Abstract
Human cytomegalovirus (HCMV) has been found in malignant gliomas at variable frequencies with efforts to date focused on characterizing the role(s) of single gene products in disease. Here, we reexamined the HCMV prevalence in malignant gliomas using different methods and began to dissect the genetics of HCMV in tumors. HCMV DNA was found in 16/17 (94%) tumor specimens. Viral DNA copy numbers were found to be low and variable, ranging from 102 to 106 copies/500 ng of total DNA. The tumor tissues had incongruences between viral DNA copy numbers and protein levels. However, nonlatent protein expression was detected in many tumors. The viral UL83 gene, encoding pp65, was found to segregate into five cancer-associated genotypes with a bias for amino acid changes in glioblastoma multiforme (GBM) in comparison to the low-grade tumors. Deep sequencing of a GBM-associated viral population resulted in 81,224 bp of genome coverage. Sequence analysis revealed the presence of intact open reading frames and higher numbers of high-frequency variations within the repeat long region compared to the unique long region, which harbors many core genes, and the unique short region (P = 0.001). This observation was in congruence with phylogenetic analyses across replication-competent viral strains in databases. The tumor-associated viral population was less variable (π = 0.1% and πAA = 0.08%) than that observed in other clinical infections. Moreover, 42/46 (91.3%) viral genes analyzed had dN/dS scores of <1, which is indicative of high amino acid sequence conservation. Taken together, these findings raise the possibility that replication-competent HCMV may exist in malignant gliomas.
INTRODUCTION
Malignant diffuse gliomas are the most common brain cancers in adults, with an annual incidence of 5 per 100,000 people and ∼14,000 new cases diagnosed each year in the United States (60). These gliomas are classified as astrocytomas, oligodendrogliomas, or ependymomas on the basis of the type of glial cell from which the cancer arises. The astrocytomas are the most common form of the tumor. According to the present World Health Organization (WHO) classification, astrocytomas are divided into four categories: grade I or pilocytic astrocytoma (PA) and subependymal giant cell astrocytoma, grade II or diffusely infiltrating astrocytoma (IAS), grade III or anaplastic astrocytoma, and grade IV or glioblastoma multiforme (GBM) (18). Among all of the categories of astrocytomas, grade IV or GBM is the most aggressive, with a median survival of 7 months (10). Despite such a thorough understanding of the disease stages, most individuals rapidly succumb to the disease irrespective of advanced therapeutics (58). Several recent studies have made the observation that a percentage of GBMs express human cytomegalovirus (HCMV) antigens (6, 31, 36, 41, 47). Determining whether HCMV influences GBM biology will increase our understanding of this cancer (5, 35, 51, 52). Moreover, the presence of HCMV in gliomas could be exploited to develop better treatments for these cancers.
HCMV is a large DNA virus with an ∼235-kbp genome that encodes approximately 165 proteins and is subdivided into three distinct regions: the repeat long and short regions (RL and RS) and the unique long (UL) and the unique short (US) regions (11). HCMV, a member of the Herpesviridae family, has several unique biological properties (1, 14, 21, 32, 34, 50, 53, 56). It infects a broad spectrum of cells in vivo, including endothelial, epithelial, hepatocytes, fibroblasts, muscle cells, leukocytes, dendritic cells, and astrocytes (48). However, irrespective of cell types or strains, there are three distinct classes of gene expression essential for a productive viral infection, immediate-early (IE), early (E), and late (L), which are expressed in a well-coordinated manner. Expression of the E genes is dependent upon the expression of the IE genes, while L gene expression occurs subsequent to IE and E gene expression (37). HCMV is a ubiquitous virus, but HCMV-related disease is rare among immunocompetent individuals. The virus is primarily a concern in immunocompromised populations, while most immunocompetent individuals maintain a reservoir of HCMV without any sign or symptom of disease (8, 17). However, in congenitally infected infants and in allograft recipients, HCMV disease is a significant cause of morbidity and mortality (19).
Evidences for the presence of HCMV in malignant gliomas have been shrouded by discrepancies. Some groups have been able to identify the presence of IE and E markers of viral infection in 93 to 100% of GBM specimens, while the percentage is lower in other grades of malignant gliomas (6, 36, 47). Others have been able to detect the viral marker in only 11 to 51% of GBMs (26, 45). The presence of viral DNA in the blood has been reported in 80% of GBM patients (36). However, this observation has been challenged in two reports containing limited numbers of samples (29, 31). In this report, we revisit the question of HCMV detection in cancer using a sensitive nested PCR approach to validate the presence of HCMV sequences, since tissue resection and chemotherapy can potentially cause damage to cellular proteins. We were also able to quantify the viral DNA copy numbers in a subset of samples. We expanded on these observations by measuring viral protein expression by immunoblotting and performed a genetic analysis across tumor tissues. In addition, we were able to study viral population genomic variability of intact 81,244-bp regions encompassing the RL region, parts of the UL region, and the US region and found all open reading frames (ORFs) to be intact and under positive selection. Although not definitive, these results leave open the possibility that unique, replication-competent virus exists within tumor tissues.
MATERIALS AND METHODS
Sample collection.
A panel of 17 snap-frozen glioma specimens (Table 1) was obtained from the tumor bank at University of Massachusetts Medical School, in accordance with the Institutional Review Board (IRB). A primary culture of a GBM tumor, GS826, was graciously provided by Alonzo Ross (UMass Medical School).
Table 1.
Description of the patient samples included in this study
| Sample | Diseasea | Age (yr) | Gender |
|---|---|---|---|
| 07T | GBM | 71 | F |
| 08T | GBM | 52 | M |
| 10T | Oligodendroglioma | 51 | F |
| 16T | GBM | 72 | F |
| 29T | Astocytoma | 74 | M |
| 40T | GBM | 64 | M |
| 41T | GBM/small-focus GSM | 68 | M |
| 54T | Ganglioglioma | 25 | M |
| 58T | GBM | 59 | M |
| 59T | Infiltrating GBM | 45 | F |
| 61T | GBM | 64 | M |
| 65T | GBM | 51 | F |
| 78T | Pilocytic astrocytoma | 3 | F |
| 80T | GBM | 75 | F |
| 95T | GBM | 54 | M |
| 161T | Astrocytoma | 53 | M |
| GS826 | GBM primary culture | 65 | M |
GBM, glioblastoma multiforme; GSM, gliosarcoma.
DNA isolation.
DNA was isolated from tumor specimens using a DNeasy Blood & Tissue kit (Qiagen, Valencia, CA) according to the manufacturer's instructions.
Nested PCR.
All of the PCR preparations were carried in an isolated laboratory where no previous work had been done on HCMV in order to prevent false positive results. A nested PCR approach was used to detect viral DNA. The external primer sequences were the forward primer 5′-CCGAAATACGCGTTTTGAGAT and the reverse primer 5′-CCAAGCCAAAAACAGTATAGC, which amplified a 1.3-kbp region of the major immediate-early promoter. This primer set recognized all of the genomes documented at the NCBI database. Human embryonic lung fibroblast (HEL) DNA was used as a negative control in each PCR run. The reaction mixture (20 μl) contained 500 ng of DNA, 20 ng of primers (IDT DNA, Coralville, IA), 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.3), 250 μM concentrations of each deoxynucleoside triphosphate (dNTP; New England Biolabs, Ipswich, MA), and 1 U of thermostable DNA Polymerase (New England Biolabs). The PCR conditions were 95°C for 5 min, followed by 50 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 10 min. The internal primers sequences were the forward primer 5′-GGCGGAGTT(G/A)TTACGACATTT and the reverse primer 5′-ATGCGGTTTTGGCAGTACAT, which amplified a 144-bp region within the 1.3-kbp amplicon generated by the external primer set. The reaction mixture (20 μl) contained 4 μl of external PCR product, 20 ng of primers, 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.3), 250 μM concentrations of each dNTP, and 1 U of thermostable Taq DNA polymerase (New England Biolabs). The PCR conditions were 95°C for 5 min, followed by 40 cycles of 95°C for 20 s, 55°C for 20 s, and 72°C for 20 s, with a final extension at 72°C for 5 min.
