Summary
Dyneins power the beating of cilia and flagella, transport various intracellular cargos and are important during mitosis. All dyneins have a ~300kDa motor domain consisting of a ring of six AAA+ domains. ATP hydrolysis in the AAA+ ring drives the cyclic relocation of a motile element, the linker domain, to generate the force necessary for movement. How the linker interacts with the ring during the ATP hydrolysis cycle is not known. Here we present a 3.3Å crystal structure of the motor domain of Saccharomyces cerevisiae cytoplasmic dynein, crystallized in the absence of nucleotides. The linker is docked to a conserved site on AAA5, confirmed by mutagenesis as functionally important. Nucleotide soaking experiments show that the main ATP hydrolysis site in dynein (AAA1) is in a low nucleotide affinity conformation and reveal the nucleotide interactions of the other three sites (AAA2-4).
Dyneins are a family of motor proteins that use the energy of ATP hydrolysis to move along microtubules. Axonemal dyneins power the beating of cilia and flagella. Intraflagellar transport (IFT) dynein is important for axoneme biogenesis. Cytoplasmic dynein is involved with a wide array of cellular processes from regulating the spindle assembly checkpoint and transport of organelles to localization of RNAs and gathering up of misfolded proteins1. Dyneins are also associated with a number of disease processes including viral transport2, failure of neuron migration in Lissencephaly1 , axonemal disorders such as primary ciliary dyskinesia 3 and neurodegeneration4.
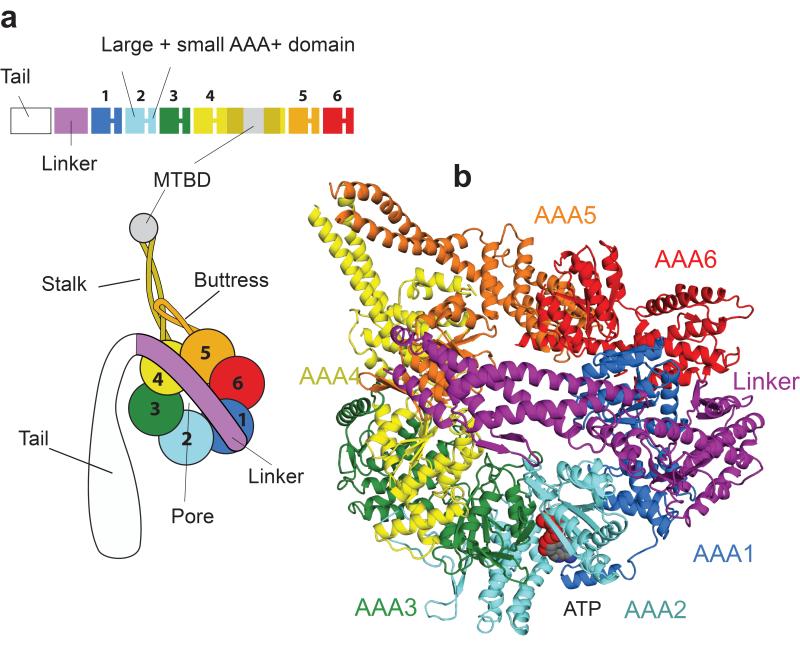
All dyneins share a conserved motor domain whose overall organization has been revealed by electron microscopy5,6 and more recently, low resolution X-ray crystallography studies7,8 (Fig. 1a). The motor domain consists of a ring of six AAA+ domains (ATPases Associated with diverse cellular Activities)9. Each AAA+ domain can be subdivided into an α/β or “large” (AAAL) and a five helix bundle or “small” subdomain (AAAS). All six AAA+ domains (AAA1-AAA6) are linked together in one polypeptide (Fig. 1a). AAA1 is the main hydrolysis site of the dynein motor domain10-12, while the role of nucleotide binding at three other sites (AAA2-AAA4) is unclear11-13. In addition to the AAA+ ring, extra structures allow dynein to act as a motor. The microtubule binding domain (MTBD) is at the end of a 15nm stalk made up of an antiparallel coiled coil emanating from AAA4S. The base of the stalk is reinforced by a coiled coil extension from AAA5S, called the buttress (or strut8), which is likely to couple ATP-binding to altered microtubule affinities of the MTBD7. Force, to move the motor along microtubules, is generated by ATP hydrolysis at AAA1 driving the cyclic relocation of a motile element called the linker. On ATP binding the linker exits the ring closer to AAA2 (pre-powerstroke position), while in the absence of nucleotide or presence of ADP it moves to exit the ring close to the base of the stalk (post-powerstroke position)5,6,11.
Figure 1.
Overview of the dynein motor domain. (a) Cartoon of the domain organization of the dynein motor. (b) Top view of a single dynein motor domain, crystallized in the absence of added nucleotide (LuAc-1 data set), showing AAA+ domains colored as in (a). ATP is firmly bound at the AAA2 nucleotide binding site (AAA2/AAA3 cleft).
