DP gene variation showed the strongest association with chronic HBV infection (8), which was validated subsequently in another Asian study (4). This variant also associated with HBV vaccination response in Asians (18). HLA-DP is an HLA class II heterodimeric molecule consisting of alpha and beta chains that are encoded by the HLA-DPA1 and -DPB1 genes, respectively. The HLA-DP molecule is expressed on antigen-presenting cells (APC), including B cells, dendritic cells, and macrophages, where it functions by presenting extracellular antigens to CD4+ T cells, initiating cellular and humoral immune responses.
In general, HLA-DP genes have been somewhat ignored in terms of their impact on human disease relative to HLA-DR and -DQ, in part because HLA-DPA1 and -DPB1 are less polymorphic and also because HLA-DP cell surface expression levels are likely to be lower than that of HLA-DR or -DQ (2). Indeed, HLA-DPA1 and -DPB1 had never been investigated for potential effects on HBV disease or outcomes prior to the GWAS findings in Asians. Here we describe the effects of HLA-DP loci on outcomes of HBV infection among individuals of European and African descent. We identified a novel 3′UTR variant termed 496A/G (rs9277534) that has a stronger association with HBV recovery/persistence than the single nucleotide polymorphisms (SNPs) identified previously in the Asian studies and than any of the individual HLA-DP alleles. The 496A/G variant correlates with levels of both HLA-DP surface protein and DPB1 transcript levels in peripheral blood cells from healthy donors, suggesting that HLA-DP expression levels may represent the functional basis for the association of this locus with outcome to HBV infection.
MATERIALS AND METHODS
Study participants.
A total of 662 subjects were included, consisting primarily of European-American and African-American participants from one of the following cohort studies: (i) Multicenter AIDS Cohort Study (MACS) (17), (ii) Multicenter Hemophilia Cohort Study (MHCS) (3), (iii) Hemophilia Growth and Development Study (HGDS) (5), and (iv) AIDS Linked to Intravenous Experience (ALIVE) (31). Details of the subjects used in this study have been described previously (26, 27).
A nested case-control design was used in which all individuals with persistent HBV infection (n = 241) were matched to one or two persons from the same cohort who recovered from HBV infection (n = 421) but were otherwise similar with regard to nongenetic factors. Matching criteria included geographic location and factors that have been associated with HBV recovery, including age within 10 years, gender, and HIV-1 status. Subjects were considered persistently infected with HBV if their serum or plasma tested positive for hepatitis B surface antigen (HBsAg) at two visits separated by a minimum of 6 months. Testing for antibodies against hepatitis B core antigen (anti-HBc) and against HBsAg (anti-HBs) was performed as needed to exclude primary HBV infection. HBV status of HIV-positive subjects was determined before antiretroviral therapy was available. The majority of subjects were European-American (n = 505; 185 with persistent infection and 320 who cleared the virus) and a smaller subset was African-American (n = 131; 46 with persistent infection and 85 who recovered). This study was approved by the institutional review boards at all participating institutions.
Peripheral blood from 75 healthy donors of European-American ancestry was obtained. This study was approved by the protocol review office of the U.S. National Cancer Institute institutional review board. A cohort of 112 healthy African-American donors was recruited at the Duke Human Vaccine Institute as part of the CHAVI 008A study. Written informed consent was obtained from all subjects at all study sites.
HLA-DP genotyping/sequencing.
HLA-DPA1 and -DPB1 alleles were characterized using sequence-based typing protocols recommended by the 13th International Histocompatibility Workshop (21, 32). Exons 2 of HLA-DPA1 and -DPB1 were selectively amplified with locus-specific primers. The amplicons were sequenced using the ABI BigDye Terminator cycle sequencing kit version 1.1 and ABI3730xl DNA analyzer (Applied Biosystems). HLA-DPA1 and -DPB1 alleles were assigned on the basis of known alleles using the ASSIGN software (Conexio Genomics).
The 3′UTR region of the HLA-DPB1 was amplified using HLA-DPB1-specific primers (forward, 5′-TTCAACGAGGATCTGCATAA-3′; reverse, 5′-GGTTAAAGTGTATTAGATTA-3′). This amplified product was then sequenced using the same primers on an ABI BigDye Terminator cycle sequencing kit version 1.1 and ABI3730xl DNA analyzer (Applied Biosystems). Data for 33 SNPs in the HLA-DPB1 3′UTR region was available for 638 patients.
Statistical analysis.
SAS 9.1 (SAS Institute) was used for data management and statistical analyses. PROC FREQ was used to compute frequencies on individual variables. Statistical significance refers to two-sided P values of <0.05. Odds ratios (OR) for persistent infection and P values were calculated by using conditional logistic regression with PROC LOGISTIC. A conditional logistic regression analysis with stepwise selection was performed in 662 patients, which included all HLA-DPB1 alleles with a frequency of greater than 5%, 496A/G, 550A/G, and other class I and II loci that were reported previously to associate with HBV recovery in our study cohort (26, 27). These variables were tested in a model using PROC LOGISTIC to determine which had independent effects. The significance level for selecting variables in the model was P values of <0.05.
