Abstract
The herpes simplex virus 1 (HSV-1) UL21 gene encodes a 62-kDa tegument protein with homologs in the alpha-, beta-, and gammaherpesvirus subfamilies. In the present study, we characterized a novel UL21-null virus and its genetic repair to determine whether this protein plays a role in early stages of the HSV-1 replication cycle. Single-step growth analyses, protein synthesis time courses, and mRNA quantifications indicated that the absence of UL21 results in a delay early in the HSV-1 replication cycle.
TEXT
Herpes simplex virus 1 (HSV-1) virions, like those of all herpesviruses, contain a proteinaceous layer, termed the tegument, located between the nucleocapsid and viral envelope. The HSV-1 tegument is composed of ∼20 different viral proteins of varying stoichiometries. Tegument proteins have been shown to play a variety of roles in infection, including the regulation of viral and host gene expression and the promotion of virus assembly and egress (2, 5, 12, 13). Tegument proteins are delivered to the host cell upon infection and, thus, can play roles at early times in infection as well as at late times when they are produced.
The UL21 protein is a 535-amino-acid component of the HSV-1 tegument (1, 17). The majority of work on UL21 has been performed with HSV-1 and pseudorabies virus (PRV), though homologs of this protein have been identified in the alpha-, beta-, and gammaherpesvirus subfamilies. Although the role of UL21 late in infection has been studied in both HSV-1 and PRV, the role this protein plays at early times in infection is unknown. In the present study, we sought to characterize the role(s) of the UL21 protein at early times in the HSV-1 replication cycle.
To identify the function(s) of UL21 at early times in HSV-1 infection, we generated both a UL21-null virus (UL21−), which lacks the entire UL21 ORF, and a repair virus (UL21R), in which the UL21 ORF was restored using the HSV-1(F) BAC pYEbac102 (18) with the “en passant” recombination system (19) and the PCR primers 5′-CCGTAGGGGGCCTCTGGGCCGTGTT-3′, ACGTCGCCGCCCGCGAAGACCCCAATAAACGTATATAGGGATAACAGGGTAATCGATTT-3′, and 5′-ACACAAGGGTGTAGTAGCGATATACGTTTATTGGGGTCTTCGCGGGCGGCGACGTAACACGCCAGTGTTACAACCAATTAACC-3′. The deletion in the UL21-null virus spanned HSV-1 bp 42074 to 43678, a region beginning with the start codon and ending with the stop codon of the UL21 open reading frame (ORF). This entire sequence was restored to its original location in the UL21R virus. Following transfection of BAC DNAs into Vero cells and subsequent virus stock production, restriction fragment length polymorphism analysis of purified viral DNAs was performed and showed the genotype was as expected (data not shown). Viral DNAs were also used for PCR amplification of the manipulated areas. Sequencing of the PCR products showed that the genetic manipulations were made as planned (data not shown). Southern blot analysis showed that the deletion in the UL21-null genome and the repair in the UL21R genome were of the correct sizes (data not shown). Immunoblot analysis showed that the UL21 protein was present in lysates of Vero cells infected with the wild-type and UL21R viruses but not the UL21-null virus (data not shown).
To determine whether the newly generated UL21-null virus possessed a defect in virus replication, single-step growth analyses were performed. Vero cells infected at a multiplicity of infection (MOI) of 5 PFU/cell with the wild-type (WT), UL21−, and UL21R viruses were collected at 0, 6, 12, 18, and 24 h postinfection and lysed, and intracellular virus was quantified by plaque assay (Fig. 1A). Medium overlying the infected cell monolayers was clarified and assayed separately (Fig. 1B). The UL21-null virus showed a 99% (two-log-unit) reduction in both intracellular and extracellular virus yields as early as 6 h postinfection compared to the wild-type and UL21 repair viruses. This reduction was less pronounced late in infection, with intracellular and extracellular virus yields being reduced by approximately 5- and 10-fold, respectively, at 18 and 24 h postinfection (hpi). These data indicate that the absence of UL21 causes a delay in the production of infectious virus.
Fig 1.
