Abstract
The ectodomain of the gD protein of herpes simplex viruses (HSVs) plays an important role in viral entry by binding to specific cellular coreceptors and mediating viral entry to the host cells. In the present study, we isolated RNA aptamers (aptamer-1 and aptamer-5) that specifically bind to the gD protein of HSV-1 with high affinity and are able to discriminate the gD protein of a different virus, HSV-2. Aptamer-1 efficiently interfered with the interaction between the gD protein and the HSV-1 target cell receptor (HVEM) in a dose-dependent manner. The 50% effective concentration (EC50) of aptamer-1 was estimated to be in the nanomolar range (60 nM). Furthermore, aptamer-1 was analyzed for anti-HSV-1 activity by using plaque assays, and it efficiently inhibited viral entry with an estimated Ki of 0.8 μM. To expand the future applications of aptamer-1, a shorter variant was designed by using both mapping and boundary analyses, resulting in the mini-1 aptamer (44-mer). Compared to the full-length aptamer, mini-1 had at least as high an affinity, specificity, and ability to interfere with gD-HVEM interactions. These studies suggest that the mini-1 aptamer could be explored further as an anti-HSV-1 topical therapy designed to prevent the risk of acquiring HSV-1 infection through physical contact.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) and HSV-2 are widespread human pathogens that infect primarily epithelial tissues before spreading to the nervous system, where they become latent. The genomes of HSV-1 and HSV-2 share 50 to 70% homology, and the two viruses also share several cross-reactive epitopes. The entry of HSV into the host cell begins with the binding of viral proteins gC and gB to proteoglycans on the host cell surface. This binding interaction is then followed by the coordinated action of four essential glycoproteins: gD, gB, gH, and gL (3, 30). Of these four glycoproteins, the gD protein is required for binding to the specific cellular receptors for viral entry. The gD protein recognizes two protein receptors, herpesvirus entry mediator (HVEM) (34) and nectin-1 (25). In addition to these receptor proteins, HSV-1 entry can also be mediated by 3-O-sulfated-modified heparan sulfate (29). The gD proteins of HSV-1 and HSV-2 are highly homologous proteins, with a sequence identity of around 86% within the ectodomain region (amino acids [aa] 1 to 319).
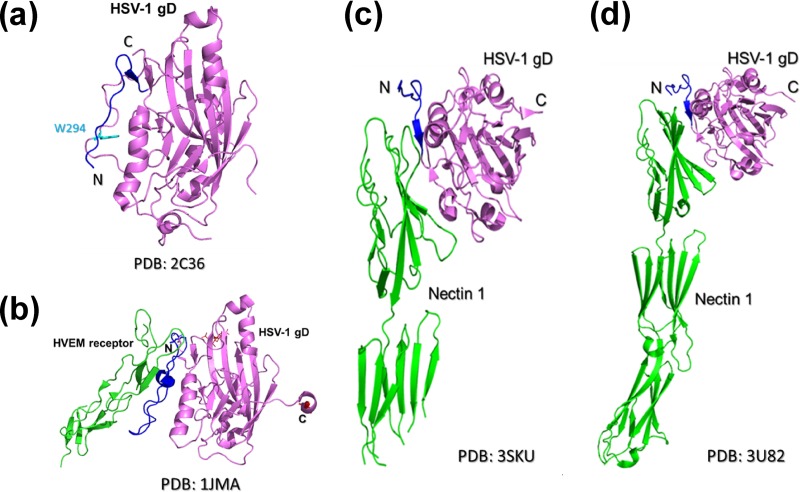
The structures of the gD protein of HSV-1 have been solved, both in the absence (Fig. 1A) (4) and in the presence (Fig. 1B) (20) of its cognate receptor, HVEM. Recently, the structure of gD bound to another receptor, nectin-1, was also determined by two groups (Fig. 1C and d) (8, 35). Those studies revealed that gD contains a V-like Ig fold between residues 56 and 184. This structural feature is commonly found in cell surface adhesion molecules. N- and C-terminal extensions flank this fold and are unfolded and disordered in the absence of the gD receptor. However, in the HVEM-bound form, the N terminus of gD folds into a hairpin structure that binds to HVEM (4). This receptor-induced conformational change is suggested to be required for the initiation of the viral entry mechanism. The C terminus of the gD ectodomain (residues 260 to 316) also plays an important role in HSV entry. Carfi et al. and Krummenacher et al. (4, 20) reported previously that the C terminus of the gD ectodomain is a flexible structural element that interferes with receptor binding. In the unliganded form of gD, the C terminus folds around the core of the protein toward the N terminus, and Trp294 anchors into a groove in the core. When HVEM is bound to gD, the C terminus moves away from the core, and the exposed N terminus folds into a hairpin to form an interface with HVEM. In contrast, when nectin-1 is bound to gD, the C terminus of gD moves away from the core, and Phe129 of nectin-1 replaces Trp294 of gD in the core groove of gD. Those authors postulated that gD Trp294 plays a key role in the controlled displacement of the gD C terminus upon receptor binding. This process is thought to be an essential feature of HSV entry, which then ensures the timely activation of membrane fusion (4). Thus, those studies suggested that both the N- and C-terminal residues of the gD protein play important roles in receptor binding and the activation of membrane fusion for viral entry, respectively.
Fig 1.
Crystal structures of the HSV-1 gD protein in free and complex forms. (a) Crystal structure of gD in the absence of receptor. The Trp294 residue of the gD protein is shown as cyan sticks. (b) Complex crystal structure of the gD-HVEM interaction. HSV-1 gD is shown in purple, its N-terminal residues (aa 1 to 37) are shown in blue, and HVEM receptor is shown green. (c and d) Complex crystal structure of the gD–nectin-1 interaction reported previously by two groups (8, 35). HSV-1 gD is shown in purple, its N-terminal residues (aa 1 to 37) are shown in blue, and the nectin-1 receptor is shown in green. N and C indicate the N-terminal and C-terminal ends of HSV-1 gD, respectively.
Aptamers are rare, functionally active molecules that are selected from a library of nucleic acids by iterative rounds of selection and amplification in a process called “SELEX” (systematic evolution of ligands and exponential enrichment). In the past, aptamers were explored on the diagnostic front, and they displayed a high discriminating ability with a high affinity for their cognate ligands. These aptamers can bind to a wide range of molecules, from simple ions to complex targets, with affinities that are at least as strong as the affinities of antibodies for cognate antigens (5, 17, 33). Several aptamers that target viral proteins have been developed, including human immunodeficiency virus type 1 proteins (Tat, Rev, reverse transcriptase, nucleocapsid protein, gp120, Gag, and integrase), human T-cell leukemia virus type 1 proteins (Rex and Tax), hepatitis B virus, hepatitis C virus proteins (internal ribosome entry site [IRES], NS3, and RNA polymerase), feline immunodeficiency virus (reverse transcriptase), whole human influenza virus, cytomegalovirus, and Rous sarcoma virus (see reference 12 and references therein). These aptamers specifically inhibit their target's functions both in vitro and in vivo (12). Previously, we selected aptamers that bound to the hemagglutinin (HA) of influenza A (H3N2) and B viruses with high specificity and affinity. These aptamers efficiently inhibited membrane fusion when challenged in vitro with whole viruses (14, 15). Interestingly, the anti-influenza A virus aptamer displayed a high level of molecular discrimination of closely related HA proteins derived from other strains within the same subtype of influenza viruses (14).
Because gD is required for viral entry to bind to specific cellular receptors, we explored an in vitro selection strategy to find specific, high-affinity aptamers that bind to the gD protein of HSV-1 from a completely random pool of nucleic acids. After completing eight rounds of selection and amplification cycles, under varied conditions for each selection cycle, we found that a gD binding population of aptamers was enriched. Upon sequencing the population, we found that the aptamer-1 and aptamer-5 clones were predominantly represented in the pool (30% and 23% frequencies, respectively). These two aptamers bind with a high affinity to the gD protein of HSV-1, with equilibrium dissociation constants (KD) of 109 and 39 nM, respectively. These aptamers displayed the ability to bind to the gD protein of HSV-1 and to discriminate between the gDs of HSV-1 and HSV-2. To improve the stability of the aptamers against endoribonucleases, we examined the incorporation of 2′-fluoro pyrimidines. After this incorporation, the affinity of aptamer-1 for gD was reduced marginally, but the affinity of aptamer-5 remained unchanged. To test whether aptamer-1 had the ability to inhibit the functions of gD, such as interacting with the target cell receptor (HVEM), we used a surface plasmon resonance (SPR)-based approach and found that aptamer-1 was able to efficiently interfere with the gD-HVEM interaction. Aptamer-1 was further analyzed for anti-HSV-1 activity by using plaque assays. The aptamer displayed anti-HSV-1 activity with a Ki value of approximately 0.8 μM. Because aptamer-1 exhibited anti-HSV-1 activity, we used mapping and boundary analyses to define the functional regions of aptamer-1 and to design an optimal length for the aptamer. A shorter aptamer, mini-1 (44-mer), efficiently bound to gD, discriminated between HSV-1 gD and HSV-2 gD, and interfered with the gD-HVEM interaction. Taken together, these findings revealed that the shorter aptamer, mini-1, was useful for blocking the gD functions of HSV-1 before the virus attached to the cognate receptors on target cells. In addition, mini-1 could also be useful in a medical diagnostic test to discriminate HSV-1 infection from HSV-2 infection.
MATERIALS AND METHODS
Viral proteins.