Measurement of HLA-DP surface protein expression.
Specificities of purified murine monoclonal antibodies (MAbs) B7/21 (AbCam), BrafB6 (Santa Cruz), Tu39, and SK10 (both Becton Dickinson) for HLA class II allotypes were tested by screening a panel of 86 HLA class II allotypes (One Lambda) using a Luminex 100 cytometer as previously described (1). For each antibody, binding was compared to purified IgG1 or IgG2a isotype controls (Sigma). HLA staining of 112 African-American donors was performed by isolation of peripheral blood lymphocytes (PBL) using Lymphoprep (Lonza) followed by incubation with human IgG (Lampire Biological) to block FcγR before staining with the unconjugated murine MAbs. Purified sheep IgG (Lampire Biological) was then added to block the nonspecific binding from the subsequent antibody, which was a sheep polyclonal phycoerythrin (PE)-conjugated anti-mouse antibody (Sigma). Free secondary-antibody-binding sites were blocked with murine IgG (Lampire Biological) before B cells were identified by staining with fluorescein isothiocyanate (FITC)-conjugated anti-CD19 antibody compared to an isotype control (both Beckman Coulter). All African-American samples were acquired using identical settings on a single FACSCalibur cytometer and analyzed using Cellquest (both Becton Dickinson). HLA staining of 75 European-American donors was performed using 0.5 ml of freshly drawn blood directly with the same antibody incubations as described above for African-Americans. Red blood cells were then removed by incubation for 30 min at 4°C in an isotonic lysis buffer (150 mM NH4Cl, 10 mM NaHCO3, 1 mM Na2EDTA, pH 7.4). Staining European-American whole blood gave greater discrimination of HLA class II binding from control MAb binding than using purified PBL for the African-American staining. Both isotype control and HLA class II staining could not be acquired using the same instrument settings for all European-Americans, as the fluorescence range exceeded that of the cytometer for some donors. We confirmed that HLA class II MAbs were staining CD19+ cells compared to isotype controls (see Fig. S1 in the supplemental material). We then used one set of cytometer settings for acquiring HLA class II MAbs and a second cytometer setting with higher PE amplification for acquiring the isotype control in all 75 European-American donors. European-American samples were acquired using a single FACScan cytometer and analyzed using CellQuest. The European- and African-American populations were acquired within separate periods of less than 15 consecutive days and showed no trends with time (data not shown). Pellets of 5 × 106 PBL or 0.5 ml of whole blood mixed with 1 ml of DNAzol (Molecular Research Center) were stored frozen from each donor for genotyping, with DNA prepared using DNeasy columns (Qiagen).
Measurement of HLA-DPB1 mRNA transcript levels.
RNA was prepared from cell suspensions of freshly isolated PBL (1 × 107 cells) using a combination of TRIzol reagent (Invitrogen) and the RNeasy microkit (Qiagen). All 15 RNA samples from European-American donors were treated with RNase-free DNase I (Qiagen) prior to cDNA synthesis. Quality and quantity of RNA was checked on an HT RNA LabChip (Caliper, Life Sciences). Each sample had two distinct peaks representing 18S and 28S rRNAs with, on average, a ratio greater than 2.0 and an RNA quality score of >8.5. Reverse transcription was performed using 1 μg of total RNA, oligo(dT), and SuperScript III reverse transcriptase (RT) (Invitrogen) in a volume of 20 ul. The gene of interest (GOI), HLA-DPB1, was then quantified using SYBR green quantitative PCR (qPCR) using the threshold cycle (CT) method (12) in an ABI 7900HT PCR machine (Applied Biosystems). Each PCR included 5 μl of power SYBR green PCR master mix (Applied Biosystems), 200 nM primers (see Table S1 in the supplemental material), and 2 μl of cDNA (5× dilution) in a total volume of 10 μl using the recommended ABI qPCR protocol. The specificity of the reaction was confirmed by melt curve analysis using the dissociation step following the qPCR protocol. Sequencing and melt curve analysis of HLA-DPB1 PCR amplicons established that the primers (see Table S1) were specific for the cDNA prepared (data not shown) and did not cross-react with other genes. All reactions were standardized to the expression of two different reference genes, RPLP0 and GAPDH (see Table S1). The amplification efficiencies of the different genes were tested by making standard curves using serial dilutions of cDNA from a sample that was also designated a calibrator. Additionally, quantitative PCR using a relative quantification method (16) was confirmed by using two primer pairs that targeted different regions of the HLA-DPB1 transcript. The quantity of HLA-DPB1 in the 15 samples and calibrator were normalized to the quantity of three different reference genes (the ACTB, RPLP0, and GAPDH genes).
RESULTS
HLA-DPB1 alleles associate with HBV recovery.