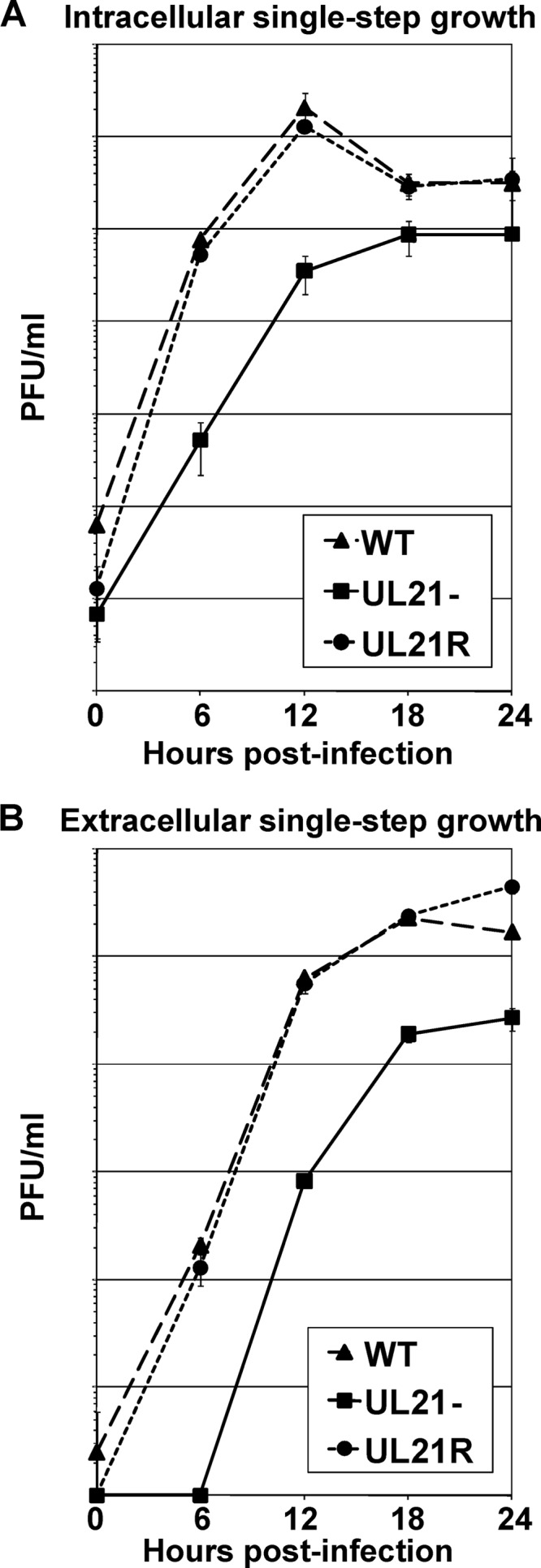
Single-step growth analyses of the WT, UL21−, and UL21R viruses. Vero cell monolayers were infected with the WT, UL21−, and UL21R viruses at an MOI of 5 PFU/cell for 1 h to allow virus adsorption. The cells were then washed extensively with citrate buffer to neutralize and remove unbound virus. The cells were overlaid with medium and held at 37°C. At the indicated times postinfection, the infected cells (A) and the overlying medium (B) were analyzed separately by plaque assay to determine intracellular and extracellular virus yields, respectively. Data points are the arithmetic means from three independent experiments; error bars represent one standard deviation.
A delay in virus production could result from a delay in the initiation of virus infection. To measure the kinetics and efficiency of UL21-null virus attachment, 25-cm2 flasks of Vero cells were infected at room temperature with 100 PFU of WT, UL21−, or UL21R viruses for 5, 10, 20, 30, or 45 min. At each time point, infected cells were washed to remove unattached virus and overlaid with medium containing human gamma globulins to neutralize any remaining unattached virus. Infected cells were then incubated at 37°C for ∼2 days, at which time viral plaques were counted to determine the percentage of the original infecting dose that had initiated viral infection by each time point. We found no difference in the number of viral plaques produced by the wild-type, UL21-null, and UL21 repair viruses (data not shown).