Despite recent advances7,8 little is known about how ATP hydrolysis is coupled to generation of force in dynein. Are there defined docking sites for the linker in the pre- and post-powerstroke conformation and if so what do they look like? Is the linker in the same position in the nucleotide free (apo) and ADP bound form of the motor and how is ATP-turnover at AAA1 connected to changes in the position of the linker? Also compared to AAA1, the roles of the other ATPase domains (AAA2-AAA4) are less clear as there is contradictory evidence whether one13-15 or two16,17 ATPs are hydrolysed per dynein step. In addition mutations in these sites reduce but do not abolish dynein motility11-13, raising the question of what function they might have during the dynein mechanochemical cycle. Here we present the first high-resolution X-ray crystal structure of a dynein motor domain and provide insights into these questions.
RESULTS
Structure Determination
To solve the dynein motor domain at 3.3Å we used an under-oil batch crystallization to improve the diffraction and reproducibility of crystals grown using a GST-dimerized dynein motor domain construct with a truncated stalk (GST-Dyn1-314kDΔ3039-3291)7. The related construct (GST-Dyn1-314kD), containing a full length stalk, walks normally along microtubule with velocities comparable to wildtype yeast dynein12. The truncated stalk version shows ATPase activity (25.9μM Pi/s/μM dimer) and undergoes ATP dependent vanadate-mediated UV cleavage13 (Supplementary Fig. 1) in AAA1 showing the main dynein site of hydrolysis responds to nucleotide. These experiments indicate that the construct is an active ATPase and can thus provide important insights into the dynein ATPase cycle.
We obtained phases by collecting single wavelength anomalous diffraction (SAD) datasets from LuAc and Na2WO4 derivatives. Several rounds of multicrystal averaging, phase extension and model building were used to produce maps of sufficient quality to build a complete model, which could be refined against a non-nucleotide soaked data set and datasets from crystals soaked with different nucleotides (Table 1). The finished structure (Fig. 1b) reveals a wealth of new data compared to a previous low resolution structure7 (PDB code: 3QMZ). It is at a high enough resolution to build side chains and nucleotides (Supplementary Fig. 2a-f) and previously unseen loop regions, including the long connector peptides that link AAA+ domains together (Supplementary Fig. 2g,h).
Table 1.
Data collection, phasing and refinement statistics
| ATP | ADP1 | AMPPNP | LuAc-12 | LuAc-2 | Na2WO4 | |
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space group | P21 | P21 | P21 | P21 | P21 | P21 |
| Cell dimensions | ||||||
| a, b, c (Å) | 175.3, 117.9,202.8 | 174.9, 119.2, 194.0 | 175.6, 118.1, 201.0 | 175.8, 118.2,202.7 | 174.9, 118.3, 204.0 | 175.9, 118.1, 199.8 |
| α, β, γ (°) | 90, 90.2, 90 | 90.0, 90.2, 90.0 | 90.0, 90.3, 90.0 | 90.0, 90.9, 90.0 | 90.0, 90.6, 90.0 | 90.0, 91.1, 90.0 |
| Resolution (Å) | 87.7 (3.3) | 49.2 (3.4) | 80.3 (3.6) | 49.1 (3.7) | 66.2 (3.5) | 81.1 (3.9) |
| Rsym or Rmerge | 8.6 (101.5) | 12.1 (187.5) | 10.2 (68.8) | 15.4 (175.6) | 8.4 (82.1) | 10.8 (107.7) |
| I / σI | 10.1 (1.7) | 19.4 (2.6) | 7.6 (2.2) | 9.4 (2.0) | 6.5 (1.4) | 9.0 (2.1) |
| Completeness (%) | 96.1 (86.5) | 99.9 (100.0) | 99.2 (97.5) | 99.9 (99.7) | 98.1 (94.7) | 99.9 (99.9) |
| Redundancy | 5.5 (5.2) | 21.8 (21.8) | 5.1 (4.6) | 9.9 (8.1) | 3.2 (3.0) | 7.4 (7.5) |
| Refinement | ||||||
| Resolution (Å) | 50.0 - 3.3 | 49.3 - 3.4 | 50.0 - 3.6 | 49.1 – 3.7 | ||
| No. reflections | 113780 | 104357 | 90155 | 84466 | ||
| Rwork / Rfree | 23.9/30.5 | 24.1/30.3 | 24.1/30.2 | 23.1/28.9 | ||
| No. atoms | 41628 | 41679 | 41642 | 41590 | ||
| Protein | 41490 | 41497 | 41469 | 41496 | ||
| Ligand/ion | 138 | 182 | 146 | 94 | ||
| Water | - | - | - | - | ||
| B-factors | ||||||
| Protein | 164.1 | 184.7 | 176.5 | 192.2 | ||
| Ligand/ion | 119.2 | 154.8 | 145.7 | 159.5 | ||
| Water | - | - | - | |||
| R.m.s deviations | ||||||
| Bond lengths (Å) | 0.011 | 0.010 | 0.010 | 0.010 | ||
| Bond angles (°) | 1.61 | 1.56 | 1.49 | 1.51 |
four crystals,
two crystals.
Values in parentheses are for highest-resolution shell.