Given that variants near HLA-DP showed association with chronic HBV infection and outcome after infection in Asian cohorts (4, 8), we genotyped the HLA-DPA1 and HLA-DPB1 genes in 662 individuals, primarily of European and African ancestry, to determine potential associations with HBV recovery/persistence. While no HLA-DPA1 alleles associated significantly (P ≤ 0.01) with HBV outcome, HLA-DPB1*04:01 and HLA-DPB1*01:01 associated significantly with recovery and persistence of HBV, respectively, in all ethnic groups combined (Table 1). The association with these alleles was consistent across the two ethnic groups (Table 1), and no other HLA-DPB1 alleles reached significance in the combined or the stratified groups.
Table 1.
Significant HLA-DPB1 associations in the study cohortsa
| Samplea | Clearance |
Persistence |
Association | P value | OR | 95% CI | ||
|---|---|---|---|---|---|---|---|---|
| No. of samples | % of samples | No. of samples | % of samples | |||||
| All samples | 421 | 241 | ||||||
| 01:01 | 60 | 14.3 | 51 | 21.2 | 01:01 vs others | 0.01 | 1.86 | 1.16–2.99 |
| Others | 361 | 85.8 | 190 | 78.8 | ||||
| 04:01 | 249 | 59.1 | 117 | 48.6 | 04:01 vs others | 0.003 | 0.56 | 0.38–0.82 |
| Others | 172 | 40.9 | 124 | 51.5 | ||||
| European-Americans | 320 | 185 | ||||||
| 01:01 | 26 | 8.1 | 22 | 11.9 | 01:01 vs others | 0.18 | 1.55 | 0.81–2.96 |
| Others | 294 | 91.9 | 163 | 88.1 | ||||
| 04:01 | 225 | 70.3 | 108 | 58.4 | 04:01 vs others | 0.01 | 0.55 | 0.36–0.84 |
| Others | 95 | 29.7 | 77 | 41.6 | ||||
| African-Americans | 85 | 46 | ||||||
| 01:01 | 32 | 37.7 | 28 | 60.9 | 01:01 vs others | 0.01 | 2.7 | 1.24–5.75 |
| Others | 53 | 62.4 | 18 | 39.1 | ||||
| 04:01 | 19 | 22.4 | 4 | 8.7 | 04:01 vs others | 0.05 | 0.27 | 0.08–0.97 |
| Others | 66 | 77.7 | 42 | 91.3 | ||||
Data were adjusted by race and cohort.
A novel SNP in the 3′UTR region of HLA-DPB1 has the strongest association with HBV recovery.
The variant that showed the strongest association with chronic HBV infection in Asians, which we term 550A/G (rs9277535), is located in the 3′UTR of the HLA-DPB1 gene (8). To determine whether this or another variant showed a similar effect in European- and African-Americans, we sequenced the entire 3′UTR region of HLA-DPB1 in all subjects, identifying 33 variants in this region. The 550A/G variant associated weakly with HBV outcome in our cohort (OR = 0.39, P = 0.01; see Table S2 in the supplemental material) relative to another variant in the 3′UTR, rs9277534 (496A/G; OR = 0.37, P = 0.0001; see Table S2). The 496GG genotype variant had a significantly higher frequency in the persistence group (19.83%) than in the recovery group (9.85%), whereas the frequency distribution of 496AA and 496AG genotypes (these combined genotypes are termed 496AX) did not differ significantly between outcome groups. The recessive susceptible association of 496GG was consistent across the stratified European- and African-American cohorts (Fig. 1). This association was also consistent in HIV-positive and HIV-negative subjects (data not shown). The 496A/G SNP is in perfect linkage disequilibrium (LD) with four other SNPs (rs9277530, rs9277531, rs9277533, and rs9277536) in the 3′UTR region of HLA-DPB1 (see Table S2), any of which could potentially be causal. None of the other 28 SNPs in the 3′UTR region of HLA-DPB1 had as strong an association as position 496, suggesting that they are not as likely to be directly involved in the mechanism underlying viral recovery.
Fig 1.
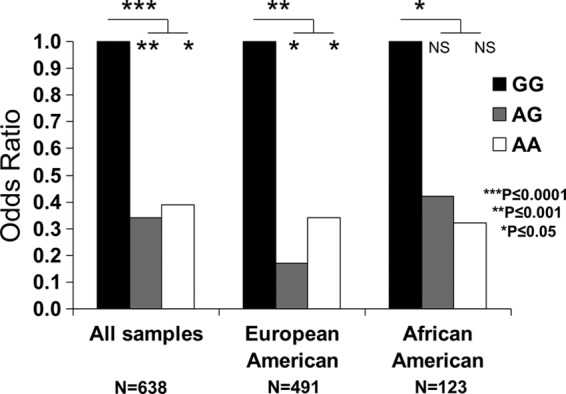
The 496A variant in the 3′UTR region of HLA-DPB1 confers dominant protection against HBV. The odds ratios are based on the frequency distribution of the three genotypes (GG, AG, and AA) of the 496A/G SNP in the persistence group versus the clearance group in the study cohort with all samples, European-Americans and African-Americans. 496GG is the reference genotype set at an OR of 1.0.