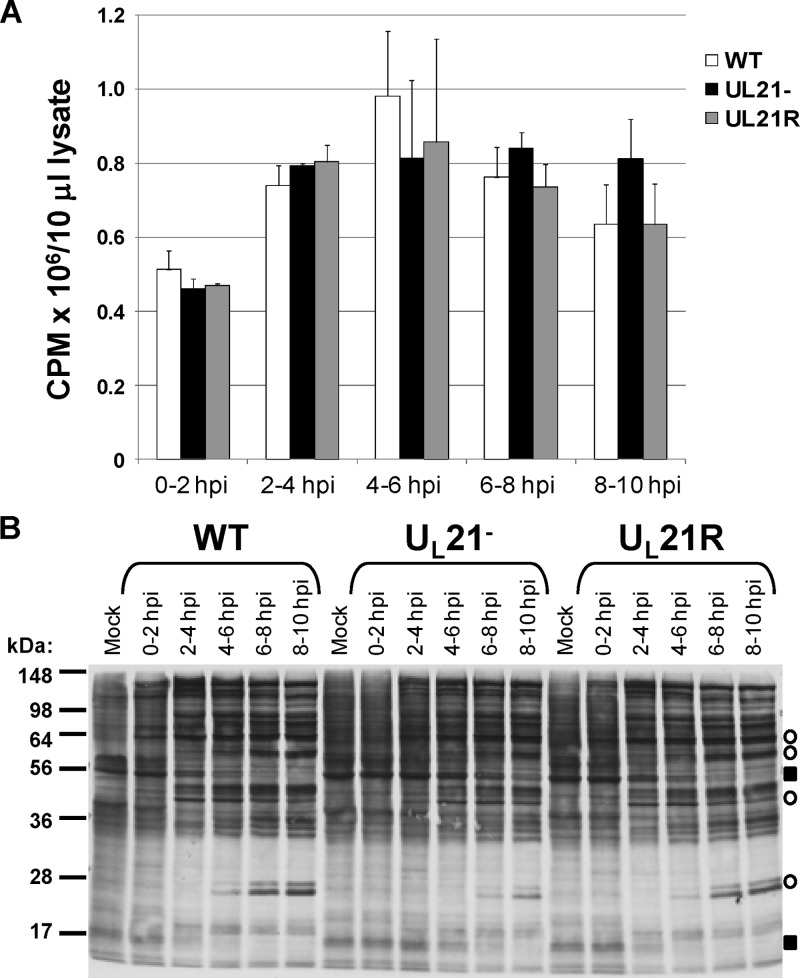
The reduced intra- and extracellular virus yields observed early in UL21-null virus infections could be caused by a defect or delay in viral protein synthesis. We examined whether total protein synthesis was decreased at various times early in UL21− virus infections by performing 35S-labeling time courses. Vero cells were either mock infected or infected with WT, UL21−, or UL21R viruses at an MOI of 10 PFU/cell by slow rocking at 4°C for 1 h, followed by removal of unattached virus and incubation at 37°C to synchronize virus entry. At 0, 2, 4, 6, or 8 h postinfection, medium overlying the infected cells was replaced with medium containing 200 μCi [35S]methionine and [35S]cysteine, and cells were returned to 37°C for 2 h. Following each 2-h labeling period, the infected, labeled cells were washed, collected, and boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Ten-microliter samples were assayed for incorporated radiolabel via scintillation counting (Fig. 2A). We found no difference in total protein synthesis between cells infected with the wild-type, UL21-null, or UL21-repair viruses for any of the time periods assayed. To examine the relative abundance of individual proteins synthesized between 0 and 2, 2 and 4, 4 and 6, 6 and 8, and 8 and 10 h postinfection, we performed autoradiography following SDS-PAGE separation of the radiolabeled cell lysates (Fig. 2B). The absence of UL21 caused a 2- to 4-h delay in the induction of viral protein synthesis, inasmuch as some proteins synthesized in infected cells, but not in mock-infected cells, began accumulating 2 to 4 h later in UL21− infections than in WT and UL21R infections. The absence of UL21 also corresponded to a 2- to 4-h delay in the shutoff host protein synthesis, inasmuch as proteins present in mock-infected cells were synthesized in UL21−-infected cells for 2 to 4 h longer than in either WT- or UL21R-infected cells.
Fig 2.