Interaction of the linker with the AAA+ ring
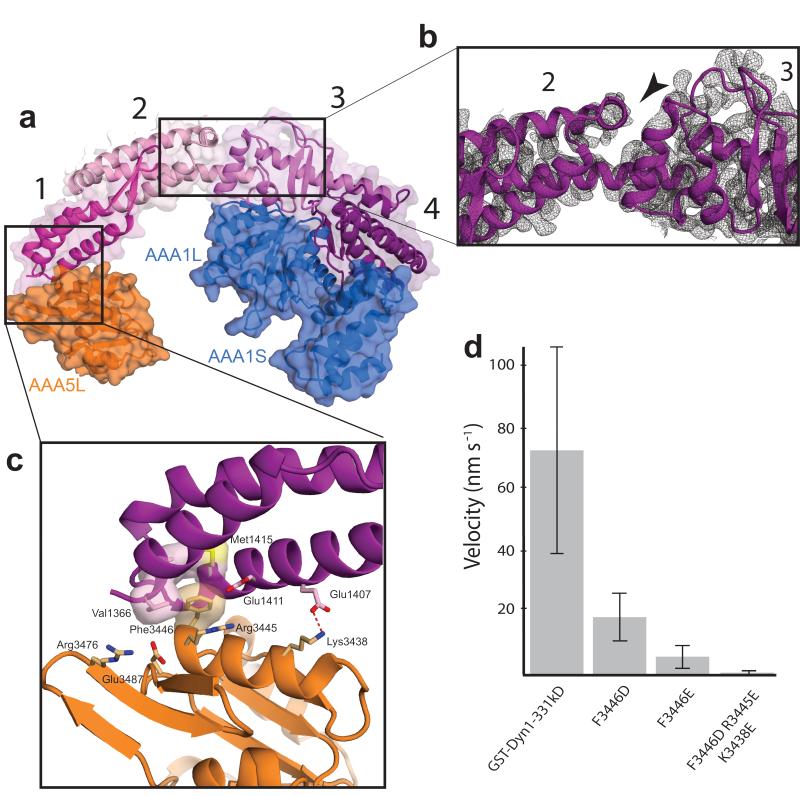
The linker is divided into four subdomains (Fig. 2a). The N-terminal subdomain 1 contacts AAA5L, subdomain 2 spans over the pore of the AAA+ ring. The C-terminal subdomains 3 and 4 interact with AAA1L via extensive contacts suggesting the linker is tightly connected to the AAA+ ring at this point. The middle of the linker consists of the junction between subdomain 2 and subdomain 3. Interestingly, unlike the rest of the linker where there are multiple interactions between subdomains, this hinge-like junction appears to form a deep cleft (Fig. 2b) with only a single helix at its base.
Figure 2.
Interaction of the linker with the AAA+ ring. (a) The four subdomains of the linker are colored different shades of purple and numbered. Subdomain 1 interacts with AAA5, while subdomains 3 and 4 interact with AAA1. (b) Cleft in the linker domain. The black arrow head points to the cleft, separating the linker into two halves between subdomains 2 and 3. The 2Fo-Fc electron density of the ATP-soaked crystal structure is contoured at 1σ. (c) Interaction site between the linker and AAA5L. The highly conserved Phe3446 of AAA5 contacts hydrophobic residues of the linker, while Arg3445 and Lys3438 mediate electrostatic interactions between AAA5L and the linker. (d) Velocity of GST-dimerized truncated dynein motors in a single molecule assay. GST-Dyn1-331kD (= wildtype), other mutations as indicated. 600 measurements were carried out for the wildtype and 400 measurements for each of the mutants, the error bars represent the s.d.. More severe mutation of the linker-AAA5 interface leads to a greater reduction in velocity.
The contact site between subdomain 1 and AAA5L consists of a hydrophobic pocket on subdomain 1 that accommodates a phenylalanine (Phe3446) on AAA5L as well as a number of surrounding electrostatic interactions (Fig. 2c). To directly test its functional relevance we mutated Phe3446 and the surrounding residues in the background of a GST dimerized, truncated dynein motor construct (GST-Dyn1-331kD), which moves similarly to full length native dynein14. The mutations resulted in severe motility defects (Fig. 2d), abnormally strong microtubule binding states and impaired ATPase activities (Supplementary Fig. 3a,b), consistent with the notion that linker binding to the AAA5 site is required for the dynein ATP hydrolysis cycle. The AAA5L and AAA1L linker contact sites are highly conserved among cytoplasmic dyneins (Fig. 3a-c) also underscoring the importance of these sites.
Figure 3.
