Abstract
Anthrax toxin is a three-part toxin secreted by Bacillus anthracis, consisting of protective antigen (PrAg), edema factor (EF), and lethal factor (LF). To intoxicate host mammalian cells, PrAg, the cell-binding moiety of the toxin, binds to cells and is then proteolytically activated by furin on the cell surface, resulting in the active heptameric form of PrAg. This heptamer serves as a protein-conducting channel that translocates EF and LF, the two enzymatic moieties of the toxin, into the cytosol of the cells where they exert cytotoxic effects. The anthrax toxin delivery system has been well characterized. The amino-terminal PrAg-binding domain of LF (residues 1–254, LFn) is sufficient to allow translocation of fused “passenger” polypeptides, such as the ADP-ribosylation domain of Pseudomonas exotoxin A, to the cytosol of the cells in a PrAg-dependent process. The protease specificity of the anthrax toxin delivery system can also be reengineered by replacing the furin cleavage target sequence of PrAg with other protease substrate sequences. PrAg-U2 is such a PrAg variant, one that is selectively activated by urokinase plasminogen activator (uPA). The uPA-dependent proteolytic activation of PrAg-U2 on the cell surface is readily detected by Western blotting analysis of cell lysates in vitro, or cell or animal death in vivo. Here we describe the use of PrAg-U2 as a molecular reporter tool to test the controversial question of what components are required for uPAR-mediated cell surface pro-uPA activation. The results demonstrate that both uPAR and plasminogen play critical roles in pro-uPA activation both in vitro and in vivo.
Keywords: Anthrax toxin, plasminogen, protective antigen, urokinase plasminogen activator, urokinase plasminogen activator receptor
1. Introduction
Anthrax toxin is a major virulence factor secreted by Bacillus anthracis, consisting of three polypeptides: a cellular receptor binding component - protective antigen (PrAg), and two enzymatic moieties - edema factor (EF) and lethal factor (LF) (1). These three proteins are individually non-toxic. To intoxicate host mammalian cells, PrAg binds to cell surface tumor endothelial marker 8 (TEM8) or capillary morphogenesis gene 2 product (CMG2), the two widely expressed anthrax toxin receptors (2, 3), and is then proteolytically activated by cell surface furin, releasing the amino-terminal 20-kDa peptide (PrAg20), thereby allowing the cell bound carboxyl-terminal 63-kDa peptide PrAg63 to form a heptamer (1). Oligomerization of PrAg63 also generates EF and LF binding sites, which span the subunit-subunit interfaces on the PrAg heptamer (4, 5). Thus, EF and LF can only bind to the oligomeric form, not the monomeric form of PrAg63. Under saturating conditions, one PrAg63 heptamer can bind a maximum of 3 molecules of LF or EF. Oligomerization of PrAg63 not only provides the binding site for LF and EF, but also triggers internalization of the toxin complex into endosomes via a lipid raft-mediated clathrin-dependent process (6, 7). A decrease in the pH in endosomes causes the PrAg63 heptamer to insert in endosomal membranes to form a channel, and through this channel LF and EF translocate to the cytosol to exert their cytotoxic effects. Therefore, PrAg is the central part of anthrax toxin, serving as the delivery vehicle for binding and translocation of LF and EF into the cytosol of the cells. The combination of PrAg and LF, termed lethal toxin (LeTx), kills animals (8, 9) and certain cells, including some murine macrophages (10), but shows no evident cytotoxicity to most other cell types. LF is a zinc-dependent metalloprotease that cleaves several mitogen-activated protein kinase kinases (MAPKKs) in their amino-terminal regions (11, 12). The combination of PrAg and EF, called edema toxin, causes edema when injected subcutaneously and death when injected systemically in experimental animals (13). EF is a calcium- and calmodulin-dependent adenylate cyclase which elevates intracellular cAMP concentrations (14), thereby causing diverse effects in cells including the impairment of phagocytosis (15).
The amino-terminal sequence of LF (residues 1–254, LFn) has substantial sequence homology to the amino-terminal sequence of EF (1). This region constitutes the PrAg hetpamer-binding domain, and is sufficient to allow translocation of fused “passenger” polypeptides to the cytosol of cells in a PrAg-dependent process (16–18). Thus, LFn fused to other bacterial toxin enzymatic domains such as the ADP-ribosylation domain of Pseudomonas exotoxin A (fusion protein 59, or in short, FP59) (17), or to reporter enzymes, such as β-lactamase (LFnLac) (19), have been generated. FP59 can be used as a potent anti-tumor agent when delivered to tumor cells using a tumor-specific PrAg (20–22). LFnLac was successfully used to image cells expressing various proteases when combined with the protease-specific PrAg proteins (19).
The unique requirement for PrAg proteolytic activation on the target cell surface provides a way to re-engineer this protein to make its activation dependent on proteases other than furin that are present on the surface of the target cell. To this end, we have generated PrAg variants that are selectively activated by urokinase plasminogen activator (uPA) (21), a serine protease that is overproduced along with its cognate receptor (uPAR) by a variety of tumor tissues and tumor cell lines. Another physiological plasminogen activator, tissue plasminogen activator (tPA), shares with uPA an extremely high degree of structural similarity and the same primary physiological substrate (plasminogen) and inhibitors (PAI-1 and PAI-2). Unlike uPA, which normally functions in tumor cells, tPA is expressed and secreted mostly by vascular endothelial cells and is primarily involved in clot dissolution. Therefore, one concern in the design of uPA-dependent PrAg proteins is to avoid cross-activation by tPA in order to minimize the potential toxicity to blood vessels. Successful discrimination of substrate sequences between uPA and tPA was made possible by the work of Madison and colleagues (23, 24) who used phage display to identify peptide sequences that are cleaved with high efficiency and selectivity by either uPA or tPA. Thus, using an optimized uPA substrate sequences GSGRSA to replace the furin site RKKR in PrAg yielded PrAg-U2, a PrAg variant that is efficiently and preferentially activated by uPA (21, 22). In contrast, when the furin site was replaced by a tPA-preferred recognition sequences, QRGRSA, the resulting PrAg-U4 was preferentially activated by tPA (21). In theory, the PrAg furin cleavage site RKKR can be changed to any other protease cleavage site to generate a PrAg cleaved by a particular protease, provided a specific substrate sequence is known. In this chapter, we will focus on PrAg-U2 protein purification and its usefulness in dissecting urokinase activation pathway both in vitro and in vivo. Using the modified anthrax toxin to image cell surface protease activities is described in the following chapter (Bugge et al.). For other applications of the modified anthrax toxins, please refer to (25).