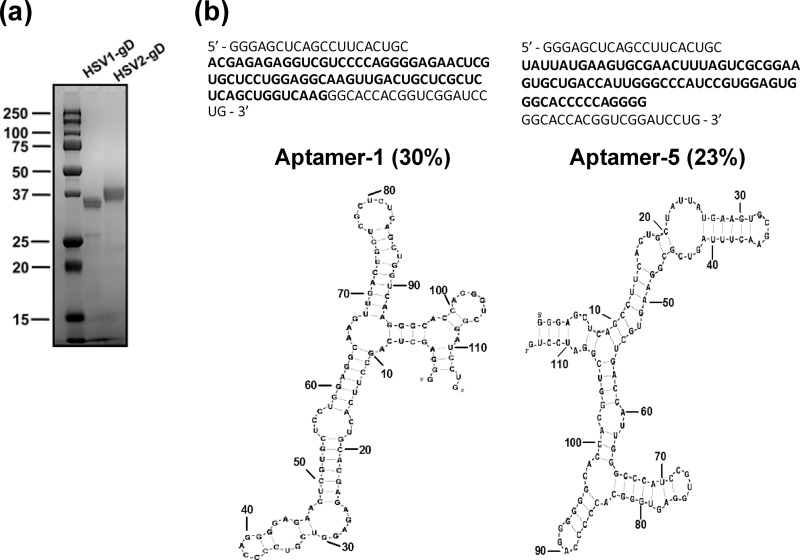
Recombinant HSV-1 gD prepared from Pichia pastoris, containing amino acids 1 to 319, was obtained from either Cortex Biochem or Vybion Inc. (New York, NY) or was expressed as a C-terminal His tag fusion protein (1 to 285 amino acids) in baculovirus, as described previously by Zhang et al. (35). We purchased the purified HSV-2 gD protein from Vybion Inc. (New York, NY) (Fig. 2A). This protein was glycosylated as it was expressed in Pichia pastoris. Both gD proteins derived from HSV-1 and HSV-2 used for the filter binding analyses were glycosylated and expressed in Pichia pastoris. A C-terminal His tag fusion protein (amino acids 1 to 285) of HSV-1 gD (expressed in baculovirus cells) was used for the gD binding analyses with HVEM and nectin-1 interactions (Fig. 2A). Both HVEM and nectin-1 proteins were purchased from Sino Biological Inc. (Beijing, China). HVEM and nectin-1 were expressed in human cells as fusion proteins with the Fc region of human IgG and a polyhistidine tag, respectively, at the C-terminal end.
Fig 2.
Purified gD proteins of HSV-1 and HSV-2 and in vitro-selected aptamers. (a) Representative Coomassie blue-stained PAGE gel showing gD proteins of HSV-1 (amino acids 1 to 285) and HSV-2 (amino acids 1 to 319). (b) Oligonucleotide sequences of the predominant aptamers found in the selected pool of cycle 8. The predicted secondary structures of aptamer-1 and aptamer-5 are presented (BayesFold, version 1.01). Boldface type indicates the randomized region of the RNA.
Design of the random RNA pool.
SELEX libraries were designed to contain a central domain of randomized sequences flanked by invariable 5′ and 3′ sequences. The single-stranded DNA (ssDNA) library sequence TCTAATACGACTCACTATAGGAGCTCAGCCTTCACTGC-N74-GGC ACCACGGTCGGATCCTG, with 74-nucleotide contiguous random sequences, was synthesized and flanked by defined sequences, including the T7 promoter (in italics). The 5′ and 3′ defined sequences were 5′-TCTAATACGACTCACTATAGGAGCTCAGCCTTCACTGC-3′ and 5′-CAGGATCCGACCGTGGTGCC-3′, respectively. PCR was performed with the random DNA library (1 × 1014 molecules) with 0.25 μM each 5′ and 3′ primers. To preserve the abundance of the original library, the PCR was limited to eight cycles to avoid amplifying a skewed population from the random DNA library. The DNA library was converted into an RNA library by in vitro transcription using a T7 Ampliscribe kit (Epicentre Technologies). This RNA library was used for selection.
In vitro selection procedure.
Selection was performed by using 15 μg (∼1014 RNA molecules) of the original RNA library in RNA binding buffer (50 mM Tris-HCl, 50 mM KCl [pH 7.5]) at a 2:1 molar ratio of RNA to protein. During the later selection cycles, the protein concentration was reduced to select high-affinity RNA molecules. The RNA was briefly heated to 95°C and then cooled to room temperature to form stable structures before the selection steps. In the selection cycles, tRNA (total tRNA from Escherichia coli; Boehringer-Mannheim) was used as a nonspecific competitor. After the RNA was incubated with the target protein at room temperature for 10 min, the protein-RNA complexes were filtered through a prewetted nitrocellulose acetate filter (HAWP filter, 0.45 μm and 13.0 mm; Millipore) fitted in a “pop-top” filter holder (Nuclepore) and washed with 1 ml binding buffer. The RNAs that were retained on the filter were eluted and recovered as previously described (14, 15). The recovered RNAs were reverse transcribed in 20 μl of a reaction mixture containing 50 mM Tris-HCl (pH 8.0), 40 mM KCl, 6 mM MgCl2, 0.4 mM deoxynucleoside triphosphates (dNTPs), 2.5 μM primer, and 5 U avian myoblastosis virus (AMV) reverse transcriptase (Seikagaku). The nucleotides and enzymes were added after a denaturation-and-annealing step (2 min at 90°C followed by incubation at room temperature for 10 min). Reverse transcription (RT) was performed for 1 h at 42°C. The resulting cDNA was amplified by PCR and used as the template to obtain RNA for the next round of selection. For the amplification of the cDNA by PCR, a 20-μl aliquot of the reverse transcription reaction mixture (cDNA reaction mixture) was diluted in 80 μl of a PCR mixture (10 mM Tris-HCl [pH 8.8], 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 5 U of Taq DNA polymerase [TaKaRa], and 0.4 μM each primer). The reaction mixture was amplified under the following conditions: 94°C for 70 s, 55°C for 50 s, and 72°C for 70 s. This program was repeated for as many cycles as needed to produce a DNA band of the correct size. The PCR product was precipitated in ethanol and used for transcription. In vitro transcription was performed at 37°C for 3 h with a T7 Ampliscribe kit (Epicentre Technologies). After treatment with DNase I, the reaction mixture was fractionated on an 8% denaturing polyacrylamide gel. The RNA was extracted from the gel, quantitated, and used for the next cycle of selection and amplification. We manipulated each selection cycle to ensure the specificity and high affinity of the HSV-1 gD binders. For example, we modified the ratio of RNA to gD, competitor concentrations, and buffer volumes. To remove the filter binders, which increased the background, we used Xenobind plates for the third, fifth, seventh, and eighth rounds of selection.
For the Xenobind selections, we initially coated the wells with 5 μg of protein per milliliter of binding buffer and blocked the remaining sites with bovine serum albumin (BSA) (3% stock solution). The wells were then washed and used for selection. In the selection cycle, the RNA pool from the second cycle was denatured at 90°C for 2 min and allowed to cool to room temperature for 10 min to facilitate the equilibration of the different conformers. The pooled RNA (0.25 μM) and tRNAs (40 μM) were then mixed in 100 μl binding buffer, loaded twice into the BSA-coated wells, and incubated for 10 min at room temperature (25°C). The unbound RNAs were then collected and loaded into wells coated with HSV-1 gD. The reaction mixture was incubated in these wells for 10 min. The unbound molecules were discarded, and the wells were washed five times with 300 μl binding buffer. The bound RNAs were recovered by using a boiling 7 M urea solution, precipitated with ethanol and regenerated by RT, PCR, and in vitro transcription.
Analysis of aptamers.
To obtain individual aptamers, the amplified PCR products from cycle 8 were directly ligated into the pCRII vector (Invitrogen) according to the manufacturer's protocol. DNA was isolated from individual clones by the alkaline lysis method and sequenced with a Dye Terminator sequencing kit (Applied Biosystems Inc. [ABI]) on a DNA sequencer (model 373A; ABI). The secondary structures of the aptamers were predicted by the BayesFold program (version 1.01). To evaluate the gD binding of the RNA pools from different selection cycles as well as the individual aptamers, internally labeled RNA was prepared by using 0.5 mCi/ml [α-32P]ATP, and binding studies were performed by using a filter binding assay similar to that previously reported (15, 21, 22). The binding and in vitro transcription conditions were similar to those used for selection, except for the RNA and gD protein concentrations. For this analysis, we used 20 nM labeled RNA and 100 or 200 nM gD protein derived from either HSV-1 or HSV-2, respectively. The RNA-protein complexes retained on the filters were washed with 1 ml binding buffer and air dried, and the radioactivity was quantitated with an image analyzer (BAS2000; Fuji Film). To ensure that the binding was specific, we performed binding assays in the presence of a 10-fold molar excess of tRNA as a nonspecific competitor.
Partial chemical modification of aptamers.
To increase the stability of the aptamer-1 and aptamer-5 RNAs against endoribonucleases, we substituted the pyrimidine bases (U and C residues) with 2′-fluoro derivatives. These modifications are known to stabilize RNA against endoribonucleases (27). The chemical modifications in aptamer-1 and aptamer-5 were introduced by in vitro transcription using a T7 Durascribe kit (Epicentre Technologies) in the presence of four types of nucleotide triphosphates (2′ fluoro [2′F] UTP, 2′F CTP, 2′OH ATP, and 2′OH GTP) on their cognate PCR templates, which were used for the preparation of the unmodified aptamer-1 and aptamer-5 RNAs. In vitro transcription was performed overnight at 37°C, and the transcribed RNAs were purified by using the method described above. The resulting RNAs had the 2′-fluoro modification at all pyrimidine positions.
Analyses of aptamer-gD interactions using SPR analysis.