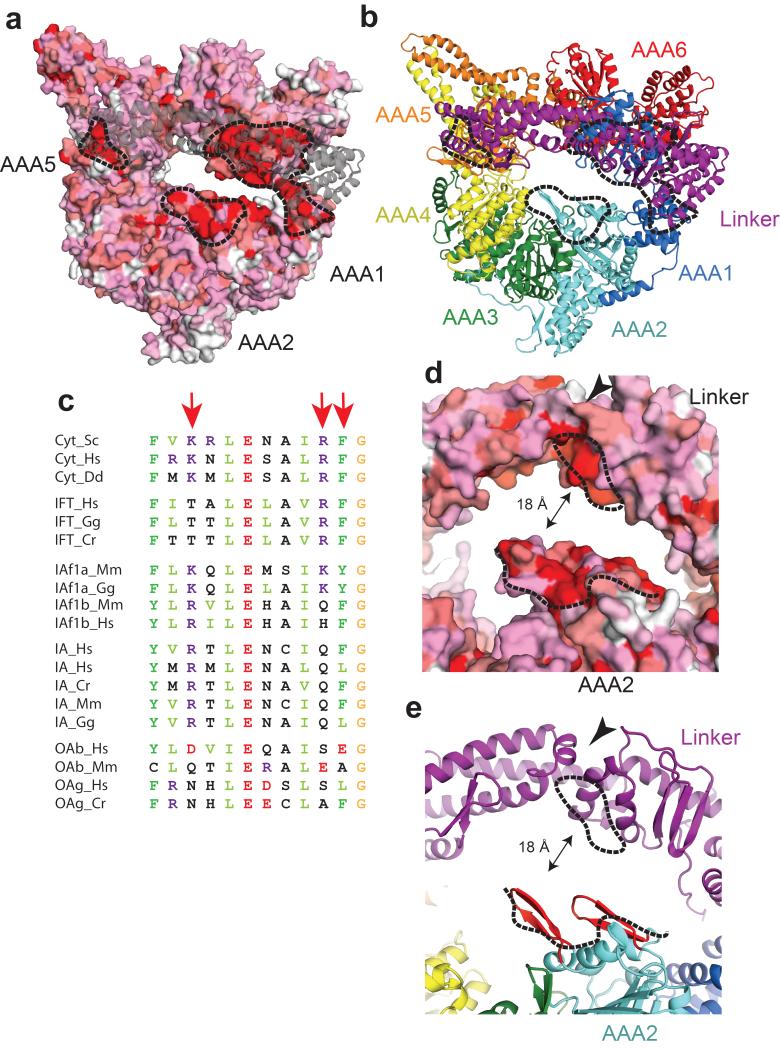
Conservation plots of the dynein motor domain. (a) Surface conservation plot of the linker proximal face of the AAA+ ring (white = not conserved, red = strictly conserved among cytoplasmic dyneins). The linker is shown in grey, transparent cartoon representation. The black dotted lines point to surface patches enriched in strictly conserved amino-acid residues (The AAA5 site corresponds to Fig 2c). (b) Color-coded cartoon representation of (a). (c) Sequence alignments of the AAA5-linker interaction site shown in Fig 2c. Cyt = cytoplasmic dynein, IFT= intraflagellar transport dynein, IAf= heterodimeric inner arm dynein IA = monomeric inner arm dynein and OA = outer arm dynein. The red arrows point to Lys3438, Arg3445 and Phe3446. Cr = Chlamydomonas reinhardtii, Dd = Dictyostelium discoideum, Gg = Gallus gallus, Hs = Homo sapiens, Mm = Mus musculus. (d) Surface conservation plot of AAA2 and the linker cleft color-coded as in (a). Conserved surface patches are indicated by black dotted lines. The surface patch of AAA2 consists of two β-hairpins. (e) Color-coded cartoon representation of (d). The β-hairpins of AAA2 are highlighted in red. The black arrowhead in (d) and (e) point to the linker cleft.
In addition to the AAA5L and AAA1L sites, two β-hairpins from AAA2 (Fig. 3d,e) are also strictly conserved and they are close to a conserved patch on the linker itself (Fig. 3d,e) which is found close to the base of the cleft between subdomains 2 and 3 (arrowhead in Figs. 2c and 3d,e). The AAA2 site forms the only highly conserved patch on the AAA+ ring not contacting the linker (Fig. 3a,b) which implies it might do so at some point of the mechanochemical cycle and hence that the linker switches between two alternate ring docking sites during the powerstroke.
Nucleotide binding to the AAA+ ring
The nucleotide binding sites sit between two neighboring AAA+ domains. The first large domain contributes the Walker A motif (P-loop), which binds phosphate groups, the Walker B motif, which contains a magnesium coordinating aspartate followed by the catalytic glutamate residue, and the sensor 1 hydrophilic residue which also contributes to catalysis. The adenine base binding pocket is between the large and small domain, and further contacts to the ATP phosphate groups are made by conserved arginine residues on the small domain (sensor 2) and the neighboring large domain (arginine finger) (Fig4a). Work on other oligomeric AAA+ domains shows that nucleotide binding closes the gap between the second and first large domain either partially, typically in the presence of ADP, or completely to bring the arginine finger into contact with the nucleotide18-20.
Figure 4.