2. Materials
2.1. Non-virulent B. anthracis Protein Expression System
Expression plasmids. pYS5 is a PrAg expressing plasmid that can shuttle between E. coli and B. anthracis (26), allowing molecular cloning to be done in E. coli and protein expression and purification in non-virulent B. anthracis strain BH450. In this plasmid, the expression of PrAg is driven by the original PrAg promoter. To make PrAg-U2, the DNA sequence encoding the PrAg furin cleavage sequence in pYS5 was changed to that encoding the uPA cleavage peptide PGSGR↓ SA (↓ indicates cleavage site), resulting in an uPA-activated PrAg expressing plasmid pYS-PrAg-U2 (21). To express FP59 in this system, the mature PrAg coding sequence in pYS5 was replaced with the FP59 coding sequence, resulting in pYS-FP59, which expresses FP59 with the PrAg signal peptide at the amino-terminus (27). All other protease-specific PrAg proteins can be efficiently made using this system.
Host strain for expression. BH450 is a protease and sporulation deficient, virulence plasmid-cured B. anthracis strain previously designated MSLL33 (28). The expression plasmids are transformed into BH450 by electroporation (the electroporation transformation protocol is available upon request).
2.2. FA Medium
FA medium is used to culture BH450 transformants for protein expression and purification.
Enriched FA medium (1 liter): mixture of 900 ml FA medium (35 g Bacto tryptone, 5 g Bacto yeast extract, autoclaved to sterilize) and 100 ml of 10×salts.
10×salts (1 liter): 60 g Na2HPO4-7H2O, 10 g KH2PO4, 55 g NaCl, 0.4 g L-tryptophan, 0.4 g L-methionine, 0.05 g thiamine-HCl, 0.25 g uracil, adjust pH to 7.5, filter to sterilize.
2.3. Protein purification
Phenyl-Sepharose Fast Flow (low substitution) resin (GE Healthcare Life Sciences). Store in 20% ethanol at 4°C. The used resin can be recycled by sequentially washing with 10 volumes of 0.1 N NaOH and large amounts of distilled water, and then stored in 20% ethanol at 4°C.
Q-Sepharose Fast Flow resin (GE Healthcare Life Sciences)
Ammonium sulfate (Sigma, St. Louis, MO). Solid ammonium sulfate is pre-cooled at −20°C before use.
Phenylmethylsulfonyl fluoride (PMSF): 30 mg/ml stock solution in isopropanol; store at −20°C.
500 mM stock solution of EDTA in H2O, pH 7.4.
Washing buffer: 1.5 M ammonium sulfate, 10 mM Tris-HCl, 1 mM EDTA, pH 8.0.
Elution buffer: 0.3 M ammonium sulfate, 10 mM Tris-HCl, pH 8.0, 0.5 mM EDTA.
Buffer A: 10 mM Tris HCl, pH 8.0, 1 mM EDTA, pH 8.0.
Buffer B: Buffer A with 0.5 M NaCl.
2.4. Reagents
Rabbit anti-PrAg serum #5308 (made in our laboratory) can recognize various PrAg species when used in Western blotting. A 1:5000 dilution can be used in Western blotting.
Human pro-uPA (single-chain uPA) (no. 107), monoclonal antibody against human uPA B-chain (no. 394), PAI-1 (no. 1094), and Glu-plasminogen (no. 410) (American Diagnostica, Inc., Greenwich, CT)
Tranexamic acid and MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (Sigma).
2.5. Cell culture and Western Blotting
HeLa cells and human 293 kidney cells obtained from American Type Culture Collection are grown in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 0.45% glucose, 2 mM glutamine, and 50 μg/ml gentamicin.
Solution of trypsin (0.25%) and EDTA (1 mM) (Invitrogen).
Hanks’ balanced salt solution (HBSS) (Biofluids, Rockville, MD).
Modified Radioimmunoprecipitation buffer (RIPA): 50 mM Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.25% sodiumdeoxycholate, 150 mM NaCl, 1 mM EDTA. May be stored up to one year if filter sterilized and stored at 4°C in the dark. RIPA buffer may be supplemented with complete protease inhibitor cocktail tablet (see below) immediately before use.
Complete protease inhibitor cocktail tablets, from Roche Diagnostics (Mannheim, Germany).
4–20% gradient Tris-glycine gels (Novex) from Invitrogen.
6×SDS sample buffer: 0.35 M Tris-HCl, pH 6.8, 10% SDS, 36% glycerol, 0.6 M dithiothreitol, 0.01% bromphenol blue. Store at −20°C in aliquots.