Aptamer-gD interactions were analyzed by using a BIAcore T100 optical biosensor at 25°C with a dextran-coated sensor chip (GE Healthcare). To determine the binding rate constants of the selected aptamers, we prepared the aptamers with 24-mer poly(A) nucleotides at their 3′ ends, which could anneal to the complementary biotinylated oligo(dT) [5′-biotin-(T)24-3′]. To prepare this RNA with the 3′-end extension, we used two primers: a forward primer similar to that used for selection and a 3′-end primer [5′-(T)24-CAGGATCCGACCGTGGTGCC-3′]. The double-stranded DNA template was generated by PCR and transcribed in vitro as described above. Similarly, the complementary sequences of the selected aptamer were prepared as the negative control by annealing the templates 5′-GGAGCTCAGCCTTCACTGCTGCTCTCTCCAGCAGGGGTCCCCTCTTGAGCACGAGGACCTCCGTTCA-3′ and 5′-CAGGATCCGACCGTGGTGCCGAACTGGTCGACTCTCGCTCGTCAGTTGAACGGAGGTCCTCGTGCT-3′, and the templates were further amplified with the sequences 5′-TCTAATACGACTCACTATAGGAGCTCAGCCTTCACTGC-3′ (the T7 promoter region is in italics) and 5′-(T)24-CAGGATCCGACCGTGGTGCC-3′. Initially, the biotinylated oligo(dT)24 was attached to streptavidin (SA chip; BIAcore) by dissolving the oligonucleotide in binding buffer (5 μM final concentration). The excess or unbound biotinylated oligonucleotide was washed away with binding buffer at a flow rate of 20 μl/min for 10 min. To analyze the binding kinetics, 20 μl aptamer was injected (100 nM final concentration) at a flow rate of 2 μl/min for 10 min. This resulted in an increase of approximately 1,200 response units (RU) upon aptamer binding to the complementary biotinylated oligonucleotide. Single-cycle kinetics were measured with various concentrations of the gD protein (10, 20, 40, 80, and 160 nM) by injecting the protein through both the experimental flow cell (flow cell 2 [FC2]) and the control flow cell (FC1) at a flow rate of 30 μl/min and with a total volume of 60 μl. The dissociation time was set for 2 min to obtain a smooth curve. Sensorgrams were corrected for nonspecific binding by subtracting the aptamer-attached curve (FC2) by the control curve (FC1), and the binding kinetics were evaluated. The sensor chip was then washed with a buffer solution, followed by the injection of 10 mM NaOH solution to strip the aptamer from the biotinylated oligo(dT)24 before the next sample analysis.
Analyses of gD-HVEM and gD–nectin-1 interactions in the presence of a selected aptamer.
To evaluate the ability of the aptamers to interfere with gD-receptor (HVEM) interactions, we developed SPR-based analyses. Initially, 100 nM HVEM was immobilized on an activated CM5 chip through an amino-coupling reaction, followed by the blocking of the unreacted carboxyl groups with 1 M ethanolamine-HCl (pH 8.5) for 7 min. The immobilized surface was washed with binding buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, and 0.005% polysorbate 20) before the HVEM and gD interactions were analyzed. Flow cell 1 was used as a control surface, and it was activated and quenched as described above for the experimental flow cell except without the addition of protein. To obtain the initial binding response, we injected 100 nM gD protein into flow cells 1 and 2. A response curve (indicating HVEM-gD interactions) was initially observed. To determine the effect of the aptamers on the HVEM-gD interactions, 100 nM gD protein was incubated with different concentrations of aptamers (0, 12.5, 25, 50, 100, and 200 nM) at 25°C for 10 min. The complexes were then injected through both the experimental flow cell (FC2) and the control flow cell (FC1) for 2 min, with a flow rate of 30 μl/min. The dissociation time was set for 2 min. Sensorgrams were corrected for nonspecific binding by subtracting the response obtained from the control flow cell (FC1) from the response obtained from the HVEM-immobilized flow cell (FC2). The sensor chip was then washed with a buffer solution, followed by washing with 0.1% SDS, until the response returned to the baseline value for the next analysis. Whether this regeneration treatment affected HVEM binding to the gD protein was verified further by injecting the gD protein alone at the end of the studies to reconfirm that HVEM maintained its activity for binding to the gD protein. The reduction in responses with increasing concentrations of the aptamer was used to calculate the 50% effective concentrations (EC50s) using Graphpad Prism 5 software, as previously described (16).
Similarly, to evaluate the ability of aptamer-1 to interfere with gD–nectin-1 interactions, 100 nM nectin-1 was immobilized on an activated CM5 chip through an amino-coupling reaction, followed by the blocking of the unreacted carboxyl groups with 1 M ethanolamine-HCl (pH 8.5), as mentioned above. The immobilized surface was washed with binding buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, and 0.005% polysorbate 20) before the nectin-1 and gD interactions were analyzed. Flow cell 1 was used as a control surface, and it was activated and quenched as described above for the experimental flow cell except without the addition of protein. To obtain the initial binding response, we injected 100 nM gD protein into flow cells 1 and 2. A response curve (indicating nectin-1–gD interactions) was initially observed (about 410 RU). To determine the effect of the aptamer-1 on the nectin-1–gD interactions, 100 nM gD protein was incubated with 12.5 nM concentrations of aptamer-1 at 25°C for 10 min. The complex was then injected through both the experimental flow cell (FC2) and the control flow cell (FC1) for 2 min, with a flow rate of 30 μl/min. The dissociation time was set for 2 min. Sensorgrams were corrected for nonspecific binding by subtracting the response obtained from the control flow cell (FC1) from the response obtained from the nectin-1-immobilized flow cell (FC2).
In vitro antiviral assay.
Vero (Africa green monkey kidney) cells were grown in minimal essential medium (MEM) containing 5% fetal bovine serum (FBS). HSV-1 (KOS strain) and HSV-2 (UW268 strain) were propagated by using Vero cells. To assess the cytotoxic effects of aptamers on uninfected cells, sample dilutions were prepared and added to Vero cell monolayers in triplicate. After incubation at 37°C for 72 h, viable cells were counted by using the trypan blue exclusion test, and cytotoxicity was evaluated. Cytotoxicity was determined by calculating the 50% cytotoxic concentration (CC50) from the concentration-response curves. For the antiviral assay, Vero cells were infected with virus at 0.1 PFU per cell for 1 h at room temperature, washed with phosphate-buffered saline, and then incubated at 37°C for 24 h. Aptamers were added during the infection and throughout the incubation period. Virus yields were determined by plaque assays at 1 day postinfection. The 50% inhibitory concentration (IC50) was obtained from the concentration-response curves. The antiviral activities of the aptamers were estimated by the selectivity index calculated from the CC50s and IC50s.
Phosphate modification and interference analysis.
To map the gD binding sites within the selected aptamer, a phosphate modification assay was performed. In vitro-transcribed RNA was treated with calf intestine phosphatase for 1 h at 37°C, extracted with phenol, and precipitated with ethanol. The RNA was labeled with [γ-32P]ATP and T4 polynucleotide kinase, electrophoresed in an 8% polyacrylamide–7 M urea gel, and eluted from the gel. The labeled RNA was allowed to fold into a tertiary structure by heating to 95°C for 2 min and slow cooling to room temperature. The 5′-end-labeled RNA (1,100 kcpm) was dissolved in buffer (20 mM HEPES [pH 8.0], 1 mM EDTA, and 2.5 μg tRNA) and mixed with 5 μl saturated N-nitroso-N-ethyl-urea in ethanol. After modification at 90°C for 2 min, the reaction was stopped by the addition of 15 μg carrier tRNA to the mixture. The sample was then recovered by ethanol precipitation. To allow the formation of the aptamer-gD complex, 20 pmol treated sample was denatured in 25 μl of selection binding buffer at 92°C for 2 min, and it was then allowed to cool at room temperature for 10 min. The complex was separated by passage through a nitrocellulose filter and partially hydrolyzed in a 200-μl solution of 100 mM Tris-HCl (pH 9.0) at 50°C for 5 min. The RNA cleavage products were recovered by ethanol precipitation and loaded onto an 8% polyacrylamide–7 M urea gel. Aliquots of the alkaline hydrolysate of the aptamer and the sample digested with RNase T1 were coelectrophoresed for band identification. The gel was dried and exposed to X-ray film for autoradiography.
Boundary analysis.
To determine the minimal domain of aptamer-1 required for efficient binding to HSV-1 gD, we prepared a shorter version of aptamer-1 (57-mer, between 10 and 64 nucleotides with an extension of the 5′ end with two G residues) and performed boundary analyses using both 3′- and 5′-labeled RNAs. Initially, the PCR product was generated by using full-length aptamer-1 DNA as the template DNA in the presence of forward and reverse primers (5′-AGTAATACGACTCACTATAGGGCCTTCACTGCACGAGAGAGGT-3′ and 5′-GCCTCCAGGAGCACGAGTTC-3′, respectively). The shorter aptamer-1 RNA was synthesized by the in vitro transcription of the PCR templates generated as described above, labeled, and purified in a manner similar to that used in the case of full-length aptamer-1. To generate the different-length RNAs, the labeled RNAs were partially hydrolyzed by incubation with alkaline hydrolysis buffer (Ambion) in the presence of 3 μg yeast RNA at 95°C for 15 min. The reaction mixture was then neutralized with 3 M sodium acetate (pH 5.2) and recovered by ethanol precipitation. These hydrolyzed, labeled RNAs (∼100 kcpm) were incubated with 500 nM HSV-1 gD at room temperature for 10 min in binding buffer. The resulting complexes were captured on a nitrocellulose filter similar to that used for the filter binding assays described above, and the bound RNAs were eluted from the filter by boiling the filter papers at 95°C for 2 min in 200 μl binding buffer containing 7 M urea. The eluted RNAs were recovered by ethanol precipitation. The recovered RNAs were coelectrophoresed on a 10% denaturing gel (polyacrylamide containing 7 M urea) along with alkaline-hydrolyzed and RNase T1 (Ambion)-digested RNAs, which served as markers for the cleaved products. Upon the completion of electrophoresis, the polyacrylamide gel was dried and exposed to X-ray film for autoradiography.
Preparation of the mini-1 aptamer and complementary sequences.