Analysis of global protein synthesis and accumulation at early times in WT, UL21−, and UL21R infections. Vero cells synchronously infected with the WT, UL21−, and UL21R viruses were incubated in the presence of [35S]methionine and [35S]cysteine from 0 to 2, 2 to 4, 4 to 6, 6 to 8, or 8 to 10 h postinfection. (A) Following cell washing and lysis, labeled proteins were detected by scintillation counting. The values are arithmetic means from three independent experiments; error bars represent one standard deviation. (B) Labeled proteins were also detected by autoradiography following SDS-PAGE separation. The experiment was performed three times with similar results. Examples of viral proteins that undergo delayed induction in UL21−-infected cells are indicated by circles, and examples of cellular proteins that undergo delayed shutoff in UL21−-infected cells are indicated by squares.
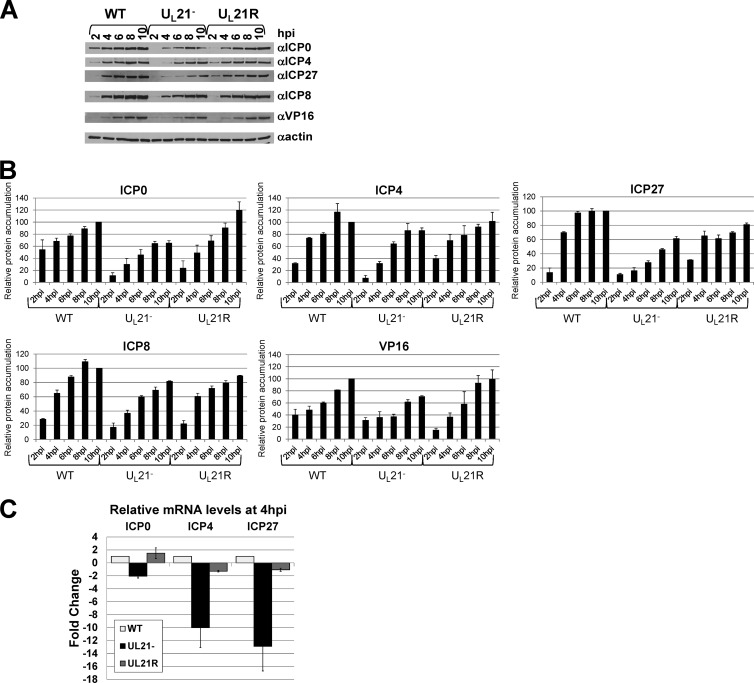
We next examined how the absence of UL21 affects the accumulation of specific immediate-early, early, and late viral proteins. We analyzed the accumulation, over time, of ICP0, ICP4, ICP27, ICP8, and VP16 by performing immunoblotting of lysates from Vero cells infected with WT, UL21−, or UL21R viruses at an MOI of 10 PFU/cell for 2, 4, 6, 8, or 10 h (Fig. 3A). For each experiment, immunoblots were quantified, and the signal from each sample was normalized to the signal obtained from the WT 10-hpi sample. Normalized values from three experiments were used to calculate relative means and standard deviations (Fig. 3B). We found that the delay in protein synthesis observed in the absence of UL21 was more pronounced for the immediate-early proteins ICP0, ICP4, and ICP27 than for either the early protein ICP8 or the late protein VP16. We therefore focused on ICP0, ICP4, and ICP27 for the experiments examining mRNA levels. The quantifications also revealed that the largest difference between wild-type and UL21− ICP0, ICP4, and ICP27 levels occurred at 4 h postinfection. This time point was therefore chosen to examine relative mRNA levels.
Fig 3.