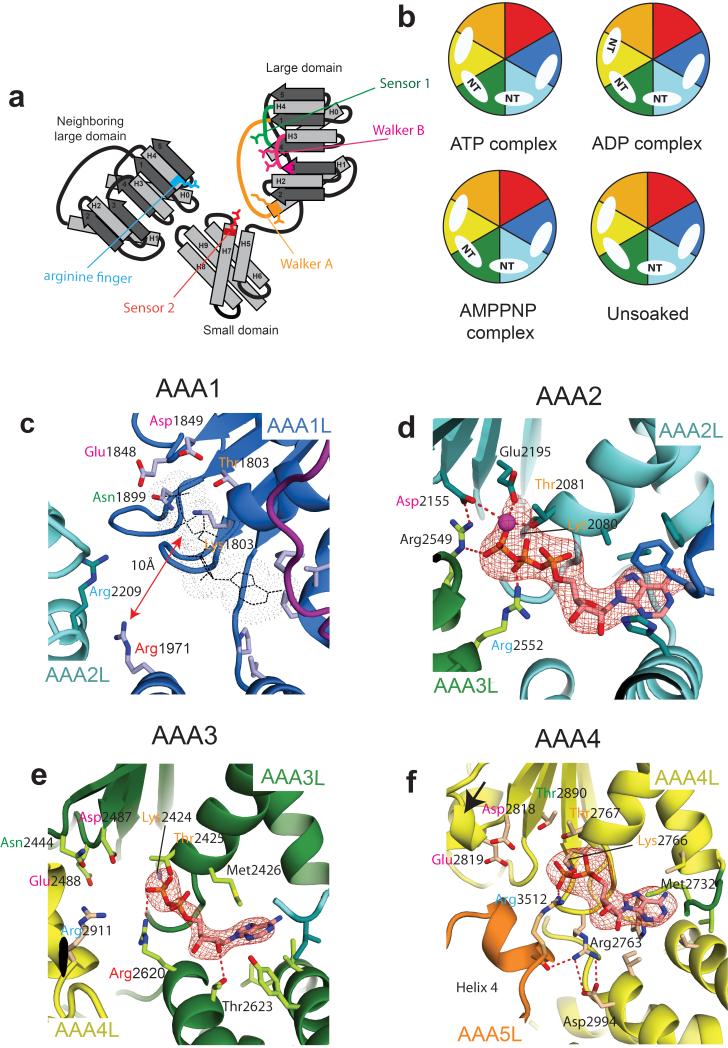
Nucleotide binding sites in the AAA+ ring. (a) Cartoon of an AAA+ nucleotide binding site showing important residues (Walker A motif in orange, Walker B motif in pink, sensor I in green, sensor II in red, arginine finger in blue. (b) Schematic representation of the nucleotide binding states of the dynein motor domain for unsoaked crystals and after soaking crystals with ADP, ATP or AMPPNP. Color-coding as in Fig. 1a, NT = nucleotide. (c) AAA1 site showing how key nucleotide binding residues are moved away from the putative nucleotide position (grey) (ATP-soaked crystal structure). (d) AAA2 site showing density for a firmly bound Mg-ATP molecule mediating multiple interactions between AAA2 and AAA3 (unsoaked crystal structure). (e) AAA3 site showing density for an exchangeable ADP molecule (ADP-soaked crystal structure). (f) AAA4 site (ADP-soaked crystal structure). The catalytic Walker B glutamate is part of a short helical fragment (black arrow) and angled away from the nucleotide. In all images the Fo-Fc difference density is contoured at 3σ and hydrogen bonding interactions are indicated as red-dotted lines. In panels (c) – (f) some residues of the nucleotide base binding pocket are shown in stick representation.
In order to investigate nucleotide binding to the different AAA+ domains in dynein we soaked crystals in ATP, ADP or AMPPNP (Fig. 4b). Compared to the nucleotide free crystal structure there are no major conformational changes. However, differences emerged between which nucleotides can bind individual AAA+ domains.
AAA1
The AAA1 site (AAA1/AAA2 interface) is devoid of nucleotide in all of our structures (Fig. 4b) and the phosphate binding Walker-A P-loop on AAA1L is occupied instead by a sulfate ion (Supplementary Fig. 4a). The AAA1 site is in a very open conformation compared to the other nucleotide binding sites (Supplementary Fig. 4a-d). It is over 10 Å wide at the bottom and nearly 20 Å wide at the top. That nucleotide binding does not occur, even though all sites are accessible and AAA3 and AAA4 are occupied, demonstrates that the AAA1 site has a very low affinity for nucleotide in the conformation of dynein in our crystals. The reason for this low affinity is that the AAA1 small domain has swung down (Supplementary Fig. 4e,f) opening up the adenine base binding pocket so that the base would be more solvent exposed than in the other nucleotide binding sites (Supplementary Figs 4g-h). In addition the small domain movement shifts the sensor 2 arginine residue so that it cannot contact the nucleotide phosphate groups (Fig. 4c).
AAA2
The AAA2 site (AAA2/AAA3 interface) contains density for a Mg-ATP molecule even when crystals were not soaked in nucleotide showing that it is tightly bound (Fig. 4d and Supplementary Fig. 4b). AAA2 lacks the catalytic glutamate in the Walker B motif. Its place is occupied by a conserved arginine (Arg2549) from AAA3 which reaches over to contact the Walker B aspartate (Asp2155) and helps anchor the ATP in place (Fig. 4d). The nucleotide is also contacted by the arginine finger (Arg2552) from AAA3 and so adds to an extensive network of salt bridges and hydrophobic interactions that hold the AAA2 and AAA3 large domains together. Thus the site is tightly closed and appears unlikely to open at any stage of the catalytic cycle.