To prepare aptamer mini-1 (44-mer) RNA by in vitro transcription, a PCR product was prepared by using aptamer-1 DNA as the template in the presence of both forward and reverse primers (5′-AGTAATACGACTCACTATAGGGCACGAGAGAGGTCGTCC-3′ and 5′-CCAGGAGCACGAGTTCT-3′, respectively). A second PCR was performed on the same template to extend the length by T24 at the 3′ end using a T extension primer (5′-T24-CCAGGAGCACGAGTTCT-3′). The resulting PCR template was used for in vitro transcription using T7 RNA polymerase. The PCR product was separately purified by denaturing PAGE similar to that used in the case of full-length aptamer-1. The resulting RNA (aptamer mini-1) has rA24 (rAAAAAAAAAAAAAAAAAAAAAAAA) at its 3′ end, which was used to immobilize the aptamers on the SA chip through biotinylated dT24. The mini-1 aptamer was analyzed for binding to HSV-1 gD by using SPR in a manner similar to that used for full-length aptamer-1.
RESULTS
Selection of high-affinity RNA ligands.
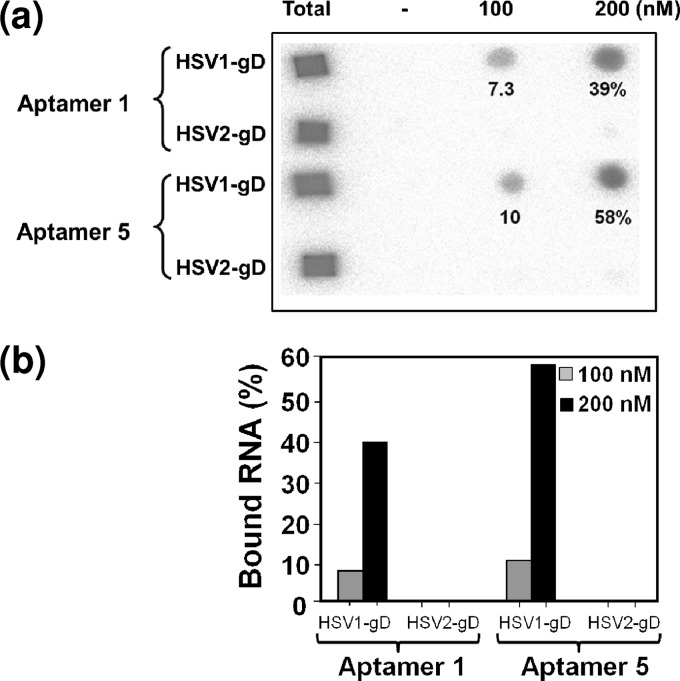
HSV infection requires the binding of the gD protein to the cell surface receptors, and this interaction is essential for receptor binding and the activation of membrane fusion. The selection of an aptamer against the gD protein of human HSV is desirable because this type of specific probe will greatly impact the development of specific diagnostic reagents and inhibitors. Previously, aptamers were demonstrated to have high affinities for several nucleic acid binding proteins. In addition, it has been revealed that aptamers can recognize various epitopes on surface proteins and that aptamers preferentially bind to active sites (15). To select an RNA aptamer against HSV-1 gD, the starting RNA repertoire was randomized over a 74-nucleotide stretch flanked by a fixed 38-nucleotide primer region at the 5′ end and a 20-nucleotide primer region at the 3′ end. This randomized pool contained approximately 1014 units of RNA species under our experimental conditions, and these species were allowed to bind to the target protein. For the initial selection, the RNA pool and gD (3.6 μM) were mixed and incubated for 10 min in the presence of buffering agents, and the free RNA was then separated from the gD-RNA complexes by filtration or by using Xenobind plates under the conditions required for the selection cycle. Bound RNAs were recovered in a hot 7 M urea solution and subsequently amplified by reverse transcription, PCR, and in vitro transcription. To allow more competition between the individual sequences during the selection cycles, an excess ratio of the RNA pool to the target protein was used for the initial selection cycles. For the subsequent selection cycles, the ratio of the gD protein to the RNAs (pool RNA and tRNA) was manipulated to increase the selection stringency (Table 1). In addition, a nonspecific competitor, tRNA, was used at different molar ratios in the selection cycles to avoid the selection of aptamers with nonspecific binding. After eight rounds of selection, both the enrichment of high-affinity gD binding aptamers and the specificity were analyzed by filter binding assays. Approximately 29% of the RNA pool was bound to gD (Table 1). No significant retention of the selected RNA pool occurred on the filter in the absence of gD, indicating that nonspecific filter binders were not coisolated during the course of selection. After the eighth cycle of amplification and selection, PCR-derived molecules from the RNA pool were subcloned and sequenced. Individual aptamers from the eighth cycle were classified based on the obtained sequences. Aptamers representing each group were analyzed directly for HSV-1 gD binding by using a filter binding assay. Upon sequencing, we found that two clones dominated the selected pool: aptamer-1 and aptamer-5. These two major sequences represented population frequencies of 30% and 23%, respectively, and combined, the aptamers represented >50% of the population in the selected pool 8 (Fig. 2B). The primary sequences of each class of aptamer were predicted, and each class assumed different secondary structures according to the BayesFold program (version 1.01) (Fig. 2B). Both of these aptamers were assayed for binding to the HSV-1 gD protein at concentrations of 100 and 200 nM in the presence of a 10-fold molar excess of tRNA to avoid nonspecific interactions with RNA. The filter binding assays showed different bindings for the interactions of gD with aptamer-1 and aptamer-5. The binding values of aptamer-1 and aptamer-5 were 39% and 58%, respectively (Fig. 3).
Table 1.
Selection cycles and binding analysis of HSV-1 gD
| Cycle | Pool RNA concn (μM) | Competitor concnb (μM) | Protein concn (μM) | Filter binding (%)c |
|---|---|---|---|---|
| 1 | 8 | 80 | 3.6 | 0 |
| 2 | 4 | 80 | 3.6 | ND |
| 3a | 0.25 | 40 | 0.2 | ND |
| 4 | 0.5 | 80 | 2.4 | ND |
| 5a | 0.25 | 40 | 0.2 | 5.3 |
| 6 | 0.5 | 80 | 1.2 | ND |
| 7a | 0.125 | 80 | 0.2 | ND |
| 8a | 0.125 | 80 | 0.2 | 29 |
Microtiter plates were used.
tRNA.
ND, not determined.
Fig 3.
Filter binding analyses of the selected aptamers. (a) Filter binding assays were performed with unmodified RNA (2′OH) in the presence of gD proteins (100 and 200 nM) from HSV-1 and HSV-2, and tRNA was used as a nonspecific competitor. (b) The amounts of labeled aptamers (unmodified) retained on the filter in the presence and absence of gD proteins were quantitated with an image analyzer (BAS2000; Fuji Film). The percentages of binding to the gD protein of HSV-1 were plotted.
Binding specificity and rate constants of the selected aptamers.
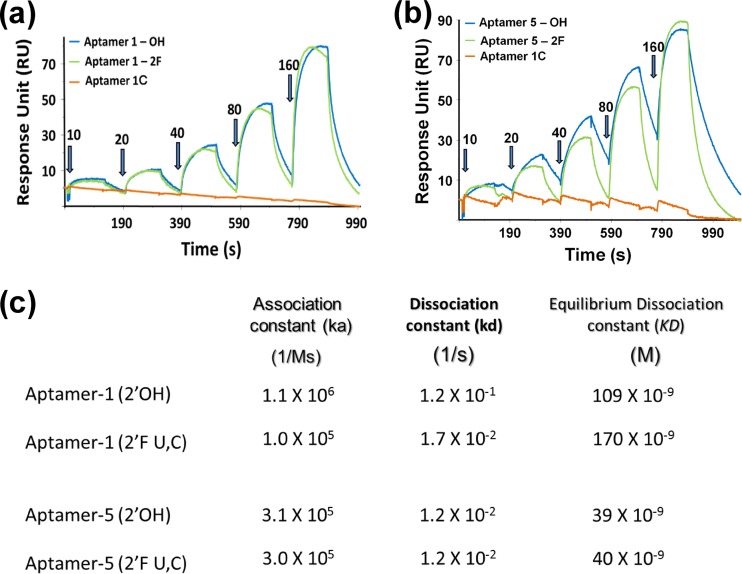
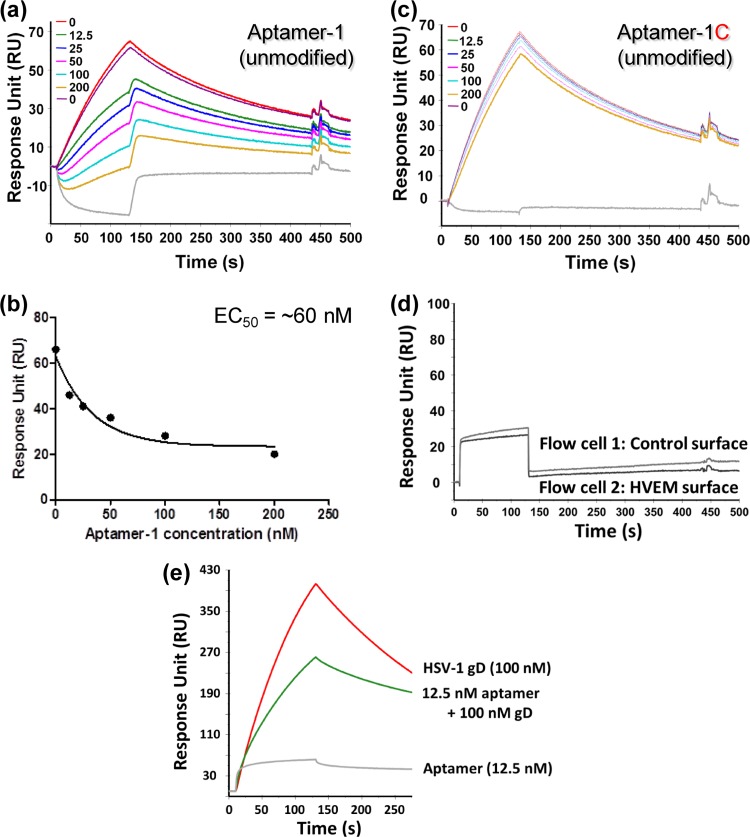
Aptamers that can discriminate between closely related species are extremely useful and aid in the development of specific diagnostic reagents. In the past, several aptamers were reported to have high abilities to discriminate between closely related molecules and species with several-thousandfold discriminatory abilities (10, 13, 17). Because the ectodomains of the gD proteins of HSV-1 and HSV-2 share 86% sequence identity, we were interested in assessing the interactions of the selected aptamers with gDs derived from HSV-1 and HSV-2. For the initial binding analyses of aptamers (aptamer-1 and aptamer-5), a bacterially expressed gD protein of HSV-2 was used. For a better comparison, for the final binding analyses, we used Pichia pastoris-expressed gD of HSV-2, since the gD protein of HSV-1 was also expressed in Pichia pastoris. The filter binding assays clearly indicated that there were no molecules that bound to the gD protein of HSV-2 retained on the filters at the two concentrations tested (Fig. 3A). This result indicated that the selected aptamers were specific for the gD protein of HSV-1 in a concentration-dependent manner, even though the two gD proteins share a high level of amino acid sequence similarity (Fig. 3). Next, to improve the stability of these aptamers against endoribonucleases, we replaced all of the pyrimidine positions with 2′-fluoro cytosine and uridine. These chemical modifications are known to enhance the stability of RNAs against ribonucleases (26). To clearly determine the binding rate constants for aptamer–HSV-1 gD complexes, we exploited a surface plasmon resonance-based analysis using a BIAcore T100 instrument. In this analysis, the use of single-cycle kinetics allowed us to continuously inject increasing concentrations of HSV-1 gD (10 to 160 nM) without regenerating the surface of the sensor chip (Fig. 4A and b). This type of analysis appears to be superior to multi-injection analyses (18, 31). These analyses revealed the equilibrium dissociation rate constants (KD), association rate constants (Ka), and dissociation rate constants (Kd) for the different aptamer-gD complexes (Fig. 4C). Both aptamer-1 and aptamer-5 have affinities for the gD protein of HSV-1 in the nanomolar range. In contrast, the complementary sequence of aptamer-1 (aptamer-1C) had no ability to bind to gD (Fig. 4A and b), suggesting that the sequences of aptamer-1 within the randomized regions are important for binding to the gD protein of HSV-1.