Real-time PCR.
The viral copy number in tumor tissues was determined by amplifying a 100-bp region within the UL123 gene in triplicate, using the forward primer 5′-TGACGCTTGTATGATGACCATGTAC, the reverse primer 5′-CAGCATCACACTAGTCTCCTCTAAG, and the hydrolysis probe P1 (6-FAM)-ACCCGACAGAACTC. The reaction mixture (20 μl) contained 500 ng of target DNA, 1 μl of the 20× primer-probe mix, and 10 μl of the TaqMan Universal master mix (Applied Biosystems, Foster City, CA). The PCR conditions were 95°C for 10 min, followed by 50 cycles of 95°C for 15 s and 60°C for 1 min. The reaction was performed with the ABI Prism 7500 HT sequence detection system. A standard curve was obtained using serial dilutions of AD169 bacterial artificial chromosome (BAC) (38) mixed with 500 ng of HEL DNA as a genomic control. The no-template controls also contained 500 ng of HEL DNA but were devoid of viral DNA.
Immunoblot analysis.
Protein lysates were made from snap-frozen tissues homogenized in cold radioimmunoprecipitation assay buffer (0.25% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 25 mM Tris [pH 7.4], 150 mM NaCl, 1 mM EDTA, and 1% Nonidet P-40 in water) supplemented with 100 mM β-mercaptoethanol (Sigma), 150 mM NaCl, and protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO) and incubated on ice for 30 min. Extracts were centrifuged at 14,000 rpm for 15 min, and soluble proteins in the supernatants were quantitated by using a Bradford assay (Bio-Rad, Hercules, CA). Protein extracts (100 μg) were separated by 10% SDS-polyacrylamide gel electrophoresis, and the resolved proteins were transferred to nitrocellulose membranes. Viral protein detection was carried out using monoclonal antibodies against pp65 (pUL83) (CA 003-100; Virusys, Randallstown, MD), IE1-72 (pUL123) (MAb 8131; Chemicon International, Temecula, CA), and gB (pUL55). The gB antibody (anti-HCMV gB 27-156) was kindly provided by William J. Britt (Department of Pediatrics, University of Alabama at Birmingham). β-Actin antibody (A5316; Sigma-Aldrich) was used as a gel loading control. Protein bands were visualized by chemiluminescence with ECL reagent (Perkin-Elmer, Waltham, MA).
Sanger sequencing analysis.
A set of overlapping primers—forward primer 5′-AAATATAGCAGCGGTGACAGGT (F1), reverse primer 5′-CCGCAAACGCAAATCAG (R1), forward primer 5-CGGATTGTGGATTTCGTTGT (F2), and reverse primer 5′-TACGGTGTTGTGTCCCAAAAATA (R2)—were used to amplify a 970-bp region of the UL83 ORF in separate reactions. The reaction mixture (25 μl) contained 250 ng of total DNA, 1× PfuUltra II PCR buffer, 25 ng of primers (IDT DNA), 250 μM concentrations of each dNTP (New England Biolabs), and 1.25 U of PfuUltra II fusion HS DNA polymerase. The PCR conditions were 95°C for 5 min, followed by 10 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, followed by 90 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 10 min. The PCR products were purified and then Sanger sequenced. Merlin (NC_006273) (11) was used as a reference sequence, and genotypes were constructed from tumor specimens as described elsewhere (44).
In order to detect errors generated by PCR amplification, 10 and 0.0001 ng of AD169 and Toledo BACs were used to amplify a region of the UL83 ORF using forward primer (F2) and reverse primer (R2). This primer set was chosen because of the larger amplicon size it generated of the two sets used. Each reaction was supplemented with 250 ng of HEL DNA. Separate sets of reactions were also run with templates comprising of a combination of AD169 and Toledo BACs in the ratios of 1:1 and 9:1. This was done to determine the sensitivity of this PCR at detecting mixed infections. The PCR conditions were the same as used for patient specimens. The PCR products were purified and assayed by Sanger sequencing. The AD169 and Toledo BAC sequences generated were aligned against the AD169 genome (GenBank accession no. BK000394.5).
Sequences of the same region of UL83 in unpassaged strains (JP [GQ221975.1], 3301 [GQ466044.1], U01, U04, and U33 [44]) obtained from productive human infections were included as controls in the genetic analysis. Sequence alignments were done using the SeqMan II application of the DNAStar package (DNAStar, Madison, WI). In order to resolve incompatibility in grouping polymorphisms during tree building due to potential parallel mutation events (homoplasy) or recombination, phylogenetic analyses were carried by the median-joining network approach using Network 4.6.0.0 software (3).
Viral genome enrichment and next-generation sequencing analysis.
The tumor specimen (58T) with the highest viral copy number was chosen for deep sequencing analysis (Table 1). Seventy-one sets of overlapping primers were designed to generate amplicons ranging in size from 500 bp to 2.6 kbp to amplify 81,244 bp of the viral genome covering the RL region, the US region, and a part of the UL region. All of the primer pairs were designed against the Merlin strain (NC_006273). In order to assess the performance of the primers against all sequenced HCMV genomes available at the National Center for Biotechnology Information (NCBI), the NCBI in silico PCR analyses tool was used, which searches for all of the sequences available within the database that can be amplified with a set of test primers. Primers were then chosen that were able to amplify HCMV genomes present at the NCBI database, without the potential to amplify human DNA. Degeneracy was introduced in primer sequences for regions with low levels of homology across strains. The reaction mixture (25 μl) contained 250 ng of genomic DNA, 1× PfuUltra II PCR buffer, 50 ng of primers (IDT DNA), 200 μM concentrations of each dNTP (New England Biolabs), 2% dimethyl sulfoxide (Fisher Scientific), and 1.25 U of PfuUltra II fusion HS DNA polymerase. The PCR conditions were 95°C for 5 min, followed by 80 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by an extension at 72°C for 10 min. The products were gel purified and confirmed to be of viral origin by Sanger sequencing before deep sequencing was performed on the Illumina GA II platform using 36-base, single-end reads. Sequence analysis was performed by using MAQ software suite (30), and Merlin (NC_006273) was used as the reference genome. Variants were identified after stringent filtering of false-positive errors using in-house scripts (44).
Phylogenetic analysis.
Phylogenetic relationships were determined among various minimally passaged strains (HAN13 [GQ221973.1], HAN20 [GQ396663.1], HAN38 [GQ396662.1], JHC [HQ380895.1], 3157 [GQ221974.1], and Merlin) and unpassaged strains (JP [GQ221975.1], 3301 [GQ466044.1], U01, U04, U33 [44]) with the tumor-associated genome. Each region of the genome, namely, the RL, UL, and the US regions, was analyzed in isolation to identify maximally evolving regions. Multisequence alignments were created using CLUSTAL W with a gap opening penalty of 15 and gap extension penalty of 6.66 (57). Phylogeny was constructed using the neighbor-joining method (46). Evolutionary distances were computed by using the maximum-composite-likelihood method (54), and the substitution rate variation among sites was modeled with a gamma distribution. Each clustering was confirmed by the bootstrap method with 1,000 replicates. The analysis was conducted using MEGA5 (55).
Statistical analyses.