2.6. Mice
3. Methods
uPA and uPAR are overexpressed by virtually all human tumors and can be considered as a hallmark of malignant conversion (33, 34). uPA and uPAR are expressed at very low levels in normal tissues, but their expression is rapidly induced in response to tissue injury, thereby providing extracellular proteolysis essential for tissue repair and remodeling (34–37). uPA is secreted as a single chain enzyme (pro-uPA) with very low intrinsic activity, and is converted to the active form, two-chain uPA, by plasmin (38). Two-chain uPA, in turn, is a potent activator of plasminogen (Plg), by cleaving the R560-V561 site in plasminogen, giving rise to active plasmin. The pivotal role of uPAR in uPA-mediated cell surface plasminogen activation is well defined biochemically (34), but the function of uPAR in vivo was recently challenged by the milder phenotype of uPAR−/− mice compared to uPA−/− mice (30, 39, 40). By taking advantage of the facts that uPA-dependent activation of PrAg-U2 is readily detected by immunoblotting in vitro, or as a cause of cell death in vivo, here we describe the use of PrAg-U2 as a molecular tool to test the established paradigms regarding uPAR-mediated cell surface uPA activation both in vitro and in vivo. The results demonstrate that both uPAR and plasminogen play critical roles in pro-uPA activation both in vitro and in vivo.
3.1. Expression and Purification of PrAg-U2
The BH450 bacteria transformed with the PrAg-U2 expressing plasmid pYS-PrAg-U2 are grown from an inoculum of several resuspended colonies in six 3-L flasks, each containing 500 ml FA medium with 10 μg/ml kanamycin for 12~15 h at 37°C with shaking at 220 rpm (see Note 1).
Place flasks in ice-water bath, and add PMSF to 10 μg/ml to the cultures. The culture supernatants are then collected by centrifugation at 4500 g for 30 min at 4°C. The supernatants are sterilized by pumping through a 0.2 μm cartridge filter (Millipak 60, Millipore Corp., Bedford, MA).. Add EDTA to 5 mM to further minimize protein degradation. All the following steps should be done in a cold room.
The proteins secreted into the culture supernatants are then precipitated on Phenyl-Sepharose Fast Flow resin in the presence of 2 M ammonium sulfate. Divide the 3 L sterile supernatant between two 3-L tissue culture roller bottles, and add pre-cooled solid ammonium sulfate (270 g per liter supernatant) and Phenyl-Sepharose Fast Flow resin (50 ml settled resin to each bottle).
Gently rotate bottles until ammonium sulfate is dissolved, and then at least 1 h more.
Collect resin on a porous plastic funnel (Bel-Art Plastics, 8 cm diameter). The porous filter should be wetted with ethanol, then washed with water, to remove air.
Wash resin on funnel with 500 ml washing buffer.
The proteins are then eluted using elution buffer. Add this initially in 10 ml portions, dropwise, to achieve laminar flow as if this were a chromatography column. Collect fractions of 10–20 ml until pigment (and protein) elute.
Collect approximately 300 ml protein elute, and place in two 200 ml-centrifuge bottles (150 ml protein elute in each) and precipitate proteins by adding solid ammonium sulfate to 70% saturation (30 g solid ammonium sulfate per 100 ml of eluate). Rotate the bottles until ammonium sulfate is dissolved, and then at least 1 h additional (It is convenient to leave this overnight).
Centrifuge in 200 ml bottles, 8000 rpm, 20 min. Pour off the supernatant. Dissolve the protein pellet in 20 ml 10 mM Tris-HCl, 1.0 mM EDTA, pH 8.0, and dialyze the solution >5 h against 10 mM Tris-HCl, 1 mM EDTA, pH 8.0.
PrAg-U2 is further purified by chromatography on a Q-Sepharose FF column using an AKTA Purifier 10 FPLC system (GE Healthcare Life Sciences) or equivalent system. The column is 1.5 cm diameter, 15 cm long. Approximately 20 ml dialyzed protein solution from last step is loaded on the column, and washed with 300 ml buffer A.
PrAg-U2 is then eluted from the column using 300 ml 0–50% gradient buffer B, pumped at 1 ml/min, and fractions are collected.
Run SDS- or native-PAGE to identify the fractions containing PrAg-U2 (see note 2). Pool the fractions with PrAg-U2, and dialyze against 5 mM Hepes, 0.5 mM EDTA. Filter sterilize, measure UV spectrum, calculate mg/ml as A280 ×1.09. Freeze aliquots (see Note 3).
FP59 can be purified to one prominent band with expected size of 53 kDa from the culture supernatant using the same procedures as described above.
3.2. Proteolytic Activation of Pro-uPA and PrAg-U2 on Cultured Cells
Cells (such as uPAR-expressing HeLa cells and uPAR-non expressing human 293 cells) are seeded in 24-well plates to allow them to grow near confluence (80~100%) the next day (see Note 4).
The cells at 80~100% confluency are washed once with HBSS, followed by incubation in 1 ml/well serum-free DMEM containing 1 μg/ml pro-uPA, 1 μg/ml Glu-plasminogen, 1 μg/mlPrAg-U2, and 2 mg/ml bovine serumalbumin (BSA) at 37°C for various lengths of times (Fig. 1).
When plasminogen activator inhibitor-1 (PAI-1) is tested, cells are preincubated with PAI-1 for 30 minprior to the addition of pro-uPA, Glu-plasminogen, and PrAg-U2. When tranexamicacid is used to strip the cell surface-bound plasminogen, cells are preincubated with serum-free DMEMcontaining 2 mg/ml BSA, 1 mM tranexamic acid, without plasminogen, for 30 min before the addition of pro-uPA and PrAg-U2 (See Note 5).
Cell culture plates are then placed on ice, and washed five times with pre-cooled (on ice) HBSS to remove unbound pro-uPA, PrAg-U2, and other additions (such as inhibitors), thenlysed in 100 μl/well of modified RIPA lysis buffer supplemented with complete protease inhibitor cocktail tablet on ice for 10 min (see Note 6).
Mix 50 μl cell lysate from each well with 10 μl 6×SDS sample buffer, heat at 95°C for 5 min, vertex vigorously to break genomic DNA before sample loading (see Note 7).