Fig 4.
Determination of kinetic parameters for the interactions of aptamer-1 and aptamer 5 with the cognate gD protein of HSV-1. (a) Single-cycle binding kinetics were measured with aptamer-1 (unmodified), aptamer-1 (chemically modified, 2′F U and C residues), and a complementary aptamer-1 (aptamer-1C) in the presence of various concentrations of the gD protein (10, 20, 40, 80, and 160 nM). (b) Single-cycle binding kinetics were measured with aptamer-5 (unmodified), aptamer-5 (chemically modified, 2′F U and C residues), and a complementary aptamer-1 (aptamer-1C) in the presence of various concentrations of the gD protein (10, 20, 40, 80, and 160 nM). (c) Rate constants (association rate constant [Ka], dissociation rate constant [Kd], and equilibrium dissociation rate constant [KD]) for each aptamer-gD complex are displayed.
Analyses of gD-HVEM interactions in the presence of aptamer-1.
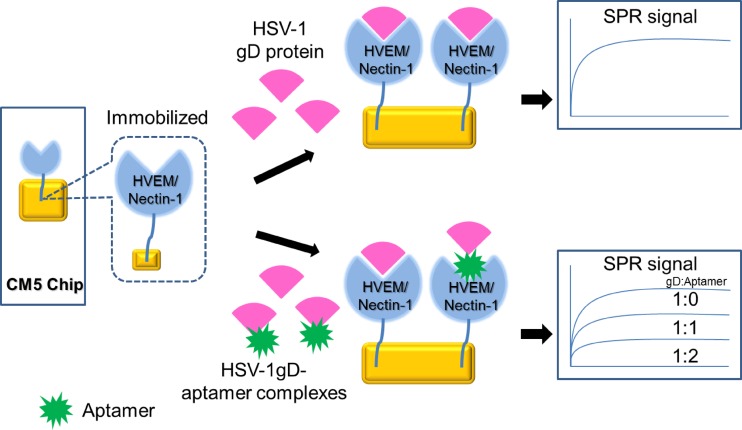
The gD protein is important for binding to specific cellular receptors, such as HVEM (34), nectin-1 (25), and modified heparan sulfate (29). HVEM belongs to the tumor necrosis factor family (6, 7), and nectin-1 is a cell adhesion molecule of the immunoglobulin superfamily (32). gD-receptor interactions have been revealed by using X-ray structural analyses (4, 8, 35). To test whether aptamer-1 has the ability to interfere with the gD-receptor (HVEM) interaction, we established an SPR-based competition assay (Fig. 5). In this assay, HVEM was immobilized by an amino-coupling reaction on a sensor chip (CM5), and 200 nM gD protein from HSV-1 was injected over the immobilized surface in the same flow cell (Fig. 6A). Upon gD binding to HVEM, a response signal was observed (∼70 response units). If aptamer-1 interfered with the gD-HVEM interaction, we assumed that the signal of this response would decrease. As predicted, by increasing the aptamer-1 concentration (12.5 to 200 nM), the signal of gD binding to HVEM decreased in a concentration-dependent manner (Fig. 6A). Using the response signals, an effector concentration of aptamer-1 required for a 50% inhibition (EC50) of HVEM-gD interactions was calculated to be in the nanomolar range (Fig. 6B). 2′F-modified aptamer-1 (2′F-modified U and C residues) gave results similar to those for unmodified aptamer-1 (data not shown). However, aptamer-1C did not interfere with gD-HVEM interactions under similar conditions (Fig. 6C). All the response signals mentioned above were calculated after deducting the aptamer response on the control and HVEM surface (Fig. 6D). Similarly, we tested the interference of aptamer-1 in nectin-1 and gD interactions qualitatively and found that aptamer-1 also interferes efficiently with the gD-nectin interactions (Fig. 6E). These analyses indicated that aptamer-1 efficiently blocks the interaction between the gD protein of HSV-1 and the cognate receptors HVEM and nectin-1, suggesting that aptamer-1 may have potent anti-HSV-1 activity.
Fig 5.
Schematic representation of the experiments used to analyze the ability of the aptamers to interfere with gD-receptor (HVEM) interactions using SPR.
Fig 6.
Ability of aptamer-1 to interfere with gD-HVEM and gD–nectin-1 interactions. (a) gD-HVEM interaction responses observed in the absence of aptamer-1 or with increasing concentrations of aptamer-1 (unmodified) ranging from 12.5 to 200 nM. The gray line represents the injection of 200 nM aptamer alone. (b) The observed responses for each concentration of aptamer-1 were plotted, and the EC50 was estimated. (c) gD-HVEM interaction responses observed with increasing concentrations of aptamer-1C (unmodified). (d) Representative raw sensorgrams observed upon aptamer injection over flow cell 1 (control surface, CM5 chip) and flow cell 2 (HVEM surface). (e) gD–nectin-1 interaction responses observed in the absence and in the presence of aptamer-1.
Aptamer-1 displayed anti-HSV-1 activity.
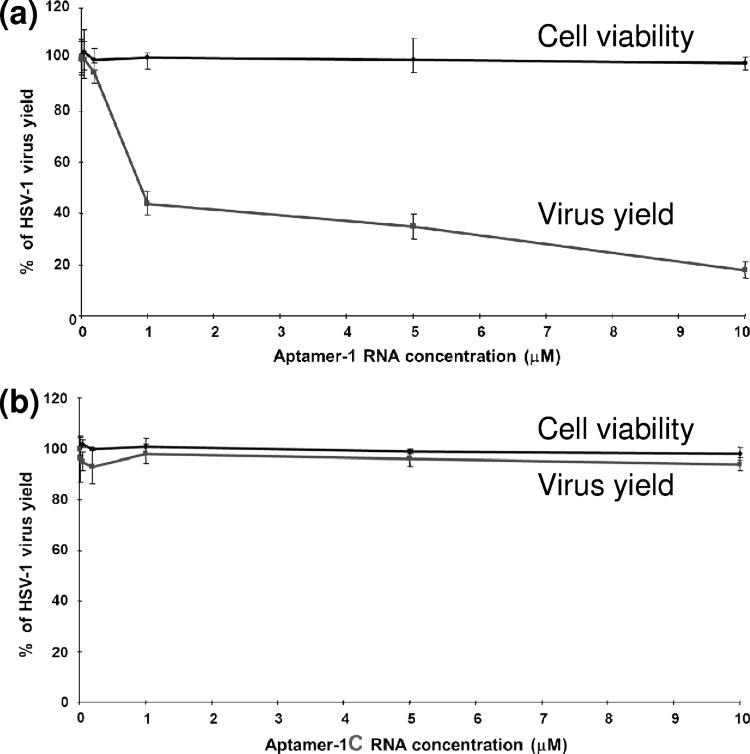
The above-described studies suggested that aptamer-1 has the ability to block gD-HVEM interactions. To determine directly whether aptamer-1 had anti-HSV-1 activity, we performed plaque assays using the KOS strain of HSV-1 in the presence of various concentrations (up to 10 μM) of aptamer-1. These assays were conducted both in the presence and in the absence of 5% fetal bovine serum (FBS), because the aptamer was partially stabilized against endoribonucleases by chemically modifying the RNA (2′F U and C residues). In this assay, we incubated 2′F-modified aptamer-1 with the viruses in binding buffer for 10 min before Vero cells were infected with the treated viruses. Our hypothesis was that if aptamer-1 interfered with HSV-1 viral binding and fusion, we should observe a reduction in the number of plaques with increasing aptamer-1 concentrations. As shown in Fig. 7A, aptamer-1 inhibited HSV-1 plaque production in a concentration-dependent manner. However, aptamer-1C exhibited no anti-HSV-1 activity (Fig. 7B). Based on this analysis, the IC50 of aptamer-1 was calculated to be 0.8 μM. Aptamer-1 had no cytotoxic effects on Vero cells and had a high selective index (>11). Aptamer-1 was also tested for anti-HSV-2 (UW268 strain) activity, and we found that the aptamer failed to possess any anti-HSV-2 activity. This result agrees with our previous binding results showing that aptamer-1 had the ability to bind specifically to the gD protein of HSV-1 and could discriminate between the gD protein derived from HSV-1 and that derived from HSV-2.