We showed previously that HLA-A*03:01, -B*08, -B*44, and -DRB1*13:02 were associated with HBV recovery/persistence in the same European-American samples used in the present study (27). In order to determine whether 496A/G in the 3′UTR of HLA-DPB1 has an effect on HBV recovery that is independent of these HLA alleles and the closely located HLA-DPB1 coding region, we used a stepwise logistic regression model that included all HLA-DPB1 alleles with a frequency of greater than 5%, the four alleles associated with HBV previously, and the 496A/G and 550A/G variants. The analysis indicates that the 496AX genotypes associate with viral recovery independently of the other variables (Table 2) in the European-Americans. A similar analysis was performed in African-American samples that included HLA-DQA1*0501 and -DQB1*0301, which were previously shown to associate with HBV persistence in these same African-Americans (26), along with all HLA-DPB1 alleles with a frequency of greater than 5%, and the 496A/G and 550A/G variants. In this subset of patients, the 496AX genotypes consistently associated with viral recovery independent of other variables. Notably, none of the HLA-DPB1 alleles remained in the model involving either ethnic group, suggesting that the mechanism underlying HLA-DPB1 association with HBV recovery does not involve presentation of a specific viral peptide by a specific HLA-DP allotype. Likewise, the 550A/G variant that was identified in the Asians to associate with outcomes of HBV infection and disease persistence did not remain in the model (neither ethnic group), showing that the association of this SNP in Asians was likely due to its strong LD with the 496A/G variant. The rs3077 SNP located in the HLA-DPA1 3′UTR region showed the second strongest association with chronic HBV infection in the Asian GWAS (8). However, this SNP associated only weakly with HBV recovery in our African-American cohorts (OR = 0.32 and P = 0.02) and did not remain significant after correcting for the effect of the 496A/G variant (data not shown). Moreover, this variant did not associate significantly with outcomes of HBV infection in European-Americans. We thus focused on the variation in the 3′UTR of HLA-DPB1 rather than that of HLA-DPA1 for the remainder of the study.
Table 2.
The effect of the 3′UTR variant at position 496 is independent of previously reported HLA associations with HBV recoverya in European- and African-Americans
| Significant independent variable | P value | OR | 95% CI |
|---|---|---|---|
| European-Americans (n = 505) | |||
| 496AX vs GG | 0.01 | 0.36 | 0.16–0.80 |
| HLA-A*03:01 vs others | 0.02 | 0.54 | 0.33–0.90 |
| HLA-B*08 vs others | 0.04 | 1.67 | 1.02–2.73 |
| HLA-B*44 vs others | 0.003 | 1.95 | 1.25–3.03 |
| African-Americans (n = 131) | |||
| 496AX vs GG | 0.01 | 0.36 | 0.16–0.81 |
| HLA-DQA*0501 vs others | 0.005 | 3.28 | 1.44–7.45 |
Data were adjusted by cohorts and analyzed using a conditional logistic regression model with stepwise selection.
HLA-DP surface expression levels correlate with 496A/G genotypes.
Previous work has shown that cell surface levels of HLA-C expression in healthy donors correlate with outcome after HIV infection (28), and a variant in the 3′UTR of HLA-C, which differentially binds a microRNA, accounts in part for the varied expression across HLA-C allotypes (10). We therefore hypothesized that the 496A/G variant in HLA-DPB1 may correlate with various levels of cell surface expression across HLA-DP allotypes, which in turn could contribute to the association of this SNP with HBV control. To test this possibility, monoclonal antibodies with reported HLA class II reactivity were subjected to extensive characterization using a panel of 86 common HLA class II allotypes (see Fig. S2 in the supplemental material). The MAbs B7/21 and BrafB6, which were previously shown to specifically immunoprecipitate HLA-DP from lymphoblastoid cell lines (6, 33), reacted strongly with all 24 HLA-DP allotypes but not with any allotypes encoded by other HLA loci tested in the panel. The MAbs Tu39 and SK10 are known to detect a range of HLA class II antigens (9, 20), and we found that a pool of these MAbs together bound all 86 HLA-DR, -DP, and -DQ allotypes tested. B7/21 and BrafB6 have a similar affinity for different HLA-DP alleles, because they show the same binding to each HLA-DP allotype relative to the level of HLA present on each bead (see Fig. S2).