10 to 15 μl samples from each well along with a protein molecular weight marker are loaded onto 4–20% gradient Tris-glycine gels to run SDS-PAGE at 120 V (see Note 8). It takes approximately 2 h for the dye to reach bottom of the gel.
Proteins on the gel are then transblotted onto nitrocellulose membranes using any method that is successful in your laboratory.
Western blottingis performed to detect pro-uPA and active form of uPA B-chain using a monoclonal antibody againsthuman uPA B-chain (no. 394, American Diagnostica, 1:1000 dilution) and goat anti-mouse IgG (HRP conjugate, pre-absorbed, Santa Cruz, 1:2000 dilution) following the universal protocols described in either Upstate Biotechnology or Santa Cruz Biotechnology Immunoblotting protocols (see Note 9).
To detect the proteolytically processed products of PrAg-U2, the same set of samples run on anther gel, are probed with 1:5000 dilution of a rabbit anti-PrAg antiserum(#5308), followed by a donkey anti-rabbit IgG (HRP conjugate, pre-absorbed, Santa Cruz, 1:2000 dilution) (see Note 9).
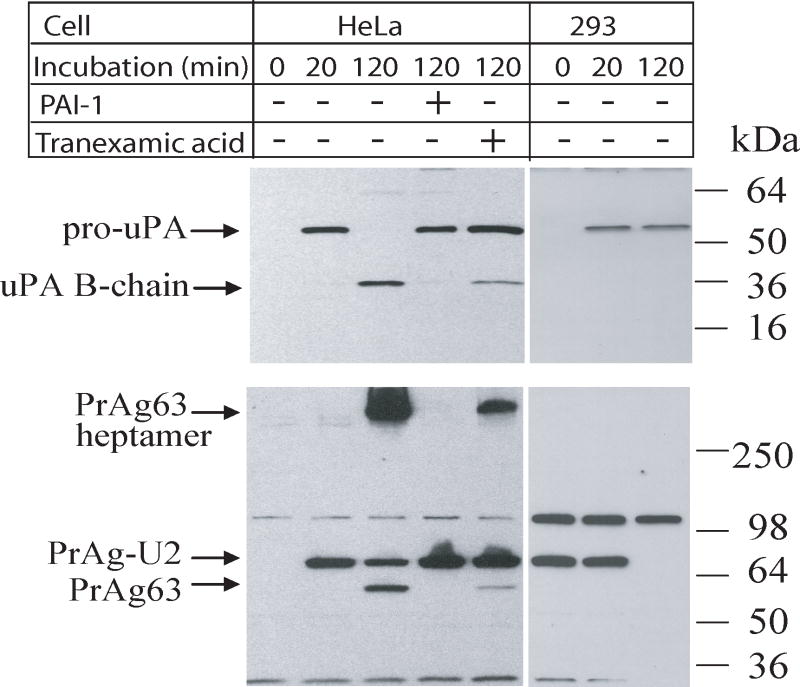
The results of an example experiment were shown in Fig. 1. HeLa cells proteolytically activated pro-uPA on the cell surface (the appearance of uPA B-chain at 120 min in SDS gel). In contrast, the uPAR non-expressing human 293 kidney cells bound weakly (probably non-specific binding) and could not proteolytically activate pro-uPA. The activation of pro-uPA by HeLa cells was completely blocked by PAI-1. Activation of PrAg-U2 on HeLa cell surface, determined by the production of the processed form PrAg63 and the formation of SDS-stable PrAg63 heptamer, exactly matched the activation profile of pro-uPA. In particular, when the activation of pro-uPA was blocked by PAI-1, or by the use of tranexamic acid, which inhibits the binding of plasminogen to the cell surface, PrAg-U2 activation was blocked in parallel. These results demonstrate that the activation of pro-uPA requires simultaneous binding of pro-uPA and plasminogen to cell surface.
Fig. 1.
Binding and processing of pro-uPA and PrAg-U2 by HeLa and 293 cells. HeLa and 293 cells were cultured to confluence in 24-well plates, and preincubated with serum-free DMEM containing 2 mg/ml BSA, 1 μg/ml of Glu-plasminogen, with or without 10 μg/ml of PAI-1 for 30 min. Some cells were preincubated with serum-free DMEM containing 2 mg/ml BSA, 1 mM tranexamic acid, without plasminogen. Then 1 μg/ml each of pro-uPA and PrAg-U2 were added to the cells and incubated for the times indicated. The cells were thoroughly washed, and the cell lysates were analyzed by Western blotting using a monoclonal antibody against the uPA B-chain (#394) (upper panel), or by using a rabbit anti-PrAg polyclonal antibody (#5308) (lower panel) to determine the binding and processing status of pro-uPA and PrAg-U2.
3.3. Cytotoxicity of PrAg-U2/FP59 to uPAR expressing cells
uPAR-expressing cells (such as HeLa cells) are seeded into 96-well plates, and grown to 30~50% confluence (see Note 10).
The cells are washed twice with serum-free DMEM to remove residual serum. Then the cells are pre-incubated for 30 min with serum-free DMEM containing 100 ng/ml pro-uPA and 1 μg/ml Glu-plasminogen with or without PAI-1 (see Note 11). Various concentrations of PrAg-U2 (0 to 1000 ng/ml) combined with FP59 (constant at 50 ng/ml) are added to the cells to give a total volume of 200 μl/well. Cells are incubated with the toxins for 6 h, the medium is replaced with fresh culture medium without toxin, and incubation continued for 48 h. (see Note 12)
Add 50 μl/well of 2.5 mg/ml MTT to the cells, incubate with the cells for 45~120 min at 37°C.