Differences among the variant frequencies across the three genomic regions sequenced were analyzed for statistical significance using a nonparametric Kruskal-Wallis test with the SPSS package (v16.0). A P value of <0.05 was considered statistically significant. Nucleotide diversity (π) (39), which measures genetic variability, and ORF amino acid diversity (πAA) were calculated using in-house algorithms. The ratios of the rates of nonsynonymous substitutions per nonsynonymous site (dN) to the rates of synonymous substitutions per synonymous site (dS), i.e., dN/dS = ω (40), were also calculated using in-house algorithms (44).
RESULTS
A broad range of viral copy numbers was detected across glioma tissues.
In order to detect the presence or absence of HCMV DNA in malignant glioma patient samples, a nested PCR approach was utilized. The reactions were initially standardized using serial dilutions of Toledo BACs (from 4.1 × 108 to 41 copies) per 500 ng of HEL DNA. Sixteen of seventeen patient samples (94%) were found to be positive for HCMV at the DNA level (Fig. 1A). The PCR products were confirmed to be HCMV sequences by Sanger sequencing (data not shown).
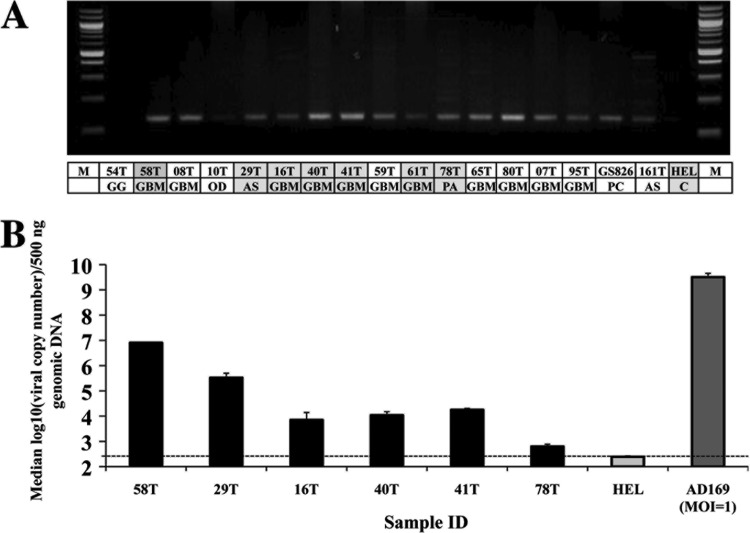
Fig 1.
Detection of HCMV DNA in tumor specimens. (A) Nested PCR products run on a 2% agarose gel. The amplicon is 144 bp in size. The first row in the table lists the sample identification, and the second row indicates the disease stage. M, 100-bp marker; GG, ganglioglioma; GBM, glioblastoma multiforme; OD, oligodendroma; AS, astrocytoma; PA, pilocytic astrocytoma; PC, primary culture of GBM tissue; C, uninfected HELs. The entries shaded in gray indicate patient specimens that were tested for viral copy number by subsequent real-time PCR. (B) Plot of median log10 (viral copy number) values from patient specimens. The dotted line denotes the lower limit of viral DNA detection.
Real-time PCR that amplified a 100-bp region from exon 4 of the UL123 ORF was used to quantify viral copy numbers in a subset of cancer specimens (n = 6) (Fig. 1B). Viral DNA copy numbers varied from a low of ∼365 copies per 500 ng of total DNA to as high as 106 copies. There was a clear correlation between band intensities from nested PCR (Fig. 1A) and viral DNA copy numbers ascertainment for the six samples assayed (Fig. 1B). The median log10 viral DNA copy number in disease was 4.033± 1.55 per 500 ng of total DNA, which is much lower than that typically found in a productive infection at a multiplicity of infection (MOI) of 1 (median log10 copy number = 9.507).
Presence of IE, E, and L proteins in glioma tissues.
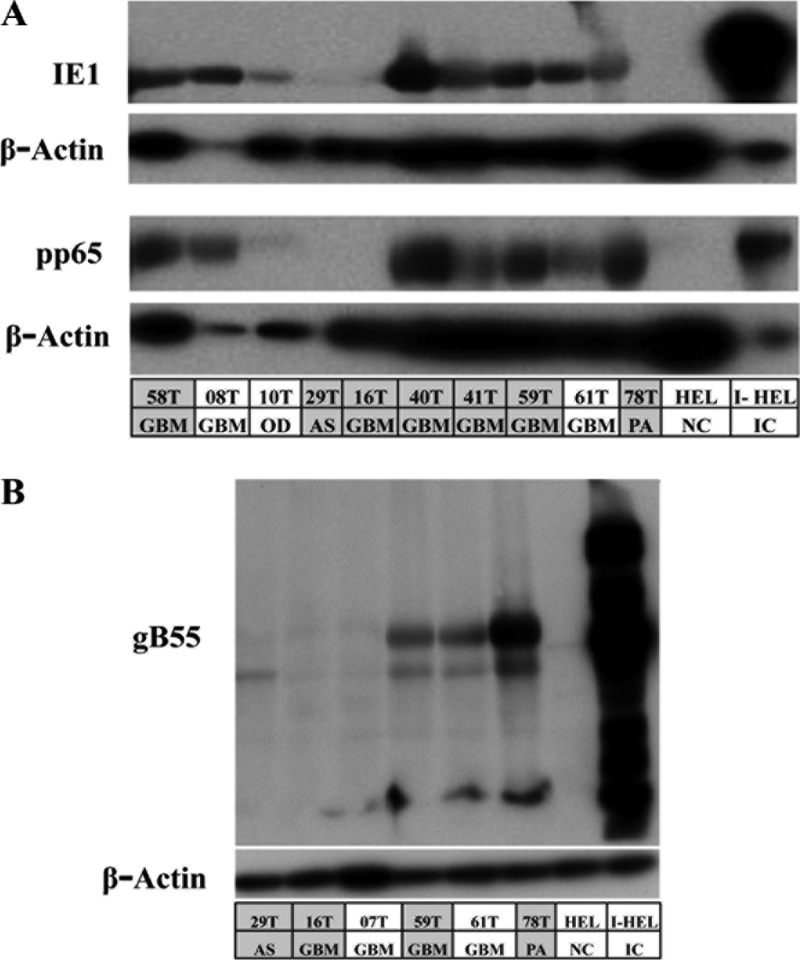
The presence of such a varied range of viral copy numbers warranted an investigation for the presence of viral proteins. Expression profiles of all three classes of viral proteins would help delineate the type of infection found in gliomas. Monoclonal antibodies against markers of immediate-early (IE1/IE2), early (pp65), and early/late (gB55) phases of the viral life cycle were used. A subset of the patient specimens (n = 12) was used for immunoblotting for pp65 and IE1/2 (Fig. 2A). The rest of the specimens could not be included because of inadequate tumor material. Nine of the twelve samples (75%) were positive for IE1 and pp65 expression. IE2 was undetectable in all of the tumor specimens. Three samples were randomly selected from among the nine tumor specimens with IE1 and pp65 protein expression and were further probed for gB55 protein expression (Fig. 2B) to compare with the three specimens with lower viral proteins. There was no clear correlation between viral protein expression and the viral DNA copy numbers, but the samples which had little to no IE1 protein expression also had little to no gB55 expression.
Fig 2.
Detection of IE, E, and L viral protein markers in gliomas. Immunoblots show the presence of viral proteins in patient specimens. The first row in the table lists the sample identification, and the second row indicates the disease stage. GBM, glioblastoma multiforme; OD, oligodendroma; AS, astrocytoma; PA, pilocytic astrocytoma. HEL, human embryonic lung fibroblasts; I-HEL, infected HEL cells (IC, infected cells). The entries shaded in gray are patient specimens that were tested for viral copy numbers in Fig. 1B. (A) Nine of twelve (75%) patient specimens were found to exhibit IE (IE1) and E (pp65) protein expression. (B) E/L (gB55) protein expression was found in the subset of tumors expressing IE and E proteins.