Analysis of individual protein and mRNA levels. (A and B) Immunoblot analysis of steady-state ICP0, ICP4, ICP27, ICP8, VP16, and β-actin protein levels at early times in WT, UL21−, and UL21R infections. Lysates of Vero cells infected with the WT, UL21− and UL21R viruses for 2, 4, 6, 8, or 10 h were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with antibodies specific to ICP0 (Abcam), ICP4 (Rumbaugh-Goodwin Institute for Cancer Research), ICP27 (Virusys), ICP8 (16), VP16 (Santa Cruz Biotechnology), and β-actin (Santa Cruz Biotechnology). The experiment was performed three times. (A) Autoradiographic visualization of immunoblots. Similar results were obtained in three experiments. (B) Quantification of immunoblots. For each experiment, bands were normalized to the signal obtained from the WT 10-hpi band. Normalized values from each experiment were used to calculate relative means. Error bars represent one standard deviation. (C) Relative quantitative real-time RT-PCR analysis of ICP0, ICP4, and ICP27 mRNA levels in Vero cells infected with the WT, UL21−, and UL21R viruses for 4 h. Primer pair sequences were as follows: ICP0, 5′-CCTCTCCGCATCACCACAGAAGCC-3′ and 5′-CAGGTCTCGGTCGCAGGGAAACAC-3′; ICP4, 5′-CCGCCGTCGCAGCCGTATC-3′ and 5′-CCCGCCCTCCTCCGTCTCC-3′; ICP27, 5′-GGTGTCGGGGTCGGAGAGAAGATG-3′ and 5′-GGCGGTGCGTGTCTAGGATTTCG-3′; 18S rRNA, 5′-CCAGTAAGTGCGGGTCATAAGC-3′ and 5′-GCCTCACTAAACCATCCAATCGG-3′. The values are arithmetic means from two experiments, each performed in triplicate. Error bars represent one standard deviation.
To determine whether the delay in accumulation of ICP0, ICP4, and ICP27 proteins in UL21− infections reflected a delay in accumulation of their respective mRNAs, we performed quantitative real-time reverse transcription-PCR (qRT-PCR). Total RNA was collected from cells infected at an MOI of 10 PFU/cell with the WT, UL21−, and UL21R viruses for 4 h. The RNAs were treated with DNase to remove contaminating DNA and then used as templates to synthesize cDNAs using random primers. Equal amounts of cDNA were used in qRT-PCR mixtures containing primers specific to the gene of interest and SYBR green for product quantification. Parallel qRT-PCR mixtures contained primers specific to 18S rRNA as a control to normalize template input. The 18S rRNA cycle threshold (CT) value for a given template was subtracted from the CT obtained for each mRNA of interest (ICP0, ICP4, and ICP27) from the same template to obtain the normalized CT value (ΔCT) for each template. The change (fold, relative to the wild type) was then calculated using the ΔΔCT method. At 4 h postinfection, steady-state ICP4 and ICP27 mRNA levels were decreased ∼10-fold and ∼13-fold, respectively, in UL21−-infected cells compared to WT- and UL21R-infected cells (Fig. 3C). ICP0 mRNA levels were also affected by the absence of UL21, although to a lesser extent, showing a 2-fold decrease in UL21−-infected cells compared to WT- and UL21R-infected cells.
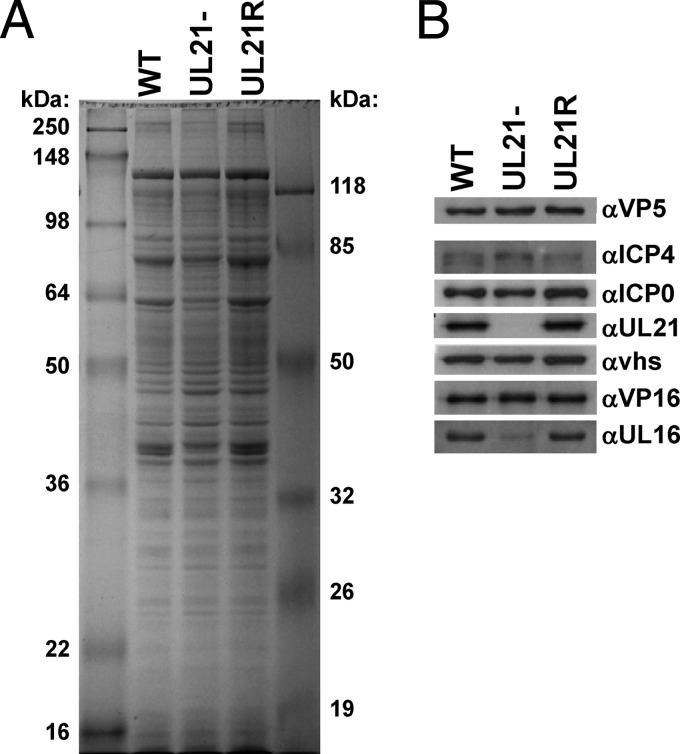
These findings show that the absence of UL21 leads to a delay in the accumulation of ICP0, ICP4, and ICP27 mRNAs, their corresponding proteins, and infectious intra- and extracellular virus. UL21 could promote timely progression through the HSV-1 replication cycle by promoting efficient packaging of tegument proteins into virions. For example, if the absence of UL21 leads to decreased levels in virions of tegument proteins with known regulatory roles, such as VP16, a delay in IE transcription could result during subsequent infection with these virions. To test this hypothesis, we purified virions from WT-, UL21−-, and UL21R-infected Vero cells as described previously (4) and examined their relative protein compositions. Figure 4A shows WT, UL21−, and UL21R purified virions separated by SDS-PAGE and stained with Coomassie brilliant blue. Figure 4B shows a panel of immunoblots of the purified virions. VP5 was used as a loading control. As expected, the UL21 protein (65 kDa) was absent in UL21− virions. Of note, VP16, vhs, ICP0, and ICP4 were present in approximately equimolar amounts in WT, UL21−, and UL21R virions. As shown previously (9), levels of the UL16 protein (41 kDa) were reduced in UL21− virions. These data show the delay in the HSV-1 replication cycle observed in the absence of UL21 is not caused by reduced virion packaging of the regulatory proteins VP16, vhs, ICP0, or ICP4.