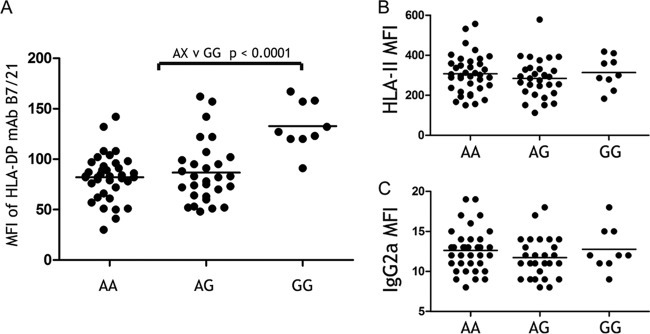
We then compared staining of CD19+ cells with these MAbs across 75 European-American donors that were randomly selected from a healthy donor pool. Fluorescence intensity of HLA-DP staining correlated significantly with the genotype of 496A/G (P < 0.0001) (Fig. 2A), whereas that for the pool of MAbs recognizing all HLA class II allotypes or an isotype control showed no correlation (Fig. 2B and C). Surface HLA-DP protein expression of donors with the GG genotype was significantly higher than that of either AG or AA, but expression on cells from individuals with the AG and AA genotypes did not differ from one another. Thus, consistent with the recessive susceptible effect of 496GG, this variant also correlated recessively with high expression of HLA-DP on B cell surfaces. B7/21 fluorescence intensity varied by up to 5.6-fold across the donor pool, with a difference of 1.6-fold on average between donors with the 496GG versus AX genotypes (see Fig. S3A and B in the supplemental material). This pattern of HLA-DP staining on CD19+ cells was replicated in a group of 112 healthy African-American donors. Once again, both of the HLA-DP-specific MAbs, B7/21 and BrafB6, bound to CD19+ cells from donors with the GG genotype at significantly higher levels than to cells from donors with the AX genotypes (see Fig. S4A and B in the supplemental material). CD19 staining on the same cells showed no difference between donors of GG versus AX genotypes (see Fig. S4C in the supplemental material). HLA-DP surface protein expression levels in the European- and African-American groups correlated only weakly with the genotype that associated with HBV recovery in Asians (i.e., 550A/G) (see Fig. S5A and B in the supplemental material).
Fig 2.
HLA-DP surface protein levels correlate with the 496A/G genotype in the 3′UTR region of HLA-DPB1. HLA-DP molecules were detected on peripheral blood CD19+ cells by flow cytometry using the HLA-DP-specific MAb B7/21. Median fluorescence intensity (MFI) of B7/21 staining was compared across 75 normal European-American donors, as grouped by their HLA-DPB1 genotype (A). MFI of staining to the same cells by a pool of Tu39 and SK10 (HLA-II) MAbs that together bind all HLA-DR/DQ/DP allotypes (B) or an isotype-negative control (C) showed no correlation with the 496A/G genotype. P values were generated by unpaired Mann-Whitney t test.
HLA-DPB1 mRNA transcript levels correlate with 496A/G genotypes.
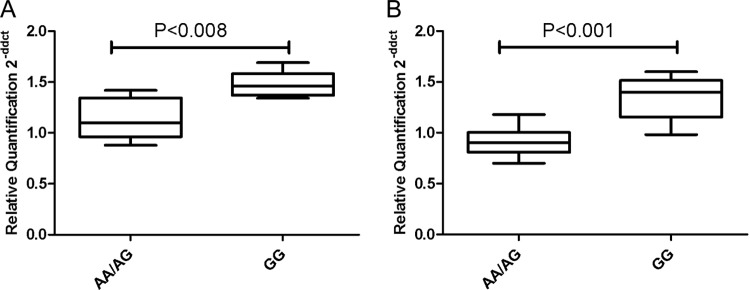
In order to validate the differential cell surface expression levels between 496GG and AX genotypes, we determined the differences in levels of HLA-DPB1 mRNA by quantitative PCR (qPCR) using the comparative CT method (12). HLA-DPB1 transcripts were amplified using primers specific for exon 3 of the HLA-DPB1 gene. Levels of HLA-DPB1 transcripts for each sample were compared to that of a calibrator (donor with 496AA genotype) and were measured by comparing the CT values of the gene of interest (GOI) (i.e., HLA-DPB1) normalized to CT values of a reference gene. The RPLP0 and GAPDH reference genes had amplification efficiencies that matched the GOI. Significant differences in HLA-DPB1 mRNA expression in cells from 15 European-American donors correlated with the 496A/G genotypes in a manner that mirrored the cell surface expression (Fig. 3). Ten samples with 496AX genotypes had significantly lower HLA-DPB1 mRNA expression than did 5 samples with the 496GG genotype after normalization to the RPLP0 (Fig. 3A) and GAPDH (Fig. 3B) reference genes. Quantitative PCR using the relative quantification method (16) produced results very similar to those of the comparative qPCR method (see Fig. S6A to F in the supplemental material). We conclude that the 3′UTR variant 496A/G associates with variation in HLA-DPB1 mRNA expression levels in a manner that reflects our surface protein flow cytometry results.
Fig 3.