Presence of cancer-associated pp65 genotypes in gliomas.
It is known that pp65 is one of the major targets of the cellular immune response (16, 17, 23, 24, 49, 59, 61), and a long-term GBM survivor who underwent autologous dendritic cell vaccination was also found to mount a strong CD8+ T-cell response to a pp65 immunodominant epitope (42). To assess both nucleotide and amino acid variability across the major pp65 epitope-producing domains, a 970-bp region of UL83 coding for amino acids 273 to 561 of pp65 was amplified and sequenced from seven tumor specimens that produced an amplification product (Table 2). This was done using two pairs of overlapping primers: F1/R1 (484 bp) and F2/R2 (622 bp). The lack of amplification in some specimens may be attributed to variability within the primer binding sites or insufficient sensitivity at low viral DNA copy number.
Table 2.
Patient samples used for assessing pp65 coding region variability
| Sample | Disease | Amplificationa using the primers: |
|
|---|---|---|---|
| F1/R1 (aa 435–561) | F2/R2 (aa 273–479) | ||
| 10T | Oligodendroglioma | + | + |
| 16T | GBM | + | + |
| 40T | GBM | + | + |
| 58T | GBM | + | + |
| 59T | GBM | + | + |
| 78T | Pilocytic astrocytoma | + | + |
| 95T | GBM | + | + |
| 07T | GBM | – | – |
| 08T | GBM | – | – |
| 29T | Astrocytoma | – | – |
| 41T | GBM/small-focus GSM | + | – |
| 54T | Ganglioglioma | – | – |
| 61T | GBM | – | – |
| 65T | GBM | – | – |
| 80T | GBM | – | – |
| 161T | Astrocytoma | – | – |
| GS826 | GBM primary culture | + | – |
aa, amino acids. A “+” indicates a positive amplification.
PCR amplification cycles can result in the introduction of errors in amplified sequences. These errors could be mistaken for polymorphisms; hence, an effort was made to ascertain the number of errors generated by PCR amplification of the pp65 coding region using the primer set F2/R2. AD169 and Toledo BACs with published genomic sequences were used as viral DNA templates, and 250 ng of HEL DNA was added to each reaction to mimic tumor tissue conditions. Both a high viral DNA copy number (107) and a low viral DNA copy number (102) were used as a template. For each concentration of template DNA, two reactions were run with individual BACs and two different combinations of AD169 and Toledo BACs at ratios of 1:1 and 9:1, respectively. The combination reactions were set up to validate the ability of the primers to detect mixed infections. At both high and low concentrations of template, no errors were detectable. Mixed infections were detectable at both ratios in the reactions containing 107 copies of viral DNA, while with 102 copies both of the BACs were detectable only at the 1:1 ratio. Taken together, the BAC resequencing data indicated the absence of PCR induced errors and also highlighted the sensitivity of the primers in detecting mixed infections at 9:1 and 1:1 ratios at high copy numbers and at a 1:1 ratio at low copy numbers (data not shown). Thus, we assumed that differences in sequence information obtained from patient samples represent true polymorphisms.
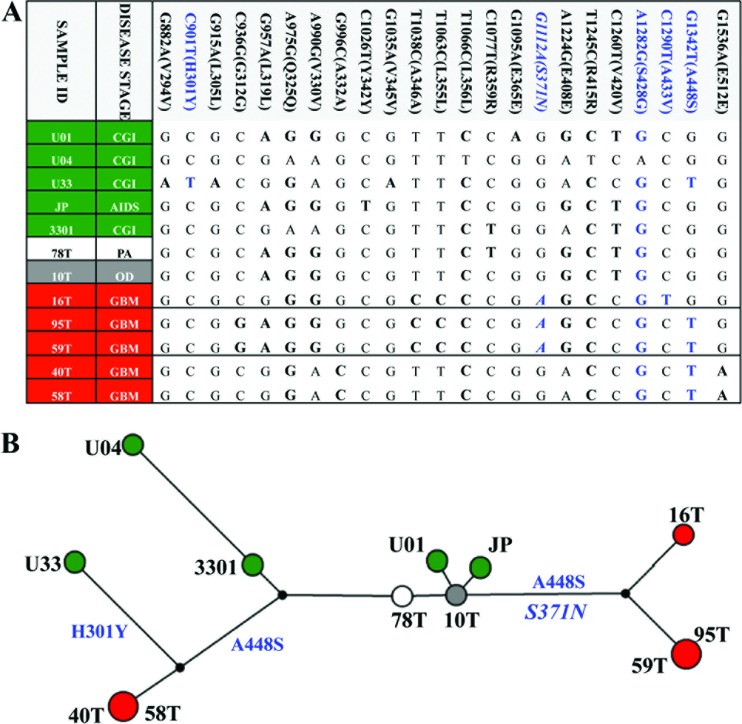
Seven cancer-associated and five noncancerous viral sequences were compared across the 970-bp region of UL83 gene, and 23 polymorphisms were identified. Eighteen of these variations were synonymous, and five were nonsynonymous (Fig. 3A, nonsynonymous variants are indicated in blue). Only one of the nonsynonymous changes, S371N, was found within a known HLA-restricted epitope (B*4006; 364-SEHPTFTSQY-373), and this was observed in two of the three GBM genotypes (25). There was no evidence of clustering among the control specimens, while four of the GBM specimens clustered into two clades, and five cancer-associated genotypes were identified (Fig. 3B). An absence of reticulations was observed which negated the presence of homoplasy or recombination within this region.
Fig 3.
Viral pp65 genotype network of viral genotypes found in cancer specimens. (A) Genotypes were constructed from 23 variations. Nucleotides in boldface are variants relative to the Merlin reference genome, with nonsynonymous variants indicated in blue. Five genotypes were identified, and the sequence of each is shown in the table. An HLA-restricted polymorphism (G1112A) is indicated in italics. CGI, congenitical CMV infection. (B) A genotype network was inferred by the median-joining method with seven cancer-associated sequences using Merlin as the reference sequence. The size of each circle represents the frequencies of each genotype, with each color showing the disease stage of the patient as noted in panel A.
Differences in sequence conservation among the UL, US, and RL regions within a cancer-associated HCMV population.
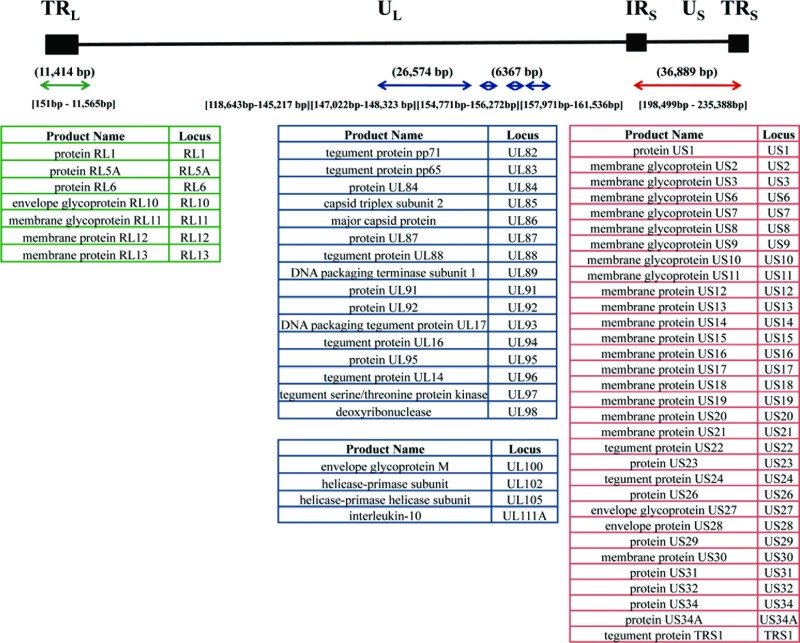
The contribution of HCMV to cancer causation or augmentation remains an open question. We reasoned that insight into the relation between HCMV and cancer can be deduced by analysis of the genomic variability of virus present within a cancer tissue relative to sequences not associated with cancer. In an attempt to understand the intrinsic changes within a glioma-associated viral genome, a patient sample was chosen with the highest viral copy number (58T). Seventy-one pairs of primers were used to amplify 81,244 bp of HCMV genome, which included the RL region (11,414 bp), parts of the UL region (26,574 and 6,367 bp), and the entire US region (36,889 bp) (Fig. 4). These PCR products were then subjected to next-generation (i.e., deep) sequencing because a drawback of traditional Sanger sequencing is the inability to detect minor variants within a sample. Using next-generation sequencing, thousands of minor HCMV sequence variants within the tumor tissue were identified, along with the major genome type or consensus sequence.