AAA3
The AAA3 nucleotide binding site (AAA3/AAA4 interface) contains all the nucleotide binding residues expected for an active AAA+ domain arranged in the correct orientation expected for an active AAA+ ATPase (Fig. 4e). The AAA3 site is empty in the nucleotide free structure but is occupied in the presence of ATP, ADP and AMPPNP (Fig. 4b). The nucleotide is more accessible than in the AAA2 site (61Å2 are solvent accessible compared to 44Å2 - Supplementary Fig. 4b,c) consistent with the AAA3 site having an intermediate affinity. The adenine fits into the base binding pocket (Fig. S4h) and packs against a methionine residue (Met2426) which moves in to contact it (Fig. 4e). The small domain of AAA3 contributes contacts to the nucleotide ribose from a conserved threonine residue (Thr2623) and to the nucleotide phosphate groups from the sensor arginine (Arg2620). The AAA3 site is in a semi-closed conformation (Supplementary Fig. 4c) similar to other AAA+ proteins in their ADP state. The arginine finger from AAA4L (Arg2911) is still over 4.5Å away from the nearest phosphate group which suggests the AAA3 site will close further for this residue to contact the nucleotide.
AAA4
In the case of the AAA4 site (AAA4/AAA5 interface) only ADP-soaks lead to clear nucleotide binding (Fig. 4b and Supplementary Fig. 4d). The AAA4 site shows a number of unusual features in our structure. The proposed catalytic glutamate (Glu2819) is angled away from the nucleotide by the presence of a short alpha helix after it (Fig. 4f) suggesting it is inactive. The relative position of the two large domains is reminiscent of a fully closed nucleotide binding site, with the arginine finger (Arg3512) in position to contact the nucleotide. Despite this position of the large domains the nucleotide is not tightly buried (84Å2 solvent accessible – Supplementary Fig. 4d). This is due to exposure of the phosphate and ribose groups as the base is buried to a similar extent as the nucleotide in AAA3 (Supplementary Fig. 4h,i) and also interacts with a methionine residue (Met2732) that moves to contact it. The position of the large domains is stabilized in part by an arginine residue (Arg2763) next to the AAA4 Walker A P-loop forming a hydrogen-bond with helix 4 of the AAA5 domain and a salt bridge with the AAA4 small domain (Fig. 4f). _ENREF_8
DISCUSSION
During the dynein mechanochemical cycle, ATP binding and hydrolysis at AAA1 drives the linker into a pre-powerstroke position close to AAA25,11 whereas phosphate release allows return to the post-powerstroke conformation. After the powerstroke, ADP is released to reach the nucleotide free (apo) state before ATP rebinding occurs12. Our structure was crystallized in the absence of nucleotide and the linker is in a post-powerstroke position with its N-terminus near the base of the stalk, suggesting it resembles the final, apo state of the dynein ATP cycle.
Both copies of the motor domain in the GST dimerized construct are in a similar conformation (mean rmsd = 1.3Å), despite making different crystal packing contacts (Supplementary Fig. 5a,b), which indicates that crystallization doesn’t distort the motor domain. Furthermore the conserved nature of the AAA5 linker binding site, together with our mutagenesis data, suggests that the post-powerstroke state we observe is relevant to understanding the dynein ATPase cycle.
The low affinity of AAA1 for nucleotide in our structure is due to the movement of the small domain both removing the sensor arginine and opening up the base binding pocket. The presence of low affinity nucleotide binding sites is a known feature of AAA+ proteins and has been demonstrated structurally20,21 and biochemically22. In the AAA+ family member ClpX, movement of the small domain is also responsible for the low affinity nucleotide binding site, although in this case it is rotated in to sterically block access to the adenine binding pocket20. The large nature of the changes between low affinity and nucleotide bound states in AAA+ proteins are almost certainly incompatible with the crystalline state of the dynein motor domain, which explains why we could not observe nucleotide binding to AAA1 in our structure. Interestingly crystals of the low nucleotide affinity (rigor) conformation of myosin, a motor protein unrelated to the AAA+ family, can bind nucleotide when soaked with ADP23. However in this case the nucleotide binding pocket is not distorted and nucleotide binding can occur with much smaller conformational changes than need to occur at AAA1.
The recent low resolution structure of a Dictyostelium dynein motor domain, crystallized in the presence of ADP8 (PDB code: 3AY1), also has its linker in a post-powerstroke conformation but with the N-terminus of the linker closer to AAA4 and not making contact with the AAA5 site. Comparison of the Dictyostelium and yeast structures suggests there are two different post-powerstroke conformations of dynein. It is possible that the presence or absence of ADP is responsible for the differences in linker position or in other words that the interaction of the linker with AAA5 is associated with ejection of the nucleotide. Previous work has not considered the possibility of multiple post-powerstroke conformations, but this may be because the difference in the linker positions is too small to be detected by the FRET11,16,24 and the negative stain EM techniques used6.
The gaps between AAA1L/AAA2L and AAA5L/AAA6L7 appear to divide the AAA+ ring into two halves. We propose that the linker, when bound to AAA5L, acts as a spacer that pushes apart these two halves, and opens up the AAA1 site to create a low nucleotide affinity state. The result is an asymmetrical, non-planar overall conformation for the AAA+ ring. Consistent with this interpretation, the AAA+ ring in the Dictyostelium crystal structure8, where the linker does not contact AAA5L, shows a more symmetrical and planar arrangement of its building units. The effect of the AAA5L site mutations may thus be explained by reducing the ability of the motor domain to enter a low affinity AAA1 site, so that the motor gets stuck with nucleotide in its main hydrolysis site. Interestingly the AAA5L site residues (Lys3438, Arg3445 and Phe3446) are very highly conserved only in cytoplasmic dyneins. In the IFT and inner-arm axonemal dyneins there is sequence similarity at these positions (Fig. 3c) suggesting they also use the AAA5 site for docking the linker to the ring. In the outer-arm dyneins this site is not well conserved (Fig. 3c) and so these motors may use a different strategy to release nucleotide from AAA1.