Remove the medium. The dark blue oxidized MTT pigment produced by viable cells is dissolved in 100 μl/well of the solvent [0.5% (w/v) SDS, 25 mM HCl, in 90% (v/v) isopropanol] by vortexing the plates, and the oxidized MTT, which is proportional to cell viability is measured as A570 using a microplate reader (see Note 13).
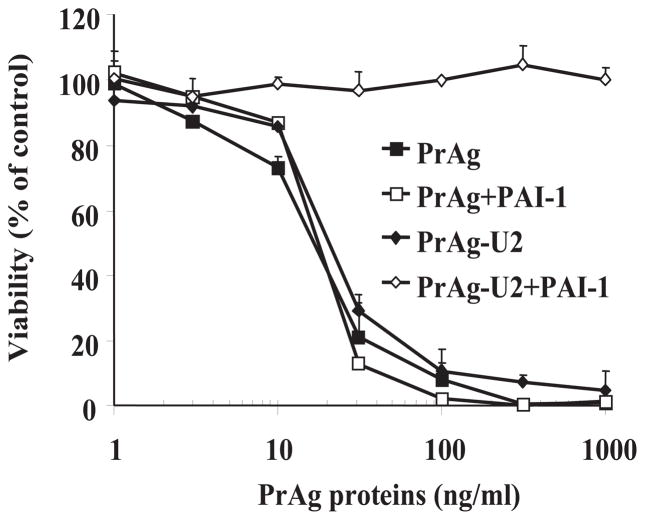
The results of an illustrative experiment are shown in Fig. 2. PrAg-U2 efficiently killed the uPA-expressing HeLa cells in a dose-depend manner, and this cytotoxicity was uPA-dependent because it was blocked by the addition of PAI-1 (see Note 14).
Fig. 2.
The cytotoxicity of PrAg-U2 to uPAR-expressing tumor cells is blocked by PAI-1. HeLa cells were cultured to 50% confluence, preincubated with serum-free DMEM containing 100 ng/ml of pro-uPA and 1 μg/ml of Glu-plasminogen with or without 2 μg/ml of PAI-1 for 30 min. Then PrAg and PrAg-U2 combined with FP59 (50 ng/ml) were added to the cells and incubated for 6 h. The toxins were removed and replaced with fresh serum-containing DMEM. MTT was added to determine cell viability at 48 h.
3.4. Activation of PrAg-U2 in vivo is dependent on the presence of uPA, uPAR, and plasminogen
The roles of uPAR, plasminogen, and PAI-1 in activation of pro-uPA can be genetically analyzed in vivo by measuring the sensitivity to PrAg-U2/FP59 of mice deficient in these plasminogen activation system components.
6–8 week-old C57BL mice deficient in uPA, uPAR, plasminogen, and control wild-type mice are injected intraperitoneally with 200 μg PrAg-U2 and 10 μg FP59 in 500 μl PBS (see Note 15).
6–8 week-old mice deficient in PAI-1 are challenged with various doses of PrAg-U2 (6, 10, 15, and 30 μg) in the presence of 10 μg FP59 in 500 μl PBS.
The mice are monitored closely (checking twice a day) for signs of toxicity for a period of 14 days after injection, by assessing weight loss, inactivity, loss of appetite, inability to groom, ruffling of fur, and shortness of breath. The mice are euthanized by CO2 inhalation at the onset of obvious malaise.
Histological analysis. Mice injected with 200 μg PrAg-U2 and 10 μg FP59 in PBS are euthanized by CO2-inhalation at the onset of malaise. The control mice injected with PBS alone are euthanized after 24–36 h by CO2-inhalation. The mice then are perfused intracardially with cold PBS, followed by 4% paraformaldehyde. The organs are post-fixed for 24 h in 4% paraformaldehyde, embedded in paraffin, sectioned, and stained with hematoxylin/eosin and subjected to microscopic analysis by a pathologist unaware of treatment or animal genotype (2–8 mice per treatment group and genotype).
Immunostaining of spleen and lymph nodes is performed with a Vectastain ABC peroxidase kit (Vector Laboratories, Inc., Burlingame, CA) with diaminobenzidine as chromogenic substrate, using rat anti-mouse CD45R/B220 antibodies (Pharmingen, San Diego, CA) to detect B lymphocytes and rabbit anti-human T cell antibodies (DAKO, Carpinteria, CA) to detect T-cells. Apoptotic cells are visualized by terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling (TUNEL) using an Apotag kit (Intergen, Gaithersburg, MD).
The results of an illustrative experiment are shown in Fig. 3. All wild type mice became terminally ill when challenged with 200 μg PrAg-U2 with FP59, with cytotoxicity observed in bone marrow, adrenal cortex, osteogenic tissues, T-cell areas of the spleen and lymph nodes (Fig. 3C–G, and data not shown). In contrast, uPA−/−, uPAR−/−, and Plg−/− mice remained completely healthy, demonstrating that both uPAR and Plg are essential cofactors in the generation of uPA activity in vivo. Microscopic examination of tissues from uPA−/−, uPAR−/−, and Plg−/− mice challenged with 200 μg PrAg-U2 with FP59 failed to demonstrate any signs of cytotoxicity to T-cell areas of the spleen and lymph nodes, bone marrow, adrenal cortex, and osteogenic tissues (Fig. 3H–J, and data not shown), providing further evidence that PrAg-U2 is activated by cell surface uPA, and demonstrating that these anatomical locations are principal sites of cell surface uPA activity in vivo. Conversely, PAI-1−/− mice were hypersensitive to PrAg-U2 combined with FP59, with a maximum tolerated dose of about 6 μg (Fig. 3B). Microscopic analysis of tissues from PAI-1−/− mice treated with just 20 μg PrAg-U2 with FP59 demonstrated bone marrow, T-cell, osteoblast, and adrenal cytotoxicity, similar to wild type mice treated with much higher concentrations of the engineered toxin (data not shown). All PrAg-U2-treated PAI-1−/− mice also presented profound edema of the small intestine frequently associated with hemorrhaging into the intestinal lumen (data not shown). This condition was never observed in wild type mice, even when treated with a 10-fold higher concentration of the engineered toxin. Taken together, these experiments unequivocally demonstrate that uPA, the binding of uPA to uPAR, and the activation of pro-uPA by plasmin are critical events in the activation of PrAg-U2 in vivo.