HLA-DPB1 mRNA levels correlate significantly with the 496A/G variant in the 3′UTR region of HLA-DPB1. HLA-DPB1 mRNA levels were detected by qPCR amplification of cDNA derived from fresh PBL of 15 healthy European-American donors. HLA-DPB1 mRNA levels were detected using a SYBR green method with primers specific for exon 3 of HLA-DPB1. Levels of HLA-DPB1 mRNA are shown relative to a calibrator using two different reference genes, RPLP0 (A) and GAPDH (B). HLA-DPB1 mRNA levels in the group of individuals with 496AX (n = 10) were significantly lower than that from 496GG (n = 5) donors. These data are representative of three independent experiments. P values were generated by unpaired Mann-Whitney t test.
Taken together with the observation that a novel 3′UTR variant, 496A/G, has the most significant overall HLA effect on HBV recovery in our European-American cohorts and the second most significant effect in our African-American cohorts, we propose that differential HLA-DP expression is one of the key determinants to outcome after HBV infection.
DISCUSSION
A number of host genes contain polymorphisms that associate with outcome to HBV infection, including CCR5, RANTES, and, most notably, HLA (24–27). A recent GWAS confirmed the primary involvement of HLA class II with chronic HBV infection, but surprisingly, the most significant genetic marker correlating with HBV outcome resided in the HLA-DP region (8) rather than in the commonly investigated HLA-DR and -DQ genes. Based on a GWAS, SNPs in the 3′UTR of HLA-DPB1 (rs9277535) and -DPA1 (rs3077) had the strongest and second strongest associations with chronic HBV infection, respectively, in two independent Asian populations. They also associated with HBV recovery/persistence in another Asian study (4). However, these SNPs associated weakly with HBV recovery/persistence in our cohorts consisting of African-American and European-American patients. Through sequence analysis of the entire 665 bp of the 3′UTR region of HLA-DPB1, we identified five variants, which are in perfect two-way LD with one another, that associate significantly with HBV recovery in both of the ethnic groups examined in our study. Since we do not know which of these variants might be causal, we have used the SNP at position 496 (rs9277534) as the representative for rs9277530, rs9277531, rs9277533, and rs9277536 SNPs in all our analyses. The 496G allele has a recessive susceptibility effect compared to that of the 496AX genotype; this association is stronger and more significant than any individual HLA-DPB1 allele or any other HLA allele previously associated with HBV recovery in our European-American cohort. Importantly, the most significant SNP (550A/G) in the GWAS report (8) did not remain significant in a stepwise multivariate model that considered 496A/G, HLA-DPB1 alleles, and other HLA class I and II alleles that had been previously associated with HBV recovery in these same cohorts. In the smaller African-American cohort, the 496A variant also confers independent protection, though to a slightly lesser extent than DQA1*0501. In fact, a recent study in a Japanese population attributes both HLA-DP and HLA-DQ loci with risk of persistent HBV infection (14). The HLA-DPA1 SNP rs3077 had only a weak association in African-Americans that was no longer significant after correcting for the association with 496A/G (data not shown). We thus concentrated our efforts on deciphering the effect of SNPs in the HLA-DPB1 region.
We surmise that the rs927753 SNP (550A/G) does not have a strong effect with HBV recovery in our cohorts, because the protective 550A allele, which marks the DPB1*04:01 protective allele, also marks the DPB1*01:01 risk allele, diminishing the association of position 550 with HBV outcome (see Tables S3 and S4 in the supplemental material). This was not a confounding factor in the Asian population, because DPB1*01:01 is uncommon in Asians and thus the 550A allele proved to be a suitable marker for protection. In contrast, the 496A/G variant distinguishes HLA-DPB1*01:01 (496G) from *04:01 (496A) and therefore shows a much stronger association in subjects of African and European descent. 496A/G is likely to be a better marker in Asians, as well, since 496G is in LD with the risk HLA-DPB1 alleles *05:01 and *03:01, whereas 496A is in LD with the protective HLA-DPB1 alleles *04:01 and *04:02 in Asians.
Evidence reported herein suggests that the influence of the HLA-DP region on HBV recovery is due to levels of HLA-DPB1 expression and less likely to differences in the peptides presented by different HLA-DPB1 alleles. All five of the common HLA-DP alleles, including HLA-DPB1*04:01, *04:02, *02:01, *01:01, and *05:01, share largely overlapping peptide binding repertoires (22) and comprise both the protective as well as the susceptible HLA-DPB1 alleles. Thus, the effect of HLA-DP on HBV may be due to differences in levels of expression rather than differences in peptide presentation. Differences in expression levels of HLA-C class I molecules have previously been implicated in HIV disease (28), and this appears to be due in part to variation in the 3′UTR of the HLA-C locus, which determines binding of a microRNA (10). We show here that HLA-DP is expressed at differential levels on freshly isolated B lymphocytes from normal donors of African and European descent, and this cell surface expression correlates with 496A/G genotypes. Individuals with the GG genotype have higher levels of HLA-DP expression than do individuals with the AA/AG genotypes. This recessive effect of 496G (alternatively the dominant effect of 496A) suggests that the mechanism regulating HLA-DP expression probably has to do with some sort of event that is sensitive to a threshold. Thus, the 496GG genotype associates with both HBV persistence and higher HLA-DP expression levels, suggesting that high HLA-DP expression may promote HBV persistence.