Fig 4.
Schematic overview of the HCMV genomic regions included for deep sequencing. The black line is a schematic of the HCMV genome, which is organized into two unique regions—UL (unique long) and US (unique short)—flanked by three repeat regions: TRL, terminal repeat long; IRS, internal repeat short; and TRS, terminal repeat short. The arrows below the line indicate the regions amplified using overlapping primers. All of the nucleotide position numbers are given in reference to the Merlin genome. The numbers above the arrows are the total numbers of base pairs amplified. The numbers below the arrows indicate the nucleotide positions covered. The tables list the ORFs sequenced within each region. The RL region is colored green, the UL region is colored blue, and the US region is colored red.
The cancer-associated sequence data was compared to the HCMV reference sequence (Merlin). The average sequencing depth was 588. A total of 1,983 segregating sites or single nucleotide variants were detected and 93.34% (1,851/1,983) were bi-allelic (two alleles at each nucleotide position), while the rest of the sites were tri-allelic 4.69% (93/1,983) and tetra-allelic 0.71% (14/1,983). The presence of tri-allelic and tetra-allelic sites was much higher than that found within noncancerous human genomes (22) and could be a result of higher mutation rates within the viral genome attributed to the viral DNA polymerase and/or because of the existence of a hypermutable cancer cell milieu.
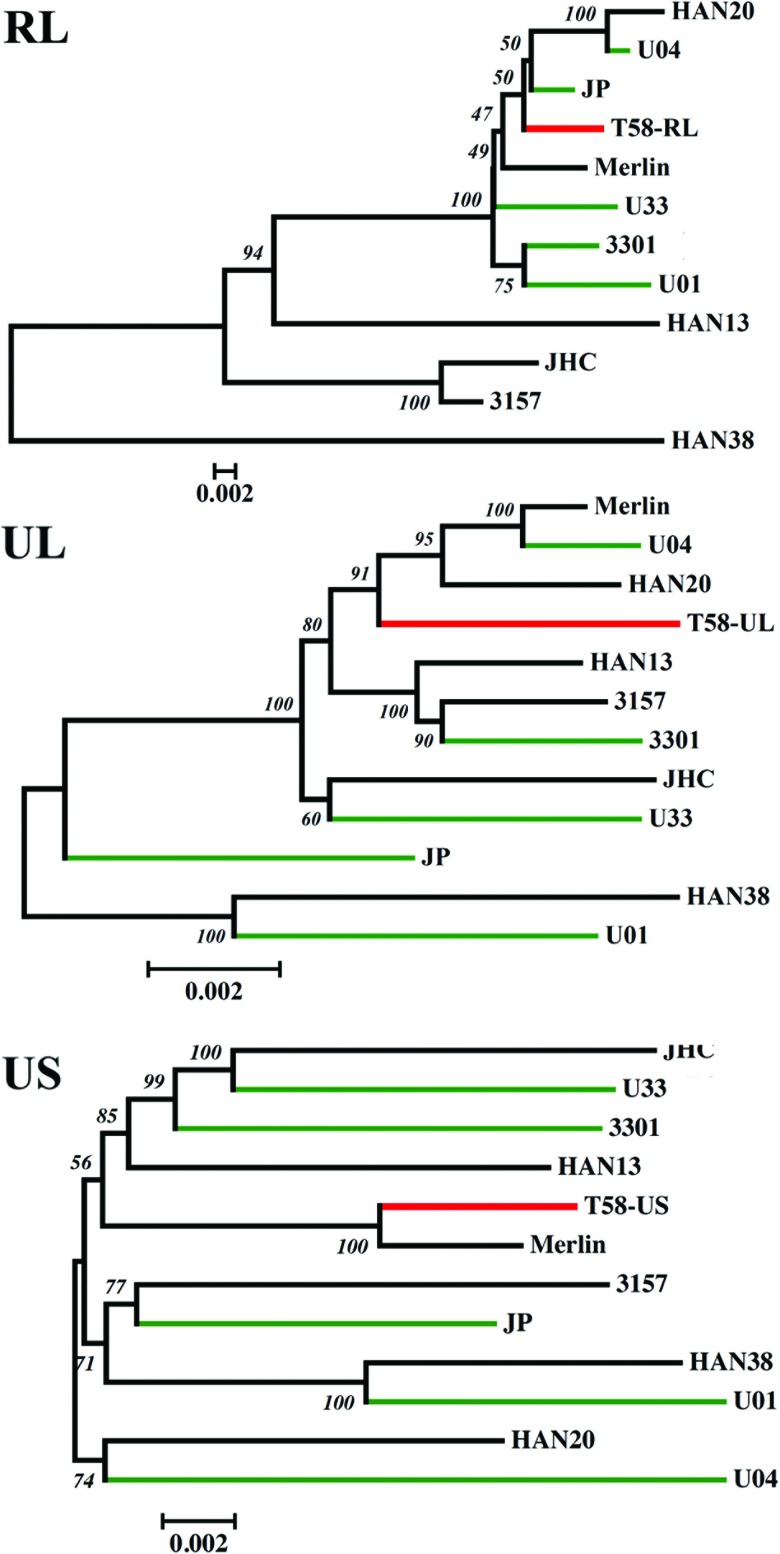
Fifty-six ORFs were completely sequenced, while three additional ORFs (UL100, UL102, and UL105) had incomplete coverage. All of the sequenced ORFs were intact. The major genome type, the consensus cancer-associated genome, was constructed using polymorphisms, which were defined as variants present at a frequency of >0.5. To better understand the taxonomic identity of the cancer-associated viral genome, a phylogenetic analysis was performed with other known minimally passaged and unpassaged strains isolated from productive infections (Fig. 5). There was a lack of congruence in the topologies generated from difference segments of the genome across various strains. The RL region was found to be the most distant among all of the strains, including the cancer-associated genome with a total branch length of 0.245 substitutions per site. The low bootstrap values further highlighted the incongruence of phylogenetic relationships within the RL region, which could be due to higher mutation rates or repeated recombination events. The RL region is still the most variable region among all productively infecting viruses, and it was also found to be most variable in the cancer-associated genome. The US region exhibited intermediate variability, with a total branch length of 0.146 substitutions per site. The UL region was found to be the least variable, with a total branch length of 0.058 substitutions per site. The cancer-associated genome, like all other HCMV strains, had a distinct genome type, but the trend toward maintaining low variability within the UL region, which harbors the core genes, was also evident. This pattern of conservation and the retention of ORFs within the cancer-associated HCMV genome is consistent with the possibility of replication-competent virus within this cancer tissue.
Fig 5.