Fig. 3.
uPA-dependent activation of PrAg-U2/FP59 requires the presence of uPA, uPAR, and plasminogen in vivo. (A) Plg, uPA, and uPAR-deficient mice are hyperresistant to uPA-activated anthrax toxin. Wild type mice and mice deficient in uPA, uPAR, and Plg were challenged with 200 μg PrAg-U2 with 10 μg FP59 intraperitoneally, and were monitored for disease. All wild type mice became terminally ill within 24 h of toxin administration, whereas no outwards or histological signs of toxicity were detected in uPA, uPAR, and Plg-deficient mice (P<0.01). (B) PAI-1-deficient mice are hypersensitive to PrAg-U2. PAI-1−/− (open bars) or wild type control (solid bars) mice were challenged with varying concentrations of PrAg-U2 with 10 μg FP59, and monitored for disease. All PAI-1−/− mice treated with 15 to 30 μg PrAg-U2 became terminally ill within 24 h of toxin administration, whereas no outwards or histological signs of toxicity were detected in wild type mice challenged with 30 μg PrAg-U2 (P<0.001). (C–J) Cell-surface uPA-dependent T-cell toxicity of PrAg-U2. Histological appearance of T cell regions of the spleen of wild type (C–G), uPA−/− (H), uPAR−/− (I), and Plg−/− (J) mice 24 h after intraperitoneal injection of PBS (C) or 200 μg PrAg-U2 with 10 μg FP59 (D–J). Scattered clusters (examples indicated with arrows) of degenerating lymphocytes in wild type mice (D), absent in PBS-treated wild type mice (C), are identified as subpopulations of T-cells, by immunostaining with T-cell (E) and B-cell (F) antibodies, undergoing apoptosis as visualized by TUNEL-staining (G). (H–J) shows the absence of T-cell pathology in the spleens of uPA−/− (H), uPAR−/− (I) and Plg−/− (J) mice. (C, D, and H–J) Hematoxylin/eosin staining. (Bars = 10 μm).
Acknowledgments
This work was supported by the intramural research program of the National Institute of Allergy and Infectious Diseases and the National Institute of Dental and Craniofacial Research, National Institutes of Health. We thank Andrei Pomerantsev for providing strain BH450, and Rasem Fattah and Dana Hsu in assistance in protein purification.
Footnotes
It is convenient to handle six 3-L flasks of culture each with 500 ml enriched FA medium at one time. PrAg and FP59 proteins are secreted into culture medium as major secreted proteins, and can usually reach to expression levels of 30 to 50 mg/L.
PrAg proteins usually elute at 28% buffer B. Note that culture supernates contain the surface array proteins EA1 and SAP which have masses like that of PrAg, which sometimes leads to confusion when only SDS gels are used for analysis.
At this point, the purity of PrAg proteins usually approaches 95%, with one prominent band evident on gels at the expected molecular mass of 83 kDa.
The cell density does not significantly affect the levels of anthrax receptors. Thus, cells near to 100% confluence are usually used for PrAg protein binding and processing analyses.
PAI-1 is a major physiological inhibitor of plasminogen activators. Tranexamic acid can compete with plasminogen for cell surface binding sites, and thus blocks binding of plasminogen to cells. Other inhibitors, such as uPAR blocking antibodies (21), can also be used by preincubation with cells for 30 min before the addition of pro-uPA and toxin.
The complete protease inhibitor cocktail tablets are expensive, and thus a portion of a tablet can be used, such as, cut one quarter of a tablet using a clean blade, and dissolve it in 5 to 10 ml of RIPA buffer. Pre-cool RIPA lysis buffer on ice before use.
After heating, samples are usually very sticky, and vigorously vortexing to shear the cellular DNA is crucial for successful sample loading on PAGE gel.
Thoroughly washing each well of gels using distilled water is crucial to get sharp protein bands.
5% (w/v) milk (dry milk from Biorad) in TPBS (PBS containing 0.05% Tween 20) is excellent blocking solution for these antibodies.
In the 48 h-cytotoxicity assay, cells with initial 30–40% confluency are used to avoid the control untreated wells reaching confluency at 48 h when the data are collected.
Other inhibitors, such as aprotinin, α2-antiplasmin, amino-terminal fragment of uPA, or the uPAR blocking antibodies can also be tested.
This cytotoxicity assay can also be performed in regular serum containing DMEM without addition of Glu-plasminogen. In this case, it is not necessary to replace toxin-containing medium with fresh routine culture medium. Similar results can be obtained using either serum-free or serum-containing medium. Fetal bovine serum is a good source for plasminogen.
MTT is dissolved in routine cell culture medium.
Evidence that the cytotoxicity of PrAg-U2/FP59 to cells is also dependent on cell-surface bound plasminogen and functional uPAR can be found in (21).
The maximum tolerated dose of wild-type C57BL mice to PA-U2 is 30 μg in the presence of 10 μg of FP59. The maximum tolerated dose is determined as the highest dose in which outward disease or histological tissue damage is not observed in any mice within a 14-day period of observation.