In contrast to the 496A/G genotypes, HLA-DP surface expression correlated relatively poorly with 550A/G genotypes. The residual correlation of position 550 with HLA-DP expression is most likely due to the significant but incomplete LD with position 496. Based on an association between 550A/G genotype and mRNA levels obtained from microarray data from liver cells and qPCR of mRNA from macrophages, O'Brien et al. (15) concluded that lower HLA-DP mRNA expression correlates with HBV disease pathogenesis. We examined HLA-DPB1 transcript levels using HLA-DPB1-specific qPCRs that targeted two different regions of HLA-DPB1 and normalized these results with three independent reference genes. The primers were specifically designed to recognize all alleles of the HLA-DPB1 locus but no alleles of any other HLA locus. Our results show that HLA-DPB1 mRNA levels were higher in total PBL from donors with the risk 496GG genotype than in donors with 496AA/AG genotypes. Thus, our findings are in complete contrast to the previous study (15) in that higher levels of HLA-DP at both the cell surface protein level and mRNA level associate with the 496GG genotype, which in turn associates with HBV persistence.
Clinical studies find that Th1 responses are more often observed in patients that recover from an HBV infection, while Th2 responses are a hallmark of chronically infected patients (13). Perhaps high HLA-DP expression favors a Th2 response characterized by vigorous antibody production along with poor CTL activity leading to HBV persistence. A recent multicenter genetic study implicates the HLA-DPB1 locus in risk of the autoimmune disease rheumatoid arthritis (RA) (19). HLA-DPB1 alleles that associated with risk of RA are the same alleles that are protective against HBV and are expressed at low levels. Taken together, this is consistent with our model of lower HLA-DP expression favoring Th1 responses that are effective in HBV recovery but are known to be disadvantageous in Th1-mediated RA (30).
Our results strongly implicate the 3′UTR region of HLA-DP with HLA-DP expression and outcomes of HBV infection. It is now necessary to determine the mechanism for varied expression of HLA-DP, which may involve regulation through differential microRNA activity, similar to HLA-C (10). If HLA-DP expression levels do indeed explain the association between HLA-DP and HBV disease pathogenesis, further work will be required to determine how varied expression of this molecule at the site of infection in the liver is affecting the immune response to HBV.
Supplementary Material
ACKNOWLEDGMENTS
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E. C.L.T. was supported by the Investigators in the Pathogenesis of Infectious Diseases Award from the Burroughs Wellcome Fund. The ALIVE study was supported by National Institutes of Health grants R01-DA-04334 and R01-DA-12568. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The MACS is funded by the National Institute of Allergy and Infectious Diseases, with additional supplemental funding from the National Cancer Institute and National Heart, Lung and Blood Institute grants UO1-AI-35042, UO1-AI-35043, UO1-AI-35039, UO1-AI-35040, and UO1-AI-35041. This work was supported by the NIH, NIAID, Division of AIDS, with the Center for HIV/AIDS Vaccine Immunology (CHAVI) (grant U19 AI067854).
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
We thank Russell Hanson and the Occupational Health Services at NCI-Frederick for sample collection from healthy donors, Bailey Kessing and Carl Macintosh for bioinformatics support, Ligia Pinto and Troy Kemp for the use of their Luminex instrument, and Arman Bashirova for helpful advice on RNA quantitation.