An important question is how the N-terminus of the linker moves to its pre-powerstroke state exiting the ring near to AAA26. Our structure shows a conserved site on AAA2 consisting of a pair of β-hairpins. This site is adjacent to a conserved patch on the underside of the linker at the base of a hinge like cleft and modelling of the closed AAA1 conformation based on the ATP-bound crystal structure of the AAA+ protein NtrC would bring these two sites in direct contact (Supplementary Fig. 5c-e). We, therefore, propose that nucleotide binding detaches the linker from AAA5 and allows it to attach via a different site to AAA2. How this interaction results in the change in linker position observed by EM and whether the cleft in the linker is acting as a pivot point are fascinating questions, but will require further studies to elucidate.
Dyneins are unusual, among motor proteins, in having multiple nucleotide binding10 sites. Mutagenesis of residues at AAA2, AAA3 and AAA4 all lead to a decrease in velocity12,13, but do not distinguish between a structural, regulatory or catalytic role. Our structure now builds on this work to provide further insight into the roles of these additional nucleotide binding sites. Despite only containing a conserved catalytic glutamate in cytoplasmic dyneins, our structure shows that the AAA3 site contains all the residues of an active AAA+ site, suggesting that it can hydrolyse ATP. Interestingly while mutagenesis of the Walker B catalytic glutamate impaired dynein velocity it did not completely abrogate it, unlike the equivalent mutation at AAA1. The conformational changes in the AAA3 cleft must in some way contribute to the catalytic cycle without being essential. The tightly bound nucleotide at AAA2 suggests it has a purely structural role and mutations at this site are acting by disrupting the rigid AAA2/AAA3 interface and hence interfering with communication between AAA1 and AAA3.
In our structure the AAA4 site appears to be in an inactive conformation. However mutagenesis of the Walker A motif in AAA1 allows UV mediated vanadate cleavage to occur at AAA412, implying that the conformation of AAA4 may respond to events at AAA1. Therefore, we cannot rule out a conformational rearrangement that activates the AAA4 site at other points of dynein’s catalytic cycle. It is interesting, however, that the Walker B motif in AAA4 is not well conserved in IFT and axonemal dyneins and mutation in cytoplasmic dyneins has no effect on velocity13_ENREF_14_ENREF_14. This suggests that whatever the role of AAA4 it is not required in all dyneins.
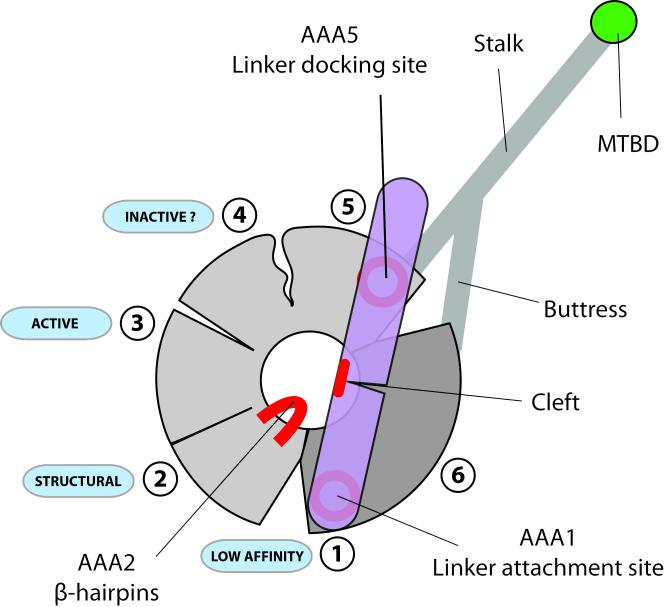
The main features of dynein revealed by our structure are summarized in figure 5. In the absence of nucleotide the linker spans the AAA+ ring contacting the conserved patches at AAA5 and AAA1. The AAA1 site is wide open and has a low affinity for nucleotide. AAA2 binds nucleotide tightly and has a structural role. AAA3 appears active for ATP hydrolysis, while in this conformation AAA4 does not. There is a cluster of conserved residues on AAA2 that is directly adjacent to a conserved patch on the underside of the linker at the base of a cleft. This may represent another interaction site with the linker. ATP binding to AAA1 would necessitate closure of the gap between AAA1 and AAA2 and could bring these two conserved sites into proximity with each other.
Figure 5.