Reference List
- 1.Leppla SH. Bacillus anthracis toxins. In: Alouf JE, Popoff MR, editors. The Comprehensive Sourcebook of Bacterial Protein Toxins. Academic Press; Burlington, MA: 2006. pp. 323–347. [Google Scholar]
- 2.Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA. Identification of the cellular receptor for anthrax toxin. Nature. 2001;414:225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- 3.Scobie HM, Rainey GJ, Bradley KA, Young JA. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc Natl Acad Sci USA. 2003;100:5170–5174. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunningham K, Lacy DB, Mogridge J, Collier RJ. Mapping the lethal factor and edema factor binding sites on oligomeric anthrax protective antigen. Proc Natl Acad Sci USA. 2002;99:7049–7053. doi: 10.1073/pnas.062160399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mogridge J, Cunningham K, Lacy DB, Mourez M, Collier RJ. The lethal and edema factors of anthrax toxin bind only to oligomeric forms of the protective antigen. Proc Natl Acad Sci USA. 2002;99:7045–7048. doi: 10.1073/pnas.052160199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abrami L, Liu S, Cosson P, Leppla SH, van der Goot FG. Anthrax toxin triggers endocytosis of its receptor via a lipid raft-mediated clathrin-dependent process. J Cell Biol. 2003;160:321–328. doi: 10.1083/jcb.200211018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu S, Leppla SH. Cell surface tumor endothelium marker 8 cytoplasmic tail-independent anthrax toxin binding, proteolytic processing, oligomer formation, and internalization. J Biol Chem. 2003;278:5227–5234. doi: 10.1074/jbc.M210321200. [DOI] [PubMed] [Google Scholar]
- 8.Ezzell JW, Ivins BE, Leppla SH. Immunoelectrophoretic analysis, toxicity, and kinetics of in vitro production of the protective antigen and lethal factor components of Bacillus anthracis toxin. Infect Immun. 1984;45:761–767. doi: 10.1128/iai.45.3.761-767.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beall FA, Taylor MJ, Thorne CB. Rapid lethal effect in rats of a third component found upon fractionating the toxin of Bacillus anthracis. J Bacteriol. 1962;83:1274–1280. doi: 10.1128/jb.83.6.1274-1280.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedlander AM. Macrophages are sensitive to anthrax lethal toxin through an acid-dependent process. J Biol Chem. 1986;261:7123–7126. [PubMed] [Google Scholar]
- 11.Duesbery NS, Webb CP, Leppla SH, Gordon VM, Klimpel KR, Copeland TD, Ahn NG, Oskarsson MK, Fukasawa K, Paull KD, Vande Woude GF. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 12.Vitale G, Pellizzari R, Recchi C, Napolitani G, Mock M, Montecucco C. Anthrax lethal factor cleaves the N-terminus of MAPKKs and induces tyrosine/threonine phosphorylation of MAPKs in cultured macrophages, Biochem. Biophys Res Commun. 1998;248:706–711. doi: 10.1006/bbrc.1998.9040. [DOI] [PubMed] [Google Scholar]
- 13.Firoved AM, Miller GF, Moayeri M, Kakkar R, Shen Y, Wiggins JF, McNally EM, Tang WJ, Leppla SH. Bacillus anthracis edema toxin causes extensive tissue lesions and rapid lethality in mice. Am J Pathol. 2005;167:1309–1320. doi: 10.1016/S0002-9440(10)61218-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leppla SH. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc Natl Acad Sci USA. 1982;79:3162–3166. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Brien J, Friedlander A, Dreier T, Ezzell J, Leppla S. Effects of anthrax toxin components on human neutrophils. Infect Immun. 1985;47:306–310. doi: 10.1128/iai.47.1.306-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arora N, Klimpel KR, Singh Y, Leppla SH. Fusions of anthrax toxin lethal factor to the ADP-ribosylation domain of Pseudomonas exotoxin A are potent cytotoxins which are translocated to the cytosol of mammalian cells. J Biol Chem. 1992;267:15542–15548. [PubMed] [Google Scholar]
- 17.Arora N, Leppla SH. Residues 1–254 of anthrax toxin lethal factor are sufficient to cause cellular uptake of fused polypeptides. J Biol Chem. 1993;268:3334–3341. [PubMed] [Google Scholar]
- 18.Milne JC, Blanke SR, Hanna PC, Collier RJ. Protective antigen-binding domain of anthrax lethal factor mediates translocation of a heterologous protein fused to its amino- or carboxy- terminus. Mol Microbiol. 1995;15:661–666. doi: 10.1111/j.1365-2958.1995.tb02375.x. [DOI] [PubMed] [Google Scholar]
- 19.Hobson JP, Liu S, Rono B, Leppla SH, Bugge TH. Imaging specific cell-surface proteolytic activity in single living cells. Nat Methods. 2006;3:259–261. doi: 10.1038/nmeth862. [DOI] [PubMed] [Google Scholar]
- 20.Liu S, Netzel-Arnett S, Birkedal-Hansen H, Leppla SH. Tumor cell-selective cytotoxicity of matrix metalloproteinase-activated anthrax toxin. Cancer Res. 2000;60:6061–6067. [PubMed] [Google Scholar]
- 21.Liu S, Bugge TH, Leppla SH. Targeting of tumor cells by cell surface urokinase plasminogen activator-dependent anthrax toxin. J Biol Chem. 2001;276:17976–17984. doi: 10.1074/jbc.M011085200. [DOI] [PubMed] [Google Scholar]