Fig 7.
Cytotoxic and anti-HSV-1 effects of aptamer-1. Uninfected and HSV-1-infected Vero cells in MEM containing 5% FBS were incubated for 72 and 24 h, respectively, in the presence of aptamer-1 (a) or aptamer-1C (b). Each value is the mean ± standard deviation (SD) from triplicate assays.
Interference of phosphates in the binding of HSV-1 gD and aptamer.
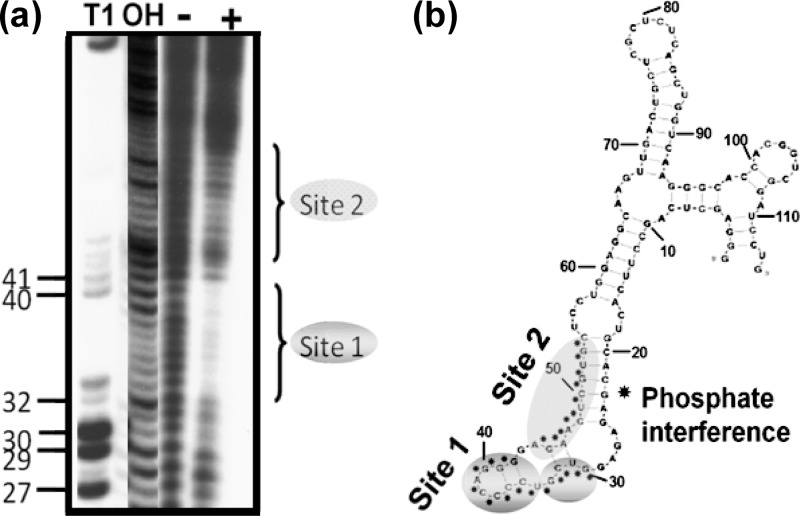
Although our previous experiments suggested that aptamer-1 had the potential to become an anti-HSV-1 drug candidate, aptamer-1 may pose limitations for future applications because of the nucleotide length of the RNA molecule. Previously commercialized and preclinically studied aptamers possess a typical length of between 30 and 40 nucleotides (1, 2). The length of aptamer-1 used in all of the above-described analyses was 113 nucleotides. Therefore, it was important to shorten the length of the aptamer to approximately 40 nucleotides. For this purpose, we undertook mapping studies using previously described modification-interference analyses (14, 15, 19) to determine the gD binding sites within aptamer-1. To identify the important phosphates of aptamer-1, we used an ethylnitrosourea (ENU) modification-and-interference analysis. Initially, aptamer-1 was labeled at the 5′ end and modified under denaturing conditions (92°C for 2 min) with ENU. Under these conditions, ethylnitrosourea modified the phosphates of the RNA at approximately 1 site/molecule. Because ethylnitrosourea reacts with phosphates and generates a phosphotriester in the RNA, the RNA could then be easily hydrolyzed by a mild alkaline treatment. The alkylated RNA was allowed to bind to HSV-1 gD, and the complexed RNAs were separated from the free RNA by filtration. The gD binding RNAs were then cleaved at the modified sites. RNAs retained on the filter were eluted from the nitrocellulose filters and loaded onto an 8% polyacrylamide gel to separate the cleaved products. In this process, the molecules that were modified at phosphates that were necessary for binding to the target were lost, and these important cleaved phosphate regions could be visualized as a footprint on the sequencing gel. Comparisons of the band intensities of the samples of complexed and free RNAs revealed the sites that are important for gD binding. Specifically, the phosphates at positions 29 to 41 were found to interfere strongly with gD binding, suggesting their importance for this interaction. In addition, the phosphate region of positions 44 to 53 was also involved in these interactions (Fig. 8). Although the nucleotide sequences were optimized during the selection process, the phosphates in random regions may also play an important role in efficient gD binding. When the gD binding sites within the predicted secondary structures of aptamer-1 were examined, it appeared that a shorter aptamer (55-mer, between positions 10 and 64) could be sufficient for efficient binding to the gD protein of HSV-1. To test whether this shorter aptamer-1 (55-mer RNA) had the ability to bind to gD, we prepared RNA by in vitro transcription and analyzed gD binding using a filter binding assay. The shorter aptamer-1 (55-mer with two “G” residues at the 5′ end to facilitate efficient transcription by T7 polymerase, resulting in a 57-mer RNA) bound gD as efficiently as full-length aptamer-1 (data not shown).
Fig 8.
Ethylnitrosourea mapping of interfering phosphates of aptamer-1 necessary for HSV-1 gD binding. (a) The labeled ethylnitrosourea-modified aptamer-protein complexes were trapped on a nitrocellulose filter. The filter was treated with a mild alkaline solution. The RNA was recovered and loaded onto the gel. The control RNA markers were prepared by partial nuclease digestion with RNase T1 (lane T1) or alkaline digestion (lane OH). Lane − denotes ENU RNA in the absence of gD. Lane + denotes ENU RNA in the presence of gD. (b) The ENU-mapped positions in the predicted secondary structure of aptamer-1 are shown, and potential interfering phosphates are indicted by asterisks.
Boundary analyses.
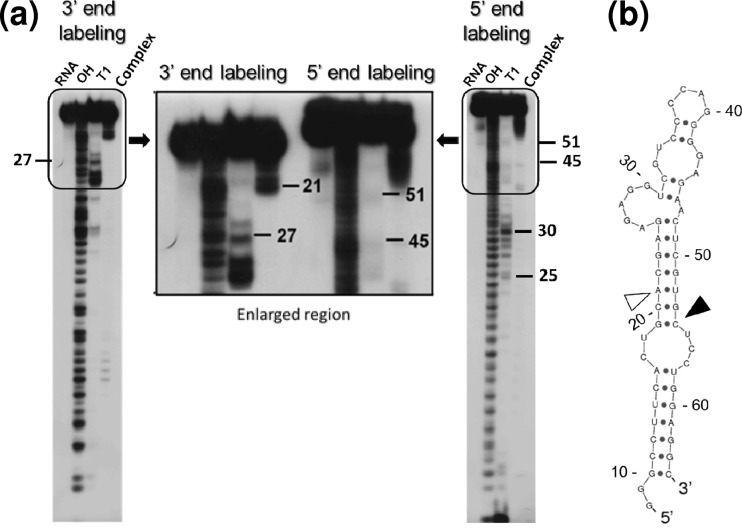
In an attempt to further shorten the length of aptamer-1 to approximately 40 nucleotides, we performed a boundary analysis to define the 3′- and 5′-end boundaries of the shorter aptamer-1 (57-mer) that were necessary for binding to gD. Both 5′- and 3′-end-labeled products of the shorter aptamer-1 (57-mer) were prepared. These labeled RNAs were partially hydrolyzed by alkaline treatment. After recovery from hydrolysis, these RNAs were incubated with HSV-1 gD. These 3′- and 5′-end-labeled RNAs were filtered through a nitrocellulose membrane after binding to the gD protein of HSV-1. In this procedure, fragments that were retained on the filter were assumed to have a desired length of RNA that possesses all of the functional groups required for binding to gD. To identify the positions and lengths of these functional groups, we recovered the RNAs that were retained on the filter after either 3′- or 5′-end labeling and loaded these RNAs onto a denaturing gel. For the 3′-end labeling of the shorter aptamer-1, products of >45 nucleotides (14 nucleotides shorter from the 5′ end) appeared to retain the ability to bind to the gD protein of HSV-1 (Fig. 9A, left). The minimally required length of RNA based on 3′-end labeling is indicated on the secondary structure by an open arrowhead in Fig. 9B. Predictions of the secondary-structure elements, single-stranded and double-stranded regions, were similar using either BayesFold or Sfold (9). In contrast, for the 5′-end labeling of the shorter aptamer-1, products of >47 nucleotides (11 nucleotides shorter from the 3′ end) appeared to retain the ability to bind to the gD protein of HSV-1 (Fig. 9A, right). The minimally required length of RNA based on 5′-end labeling is indicated on the secondary structure by a filled arrowhead in Fig. 9B. The lengths of RNAs recovered from the end-labeled RNAs suggested that RNAs that possessed the sequence between positions 19 and 54 were able to bind gD and be retained on the filter. These findings suggested that this region (nucleotides 19 to 54) is sufficient for binding to the gD protein (Fig. 9). These results agreed with data from the ENU mapping studies that indicated that nucleotides 20 to 53 of aptamer-1 possess the binding sites for HSV-1 gD.
Fig 9.
Boundary analysis of the shorter aptamer-1 (57-mer), labeled at the 3′ and 5′ ends of the RNA and complexed with HSV-1 gD. (a) The labeled alkali-hydrolyzed aptamer-protein complex was trapped on a nitrocellulose filter. The RNA was recovered and loaded onto the gel. The control RNA markers were prepared by partial nuclease digestion with RNase T1 (lane T1) or alkaline digestion (lane OH). (b) The 3′- and 5′-end boundaries that were identified by boundary analyses are indicated by open and closed arrowheads, respectively, on the predicted secondary structure of the shorter aptamer-1 (57-mer). The secondary structure was predicted by using Sfold (9; http://sfold.wadsworth.org/cgi-bin/index.pl).