Fig 4.
Immunoblot analysis of purified WT, UL21−, and UL21R virions. Purified virions were separated by SDS-PAGE and either stained with Coomassie brilliant blue (A) or transferred to a nitrocellulose membrane for immunoblot analysis (B) using antibodies against VP5 (3), ICP4 (Rumbaugh-Goodwin Institute for Cancer Research), ICP0 (Abcam), UL21 (1), vhs (8), VP16 (Santa Cruz Biotechnology), and UL16 (10). The experiment was performed twice, with similar results.
By specifically studying the role of UL21 in early stages of HSV-1 replication, we made the novel finding that deletion of UL21 causes a delay early in the virus replication cycle. Single-step growth analyses showed that the deletion of UL21 results in a delay in the production of infectious virions. This was previously demonstrated in a study by Baines et al. in which an HSV-1 mutant virus missing the first 484 codons of UL21 was generated (1). Compared to the wild-type and UL21 repair viruses, the UL21 mutant produced virus yields that were reduced by 1.5 log units on Vero cells at 8 h postinfection. However, by 20 and 28 h postinfection, the difference between wild-type and UL21-null virus yields decreased to approximately 0.5 log units. In the present study, the delay in the virus replication cycle was also observed in 35S-labeling protein synthesis time courses (Fig. 2B). By monitoring the accumulation of individual proteins over time via immunoblots (Fig. 3A and B), we found that the UL21−-associated delay in viral protein synthesis was more pronounced for the immediate-early proteins ICP0, ICP4, and ICP27 than for either the early protein ICP8 or the late protein VP16. Similarly, steady-state ICP4, ICP27, and, to a lesser extent, ICP0 mRNA levels were decreased at 4 h postinfection in UL21−-infected cells compared to WT- and UL21R-infected cells (Fig. 3C).
Interestingly, virus-induced shutoff of several host proteins was also delayed in UL21−-infected cells compared to WT- and UL21R-infected cells (Fig. 2B). The effect of UL21 on host protein shutoff is likely an indirect consequence of its role in promoting the synthesis of ICP27, since several studies have shown that ICP27 inhibits host cell pre-mRNA splicing, resulting in the shutoff of host protein synthesis (6, 7, 14, 15).
There are a number of possible roles for UL21 in promoting timely progression through early stages of the HSV-1 replication cycle. For example, UL21 may play a role in ensuring timely transport of the DNA-filled capsid to the nucleus and/or in nuclear genome delivery. Alternatively, UL21 delivered to infected cells may act directly or indirectly to aid in guiding the cellular transcription machinery toward the synthesis of viral mRNAs and away from the synthesis of cellular mRNAs. Finally, the delay in HSV-1 replication we observed in the absence of UL21 may be an indirect consequence of reduced UL16 quantities present in UL21− virions (Fig. 4B) (9). Sequence analysis of UL16 homologs from several herpesviruses identified a possible zinc-binding domain in the C terminus (20), and studies with the UL16 protein from HSV-2 have shown that this protein may possess DNA binding activity (11). It is not known whether UL16 is involved in transcriptional regulation, but if so, UL21 could act indirectly to promote timely viral protein synthesis by promoting the incorporation of UL16 into virions. In any case, future studies that further define the role of UL21 in promoting timely progression through early stages of the HSV-1 replication cycle will be of interest.