Schematic representation of the nucleotide free dynein motor domain. The linker is in a post-powerstroke position conformation and interacts with the AAA+ via conserved surface patches on AAA5 and AAA1. Conserved amino-acid residues on the β-hairpins of AAA2 are in close proximity to a likewise conserved region near the linker cleft. Nucleotide induced gap closure of the AAA1 site may bring these two areas into contact. The linker, in interacting with AAA5 and AAA1 at the same time, might act as a spacer pushing apart the two halves of the motor domain (light and dark grey) and causing gap opening between AAA1/AAA2 and AAA5/AAA6. The gap at AAA1 leads to a low nucleotide affinity state for this site. The AAA2 site contains a tightly bound ATP and is likely to play a purely structural role. Sites AAA3 and AAA4 appear to adopt a semi-closed conformation and can bind and release nucleotide. While the AAA3 ATP hydrolysis site seems to be in a catalytically competent conformation, important catalytic residues of the AAA4 site might not be able to support ATP hydrolysis.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Carol Cho, for her work helping to identify heavy-atom derivatives and suitable crystallisation conditions. We also thank M. Schlager, A. Diamant, C. Cho, R. Vale, K. Nagai and Lori Passmore for helpful discussions and their comments on the manuscript. This work was supported by the Medical Research Council (MC_UP_A025_1011).
Footnotes
ACCESSION CODES The coordinates and structure factors for the ATP, ADP, AMPPNP and LuAc complexes have been deposited in the protein data bank with deposition codes 4AKG, 4AI6, 4AKH and 4AKI.
AUTHOR CONTRIBUTIONS E.S.G., H.S. and A.P.C. produced, purified and crystallized the protein. H.S. and E.S.G. prepared heavy-atom derivatives. H.S. and A.P.C. collected data on crystals and determined the structure. H.S. carried out phasing. All authors built the model. E.S.G. and A.P.C. conducted the in-vitro experiments. A.P.C. and H.S. wrote the paper.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
REFERENCES
- 1.Vallee RB, Williams JC, Varma D, Barnhart LE. Dynein: An ancient motor protein involved in multiple modes of transport. J Neurobiol. 2004;58:189–200. doi: 10.1002/neu.10314. [DOI] [PubMed] [Google Scholar]
- 2.Dodding MP, Way M. Coupling viruses to dynein and kinesin-1. EMBO J. 2011;30:3527–3539. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leigh MW, et al. Clinical and genetic aspects of primary ciliary dyskinesia/Kartagener syndrome. Genet Med. 2009;11:473–487. doi: 10.1097/GIM.0b013e3181a53562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hafezparast M, et al. Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science. 2003;300:808–812. doi: 10.1126/science.1083129. [DOI] [PubMed] [Google Scholar]
- 5.Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 6.Roberts AJ, et al. AAA+ Ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kon T, Sutoh K, Kurisu G. X-ray structure of a functional full-length dynein motor domain. Nat Struct Mol Biol. 2011;18:638–642. doi: 10.1038/nsmb.2074. [DOI] [PubMed] [Google Scholar]
- 9.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 10.Gibbons IR, Gibbons BH, Mocz G, Asai DJ. Multiple nucleotide-binding sites in the sequence of dynein beta heavy chain. Nature. 1991;352:640–643. doi: 10.1038/352640a0. [DOI] [PubMed] [Google Scholar]
- 11.Kon T, Mogami T, Ohkura R, Nishiura M, Sutoh K. ATP hydrolysis cycle-dependent tail motions in cytoplasmic dynein. Nat Struct Mol Biol. 2005;12:513–519. doi: 10.1038/nsmb930. [DOI] [PubMed] [Google Scholar]
- 12.Kon T, Nishiura M, Ohkura R, Toyoshima YY, Sutoh K. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry. 2004;43:11266–11274. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- 13.Cho C, Reck-Peterson SL, Vale RD. Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. J Biol Chem. 2008;283:25839–25845. doi: 10.1074/jbc.M802951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reck-Peterson SL, et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimizu T, Johnson KA. Kinetic evidence for multiple dynein ATPase sites. J Biol Chem. 1983;258:13841–13846. [PubMed] [Google Scholar]
- 16.Mogami T, Kon T, Ito K, Sutoh K. Kinetic characterization of tail swing steps in the ATPase cycle of Dictyostelium cytoplasmic dynein. J Biol Chem. 2007;282:21639–21644. doi: 10.1074/jbc.M701914200. [DOI] [PubMed] [Google Scholar]
- 17.Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- 18.Chen B, et al. Engagement of arginine finger to ATP triggers large conformational changes in NtrC1 AAA+ ATPase for remodeling bacterial RNA polymerase. Structure. 2010;18:1420–1430. doi: 10.1016/j.str.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies JM, Brunger AT, Weis WI. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 22.Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144:526–538. doi: 10.1016/j.cell.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coureux PD, Sweeney HL, Houdusse A. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 2004;23:4527–4537. doi: 10.1038/sj.emboj.7600458. doi:10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imamula K, Kon T, Ohkura R, Sutoh K. The coordination of cyclic microtubule association/dissociation and tail swing of cytoplasmic dynein. Proc Natl Acad Sci U S A. 2007;104:16134–16139. doi: 10.1073/pnas.0702370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Kabsch W. Xds. Acta Crystallogr D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leslie AGW, Powell HR. Processing diffraction data with MOSFLM. Nato Sci Ser Ii Math. 2007;245:41–51. [Google Scholar]
- 28.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Mccoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowtan K. DM: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.