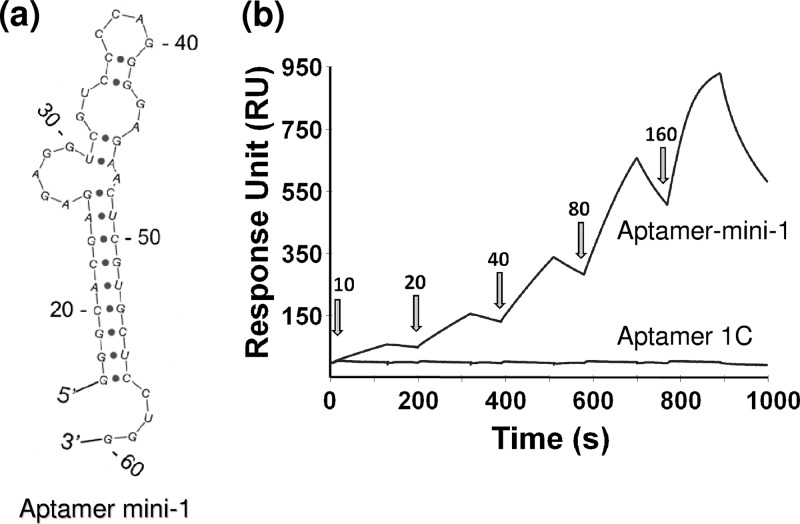
Minimal aptamer-1 that binds efficiently to gD of HSV-1.
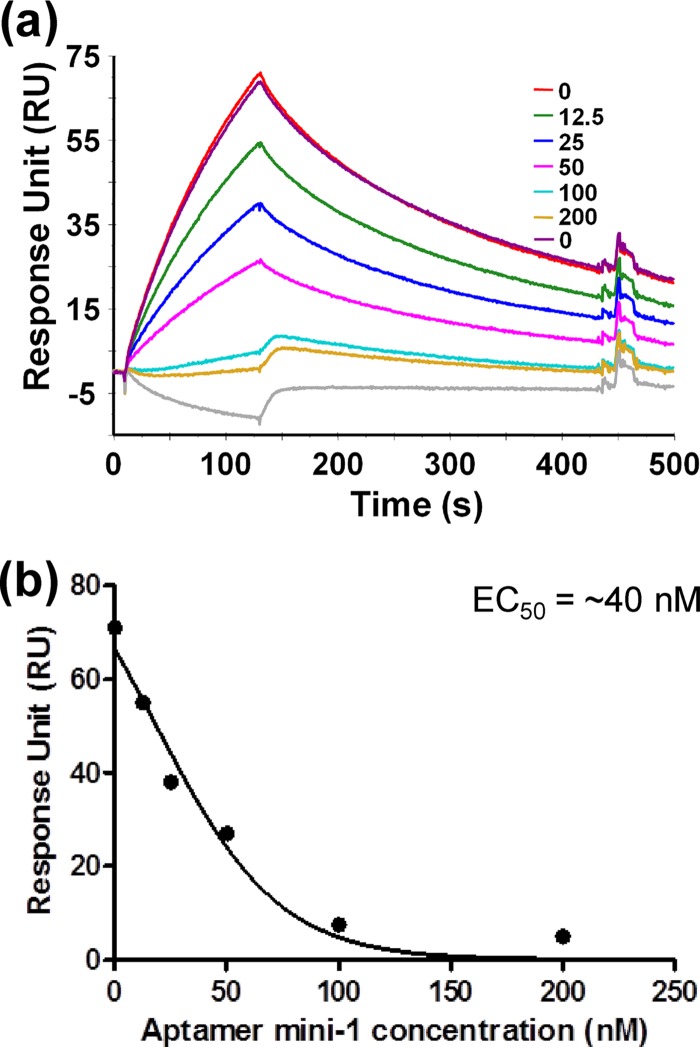
To confirm the finding that the region between positions 19 and 54 was sufficient for aptamer-1 binding to gD, we prepared an even shorter variant, aptamer mini-1 (44-mer, representing nucleotides 20 to 60 of full-length aptamer-1 with the addition of GGG at the 5′ end to facilitate efficient in vitro transcription by T7 polymerase) (Fig. 10A). The secondary structure of aptamer mini-1 was predicted by using the Sfold program (9). We analyzed the binding of aptamer mini-1 to HSV-1 gD using an SPR-based analysis (Fig. 10B). SPR-based analyses were performed in a manner similar to that used for full-length aptamer-1 to derive the rate constants of the aptamer mini-1–gD complex. Aptamer mini-1 bound to the gD protein with an association constant (Ka), a dissociation constant (Kd), and an equilibrium dissociation constant (KD) of 3.4 × 105 l/Ms, 1.1 × 10−2 1/s, and 32 × 10−9 M, respectively. Taken together, the mapping and direct binding studies revealed that the minimal motif required for the binding of an aptamer to HSV-1 gD was retained within positions 20 to 60 of full-length aptamer-1 (Fig. 10A). Thus, aptamer mini-1 represents the shortest aptamer that efficiently binds to the gD protein of HSV-1. To determine whether aptamer mini-1 (44-mer) retained the ability to inhibit gD-HVEM interactions, we performed the same competition assay as that used for full-length aptamer-1 (SPR-based assay) (Fig. 5). As shown in Fig. 11, aptamer mini-1 also interfered with HVEM-gD interactions in a concentration-dependent manner, similarly to full-length aptamer-1. By using the SPR response signals, the effector concentration of aptamer mini-1 required for a 50% inhibition (EC50) of HVEM-gD interactions was calculated and found to be in the nanomolar range (40 nM). However, a control experiment using the complementary sequence of aptamer-1 did not show any interference in the binding reaction between gD and HVEM (data not shown).
Fig 10.
Aptamer mini-1 and its ability to bind to gD of HSV-1. (a) Predicted secondary structure of aptamer mini-1 (9). (b) Single-cycle binding kinetics measured with aptamer mini-1 (unmodified) and a complementary full-length aptamer-1 in the presence of various concentrations of the gD protein (10, 20, 40, 80, and 160 nM). Blue and red lines indicate aptamer mini-1 and full-length aptamer-1C, respectively.
Fig 11.
Ability of aptamer mini-1 (44-mer) to interfere with gD-HVEM interactions. (a) gD-HVEM interaction responses observed in the absence of or with increasing concentrations of aptamer mini-1 (unmodified) ranging from 12.5 to 200 nM. The gray line represents the injection of 200 nM aptamer alone. (b) The observed responses for each concentration of aptamer mini-1 were plotted, and the EC50 was estimated.
DISCUSSION
The interaction of the HSV-1 gD protein with three cellular receptors, HVEM, nectin-1, and modified heparin sulfate, is essential to initiate cell entry through the cooperative fusogenic action of the gD, gB, and gH/gL proteins (3, 30). Therefore, it is desirable to design molecules that specifically bind to and interfere with the function of the gD protein of HSV-1. In the current study, we isolated two aptamers that bind to HSV-1 gD. Aptamer-1 and aptamer-5 displayed high affinities for the HSV-1 gD protein in the nanomolar range, and both aptamers had the ability to discriminate between the gDs of HSV-1 and HSV-2. Both aptamers were also predicted to fold into stem-loop-like structures, as shown in Fig. 2B. Subsequently, these aptamers were partially stabilized by the incorporation of 2′-fluoro pyrimidines, because such modifications can increase the stability of RNA against endoribonucleases (27). By including these chemical modifications, the affinity of aptamer-1 for gD was marginally affected, whereas the affinity of aptamer-5 remained unchanged compared to the unmodified aptamers (2′OH).
To evaluate the ability of an aptamer to block the function of the gD protein (for example, the interaction with the HVEM receptor), we developed an SPR-based assay to evaluate whether an aptamer could interfere with gD-HVEM interactions. For this assay, the HVEM receptor was immobilized on a CM5 chip (GE Healthcare), and we used gD as a ligand to flow over the HVEM molecular surface. The gD protein of HSV-1 binds to HVEM with an equilibrium dissociation constant (KD) in the nanomolar range (28). In the absence of aptamer-1, gD binds to HVEM. However, with increasing aptamer concentrations (12.5 to 200 nM), the interaction between gD and HVEM decreased. The effective concentration of the aptamer required to inhibit 50% of the gD-HVEM interaction was in the low-nanomolar range (Fig. 6A and b). Aptamer-1C, a control with the complementary sequence of aptamer-1, failed to interfere with the gD-HVEM interaction (Fig. 6C). This result was in agreement with the SPR binding data for aptamer-1C, in which aptamer-1C failed to bind to the gD protein of HSV-1 (Fig. 4A). Both the unmodified and partially modified (2′F) aptamers displayed similar levels of inhibition (data not shown), indicating that the binding sites of these aptamers on the gD protein could be the same. These results suggested that the aptamer-1 sequence is important for binding and for interfering with gD-HVEM interactions. It is interesting to consider how the aptamer is able to discriminate between the gD proteins of HSV-1 and HSV-2, despite their high sequence similarity. The aptamer binding site within the gD protein may favor the discrimination of the gD protein of HSV-1 from that of HSV-2. Aptamer-1 was able to interfere with HVEM binding to gD. In the gD-HVEM complex (4), the N-terminal region of gD is the binding site for HVEM. However, the sequence in this region is well conserved between HSV-1 and HSV-2. One possibility is that part of the aptamer binds to the N-terminal region of gD and that another part of the aptamer binds to a different site that differs between HSV-1 and HSV-2. For example, the Glu86-Asp87-Val88 and Gln91-Pro92 regions of gD, which are located on the protein surface and are possibly spatially close to the N-terminal region, may be the other binding site of the aptamer. Another possibility is that the aptamer binds to a separate region of gD and induces a conformational change at the N-terminal end, which may lead to nonproductive interactions with HVEM. These types of binding effects have been observed frequently for several RNA-ligand complexes (23, 24). In addition to the above-mentioned observation, we also found that aptamer-1 interferes with interactions between the gD protein of HSV-1 and nectin-1. Nevertheless, direct structural analyses of the aptamer-gD complex may both reveal the molecular basis of aptamer discrimination and shed light on the mechanism of how the aptamer interferes with interactions between gD and its cognate receptors HVEM and nectin-1.