The evolutionary relationship of the cancer-associated genome was inferred using the neighbor-joining method. Optimal unrooted trees with the sum of branch lengths for RL = 0.24579274, UL = 0.05801844, and US = 0.14616272 are shown. The percentages of replicate trees in which the associated strains clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The trees are drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances are in the units of the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). The analyses involved 12 HCMV genomic sequences. The viral strains with green branches are from congenital infections sequenced previously in the laboratory (44). The 58T viral strain with red branches is the GBM-associated virus.
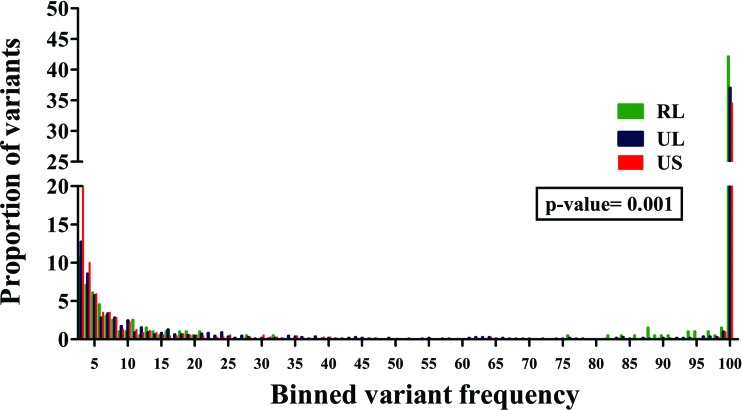
The total viral population present within the tumor tissue was also characterized. The average nucleotide diversity (π) was 0.1%. The nucleotide variations were classified on the basis of frequency (site frequency spectrum) across the three genomic regions sequenced, and the proportion of variations in each frequency class (1 to 100%) were calculated to form a distribution (Fig. 6). A significant difference was found among the three genomic regions (P = 0.001), and the UL and US regions were found to be less variable than the RL region within the population. High-frequency variations (>50%) were also highest in the RL region. None of the nucleotide changes resulted in any premature stop codons, and there was no evidence of any insertion or deletion events. Thus, all of the ORFs analyzed from the viral population of this tumor were found to be intact. The average πAA was lower than the nucleotide diversity at 0.08% and was due to the fact that not all nucleotide changes result in an amino acid change within an ORF. The RL ORFs were found to have the highest πAA (0.1%), followed by the UL ORFs (0.09%), while the US region had the lowest πAA (0.07%), although this difference was not statistically significant (P = 0.611). Thus, at the population level, there was a trend toward less variability within the UL and US regions, while the RL region remained the most variable, as was observed in non-cancer-associated HCMV genomes upon phylogenetic analysis.
Fig 6.
A significant difference was found in the frequency distribution of variants across the different regions of viral genomic sequences within a GBM specimen. A frequency distribution of all of the variants found within the 58T GBM specimen across the RL (green), UL (blue), and US (red) regions. Kruskal-Wallis testing revealed a significant difference among the distributions, with a P value of 0.001.
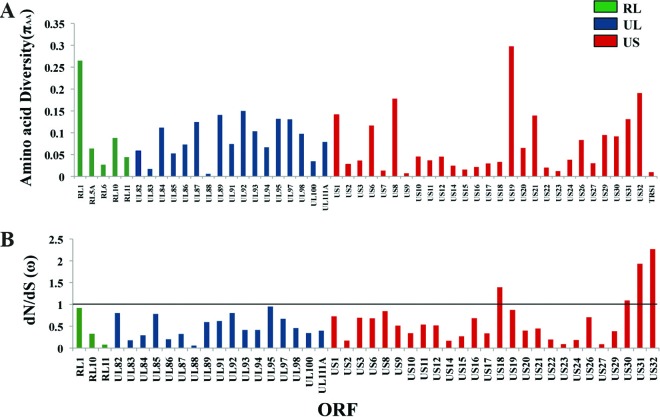
Each of the viral ORFs sequenced from the cancer-associated population was examined to identify targets, if any, of selection or sequence conservation (Fig. 7). All of the ORFs displayed some degree of amino acid variability. The ORFs that exhibited the highest amino acid diversity (πAA) values were RL1 (0.3%) and US19 (0.3%), US32 (0.19%), and US8 (0.18%) (Fig. 7A). On anchoring the nonsynonymous changes with the synonymous changes (dN/dS or ω), the genome-wide average was 0.68, and all but four ORFs had a ω of <1, which is indicative of amino acid conservation or selection. The late gene, US32, whose predicted protein product exhibited a high πAA value, also had the highest ω value of 2.27, followed by US31 (ω = 1.93) and US18 (ω = 1.39) (Fig. 7B). US32 is dispensable for growth in culture (12), but its protein product might be important in vivo, since it has been observed to be immunogenic in an HLA-transgenic mouse model (26). The UL83 ORF, encoding pp65, a part of which had been sequenced from different tumor specimens (Fig. 3A and B), was found to have a low πAA value of 0.02% and an ω value of 0.18 in this GBM specimen, a finding which could be indicative of functional selective constraints.
Fig 7.
Deep sequencing revealed amino acid variability across the viral ORFs within the 58T GBM specimen. (A) Plot showing amino acid diversity (πAA) across different ORFs. (B) Plot of variable dN/dS (ω) values across different ORFs. The RL region is indicated in green, the UL region is indicated in blue, and the US region is indicated in red. The straight line denotes a cutoff of ω = 1 under an assumption of neutrality. ORFs containing only nonsynonymous changes were not included in the dN/dS analysis.
DISCUSSION
A clear understanding of the role of HCMV in malignant gliomas requires a combinatory approach with dissection of viral DNA copy numbers, ORF and protein expression profiles, genomic status, and genome variability found in tumor specimens. This effort was geared at beginning to understand these different aspects of HCMV infection in gliomas.
Limitations of screening strategies.
Other studies have largely relied upon techniques such as immunohistochemistry and in situ hybridizations to ascertain the presence of HCMV in cancer (6, 28, 31, 36, 47). These methods are adept at elucidating the location of viral proteins and RNA within cancer tissues but are not cost-effective and can be prone to user errors (41). DNA remains as the most stable molecule within a cell upon resection of tissues from patients, and thus the efficacy of a sensitive nested PCR approach remains unparalleled for screening purposes. Our nested PCR protocol was able to detect very low levels of viral DNA beyond the limits of quantitative real-time PCR. Therefore, we suggest that a better approach for screening would be to probe for HCMV DNA in tumor tissues using robust amplification strategies.
Variable viral DNA copy numbers.
High viral DNA copy numbers have often been linked to poor prognoses in cancers associated with viral infections (2, 4, 7, 13, 15, 20, 33, 62). Exceptions to this paradigm include infections that result in integrated viral genomes (9). Our attempt at quantifying HCMV DNA copy numbers was successful in only 37.5% of the glioma specimens. This could be attributed to variations at primer/probe binding sites, as well as low viral DNA copy numbers. However, extensive viral DNA copy number variability was observed in the seven tumor specimens analyzed. Why there is such variability in DNA loads is unclear, but it could be due to differences in the various amounts of nontumor tissue affecting the ratio of viral to total DNA. Another possibility is that nontumor cells, such as endothelial cells or tissue macrophages infected with replicating viral genomes, skewed viral DNA loads to the high end of the spectrum for certain tumors. Lastly, we cannot rule out the possibility that viral DNA replication may have occurred in the tumor tissue of some samples studied.
Viral protein expression profile and incongruence with viral DNA copy numbers.