- 22.Liu S, Aaronson H, Mitola DJ, Leppla SH, Bugge TH. Potent antitumor activity of a urokinase-activated engineered anthrax toxin. Proc Natl Acad Sci USA. 2003;100:657–662. doi: 10.1073/pnas.0236849100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ke SH, Coombs GS, Tachias K, Navre M, Corey DR, Madison EL. Distinguishing the specificities of closely related proteases. Role of P3 in substrate and inhibitor discrimination between tissue-type plasminogen activator and urokinase. J Biol Chem. 1997;272:16603–16609. doi: 10.1074/jbc.272.26.16603. [DOI] [PubMed] [Google Scholar]
- 24.Ke SH, Coombs GS, Tachias K, Corey DR, Madison EL. Optimal subsite occupancy and design of a selective inhibitor of urokinase. J Biol Chem. 1997;272:20456–20462. doi: 10.1074/jbc.272.33.20456. [DOI] [PubMed] [Google Scholar]
- 25.Liu S, Schubert RL, Bugge TH, Leppla SH. Anthrax toxin: structures, functions and tumour targeting. Expert Opin Biol Ther. 2003;3:843–853. doi: 10.1517/14712598.3.5.843. [DOI] [PubMed] [Google Scholar]
- 26.Singh Y, Chaudhary VK, Leppla SH. A deleted variant of Bacillus anthracis protective antigen is non-toxic and blocks anthrax toxin action in vivo. J Biol Chem. 1989;264:19103–19107. [PubMed] [Google Scholar]
- 27.Liu S, Leung HJ, Leppla SH. Characterization of the interaction between anthrax toxin and its cellular receptors. Cell Microbiol. 2007;9:977–987. doi: 10.1111/j.1462-5822.2006.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pomerantsev AP, Sitaraman R, Galloway CR, Kivovich V, Leppla SH. Genome engineering in Bacillus anthracis using Cre recombinase. Infect Immun. 2006;74:682–693. doi: 10.1128/IAI.74.1.682-693.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carmeliet P, Schoonjans L, Kieckens L, Ream B, Degen J, Bronson R, De VR, van den Oord JJ, Collen D, Mulligan RC. Physiological consequences of loss of plasminogen activator gene function in mice. Nature. 1994;368:419–424. doi: 10.1038/368419a0. [DOI] [PubMed] [Google Scholar]
- 30.Bugge TH, Suh TT, Flick MJ, Daugherty CC, Romer J, Solberg H, Ellis V, Dano K, Degen JL. The receptor for urokinase-type plasminogen activator is not essential for mouse development or fertility. J Biol Chem. 1995;270:16886–16894. doi: 10.1074/jbc.270.28.16886. [DOI] [PubMed] [Google Scholar]
- 31.Bugge TH, Flick MJ, Daugherty CC, Degen JL. Plasminogen deficiency causes severe thrombosis but is compatible with development and reproduction. Genes Dev. 1995;9:794–807. doi: 10.1101/gad.9.7.794. [DOI] [PubMed] [Google Scholar]
- 32.Carmeliet P, Kieckens L, Schoonjans L, Ream B, van NA, Prendergast G, Cole M, Bronson R, Collen D, Mulligan RC. Plasminogen activator inhibitor-1 gene-deficient mice. I Generation by homologous recombination and characterization. J Clin Invest. 1993;92:2746–2755. doi: 10.1172/JCI116892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andreasen PA, Kjoller L, Christensen L, Duffy MJ. The urokinase-type plasminogen activator system in cancer metastasis: a review. Int J Cancer. 1997;72:1–22. doi: 10.1002/(sici)1097-0215(19970703)72:1<1::aid-ijc1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 34.Dano K, Romer J, Nielsen BS, Bjorn S, Pyke C, Rygaard J, Lund LR. Cancer invasion and tissue remodeling--cooperation of protease systems and cell types. APMIS. 1999;107:120–127. doi: 10.1111/j.1699-0463.1999.tb01534.x. [DOI] [PubMed] [Google Scholar]
- 35.Romer J, Bugge TH, Pyke C, Lund LR, Flick MJ, Degen JL, Dano K. Impaired wound healing in mice with a disrupted plasminogen gene. Nat Med. 1996;2:287–292. doi: 10.1038/nm0396-287. [DOI] [PubMed] [Google Scholar]
- 36.Lund LR, Bjorn SF, Sternlicht MD, Nielsen BS, Solberg H, Usher PA, Osterby R, Christensen IJ, Stephens RW, Bugge TH, Dano K, Werb Z. Lactational competence and involution of the mouse mammary gland require plasminogen. Development. 2000;127:4481–4492. doi: 10.1242/dev.127.20.4481. [DOI] [PubMed] [Google Scholar]
- 37.Heymans S, Luttun A, Nuyens D, Theilmeier G, Creemers E, Moons L, Dyspersin GD, Cleutjens JP, Shipley M, Angellilo A, Levi M, Nube O, Baker A, Keshet E, Lupu F, Herbert JM, Smits JF, Shapiro SD, Baes M, Borgers M, Collen D, Daemen MJ, Carmeliet P. Inhibition of plasminogen activators or matrix metalloproteinases prevents cardiac rupture but impairs therapeutic angiogenesis and causes cardiac failure. Nat Med. 1999;5:1135–1142. doi: 10.1038/13459. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen LS, Hansen JG, Skriver L, Wilson EL, Kaltoft K, Zeuthen J, Dano K. Purification of zymogen to plasminogen activator from human glioblastoma cells by affinity chromatography with monoclonal antibody. Biochemistry. 1982;21:6410–6415. doi: 10.1021/bi00268a014. [DOI] [PubMed] [Google Scholar]
- 39.Bugge TH, Flick MJ, Danton MJ, Daugherty CC, Romer J, Dano K, Carmeliet P, Collen D, Degen JL. Urokinase-type plasminogen activator is effective in fibrin clearance in the absence of its receptor or tissue-type plasminogen activator. Proc Natl Acad Sci USA. 1996;93:5899–5904. doi: 10.1073/pnas.93.12.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carmeliet P, Moons L, Dewerchin M, Rosenberg S, Herbert JM, Lupu F, Collen D. Receptor-independent role of urokinase-type plasminogen activator in pericellular plasmin and matrix metalloproteinase proteolysis during vascular wound healing in mice. J Cell Biol. 1998;140:233–245. doi: 10.1083/jcb.140.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]