Because aptamer-1 efficiently blocked the gD-HVEM and gD–nectin-1 interactions, which are essential for HSV-1 infection, we directly tested aptamer-1 for antiviral activity using a plaque assay. For this assay, we used a partially modified aptamer-1 (2′F U and C incorporated RNA) to partially protect the RNA against endoribonucleases. The plaque assays clearly demonstrated that aptamer-1 had the ability to efficiently block HSV-1 (Fig. 7A) but not HSV-2 infection. The cytotoxicity index of this RNA suggested that aptamer-1 had minimally toxic effects on the target cells. However, by a comparison of the inhibitory effects of aptamer-1 on gD-HVEM interactions by the SPR-based and anti-HSV-1 assays, aptamer-1 was found to be less effective in the latter assay. We believe that in the anti-HSV-1 assay used here, the stability of the aptamer could be affected because the partially modified aptamer was protected only against endo-RNase but not exoribonucleases. In contrast, the analyses of aptamer interference in gD-HVEM and gD–nectin-1 interactions were performed with purified proteins in vitro.
Aptamer-1, used in all of the above-mentioned experiments, was 113 nucleotides in length. However, the aptamer length for commercial applications is typically in the range of 30 to 40 nucleotides, to facilitate the ease of synthesis as well as to reduce the cost of synthesis. Considering this point, we performed mapping studies to identify the critical region within aptamer-1 that is necessary for gD binding, using ENU and boundary analyses. Both the mapping and direct binding studies revealed that the minimal motif required for the binding of aptamer to HSV-1 gD was contained within the region of full-length aptamer-1 from positions 20 to 60 (Fig. 8 and 9). Thus, aptamer mini-1 (44-mer) (Fig. 10A) represents the smallest aptamer that could efficiently bind to the gD protein of HSV-1 and interfere with gD-HVEM interactions (Fig. 11A and b).
The results of the current study suggest that aptamer-1 has a unique ability to specifically bind to the gD protein of HSV-1 with a high affinity. This binding can also block the gD-HVEM and gD–nectin-1 interactions, giving the aptamer anti-HSV-1 activity. We believe that aptamer mini-1 now warrants further preclinical studies as an anti-HSV-1 topical product designed to prevent, or at least to significantly reduce, the risk of acquiring HSV-1 infection through physical contact. One aspect of aptamer mini-1 that must be further explored before such studies are performed is the stabilization of the RNA against exoribonucleases at the 5′ and 3′ ends. Furthermore, it appears that aptamer-1 or aptamer mini-1 could be useful both for blocking gD receptor recognition on target cells and in medical diagnostics to discriminate HSV-1 infection from HSV-2 infection.
ACKNOWLEDGMENTS
This work was supported by AIST internal grants to Penmetcha K. R. Kumar.
We thank Hiroshi Mizuno and Emi Suenaga for useful discussions.
Footnotes
Published ahead of print 18 April 2012
REFERENCES
- 1. Becker RC, Chan MY. 2009. REG-1, a regimen comprising RB-006, a factor IXa antagonist, and its oligonucleotide active control agent RB-007 for the potential treatment of arterial thrombosis. Curr. Opin. Mol. Ther. 11:707–715 [PubMed] [Google Scholar]
- 2. Becker RC, Povsic T, Cohen MG, Rusconi CP, Sullenger B. 2010. Nucleic acid aptamers as antithrombotic agents: opportunities in extracellular therapeutics. Thromb. Haemost. 103:586–595 [DOI] [PubMed] [Google Scholar]
- 3. Campadelli-Fiume G, Cocchi F, Menotti L, Lopez M. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305–319 [DOI] [PubMed] [Google Scholar]
- 4. Carfi A, et al. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169–179 [DOI] [PubMed] [Google Scholar]
- 5. Chu TC, et al. 2006. Aptamer:toxin conjugates that specifically target prostate tumor cells. Cancer Res. 66:5989–5992 [DOI] [PubMed] [Google Scholar]
- 6. Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. 1998. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 72:9992–10002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Connolly SA, et al. 2002. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM). J. Virol. 76:10894–10904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Di Giovine P, et al. 2011. Structure of herpes simplex virus glycoprotein D bound to the human receptor nectin-1. PLoS Pathog. 7:e1002277 doi:10.1371/journal.ppat.1002277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ding Y, Lawrence CE. 2003. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 31:7280–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Geiger A, Burgstaller P, Eltz HV, Roeder A, Famulok M. 1996. RNA aptamers that bind L-arginine with sub-micromolar dissociation constants and high enantio-selectivity. Nucleic Acids Res. 24:1029–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reference deleted.
- 12. Gopinath SCB. 2007. Anti-viral aptamer. Arch. Virol. 152:2137–2157 [DOI] [PubMed] [Google Scholar]
- 13. Gopinath SCB, Balasundaresan D, Akitomi J, Mizuno H. 2006. An RNA aptamer that discriminates bovine factor IX from human factor IX. J. Biochem. 140:667–676 [DOI] [PubMed] [Google Scholar]
- 14. Gopinath SCB, et al. 2006. An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits hemagglutinin-mediated membrane fusion. J. Gen. Virol. 87:479–487 [DOI] [PubMed] [Google Scholar]
- 15. Gopinath SCB, Sakamaki Y, Kawasaki K, Kumar PKR. 2006. An efficient RNA aptamer against human influenza B virus hemagglutinin. J. Biochem. 139:837–846 [DOI] [PubMed] [Google Scholar]
- 16. Gopinath SCB, Shikamoto Y, Mizuno H, Kumar PKR. 2007. Snake-venom-derived factor IX-binding protein specifically blocks the gamma-carboxyglutamic acid-rich-domain-mediated membrane binding of human factors IX and X. Biochem. J. 405:351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jenison RD, Gill SC, Pardi A, Polisky B. 1994. High-resolution molecular discrimination by RNA. Science 263:1425–1429 [DOI] [PubMed] [Google Scholar]
- 18. Karlsson R, Katsamba PS, Nordin H, Pol E, Myszka DG. 2006. Analyzing a kinetic titration series using affinity biosensors. Anal. Biochem. 349:136–147 [DOI] [PubMed] [Google Scholar]
- 19. Krol A, Carbon P. 1989. A guide for probing native small nuclear RNA and ribonucleoprotein structures. Methods Enzymol. 180:212–227 [DOI] [PubMed] [Google Scholar]
- 20. Krummenacher C, et al. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 24:4144–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kumar PKR, et al. 1997. Isolation of RNA aptamers specific to the NS3 protein of hepatitis C virus from a pool of completely random RNA. Virology 237:270–282 [DOI] [PubMed] [Google Scholar]
- 22. Kumarevel TS, Gopinath SCB, Nishikawa S, Mizuno H, Kumar PKR. 2004. Identification of important chemical groups of the hut mRNA for HutP interactions that regulate the hut operon in Bacillus subtilis. Nucleic Acids Res. 32:3904–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kumarevel TS, Mizuno H, Kumar PKR. 2005. Structural basis of HutP-mediated anti-termination and roles of the Mg2+ ion and L-histidine ligand. Nature 434:183–191 [DOI] [PubMed] [Google Scholar]
- 24. Matsugami A, et al. 2003. Structural basis of the highly efficient trapping of the HIV Tat protein by an RNA aptamer. Structure 11:533–545 [DOI] [PubMed] [Google Scholar]
- 25. Montgomery RI, Warner MS, Lum BJ, Spear PG. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427–436 [DOI] [PubMed] [Google Scholar]
- 26. Pieken WA, Olsen DB, Benseler F, Aurup H, Eckstein F. 1991. Kinetics characterization of ribonuclease-resistant 2′-modified hammerhead ribozymes. Science 253:314–317 [DOI] [PubMed] [Google Scholar]
- 27. Rusconi CP, et al. 2002. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature 419:90–94 [DOI] [PubMed] [Google Scholar]
- 28. Rux AH, et al. 1998. Functional region IV of glycoprotein D from herpes simplex virus modulates glycoprotein binding to the herpesvirus entry mediator. J. Virol. 72:7091–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shukla D, et al. 1999. A novel role for 3-O-sulfated heparin sulfate in herpes simplex virus 1 entry. Cell 99:13–22 [DOI] [PubMed] [Google Scholar]
- 30. Spear PG, Eisenberg RJ, Cohen GH. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1–8 [DOI] [PubMed] [Google Scholar]
- 31. Suenaga E, Mizuno H, Kumar PKR. 2012. Monitoring influenza hemagglutinin and glycan interactions using surface plasmon resonance. Biosens. Bioelectron. 32:195–201 [DOI] [PubMed] [Google Scholar]
- 32. Takai Y, Ikeda W, Ogita H, Rikitake Y. 2008. The immunoglobulin-like cell adhesion molecule nectin and its associated protein afadin. Annu. Rev. Cell Dev. Biol. 24:309–342 [DOI] [PubMed] [Google Scholar]
- 33. Tombelli S, Minunni M, Mascini M. 2005. Analytical applications of aptamers. Biosens. Bioelectron. 20:2424–2434 [DOI] [PubMed] [Google Scholar]
- 34. Whitbeck JC, et al. 1997. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J. Virol. 71:6083–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang N, et al. 2011. Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion. Nat. Commun. 2:577 doi:10.1038/ncomms1571 [DOI] [PubMed] [Google Scholar]