Footnotes
Published ahead of print 11 April 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Apps R, et al. 2009. Human leucocyte antigen (HLA) expression of primary trophoblast cells and placental cell lines, determined using single antigen beads to characterize allotype specificities of anti-HLA antibodies. Immunology 127:26–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Edwards JA, Durant BM, Jones DB, Evans PR, Smith JL. 1986. Differential expression of HLA class II antigens in fetal human spleen: relationship of HLA-DP, DQ, and DR to immunoglobulin expression. J. Immunol. 137:490–497 [PubMed] [Google Scholar]
- 3. Goedert JJ, et al. 1989. A prospective study of human immunodeficiency virus type 1 infection and the development of AIDS in subjects with hemophilia. N. Engl. J. Med. 321:1141–1148 [DOI] [PubMed] [Google Scholar]
- 4. Guo X, et al. 2011. Strong influence of human leukocyte antigen (HLA)-DP gene variants on development of persistent chronic hepatitis B virus carriers in the Han Chinese population. Hepatology 53:422–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hilgartner MW, et al. 1993. Hemophilia growth and development study. Design, methods, and entry data. Am. J. Pediatr. Hematol. Oncol. 15:208–218 [DOI] [PubMed] [Google Scholar]
- 6. Horejsi V, et al. 1988. Characterization of a new murine monoclonal antibody against human DP antigens. Tissue Antigens 32:6–11 [DOI] [PubMed] [Google Scholar]
- 7. Hyams KC. 1995. Risks of chronicity following acute hepatitis B virus infection: a review. Clin. Infect. Dis. 20:992–1000 [DOI] [PubMed] [Google Scholar]
- 8. Kamatani Y, et al. 2009. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat. Genet. 41:591–595 [DOI] [PubMed] [Google Scholar]
- 9. Klohe EP, et al. 1988. Analysis of the molecular specificities of anti-class II monoclonal antibodies by using L cell transfectants expressing HLA class II molecules. J. Immunol. 141:2158–2164 [PubMed] [Google Scholar]
- 10. Kulkarni S, et al. 2011. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature 472:495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee WM. 1997. Hepatitis B virus infection. New Engl. J. Med. 337:1733–1745 [DOI] [PubMed] [Google Scholar]
- 12. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 13. Locarnini S. 2004. Molecular virology of hepatitis B virus. Semin. Liver Dis. 24(Suppl 1):3–10 [DOI] [PubMed] [Google Scholar]
- 14. Mbarek H, et al. 2011. A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum. Mol. Genet. 20:3884–3892 [DOI] [PubMed] [Google Scholar]
- 15. O'Brien TR, et al. 2011. Risk alleles for chronic hepatitis B are associated with decreased mRNA expression of HLA-DPA1 and HLA-DPB1 in normal human liver. Genes Immun. 12:428–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 doi:10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Phair J, et al. 1992. Acquired immune deficiency syndrome occurring within 5 years of infection with human immunodeficiency virus type-1: the Multicenter AIDS Cohort Study. J. Acquir. Immune Defic. Syndr. 5:490–496 [PubMed] [Google Scholar]
- 18. Png E, et al. 2011. A genome-wide association study of hepatitis B vaccine response in an Indonesian population reveals multiple independent risk variants in the HLA region. Hum. Mol. Genet. 20:3893–3898 [DOI] [PubMed] [Google Scholar]
- 19. Raychaudhuri S, et al. 2012. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 44:291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robbins PA, Evans EL, Ding AH, Warner NL, Brodsky FM. 1987. Monoclonal antibodies that distinguish between class II antigens (HLA-DP, DQ, and DR) in 14 haplotypes. Hum. Immunol. 18:301–313 [DOI] [PubMed] [Google Scholar]
- 21. Rozemuller EH, van der Zwan A, Tilanus MGJ. 2006. Sequence based typing for HLA-DPA1, p 372–373 In Hansen JA. (ed), Immunology of the human MHC, Proceedings of the 13th International Histocompatibility Workshop aand Conference, vol II IHWC Press, Seattle, WA [Google Scholar]
- 22. Sidney J, et al. 2010. Five HLA-DP molecules frequently expressed in the worldwide human population share a common HLA supertypic binding specificity. J. Immunol. 184:2492–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singh R, Kaul R, Kaul A, Khan K. 2007. A comparative review of HLA associations with hepatitis B and C viral infections across global populations. World J. Gastroenterol. 13:1770–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thio CL, et al. 2007. Genetic protection against hepatitis B virus conferred by CCR5Delta32: evidence that CCR5 contributes to viral persistence. J. Virol. 81:441–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thio CL, et al. 2008. Interaction between RANTES promoter variant and CCR5Delta32 favors recovery from hepatitis B. J. Immunol. 181:7944–7947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thio CL, et al. 1999. Class II HLA alleles and hepatitis B virus persistence in African-Americans. J. Infect. Dis. 179:1004–1006 [DOI] [PubMed] [Google Scholar]
- 27. Thio CL, et al. 2003. Comprehensive analysis of class I and class II HLA antigens and chronic hepatitis B virus infection. J. Virol. 77:12083–12087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomas R, et al. 2009. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat. Genet. 41:1290–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thursz M, Yee L, Khakoo S. 2011. Understanding the host genetics of chronic hepatitis B and C. Semin. Liver Dis. 31:115–127 [DOI] [PubMed] [Google Scholar]
- 30. van Roon JA, Bijlsma JW. 2002. Th2 mediated regulation in RA and the spondyloarthropathies. Ann. Rheum. Dis. 61:951–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vlahov D, et al. 1998. Prognostic indicators for AIDS and infectious disease death in HIV-infected injection drug users: plasma viral load and CD4+ cell count. JAMA 279:35–40 [DOI] [PubMed] [Google Scholar]
- 32. Vooter CE, Kik MC, van den Berg-Loonen EM. 2006. Sequence based typing for HLA-DPB1, p 376–379 In Hansen JA. (ed), Immunology of the human MHC, Proceedings of the 13th International Histocompatibility Workshop and Conference, vol II IHWC Press, Seattle, WA [Google Scholar]
- 33. Watson AJ, DeMars R, Trowbridge IS, Bach FH. 1983. Detection of a novel human class II HLA antigen. Nature 304:358–361 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.