ACKNOWLEDGMENTS
These studies were supported by an ASM Robert D. Watkins Graduate Research Fellowship to E.F.M., NIH R01 grants GM 50740 and AI 52341 to J.D.B., NIH NRSA F32 GM067519 and NIH R21 AI 083952-01 to C.D., a Howard Hughes Medical Institute Undergraduate Science Education grant to The University of Alabama, and the Louis Stokes Alliance for Minority Participation Bridge to the Doctorate Program supported by NSF HRD-0602359.
We thank Duncan Wilson for the gift of the anti-vhs antibody and Bill Ruyechan for the gift of the anti-ICP8 antibody.
Footnotes
Published ahead of print 11 April 2012
REFERENCES
- 1. Baines JD, Koyama AH, Huang T, Roizman B. 1994. The UL 21 gene of herpes simplex virus 1 is dispensable for replication in cell culture. J. Virol. 68:2929–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Campbell ME, Palfreyman JW, Preston CM. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180:1–19 [DOI] [PubMed] [Google Scholar]
- 3. Cohen GH, et al. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34:521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Duffy C, et al. 2006. Characterization of a UL49-null mutant: VP22 of herpes simplex virus type 1 facilitates viral spread in cultured cells and the mouse cornea. J. Virol. 80:8664–8675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fuchs W, Granzow H, Klupp BG, Kopp M, Mettenleiter TC. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hardwicke MA, Sandri-Goldin RM. 1994. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J. Virol. 68:4797–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hardy WR, Sandri-Goldin RM. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 68:7790–7799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee GE, Church GA, Wilson DW. 2003. A subpopulation of tegument protein vhs localizes to detergent-insoluble lipid rafts in herpes simplex virus-infected cells. J. Virol. 77:2038–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meckes DG, Jr, Marsh JA, Wills JW. 2010. Complex mechanisms for the packaging of the UL16 tegument protein into herpes simplex virus. Virology 398:208–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nalwanga D, Rempel S, Roizman B, Baines JD. 1996. The UL 16 gene product of herpes simplex virus 1 is a virion protein that colocalizes with intranuclear capsid proteins. Virology 226:236–242 [DOI] [PubMed] [Google Scholar]
- 11. Oshima S, et al. 1998. Characterization of the UL16 gene product of herpes simplex virus type 2. Arch. Virol. 143:863–880 [DOI] [PubMed] [Google Scholar]
- 12. Pellett PE, McKnight JL, Jenkins FJ, Roizman B. 1985. Nucleotide sequence and predicted amino acid sequence of a protein encoded in a small herpes simplex virus DNA fragment capable of trans-inducing alpha genes. Proc. Natl. Acad. Sci. U. S. A. 82:5870–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Read GS, Karr BM, Knight K. 1993. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J. Virol. 67:7149–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sacks WR, Greene CC, Aschman DP, Schaffer PA. 1985. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J. Virol. 55:796–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sandri-Goldin RM, Mendoza GE. 1992. A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 6:848–863 [DOI] [PubMed] [Google Scholar]
- 16. Shelton LS, Albright AG, Ruyechan WT, Jenkins FJ. 1994. Retention of the herpes simplex virus type 1 (HSV-1) UL37 protein on single-stranded DNA columns requires the HSV-1 ICP8 protein. J. Virol. 68:521–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takakuwa H, et al. 2001. Herpes simplex virus encodes a virion-associated protein which promotes long cellular processes in over-expressing cells. Genes Cells 6:955–966 [DOI] [PubMed] [Google Scholar]
- 18. Tanaka M, Kagawa H, Yamanashi Y, Sata T, Kawaguchi Y. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 77:1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol. Biol. 634:421–430 [DOI] [PubMed] [Google Scholar]
- 20. Wing BA, Lee GCY, Huang ES. 1996. The human cytomegalovirus UL94 open reading frame encodes a conserved herpesvirus capsid/tegument-associated virion protein that is expressed with true late kinetics. J. Virol. 70:3339–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]