Although viral proteins associated with all phases of productive replication were observed in many tumors, there was no clear correlation between viral DNA copy number and viral protein expression. This incongruence could be indicative of the presence of nonfunctional genomes or persistent infections with limited foci of viral replication. Regardless of the mechanism, HCMV infection in the tumors is atypical given that viral DNA and protein levels do not correlate with what is seen classically in productive and latent infections. The absence of viral protein expression in three tumor tissues despite the presence of readily detectable viral DNA could be an effect of limited sensitivity of the antibodies used or the presence of latent infections within the subset. In contrast, the presence of IE, E, and E/L protein expression in 75% of glioma samples is supportive of replication that is consistent with persistent infections. Further studies need to be performed with larger sample sizes to conclusively define the infection status of HCMV in gliomas.
Understanding the results of the phylogenetic analyses.
Phylogenetic analysis of the 970-bp region of UL83, encoding pp65, revealed the presence of ten distinct genotypes at the nucleotide level with four of these genotypes harboring amino acid changes. Phylogenetically, there was no evidence of clustering among the productively infecting viral genotypes, while the five GBM-associated genotypes clustered into three clades or groups. Amino acid changes were also more prevalent within the GBM specimens, with the exception of the one specimen, U33, which harbored an amino acid change (H301Y). These results raise the possibility that cancer-associated genotypes exist. However, the identification of cancer-associated genotypes does not necessarily mean that cancer-specific genotypes exist in GBM. Expanding from single ORF genotyping to complete genomic analysis and analyzing patient matched samples from tumor and nontumor sources would be the ideal means to understanding the relationship between sequence and disease.
Attempts to sequence entire HCMV genomes in tumor tissues were thwarted by limited tissue amounts and difficulty in amplifying many regions of the viral genome. This latter issue may be due to the absence of these sequences in the tumor analyzed. However, a recent study has provided evidence that most of the HCMV genome is present in GBMs (43). Our data further substantiate the previous finding and expand on it with near-genome-scale sequence analysis of a viral population within tumor tissue. The values for the average π (0.1%) and πAA (0.08%) of the HCMV population in 58T tumor are less than those observed for HCMV populations analyzed from symptomatic congenital infections with π (0.22%) and πAA (0.18%) (44). The presence of low levels of π and πAA could be the result of low population size (63) rather than positive selection events. However, the 58T tumor was large and contained high levels of viral DNA copy numbers among the tumors analyzed but still displayed low nucleotide and amino acid diversity scores. Clearly, further study across a number of patient specimens is warranted to better define the population diversity of HCMV genomes in malignant gliomas.
The presence of many intact ORFs was evident within the cancer-associated viral population. Site frequency spectrum analysis of the cancer-associated viral population within the 58T GBM specimen revealed the RL region to be the most diverse at both the nucleotide and the amino acid levels. In agreement with this finding, phylogenetic analyses also showed the RL region to be the most diverse across the productively infecting and cancer-associated strain. In contrast, the sequenced UL regions, which harbor the core genes, were found to be conserved at the nucleotide and amino acid level within the cancer-associated population, as well as among viruses isolated from productive infections. All but four of the ORFs analyzed had ω values of <1, which is often indicative of sequence conservation. These observations, along with the protein expression profiles, suggest the presence of replication-competent virus within this tumor specimen. However, the lack of detectable IE2 expression in this or any tumor analyzed suggests that there was little or no virus replication, at least at the time of tumor resection. Further efforts should be geared toward expanding this knowledge base across multiple tumor-associated viral genomes to understand whether this genomic signature is the norm or an exception.
Taken together, our results emphasize the existence of HCMV in glioma tissues and the presence of a cascade of viral protein expression typical of replicative virus. There is also evidence of cancer-associated pp65 genotypes. The presence of elevated tri-allelic and tetra-allelic sites in the cancer-associated, HCMV DNA sequences also argues for the presence of distinct cancer-associated mutation rates within the viral genomes in comparison to human genomes (27). The presence of intact ORFs without premature stop codons or insertion-deletion events, although not conclusive, is consistent with an absence of viral integration within tumor tissue. The potential for the presence of replicative virus was further supported at the genomic level within the 58T tumor specimen where the UL region, encoding the core enzymes, was found to be conserved. Individual ORFs were also found to conserve amino acid signatures with ω <1, which is suggestive of functional constraints. Although far from definitive, these data provide an indication for the presence of replication-competent HCMV within malignant glioma tissues.
ACKNOWLEDGMENTS
We thank Trudy Morrison and members of the Kowalik laboratory for research advice and editorial feedback.
This study was supported by the NIH (grants R01AI076189 to T.F.K. and F32AI084437 to N.R.) and the Goldhirsch Foundation (T.F.K.).
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Published ahead of print 11 April 2012
REFERENCES
- 1. Arnoult D, et al. 2004. Cytomegalovirus cell death suppressor vMIA blocks Bax- but not Bak-mediated apoptosis by binding and sequestering Bax at mitochondria. Proc. Natl. Acad. Sci. U. S. A. 101:7988–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asito AS, et al. 2010. Elevated anti-Zta IgG levels and EBV viral load are associated with site of tumor presentation in endemic Burkitt's lymphoma patients: a case control study. Infect. Agents Cancer 5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bandelt H-J, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16:37–48 [DOI] [PubMed] [Google Scholar]
- 4. Boccardo E, Villa LL. 2007. Viral origins of human cancer. Curr. Med. Chem. 14:2526–2539 [DOI] [PubMed] [Google Scholar]
- 5. Cinatl J, Vogel JU, Kotchetkov R, Doerr WH. 2004. Oncomodulatory signals by regulatory proteins encoded by human cytomegalovirus: a novel role for viral infection in tumor progression. FEMS Microbiol. Rev. 28:59–77 [DOI] [PubMed] [Google Scholar]
- 6. Cobbs CS, et al. 2002. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 62:3347–3350 [PubMed] [Google Scholar]
- 7. Cobbs CS, Soroceanu L, Denham S, Zhang W, Kraus MH. 2008. Modulation of oncogenic phenotype in human glioma cells by cytomegalovirus IE1-mediated mitogenicity. Cancer Res. 68:724–730 [DOI] [PubMed] [Google Scholar]
- 8. Crough T, Khanna R. 2009. Immunobiology of human cytomegalovirus: from bench to bedside. Clin. Microbiol. Rev. 22:76–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Das D, Bhattacharjee B, Sen S, Mukhopadhyay I, Sengupta S. 2010. Association of viral load with HPV16 positive cervical cancer pathogenesis: causal relevance in isolates harboring intact viral E2 gene. Virology 402:197–202 [DOI] [PubMed] [Google Scholar]
- 10. Daumas-Duport C, Scheithauer B, O'Fallon J, Kelly P. 1988. Grading of astrocytomas. A simple and reproducible method. Cancer 62:2152–2165 [DOI] [PubMed] [Google Scholar]
- 11. Dolan A, et al. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85:1301–1312 [DOI] [PubMed] [Google Scholar]
- 12. Dunn W, et al. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223–14228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dourmishev LA, Dourmishev AL, Palmeri D, Schwartz RA, Lukac DM. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67:175–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dziurzynski K, et al. 2011. Glioma-associated cytomegalovirus mediates subversion of the monocyte lineage to a tumor propagating phenotype. Clin. Cancer Res. doi:10.1158/1078-0432.CCR-11-0414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fontaine J, et al. 2005. High-level of correlation of human papillomavirus-16 DNA viral load estimates generated by three real-time PCR assays applied on genital specimens. Cancer Epidemiol. Biomarkers Prev. 14:2200–2207 [DOI] [PubMed] [Google Scholar]
- 16. Gallot G, et al. 2001. Purification of Ag-specific T lymphocytes after direct peripheral blood mononuclear cell stimulation followed by CD25 selection. I. Application to CD4+ and CD8+ cytomegalovirus phosphoprotein pp65 epitope determination. J. Immunol. 167:4196–4206 [DOI] [PubMed] [Google Scholar]
- 17. Gibson L, et al. 2004. Human cytomegalovirus proteins pp65 and immediate early protein 1 are common targets for CD8 T-cell responses in children with congenital or postnatal human cytomegalovirus infection. J. Immunol. 172:2256–2264 [DOI] [PubMed] [Google Scholar]
- 18. Gladson C, Prayson LRA, Liu WM. 2010. The pathobiology of glioma tumor. Annu. Rev. Pathol. Mech. Dis. 5:33–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hassan J, Connell J. 2007. Translational mini-review series on infectious disease: congenital cytomegalovirus infection: 50 years on. Clin. Exp. Immunol. 149:205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hsieh P-P, et al. 2007. EBV viral load in tumor tissue is an important prognostic indicator for nasal NK/T-cell lymphoma. Am. J. Clin. Pathol. 128:579–584 [DOI] [PubMed] [Google Scholar]
- 21. Hsu CH, et al. 2004. HCMV IE2-mediated inhibition of HAT activity downregulates p53 function. EMBO J. 23:2269–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hüebner C, Petermann I, Browning BL, Shelling AN, Ferguson LR. 2007. Tri-allelic single nucleotide polymorphisms and genotyping error in genetic epidemiology studies: MDR1 (ABCB1) G2677/T/A as an example. Cancer Epidemiol. Biomarkers Prev. 16:1185–1192 [DOI] [PubMed] [Google Scholar]
- 23. Kern F, et al. 2002. Cytomegalovirus (CMV) phosphoprotein 65 makes a large contribution to shaping the T cell repertoire in CMV-exposed individuals. J. Infect. Dis. 185:1709–1716 [DOI] [PubMed] [Google Scholar]
- 24. Khan N, Cobbold M, Keenan R, Moss PAH. 2002. Comparative analysis of CD8+ T cell responses against human cytomegalovirus proteins pp65 and immediate early 1 shows similarities in precursor frequency, oligoclonality, and phenotype. J. Infect. Dis. 185:1025–1034 [DOI] [PubMed] [Google Scholar]
- 25. Kondo E, et al. 2004. Identification of novel CTL epitopes of CMV-pp65 presented by a variety of HLA alleles. Blood 103:630–638 [DOI] [PubMed] [Google Scholar]
- 26. Krishnan A, et al. 2008. A novel approach to evaluate the immunogenicity of viral antigens of clinical importance in HLA transgenic murine models. Immunol. Lett. 120:108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kumar S, Subramanian S. 2002. Mutation rates in mammalian genomes. Proc. Natl. Acad. Sci. U. S. A. 99:803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lau SK, et al. 2005. Lack of association of cytomegalovirus with human brain tumors. Mod. Pathol. 18:838–843 [DOI] [PubMed] [Google Scholar]
- 29. Lehrer S, et al. 2011. No circulating cytomegalovirus in five patients with glioblastoma multiforme. Anticancer Res. 31:959–960 [PubMed] [Google Scholar]
- 30. Li H, Ruan J, Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lucas K, Bao GL, Bruggeman R, Dunham K, Specht C. 2011. The detection of CMV pp65 and IE1 in glioblastoma multiforme. J. Neurooncol. 103:231–238 [DOI] [PubMed] [Google Scholar]
- 32. Luo MH, Rosenke K, Czornak K, Fortunato EA. 2007. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J. Virol. 81:1934–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martin D, Gutkind JS. 2008. Human tumor-associated viruses and new insights into the molecular mechanisms of cancer. Oncogene 27(Suppl 2):S31–S42 [DOI] [PubMed] [Google Scholar]
- 34. Maussang D, et al. 2006. Human cytomegalovirus-encoded chemokine receptor US28 promotes tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 103:13068–13073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Michaelis M, Doerr HW, Cinatl J., Jr 2009. The story of human cytomegalovirus and cancer: increasing evidence and open questions. Neoplasia 11:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mitchell DA, et al. 2007. Sensitive detection of human cytomegalovirus in tumors and peripheral blood of patients diagnosed with glioblastoma. Neuro Oncol. 10:10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mocarski ES, Shenk T, Pass RF. 2007. Cytomegalovirus, p 2701–2772 In Knipe DM, et al. (ed), Fields virology. Lippincott/The Williams & Wilkins Co, Philadelphia, PA [Google Scholar]
- 38. Murphy E, et al. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 100:14976–14981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nei M, Li WH. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U. S. A. 76:5269–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418–426 [DOI] [PubMed] [Google Scholar]
- 41. Poltermann BS, et al. 2006. Lack of association of herpesviruses with brain tumors. J. Neurovirol. 12:90–99 [DOI] [PubMed] [Google Scholar]
- 42. Prins RM, Cloughesy TF, Liau LM. 2008. Cytomegalovirus immunity after vaccination with autologous glioblastoma lysate. N. Engl. J. Med. 359:539–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ranganathan P, Clark PA, Kuo JS, Salamat MS, Kalejta RF. 2012. Significant association of multiple human cytomegalovirus genomic Loci with glioblastoma multiforme samples. J. Virol. 86:854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Renzette N, Bhattacharjee B, Jensen JD, Gibson L, Kowalik TF. 2011. Extensive genome-wide variability of human cytomegalovirus in congenitally infected infants. PLoS Pathog. 7:e1001344 doi:10.1371/journal.ppat.1001344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sabatier J, et al. 2005. Detection of human cytomegalovirus genome and gene products in central nervous system tumours. Br. J. Cancer 92:747–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 47. Scheurer ME, Bondy ML, Aldape KD, Albrecht T, El-Zein R. 2008. Detection of human cytomegalovirus in different histological types of gliomas. Acta Neuropathol. 116:79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sinzger C, Digel M, Jahn G. 2008. Cytomegalovirus cell tropism. Curr. Top. Microbiol. Immunol. 325:63–83 [DOI] [PubMed] [Google Scholar]
- 49. Slezak SL, et al. 2007. CMV pp65 and IE-1 T cell epitopes recognized by healthy subjects. J. Translational Med. 5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Slinger E, et al. 2010. HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6 STAT3 axis. Sci. Signal. 3:1–10 [DOI] [PubMed] [Google Scholar]
- 51. Soroceanu L. 2008. HCMV microinfections in inflammatory diseases and cancer. J. Clin. Virol. 41:218–223 [DOI] [PubMed] [Google Scholar]
- 52. Soroceanu L, Cobbs CS. 2010. Is HCMV a tumor promoter? Virus Res. 157:193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Strååt K, et al. 2009. Activation of telomerase by human cytomegalovirus. J. Natl. Cancer Inst. 101:441–443 [DOI] [PubMed] [Google Scholar]
- 54. Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030–11035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. c18 doi:10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Terrasson J, et al. 2005. p73-dependent apoptosis through death receptor: impairment by human cytomegalovirus infection. Cancer Res. 65:2787–2794 [DOI] [PubMed] [Google Scholar]
- 57. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wei J, et al. 2010. Glioma-associated cancer-initiating cells induce immunosuppression. Clin. Cancer Res. 16:461–473 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59. Wei J, et al. 2011. Hypoxia potentiates glioma-mediated immunosuppression. PLoS One 6:e16195 doi:10.1371/journal.pone.0016195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wen PY, Kesari S. 2008. Malignant gliomas in adults. N. Engl. J. Med. 359:492–507 [DOI] [PubMed] [Google Scholar]
- 61. Wills MR, et al. 1996. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J. Virol. 70:7569–7579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ylitalo N, et al. 2000. Consistent high viral load of human papillomavirus 16 and risk of cervical carcinoma in situ: a nested case-control study. Lancet 355:2194–2198 [DOI] [PubMed] [Google Scholar]
- 63. Yu N, et al. 2003. Low nucleotide diversity in chimpanzees and bonobos. Genetics 164:1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]